Abstract
An epigenetic trait is a stably inherited phenotype resulting from changes in a chromosome without alterations in the DNA sequence. Epigenetic modifications, such as; DNA methylation, together with covalent modification of histones, are thought to alter chromatin density and accessibility of the DNA to cellular machinery, thereby modulating the transcriptional potential of the underlying DNA sequence. As epigenetic marks under environmental influence, epigenetics provides an added layer of variation that might mediate the relationship between genotype and internal and external environmental factors. Integration of our knowledge in genetics, epigenomics and genomics with the use of systems biology tools may present investigators with new powerful tools to study many complex human diseases such as kidney disease.
EPIGENETICS
An epigenetic trait is a stably inherited phenotype resulting from changes in a chromosome without alterations in the DNA sequence. During normal development somatic cells that are descended from a single progenitor, and contain near-identical genotype, differentiate to acquire diverse biological function by expressing and repressing different set of genes. During development new epigenetic marks are established. This process is brought about by modification of the genetic material without changing the nucleotide sequence. Later on in development epigenetic marks are maintained through cell division to maintain cell identity. Recent discoveries suggest that via epigenetic reprogramming few key molecules can also change fully differentiated cells back to induced pluripotent stem cells (iPS cells)(1–3). Epigenetic changes can potentially lead to gene expression changes and disease development. The recognition of the role of epigenetic dysregulation in human disease started in oncology, but has now extended to other disciplines(4). Despite the significant advances on this field, very little is known about epigenetic changes that characterize different disease conditions and tissue fibrosis(5).
EPIGENETICS AND CHRONIC KIDNEY DISEASE
Very little is known about epigenetic changes and regulation in the context of acute or chronic kidney disease (CKD). In our view at least three key clinical observations suggest that epigenetic changes contribute to CKD development.
Firstly evidence is emerging highlighting the important role of fetal programming in the development of adult diseases, suggesting a possible common pathophysiologic cause. Hypertension and CKD are highly prevalent diseases that tend to occur more frequently among disadvantaged populations, in whom prenatal care also tends to be poor. Epidemiologic evidence accumulated over the past 2 decades has shown an association between low birth weight and subsequent adult HTN, diabetes, cardiovascular disease and CKD (6). A direct correlation of LBW have been reported with microalbuminuria in type 1 diabetics, with albuminuria in type 2 diabetic Pima Indians, with microalbuminuria in older nondiabetic persons, and with the progression of CKD(7). Animal studies and indirect evidence from human studies support the hypothesis that low birth weight, as a marker of adverse intrauterine circumstances, causes or is associated with a congenital deficit in nephron number(7). The precise mechanism of the reduction in nephron number is unknown, and based on studies performed in rodents several hypotheses have been put forward, including increased apoptosis in the developing kidney, alterations in renal renin–angiotensin system activity, and increased fetal glucocorticoid exposure(8). A reduction in nephron number is associated with compensatory glomerular hypertrophy leading to the development of hypertension and glomerulosclerosis, which then starts a vicious cycle leading to further nephron loss and ultimately to the development of chronic kidney disease(6). While the low nephron number hypothesis has been developed by Barker and Brenner in the early 1960’s, the underlying molecular mechanism of the “intrauterine programming” on later life CKD is not yet known (7). As epigenetic marks are established during development we can speculate that epigenetic differences potentially play role in this process.
In addition to this “fetal programming”, studies indicate that similar “programming” might occur during adulthood as well. Data from the Epidemiology of Diabetes Interventions and Complications (EDIC) study, which followed 1441 patients with type 1 diabetes after they completed the Diabetes Control and Complication Trial (DCCT), show that early chronic exposure to a moderately high level of hyperglycemia has prolonged effects on diabetic complications during subsequent periods of improved glycemia, a phenomenon termed "metabolic memory" (9, 10). For example, atherosclerotic changes not present at the end of the DCCT appeared subsequently in the previously higher HbA1c group, followed by a twofold increase in myocardial infarction, strokes, and cardiovascular death. These changes occurred despite the fact that the hemoglobinA1c (HbA1c) levels were identical in the two groups during the entire time. Recent in vitro studies suggest that epigenetic differences and changes in histone modification that occur during periods of hyperglycemia might play role in “hyperglycemic memory” (11, 12).
In addition to “hyperglycemic memory” recent studies indicate the presence of “uremic memory” effect as well. While traditionally we thought that acute kidney injury resolves without many long term sequelae, clinical observational studies indicate that as compared with controls, patients who suffered dialysis-dependent acute kidney injury during their hospitalization have a 28-fold increased risk of developing stage 4 CKD or end-stage renal disease, despite the initial functional recovery(13, 14). The mechanism that leads to this increase in CKD development is not fully understood and epigenetic changes could play a key role in this process.
EPIGENETIC MODIFICATIONS: DNA METHYLATION
The epigenome consists of chromatin, a protein-based structure around which the DNA is wrapped, and its post-translational modifications as well as methylation of cytosines (15). DNA methylation could displace transcription factors that normally bind to the DNA and/or could attract methyl binding-proteins, which are associated with gene silencing and chromatin compaction(16).
DNA methylation is the most widely studied epigenetic modification. In eukaryotes, it consists of the covalent addition of a methyl group at the 5-position of cytosines, and is usually associated with gene silencing(17). This methylation occurs on cytosines that are followed by guanines (CpG dinucleotides). In the human genome, CpG dinucleotides are generally concentrated in regions called CpG islands, which are preferentially located in promoter regions and usually do not contain 5-methylcytosines. However, some physiological processes require DNA methylation of CpG islands (18, 19). These include silencing of imprinted genes, in which only one allele should be expressed in a progenitor-of-origin manner (i.e. only paternal or maternal expression) and X chromosome inactivation in women, as a mechanism for dosage compensation (20).
CpG dinucleotides that are not embedded in CpG islands are mainly located in repetitive or centromeric sequences. Additionally, some are distributed in gene or intergenic regions. The CpGs located outside the CpG islands are usually methylated, a process that is associated with the maintenance of chromosomal stability, translocation prevention, and endoparasitic sequence silencing (Figure 1). Functionally, DNA methylation is mostly, but not exclusively associated with untranscribed chromatin. Thus, most methylated DNA in human beings occurs in non-coding regions of the genome, such as repetitive sequences, transposons, and endogenous retroviruses acquired throughout evolution.
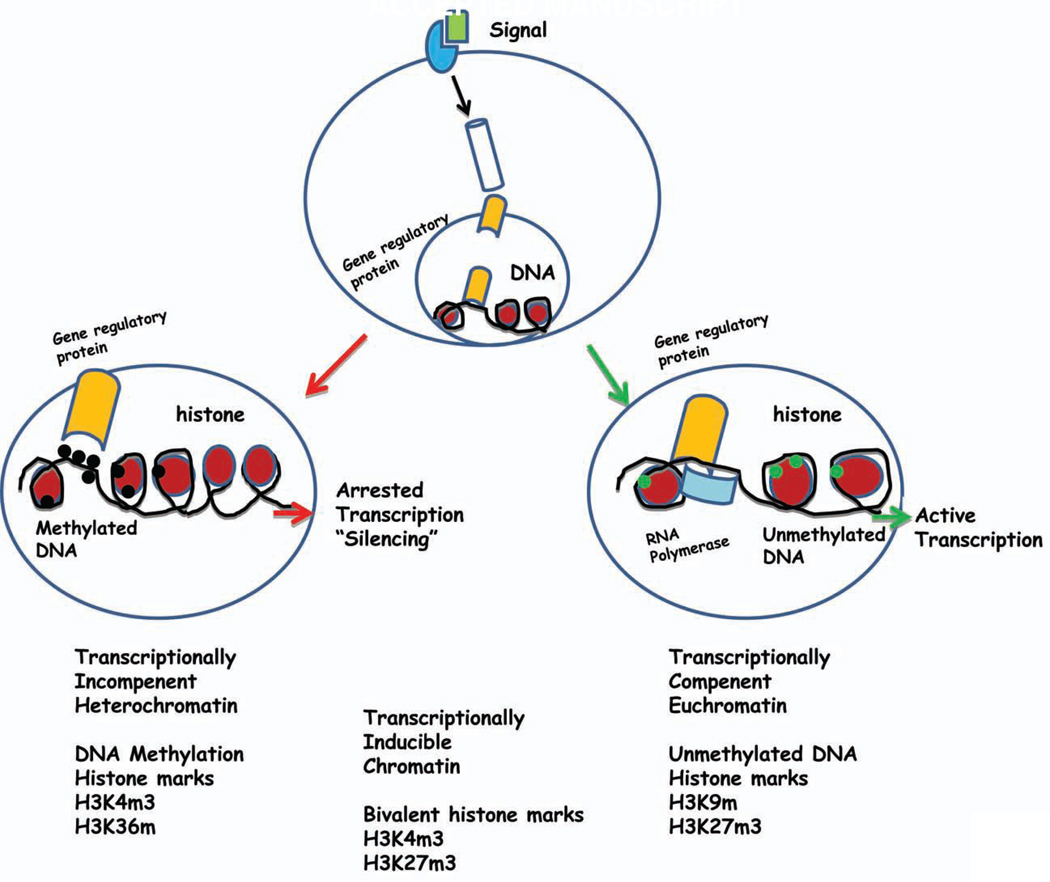
Figure 1. The effect of different external signals depends on epigenetic modification in the target cells.
The cellular response to external signals may reflect chromatin-based (epigenetic) differences superimposed on the static genetic code. In this example, the 5-regulatory region of a disease susceptibility gene is depicted in cells. While in some cells the promoter assumes an “open” chromatin architecture characterized by activating histone posttranslational marks, decreased nucleosome density, and the lack of DNA methylation. RNA polymerase II is recruited to the promoter, resulting in productive transcription. Alternatively, in other cells the genetically identical promoter assumes a “closed” chromatin configuration characterized by repressive posttranslational marks, increased nucleosome density, and prominent DNA methylation. RNA polymerase II is not recruited to the promoter, and no transcription occurs. Transcriptionally Competenent Euchromatin is usually associated with umethylated DNA, and trimethyl H3K9 and H3K23 marks. Transcriptionally Incompetent Heterochromatin is generally associated with methylated DNA and trimethyl H3K4 and H3K6 marks.
HISTONE MODIFICATION
The function of chromatin is to package DNA into smaller volume to fit in the cell nucleus. The major proteins involved in chromatin are histone proteins (H1–4). The four core histones H2A, H2B, H3 and H4 may undergo a range of post-translational modification, including acetylation, methylation, O-GlcNac modification, phosphorylation, SUMOylation, ADP-ribosylation and ubiquitination(21). Histone modifications are indicators of active or repressed chromatin, and the “histone code” hypothesis proposes that combinations of specific histone modifications create a complex defining the functional hierarchy for chromatin regulation(22–24). The key enzymes involved in the acetylation at lysine residues include histone acethyltransferase (HATs), histone deacetylases (HDACs)(15). Methylation of histones is carried out by histone methyltransferases and demethylation by histone demethylases such as the jumonji protein family(15). Histone lysine residues can be monomethylated, dimethylated, or trimethylated. The effect of each modification depends on both the identity of the modified residue and the extent of methylation. For example, methylation of histone H3 on lysines 4 and 36 (H3K4 and H3K36) is generally associated with an “open” euchromatin structure and transcriptional activation, whereas methylation of histone H3 on lysines 9 and 27 (H3K9 and H3K27) is generally associated with a “closed” heterochromatin structure and gene silencing (Figure 1). The situation, however, appears to be more complex with the recent discovery of bivalent domains (25, 26). Regions of chromatin simultaneously marked by a histone modification associated with active transcription (histone H3 lysine 4 trimethylation) and a modification associated with repression (histone H3 lysine 27 trimethylation). It is postulated that bivalent marks identify genes that are silent but poised for transcription (26, 27) (Figure 1).
It also appears that there is a functional link between DNA methylation and histone modifications. Histone deacethylases (HDACs) are recruited to methylated DNA by methyl-CpG binding proteins. DNA methylation and histone modifications might function in close interplay with nucleosome remodeling and positioning complexes that bind specific histone modifications such as trimethylated H3K4 and methyl CpG binding proteins and move nucleosomes on DNA by ATP-dependent mechanisms(23). New studies suggest an important interplay between the different epigenetic marks (histone modification, DNA methylation and microRNAs)(24) (Figure 1).
METHODS TO ANALYZE THE EPIGENOME: DNA METHYLATION
There are multiple methods available to detect DNA methylation changes. The ideal assay to test cytosine methylation would test every CG dinucleotide individually and quantitatively throughout the genome, preserving information about cis-relationships of methylation states between CGs, and allowing high sample throughput. The currently available methods can be grouped into three categories: (a) based on the use of methylation-sensitive or -insensitive restriction enzyme; (b) chemical modification of DNA by sodium bisulfite followed by sequencing; (c) complexing and precipitation with methyl-binding protein or antibodies against methylated cytosines.
One of the most commonly used methods to study DNA methylation on a whole genome scale is based on the use of restriction enzyme pairs that distinguish between methylated and unmethylated DNA. For example HpaII and MspI recognize the same nucleotide sequence, however HpaII only digests unmethylated DNA. The assay that became known as HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) is based on this principle (28). Following the digestion of the genomic DNA with HpaII and MspI that interrogates both CpG islands and other unmethylated CpG-rich genomic regions at high resolution (~50 bp) the fragments are amplified using a PCR based method. In the earlier version of the assay the HpaII digestion fragments (unmethylated DNA enrichment) and the methylation-insensitive isoschizomer (MspI) fragments were co-hybridized onto a customized high density arrays (28). Recently, instead of arrays, the massively parallel sequencing method is used to quantify fragments, which has significantly improved the sensitivity of the assay (29).
The classic method to study DNA methylation is based on bisulphite treatment of the DNA, which converts cytosine to uracil unless the base is methylated thereby differentiating methylated from unmethylated DNA (30). The converted product can be read by sequencing. The main limitation of the assay is the cost of sequencing, although emerging very high-throughput sequencing technologies may bring the cost down in future. Bisulphite sequencing is usually the method of choice for confirming genome-wide analytical studies and for the description of targeted methylation changes.
A third approach used to identify DNA methylation is based on the use of anti-methylcytosine antibodies or methyl binding proteins to isolate methylated DNA. The method, termed MeDIP, uses denatured genomic DNA of a desired fragment length (generated by restriction or sonication) that is incubated with an antibody directed against 5-methyl-cytosine (α-5mC), and methylated DNA is isolated by immunoprecipitation (IP). Enrichment of target sequences in the methylated fraction can be quantified by standard DNA detection methods such as PCR, microarrays or sequencing in order to investigate methylation changes (31).
HISTONE MODIFICATION
Modification of histones are most commonly detected by chromatin immunoprecipitation (ChIP). This method is based on cross linking of DNA and histones and using highly selective antibodies that detect modified histones(23, 27). Two slightly different approaches are commonly used in ChIP, Native-ChIP (N-ChIP) and Cross-Linking ChIP (X-ChIP). The primary difference between the N-ChIP and X-ChIP protocols is the preliminary processing step. N-ChIP protocols typically employ a micrococcal nuclease digestion to fragment chromatin, whereas X-ChIP protocols utilize formaldehyde cross-linking to stabilize protein-DNA interactions prior to sonication to fragment chromatin and immunoprecipitation to pull down the specific complexes. ChIP on Chip, the common term for applying Chromosome Immunoprecipitation to microarrays to study various histone modifications. The drawback of ChIP-chip is the inability to analyze repetitive elements as their inclusion in the array will interfere with the hybridization. In addition, bias may be introduced when amplification is used to generate large amount of DNA. Recently with the introduction of massively parallel sequencing equipments and the decreasing cost of sequencing, ChIP is being used more frequently with sequencing based methods (ChIP-Seq). The advantage of ChIP-Seq is the unbiased approach, the relative ease of analysis, and the fact that exact sequence of the pulled down DNA is known.
CANCER EPIGENOMICS
Epigenetic modifications associated with cancer have been studied extensively. Global hypomethylation was the first epigenetic change that was described in cancer cells. Most recent studies indicate that cancer involves both global and gene-specific hypomethylation and hypermethylation, as well as widespread chromatin modifications (32, 33).
For example in tumors, a global hypomethylation of the genome causes chromosome instabilities and transcriptional activation of oncogenes and prometastatic genes such as r-ras and this process has been suggested to initiate oncogenesis (34). It has been shown that many growth-promoting genes are activated through hypomethylation in tumours, such as carbonic anhydrase IX in renal-cell cancer, and S100 calcium-binding protein A4 in colon cancer (35, 36).
On the other hand, hypermethylation of CpG islands in the promoter region of a tumor suppressor is often associated with gene silencing. Some candidate genes linked to promoter specific hypermethylation include Rb, the gene associated with retinoblastoma, p16, VHL (von Hippel–Lindau), MLH1, APC (adenomatosis polyposis coli) and E-cadherin (33, 37, 38).
Studies have also shown that chromatin and histone alterations play important role in cancer development. For example, histone alterations leading to gene silencing have been found in many cancer types such as prostate, colorectal, and lymphomas (39, 40). These studies clearly establish the causative role that epigenetic modification play in regulating gene expression and phenotype development (Figure 2).
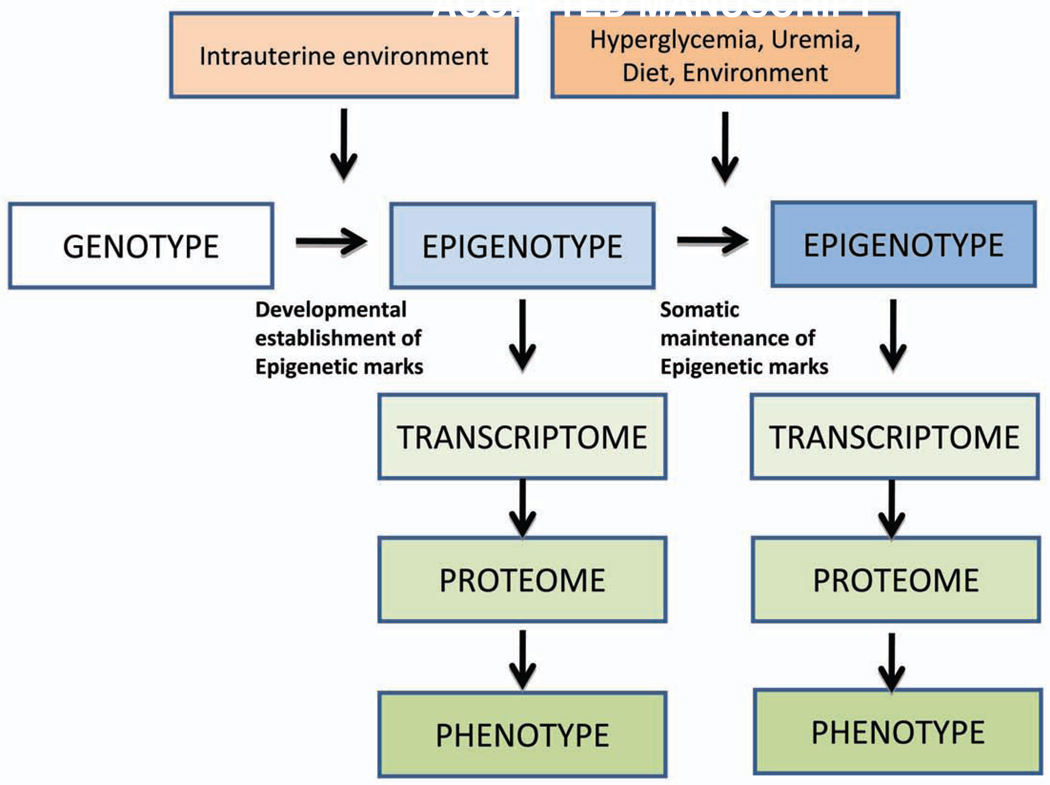
Figure 2. The complex Interaction between Genotype and Phenotype Development.
During normal development somatic cells that descended from a single progenitor, and contain similar genotype, differentiate to acquire diverse biological function by expressing and repressing different set of genes via establishing new epigenetic marks. While the genotype of an individual does not change hyperglycemia, uremia, different dietary and environmental factors might change the epigenome of cells leading to differences in gene and protein expression. Differences in the epigenotype might be response for the different (including disease) phenotype development. While the genotype is stable there is a more dynamic link between environmental factors and phenotype development.
CHALLENGES AND ADDITIONAL CONSIDERATIONS
While the current methodology is sufficient for conducting genome-wide screens of much of the epigenetic information, there are number of issues to be considered. Over the last couple of years research in the area of epigenetics has experienced and exponential growth, and a number of new assays and methods have been developed to describe epigenetic marks, however we still need a better understanding comparing the currently available different epigenenomic assays (for example for methylome analysis). New technologies are also needed to study higher order chromatin organization and function.
At present more than 20 different epigenetic marks have been identified, and we are just about to understand the functional consequences of the different epigenetic marks. It is not clear whether some marks are functionally more relevant and important than others. It is also very likely that we have not yet identified all the different epigenetic marks and we expect that new marks might be discovered in the near future.
In contrast to the DNA sequence, the epigenome varies among different normal cell types and between normal and diseased cells (Figure 2). Therefore selection of reference cells and reference tissues for analysis in the epigenome must be performed with extreme care. It is thereby appears to be critical to use the cell/organ of interest (i.e. the kidney) for the epigenomic studies. To deal with the problem of cell heterogeneity, microdissection will be essential in preparing the samples. Significant progress has been made in this area, largely through gene expression profiling and immunophenotyping, and this information needs to be taken into account in making this critical decision on sample selection(41). It would be a disastrous waste of resources to analyze the epigenome in tissue samples that subsequently prove to be poorly defined and heterogeneous. Therefore to study renal epigenomics it is essential to analyze and define marks in microdissected kidney tissue (or cells) as epigenetic marks are fundamentally different in different cells and tissues. In addition, due to the dramatic species differences it is probably also critical to analyze human kidney tissue(42).
Integration of epigenetic information into existing genome databases is essential. Computational algorithms that incorporate all levels of epigenetic information for each stretch of DNA sequence in the genome would be optimal. However, computational and statistical analysis of epigenomics data is challenging.
INTEGRATION OF EPIGENETIC STUDIES OPPORTUNITIES FOR SYSTEMS BIOLOGY
Epigenomic data promises a number of unique contributions to the study of genomic sciences and systems biology. First, epigenetic information is inherently multiplex, with hundreds of potentially methylated cytosines in a gene and dozens of known post-translational modifications of chromatin. As we discussed it also appears that the different epigenetic marks are not fully independent. Secondly, epigenetic information is quantitative, in contrast to the sequence itself, which is discrete. Tissues can maintain partial methylation at a locus, and the extent of methylation at affected sites can vary. Chromatin modifications are also inherently quantitative due to the opposing action of gene-activating (trithorax proteins) and gene-silencing (polycomb) complexes. Thirdly, epigenetics may aid in understanding the function of DNA regulatory sequence within the mammalian genome. Indeed, there is more constrained non-coding than coding sequence in most complex genomes, and much of this is methylated(16). Finally, the topological conformation of DNA within the nucleus is thought to be epigenetically controlled, and emerging studies suggest that its arrangement is important in gene regulation.
Epigenomics information adds an important layer of information that is not captured by genetics and genomics. While gene expression studies have had a great impact in the study of many different diseases including kidney disease, it is important to recognize that there are limitations associated with this technique. Gene expression studies capture a snapshot of the cell’s transcriptome, detecting genes being actively transcribed at the time of RNA extraction, but they do not capture information about the cell’s potential transcriptional response to stimuli and the genes’ regulatory state. For example by performing gene expression on developing and disease kidney samples we might find that the same or similar signaling pathways being regulated i.e. common regulation of Notch and wnt/beta catenin pathway (43, 44) (Figure 1). However, the functional targets of these pathways are fundamentally different in plastic/developing or mature/terminally differentiated cells. Epigenetic modifications might be responsible for these fundamental differences in cellular response. Therefore it is critically important to capture epigenetic information for systems biology studies as in addition to genetic variation, epigenetics provides an added layer of variation.
With the recent advances in high-throughput technologies, the need for an integrative approach using bioinformatics, computation, and statistical analysis is immense. Deciphering the epigenetic mechanisms and integrating the information obtained from genetic, genomic, and functional genomic studies will enhance our understanding of the mechanisms underlying the complex human diseases. The integration and analysis of the diverse information generated by systems biology studies will present new ways of prognosis, diagnosis, and treatments of complex human diseases such as chronic kidney disease.
SUMMARY
There are ongoing studies to characterize the role of epigenetic changes in human complex diseases. The National Institute of Health has a large program initiative to develop new epigenomic assays, generate reference epigenomic maps and describe disease specific epigenomic changes (epigenomics@nih.gov). Unlike the genetic sequence, the epigenome is under the influence of various environmental factors and the epigenome might mediate the relationship between genotype and internal and external environmental factors (45), (46) (Figure 2). Emerging evidence implicates that epigenetics plays role in the pathogenesis of chronic kidney disease which calls for the development of genome scale epigenomic and system biology studies.
Acknowledgement
We thank the members of Einstein Center for Epigenomics and the Susztak lab for the discussion. Our studies are supported by NIH 1R01DK087635 to K.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 5.Dressler GR. Epigenetics, development, and the kidney. J Am Soc Nephrol. 2008;19:2060–2067. doi: 10.1681/ASN.2008010119. [DOI] [PubMed] [Google Scholar]
- 6.Zandi-Nejad K, Luyckx VA, Brenner BM. Adult hypertension and kidney disease: the role of fetal programming. Hypertension. 2006;47:502–508. doi: 10.1161/01.HYP.0000198544.09909.1a. [DOI] [PubMed] [Google Scholar]
- 7.Luyckx VA, Brenner BM. Low birth weight, nephron number, and kidney disease. Kidney Int Suppl. 2005:S68–S77. doi: 10.1111/j.1523-1755.2005.09712.x. [DOI] [PubMed] [Google Scholar]
- 8.Keller G, Zimmer G, Mall G, Ritz E, et al. Nephron number in patients with primary hypertension. The New England journal of medicine. 2003;348:101–108. doi: 10.1056/NEJMoa020549. [DOI] [PubMed] [Google Scholar]
- 9.Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. The New England journal of medicine. 2000;342:381–389. doi: 10.1056/NEJM200002103420603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopes-Virella MF, Carter RE, Gilbert GE, Klein RL, et al. Risk factors related to inflammation and endothelial dysfunction in the DCCT/EDIC cohort and their relationship with nephropathy and macrovascular complications. Diabetes Care. 2008;31:2006–2012. doi: 10.2337/dc08-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Osta A, Brasacchio D, Yao D, Pocai A, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. The Journal of experimental medicine. 2008;205:2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, et al. Hyperglycemia induces a Dynamic Cooperativity of Histone Methylase and Demethylase Enzymes associated with Gene-Activating Epigenetic Marks that co-exist on the Lysine Tail. Diabetes. 2009 doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golestaneh L, Melamed ML, Hostetter TH. Uremic memory: the role of acute kidney injury in long-term outcomes. Kidney international. 2009;76:813–814. doi: 10.1038/ki.2009.314. [DOI] [PubMed] [Google Scholar]
- 14.Lo LJ, Go AS, Chertow GM, McCulloch CE, et al. Dialysis-requiring acute renal failure increases the risk of progressive chronic kidney disease. Kidney international. 2009;76:893–899. doi: 10.1038/ki.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. The EMBO journal. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nature reviews. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 17.Lister R, Pelizzola M, Dowen RH, Hawkins RD, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Current opinion in cell biology. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Agrelo R, Wutz A. X inactivation and disease. Seminars in cell & developmental biology. 2009 doi: 10.1016/j.semcdb.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science (New York, NY. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 21.Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochimica et biophysica acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenuwein T, Allis CD. Translating the histone code. Science (New York, NY. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 23.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 24.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. The international journal of biochemistry & cell biology. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes & development. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, Ku M, Jaffe DB, Issac B, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 28.Khulan B, Thompson RF, Ye K, Fazzari MJ, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda M, Glass JL, Thompson RF, Mo Y, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37:3829–3839. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber M, Davies JJ, Wittig D, Oakeley EJ, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 33.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 34.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 35.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochimica et biophysica acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Danesh FR, Kanwar YS. Modulatory effects of HMG-CoA reductase inhibitors in diabetic microangiopathy. Faseb J. 2004;18:805–815. doi: 10.1096/fj.03-0839rev. [DOI] [PubMed] [Google Scholar]
- 37.Greger V, Passarge E, Hopping W, Messmer E, et al. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 38.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 39.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 40.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 41.Kretzler M, Cohen CD, Doran P, Henger A, et al. Repuncturing the renal biopsy: strategies for molecular diagnosis in nephrology. J Am Soc Nephrol. 2002;13:1961–1972. doi: 10.1097/01.asn.0000020390.29418.70. [DOI] [PubMed] [Google Scholar]
- 42.Brosius FC, 3rd, Alpers CE, Bottinger EP, Breyer MD, et al. Mouse Models of Diabetic Nephropathy. J Am Soc Nephrol. 2009 doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niranjan T, Bielesz B, Gruenwald A, Ponda MP, et al. The Notch pathway in podocytes plays a role in the development of glomerular disease. Nature medicine. 2008;14:290–298. doi: 10.1038/nm1731. [DOI] [PubMed] [Google Scholar]
- 44.Si H, Banga RS, Kapitsinou P, Ramaiah M, et al. Human and Murine Kidneys show Gender- and Species-Specific Gene Expression Differences in Response to Injury. PLoS ONE. 2009 doi: 10.1371/journal.pone.0004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79:1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]