Summary
A puzzling observation in patients with oxidative phosphorylation (OXPHOS) deficiencies is the presence of combined enzyme complex defects associated with a genetic alteration in only one protein-coding gene. In particular, mutations in the mtDNA encoded cytochrome b gene are associated either with combined complex I+III deficiency or with only complex III deficiency. We have reproduced the combined complex I+III defect in mouse and human cultured cell models harboring cytochrome b mutations. In both, complex III assembly is impeded and causes a severe reduction in the amount of complex I, not observed when complex III activity was pharmacologically inhibited. Metabolic labeling in mouse cells revealed that complex I was assembled, although its stability was severely hampered. Conversely, complex III stability was not influenced by the absence of complex I. This structural dependence among complexes I and III was confirmed in a muscle biopsy of a patient harboring a nonsense cytochrome b mutation.
Introduction
Mammalian mitochondrial DNA (mtDNA) is the only cytoplasmic-located chromosome. It contains the information for 37 genes (including 2 ribosomal RNAs and 22 tRNAs) all devoted to the synthesis of 13 peptides. These are needed to build, in coordination with a far larger number of proteins encoded in nuclear chromosomes, four out of the five multiprotein complexes that constitute the oxidative phosphorylation (OXPHOS) system. Three of these complexes, I, III, and IV, are responsible for the transfer of electrons from NADH to molecular oxygen to form water. The released energy allows the generation of a proton gradient throughout the mitochondrial inner membrane, which drives the synthesis of ATP by the H+-ATPase (complex V). Although all the protein components of the different complexes have been identified, it remains unknown how the synthesis of the OXPHOS system is regulated and how the balance of the different complexes is achieved.
A source of information is the growing number of genetic defects affecting the OXPHOS system in humans, characterized in both nuclear and mtDNA genes. A particularity of many of the disease-causing mutations in OXPHOS genes is the variability in the phenotype manifestations at the symptomatic, biochemical, molecular, and genetic levels (DiMauro and Schon, 2003). In particular, the description of combined complex I + III defects in a subset of patients in which only genetic defects in complex III (ubiquinol cytochrome c reductase) could be established remains a puzzling observation (Andreu et al., 1999; Bruno et al., 2003; Lamantea et al., 2002). This phenomenon is not fully symmetric, as genetic alterations in mtDNA encoded complex I genes only cause an isolated complex I deficiency and have never been associated with combined complex I + III defects (DiMauro and Schon, 2003). Interestingly, a partial reduction of complex III and total loss of complex I activity associated with mutations in the nuclear encoded NDUFS4 complex I gene has been reported (Budde et al., 2000). The majority of the complex III genetic defects characterized to date are due to mutations in cytochrome b (Andreu et al., 1998, 1999; Keightley et al., 2000; Lamantea et al., 2002), the only mtDNA encoded gene for this complex and essential for its assembly (di Rago et al., 1993; Sidhu and Beattie, 1983).
To investigate the molecular cause of these observations we have used human and mouse cell models of complex III deficiency due to mutations in the cytochrome b gene. The mouse cell line harbors a missense mutation causing a E373K change at the carboxy terminal end of the protein and was generated and isolated in our laboratory by a novel methodology (unpublished data). The human cell line represents a unique case, since it is the only reported transmitochondrial cell line with defective CYT b (harboring a mtDNA 4-base micro-deletion) ever established from human patients (Rana et al., 2000). The human mutation causes the loss of cytochrome b and abolishes the assembly of complex III. In addition, the novel missense mutation in the mouse cytochrome b gene also impedes the assembly of complex III. Surprisingly, we have observed that the absence of assembled complex III in both the human and the mouse mutants causes a dramatic loss of complex I. We have obtained the same result in a muscle biopsy of a human patient harboring a nonsense (G142X) CYT b mutation (Bruno et al., 2003), confirming that the complex interdependence is also relevant in differentiated cells. In addition, we have investigated the consequences of the absence of complex I for the activity and assembly of complex III in a mouse cell line unable to assemble complex I due to the lack of the ND6 subunit (Acin-Perez et al., 2003; Bai and Attardi, 1998; Bayona-Bafaluy et al., 2003). We provide compelling evidence that demonstrates the requirement of fully assembled complex III, functional or not, for the stabilization of complex I. Therefore, we can provide a molecular mechanism for the occurrence of either combined complex I + III defects or isolated complex III defect induced by mutations in the cytochrome b gene. Conversely, we observed that in cultured cells the assembly and activity of complex III is not significantly affected by the lack of complex I.
Results
Isolation of a Complex III-Deficient Mouse Cell Line
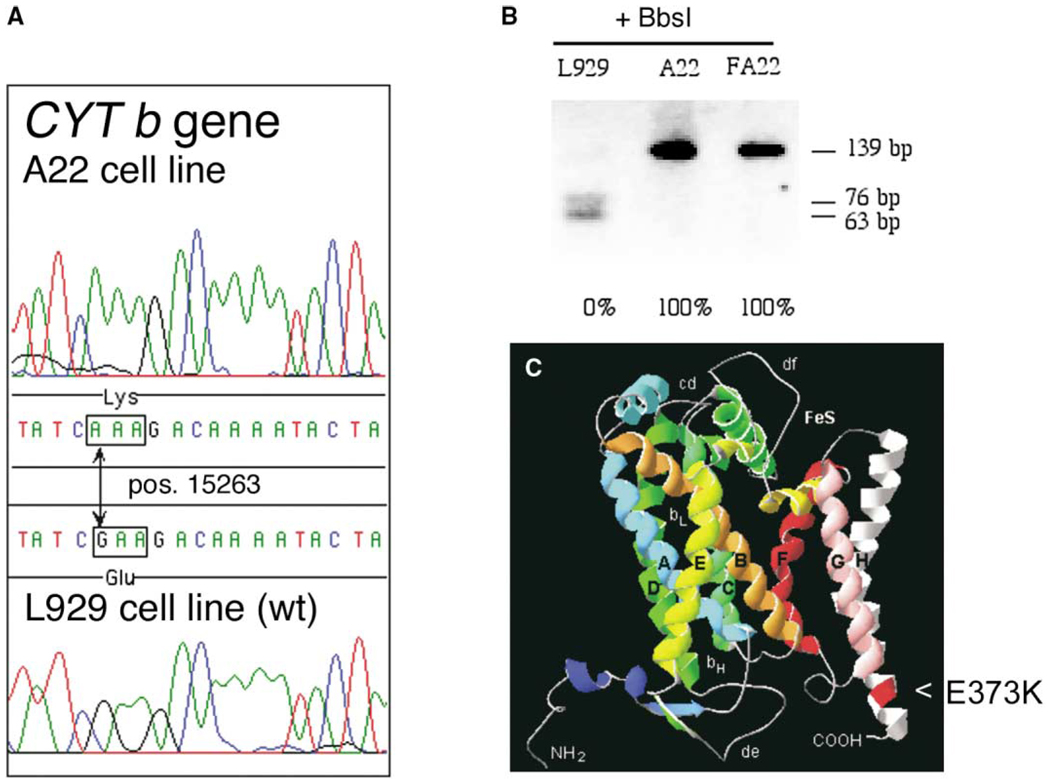
Chemical mutagenesis was used to randomly induce mutations in mouse L929 cells. Then we applied a protocol (see Experimental Procedures) to specifically select cells potentially mutated in complex III or complex IV mtDNA encoded genes. In that way a cell clone, named A22, was isolated and its biochemical analysis allowed us to conclude that complex IV was normally active whereas complex III activity was virtually undetectable (see below). Since random mutagenesis was used in the generation of the cell line, the cause of the OXPHOS deficiency could be due to alterations in either mtDNA or in nuclear encoded genes. By mitochondrial transfer from A22 to a receptor cell line lacking mtDNA (thus generating atransmitochondrial clone called FA22) and fully sequencing the A22 mtDNA, we concluded that this complex III defect was due to a single base substitution within the CYT b gene, which could be potentially deleterious: a G15263A transition that mutates the E373 to K at the carboxy terminus of the CYT b protein (see comment 1 in Experimental Procedures). This mutation was neither detected in L929 (Figure 1A) nor in three A22 isogenic control cell lines (see comment 2 in Experimental Procedures): E9, E14, or E18 mtDNAs (not shown). RFLP analysis of the G15263A mutation revealed that it was present in homoplasmic form both in A22 and in FA22 (Figure 1B). Therefore, the respiration defect observed in this cell line seems to be due to a malfunction of complex III as a consequence of the mutation found in the cytochrome b gene (Figures 2A to 2E). This mutation can also explain why A22 and FA22 become auxotrophic for uridine, since pyrimidin synthesis requires the activity of the enzyme dihydroorotate dehydrogenase (EC 1.3.99.11) that transfers electrons to ubiquinone and requires functional complexes III and IV to be recycled.
Figure 1. Characterization of the Cytochrome b Mutation.
(A) Chromatogram showing the G15263A transition within the CYT b gene of A22 cells.
(B) RFLP analysis of the presence and homoplasmic status for the G15263A transition that disrupts a restriction site for BbsI.
(C) Structural model of the mouse CYT b protein derived by analogy with the bovine crystallographic data (Tsukihara et al., 1996). The E373K substitution is located at the helix H close to the carboxy terminus end of the protein.
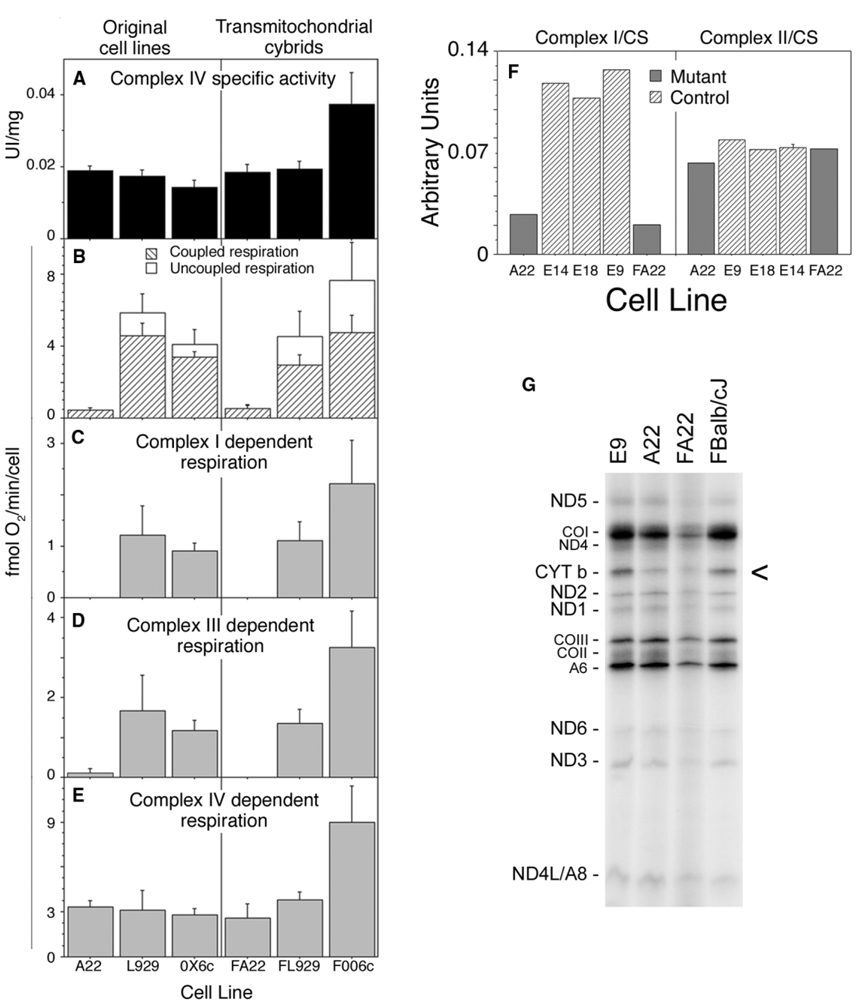
Figure 2. Functional Analysis of OXPHOS Performance and Mitochondrial Protein Synthesis.
Spectrophotometry measurements of complex IV activity in total cell extracts (A), or oxygraphic measurement of overall respiration (B), glutamate plus malate-dependent respiration (C), succinate plus glycerol-3-phosphate-dependent respiration (D), or TMPD (N,N,N′,N′-TetraMethyl-p-PhenyleneDiamine)-dependent respiration (E) of the original cell clones (left) and their transmitochondrial cell lines (right). OX6c: average values of cell lines E9, E14, and E18 isogenic with A22 but without the mutation in CYT b. FOO6c: average values of the transmitochondrial control cell lines FBalb/cJ, FC57BL/6J, and FCBA/J generated as described in Experimental Procedures. (F) Spectroscopic measurement of isolated complex I and II in mutant (A22, FA22) and control cells (E9, E14, E18) normalized by citrate synthase activity (values for Complex I-specific activities were in UI/g: E9 = 89.31 ± 11; E14 = 84.85 ± 16.9; E18 = 71.54 ± 3.9; A22 = 13.43 ± 2.7 and FA22 = 10.81 ± 2.4 and for Complex II: E9 = 91.5 ± 15; E14 = 83.5 ± 12; E18 = 89.8 ± 17; A22 = 82.8 ± 18 and FA22 = 79.2 ± 15.4). (G) Fluorogram, after electrophoresis through an SDS-polyacrylamide gradient gel, of the mitochondrial translation products of the indicated mutant and wild-type cells, labeled with [35S]-methionine for 1 hr in the presence of emetine. COI, COII, and COIII, subunits I, II, and III of cytochrome c oxidase; ND1 ND2, ND3, ND4, ND4L, ND5, and ND6, subunits 1, 2, 3, 4, 4L, 5, and 6 of the respiratory chain NADH dehydrogenase; A6 and A8, subunits 6 and 8 of H+-ATPase; CYT b, apocytochrome b.
Complex I but Not Complex II Activity Is Severely Reduced in A22 and FA22 Cells
Since we had a missense mutation in CYT b that abolished complex III activity, we investigated if complex I activity was affected by the CYT b mutation to discern which of the phenotypes observed in human patients was mimicked in A22 cells. For that purpose we measured independently the activity of complexes I and II in isolated mitochondria normalizing by citrate synthase activity. Complex I activity was severely impaired in both A22 and FA22 cells whereas isolated complex II activity was not significantly different from control cells (Figure 2F). This observation confirmed that we had reproduced the biochemical phenotype observed in those patients where the loss of function in complex III causes an associated defect in complex I.
Complex III Is Not Assembled in A22 Cells
Although the role of the C-terminal helix of cytochrome b where the mutation is located is poorly understood, the high degree of conservation of E373 pointed to a relevant structural role of this residue in the proper folding of the peptide (Iwata et al., 1998) (Figure 1C). Moreover, in vivo pulse label experiments of mtDNA-encoded proteins revealed that CYT b is indeed synthesized in the mutant cells but at a lower rate than in controls (Figure 2G). We hypothesized that this could be due to a higher rate of degradation because it is not properly folded and cannot be incorporated into assembled complexes.
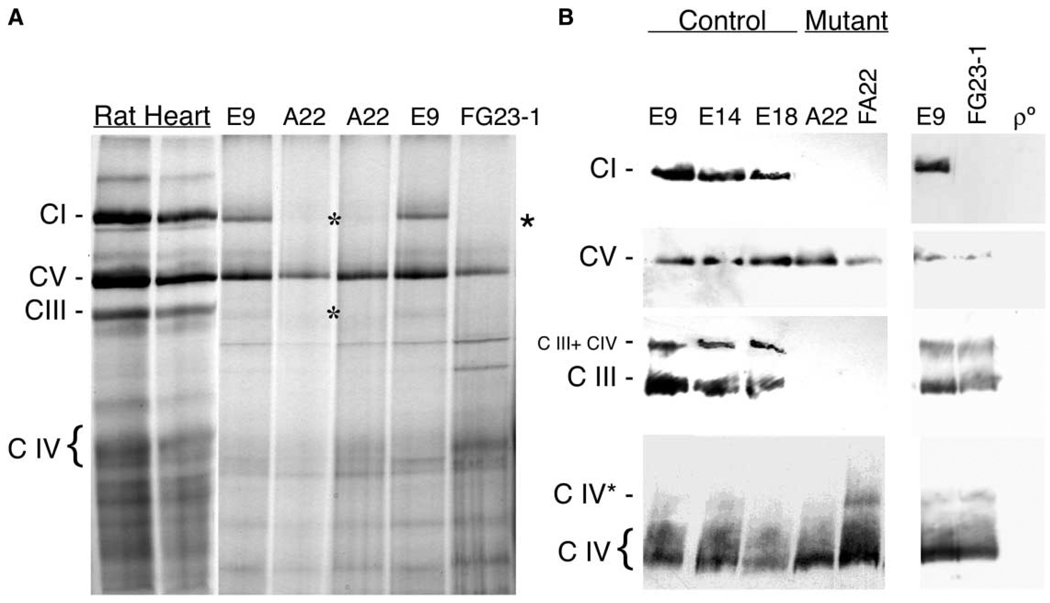
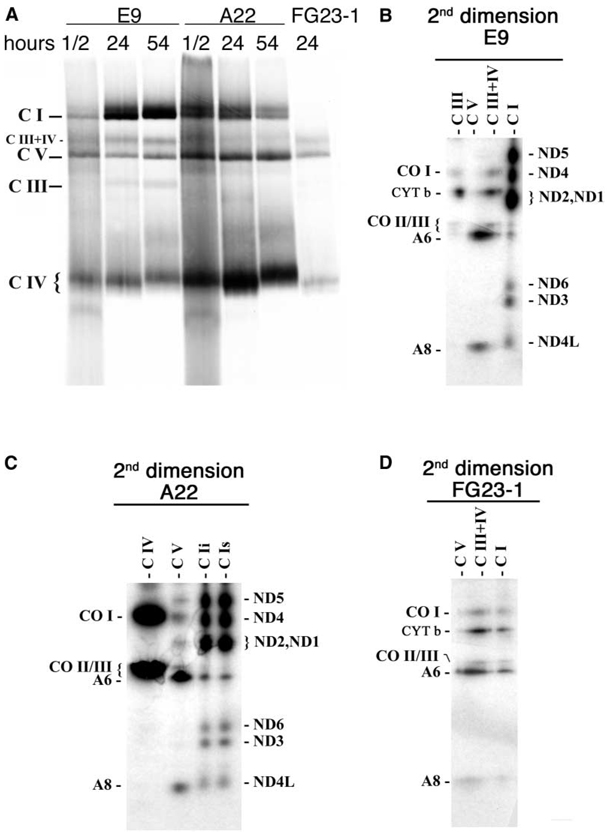
In order to test if assembly of the cytochrome bc1 complex was achieved in the mutant cell line, we performed Blue-Native gel electrophoresis (BNGE), which allows the visualization of the assembled respiratory complexes (Schägger, 1996). The identity of the different complexes was confirmed by metabolic labeling (see Experimental Procedures for an extended description) of mtDNA encoded proteins and second dimension denaturing electrophoresis (see below) and by Western blotting of the assembled complexes I, III, IV, and V, using specific antibodies (Figure 3B). The antibody against the complex III core 2 protein could detect two main assembled complexes in control cells, one that represents the complex III alone and the other, of higher molecular weight, that may represent the association of complex III and IV. Interestingly, none of the complex III-containing bands could be observed either in A22 or in FA22 cells even after overexposing the blots, confirming that complex III is not assembled in cells carrying the cytochrome b E373K mutation (Figures 3A and 3B).
Figure 3. Analysis of the Assembly Status of the Different OXPHOS Complexes.
(A) Blue-Native gel electrophoresis (BNGE) of the mitocondrial OXPHOS complexes in E9 and A22 cells using rat heart mitochondria as a control and showing also the ND6-deficient cell line FG23-1.
(B) Western blot of the different assembled complexes probed with anti NDUFS3 for complex I (C I), anti β F1-ATPase for complex V (C V), anti-Core 2 for complex III (C III), anti-CO I for complex IV (C IV), in the indicated control (E9, E14, E18), cytochrome b mutant (A22, FA22), ND6 mutant (FG23-1), and mtDNA-less cells (ρ°). CI, complex I, C III, complex III, C IV, complex IV, C III + IV, combined complex III + IV, CIV* probable complex IV dimer. Asterisks indicate the absence of CI or CIII in A22 and FG23-1 cells, respectively.
Steady State Levels of Assembled Complex I in A22 and FA22 Cells Are Significantly Reduced
Interestingly, BNGE also showed no complex I in samples obtained from A22 cells (Figure 3A). In addition, Western blots with antibodies against the nuclear encoded NDUFS3 protein also failed to detect complex I in these samples (Figure 3B). Only after loading a large amount of mitochondrial proteins and overexposing the blots, assembled complex I could be barely detected in A22 samples (not shown). This observation suggests an interdependence in assembly or stability between complex III and complex I. Since FA22 showed the same phenotype (Figure 3B), the implication of nuclear genes in the complex I defect can be excluded.
In order to reveal quantitative changes in the steady state of complex I and III proteins, we used total protein obtained from mouse cultured cells and subjected to SDS/PAGE followed by Western blot with antibodies against different respiratory complexes and, in some cases, against actin. This procedure can detect the alteration in complex I steady state levels both in cells lacking assembled complex I due to a frameshift mutation in the mtDNA encoded ND6 gene (FG23-1 cells), and in cells lacking both complex III and complex I (A22 and FA22 cells) (Table 1).
Table 1.
Relative Amount of Respiratory Complexes Estimated by SDS-PAGE/Western Blot
| Mouse | Human | ||||||
|---|---|---|---|---|---|---|---|
| Cell Line | Cell Line | Muscle Biopsy | |||||
| Control | CIII mutant | CI mutant | Control | CIII mutant | Control | Patient | |
| CI/CIV | 100 ± 3.63 | 16.41 ± 1.45* | 5.41 ± 0.82* | 100 ± 36.5 | 29 ± 10* | 100 | 25.5 |
| CII/CIV | – | – | – | 100 ± 35.6 | 171 ± 19.9 | 100 | 92.3 |
| CIII/CIV | 100 ± 3.63 | 6.15 ± 0.00* | 88.72 + 8.2 | 100 ± 34.4 | 3.6 ± 2.3* | 100 | 15.5 |
| CV/CIV | 100 ± 10.66 | 91.51 ± 1.88 | 85.29 + 5.3 | – | – | 100 | 176 |
| CI/actin | – | – | – | – | – | 100 | 11 |
| CII/actin | 100 ± 3.53 | 102 ± 19.20 | – | – | – | 100 | 39 |
| CIII/actin | 100 ± 4.20 | 10 ± 0.50* | – | – | – | 100 | 6.6 |
| CIV/actin | 100 ± 5.60 | 89 ± 7.10 | – | – | – | 100 | 43.0 |
| CV/actin | – | – | – | – | – | 100 | 75.0 |
p < 0.05.
Data from mouse cells are the average of E9 and E18 cell lines (control), A22 and FA22 (CIII mutant) and FG23-1 (CI mutant). Data from human cells are the average of 143B and 4.1 cell line (control) and 3E and 3B (mutant).
Complex I Assembly Is Not Affected by Pharmacological Inhibition of Complex III Activity
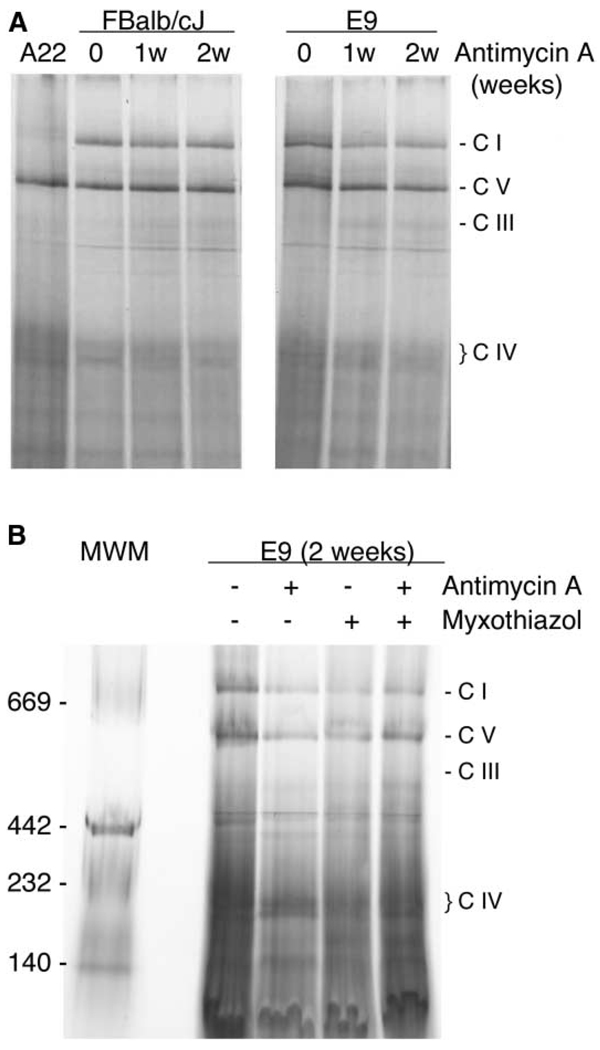
In order to establish whether the lack of function or the lack of assembled complex III triggered the reduction of assembled complex I, we treated two control cell lines (E9 and FBalb/cJ) with a complex III-specific inhibitor (antimycin A) and then followed the fate of assembled complex I and III over 2 weeks. Cells treated with antimycin A became immediately unable to grow in a galactose-containing medium. However, they were unexpectedly uridine independent (see comment 3 in Experimental Procedures). 1 or 2 weeks of continuous treatment with antimycin A did not have a significant effect in the level of assembled complex III or complex I in FBalb/cJ or E9 control cells (Figure 4A). Moreover, the use of myxothiazol alone or myxothiazol plus anti-mycin A immediately renders the cells uridine dependent and does not significantly affect the level of assembled complex I after 2 weeks of treatment (Figure 4B). Together, these results indicate that it is the physical absence of complex III rather than the lack of its activity that triggers the catastrophic loss of complex I.
Figure 4. The Absence of Complex III Activity Does Not Influence the Assembly of Complex I.
(A) Blue-Native gel electrophoresis showing the assembly status of the OXPHOS complexes in two different control cell lines after one or two weeks of treatment with antimycin A.
(B) Blue-Native gel electrophoresis showing the assembly status of the OXPHOS complexes in control cells treated for 2 weeks with the indicated complex III inhibitors.
Complex I Is Assembled in the Absence of Complex III but It Is Unstable
It remained to be determined which step of complex I biogenesis is influenced by the absence of complex III. In other words, does complex I assembly require the presence of complex III or is the assembled complex I stabilized by complex III? To address this question we selectively labeled in vivo the mtDNA encoded OXPHOS subunits by giving the cells a pulse with 35S-methionine in the presence of cycloheximide for 1 hr. Then the drug and the label were removed and the incorporation of the labeled proteins in fully assembled complexes was followed at different chase periods (Figure 5A). To confirm that only mtDNA-encoded proteins were responsible for the signal of the different complexes, the labeled bands were cut out of the first dimension gel and run on a denaturing second dimension SDS-PAGE to disaggregate the complexes into their protein components (Figures 5B to 5D). As shown, only mtDNA-encoded proteins were radioactively labeled and the components of each band in the first dimension could be unequivocally identified. Thus, by comparing the pattern of labeling of control (E9) and mutant (A22) cells at a very short chase time (half an hour), it could be established that all respiratory complexes but complex III were detected in A22 cells. This demonstrates that the primary effect of the CYT b mutation is to prevent the proper folding of the protein and therefore the assembly of complex III (Figure 5C). It should be emphasized that A22 cells were able to assemble complex I. However, complex I was observed as two clearly distinct close-migrating bands, with the faster migrating one being much more prominent in control cells, but of similar intensity to the slower-migrating band in the A22 pattern (Figure 5A). The reason for this abnormal distribution of complex I in two bands in the mutant cells is unknown. More important, the behavior of complex I during the chase was quite different between control and mutant cells. Thus, while its proportion clearly increased in E9 control cells, it decreased in A22 cells (Figure 5A). Therefore, complex I is fully assembled in A22 cells but it is unstable, very likely due to the absence of assembled complex III.
Figure 5. Metabolic Labeling of the Assembled OXPHOS Complexes.
(A) Fluorogram, after Blue-Native electrophoresis of the mitochondrial translation products of mutant and wild-type cells, pulse-labeled with [35S]-methionine for 1 hr in the presence of cycloheximide and chased for the indicated number of hours.
(B to D) Fluorograms of bidimensional electrophoresis (Blue-Native electrophoresis followed by SDS-polyacrylamide gel electrophoresis), of the mitochondrial translation products of mutant and wild-type cells shown in (A) (24 hr chase). Nomenclature over the lines indicates the putative complex that is expected to migrate in the first dimension in the area of the gel taken for the second dimension regardless if the complex is observed or not. The double band observed in the area of complex I in the first dimension from A22 cells was analyzed separately by second dimension in (C): faster migrating band (C Ii) and slower migrating band (C Is). The remaining symbols are as indicated in Figure 4.
The Presence of Complex III Is Also Required for the Stability of Complex I in Human Cultured and Differentiated Cells
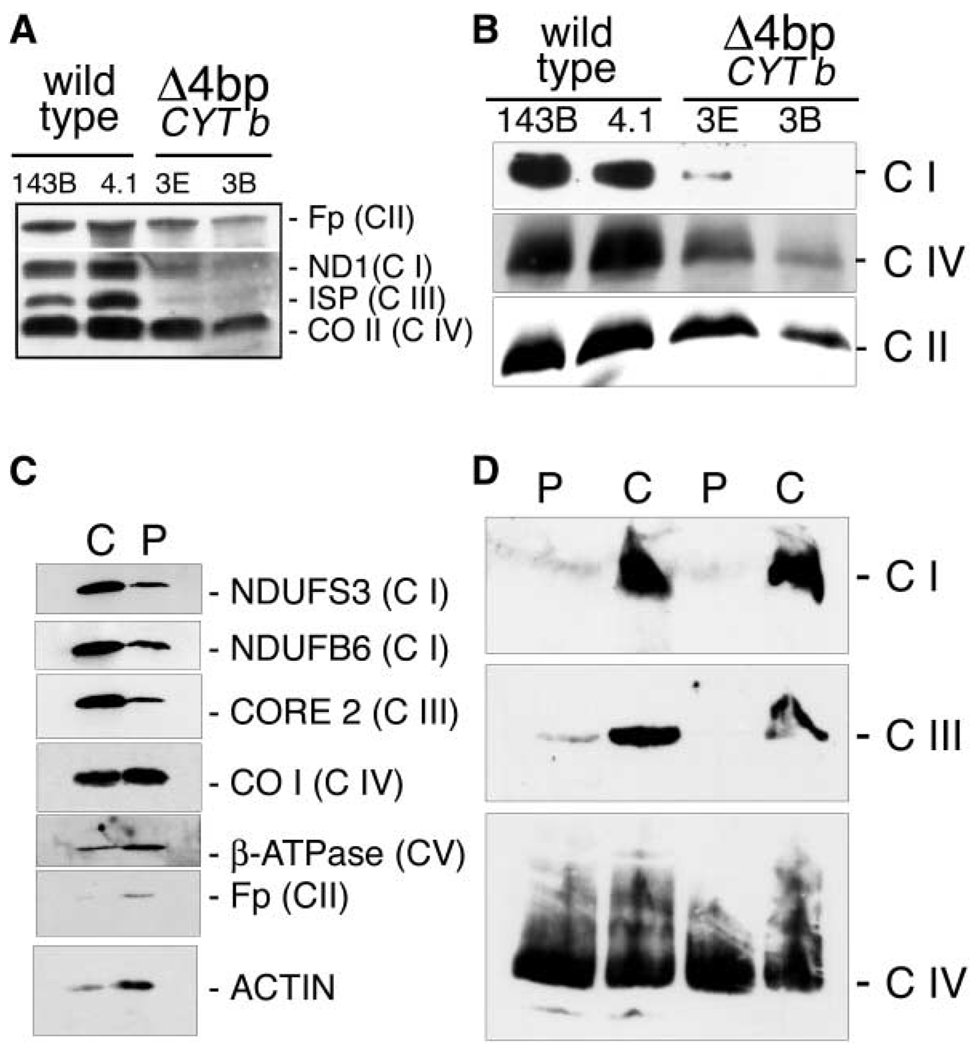
To determine whether the observed dependence of complex I stability upon the presence of complex III could be of significance in humans, we took advantage of the existence of a human transmitochondrial cell line that lacks cytochrome b and therefore does not assemble complex III (Rana et al., 2000). Since complex I is barely detected by direct staining of BNGE in samples from human cultured cells we had to determine it by BNGE followed by immunodetection. Complex I was drastically reduced in human cells lacking complex III (Figure 6B). In these cells, there was also a decrease in the levels of complex IV, but this decrease was by far less severe than the one observed for complex I.
Figure 6. Human Cells Lacking Cytochrome b Do Not Assemble Complex III and Show a Concomitant Defect in the Assembly of Complex I.
(A and C) Quantification by SDS-PAGE and Western blot of the relative level of proteins belonging to the indicated OXPHOS complex in total protein extracts from (A) human cultured control cells (143B and 4.1) and mutant cells (3E and 3B) and (C) from control (C) and patient (P) muscle biopsies. (B and D) Blue-Native gel electrophoresis of mitochondrial proteins isolated from the indicated human cultured cells (B) and muscle biopsies (D).
The reason for the very low level of assembled complex I in BNGE when using mitochondrial preparations from cultured human cells is unclear and prevents a reliable quantitative estimation of the Complex I decrease induced by the absence of complex III. To overcome this problem, we used SDS/PAGE followed by Western blot to reveal quantitative changes in the steady state levels of complex I and III proteins as we had previously determined for mouse cell lines.
We could confirm that human cells lacking cytochrome b (clones E3 and B3) showed a dramatic and significant reduction in the level of the nuclear encoded complex III protein ISP but also in the level of the mitochondrial encoded complex I protein ND1 (Figure 6A; Table 1). Therefore, we have confidently established that also in human cells the steady state level of complex I is strongly reduced in the absence of complex III.
We could have access to a muscle biopsy of a patient suffering from exercise intolerance (Bruno et al., 2003) that harbors a CYT b mutation restricted to muscle but in a high proportion (95%). The G15170A transition found in this patient is a nonsense mutation causing the transformation of the codon 142 from Gly to Stop and therefore impeding the assembly of complex III. Interestingly, analysis of muscle biopsies revealed that not only complex III but also complex I activity was severely reduced (Bruno et al., 2003). We performed Western blot analysis after BNGE or SDS-PAGE and demonstrated by the two approaches the concomitant loss of complex I and III in muscle cells (Figures 6C and 6D; Table 1). Thus, we have also confirmed, in differentiated muscle cells, the requirement of assembled Complex III for the maintenance of Complex I.
The Presence of Complex I Is Not Required for the Stability of Complex III
The observed complex I dependence on the assembled complex III prompted us to inquire if this was a reciprocal phenomenon, and to investigate if the absence of complex I could also compromise the stability of complex III. To address this question we took advantage of another mtDNA mutant cell line isolated in our laboratory, FG23-1. The mutant mtDNA of this cell line harbors an insertion of a C at the ND6 gene (ND613887iC) in more than 98% of its molecules. This mutation renders the cells incapable of assembling the mtDNA encoded subunits into complex I, as described elsewhere (Bai and Attardi, 1998). BNGE also revealed that complex I was not detected when the steady state level of respiratory complexes was investigated in our FG23-1 cells (Figures 3A and 3B). Moreover, metabolic labeling also failed to detect any level of fully assembled complex I in FG23-1 cells (Figures 5A and 5D) confirming that the presence of ND6 is required for the successful assembly of complex I. In agreement with the biochemical data (Acin-Perez et al., 2003), BNGE also revealed that complex III is assembled in FG23-1 cells (Figure 3). Therefore, the absence of complex I does not significantly impair the assembly and stability of complex III.
Discussion
In this investigation we have demonstrated that assembled complex III is required to stabilize the structure of complex I in mammalian cells. Support for the effect on complex I being exclusively due to the mutation in cytochrome b comes from the following observations. (1) mtDNA in the mouse cell line has been fully sequenced and it showed no mutations other than the CYT b E373K. (2) The replacement of the nucleus did not influence the phenotype. (3) The observation is consistent in three CYT b mutants belonging to two distant species and in cultured and differentiated cells. Support for the effect on complex I being due to the physical absence of complex III and not to the lack of activity comes from the observation that long-term inhibition of complex III activity by antimycin A, myxothiazol, or both did not significantly affect the assembly of complex I.
These findings have profound implications in our understanding of how the biogenesis of the OXPHOS system is regulated and of mtDNA linked diseases. Thus, our observation may explain why in some patients CYT b mutations are associated with an isolated complex III defect while in others they produce a combined complex I + III defect. If our proposal is correct, those mutations that disturb the assembly of complex III (type A mutations) would reveal a combined complex I + III defect. On the contrary, mutations that impair the function but not the assembly of complex III (type B mutations) would induce an isolated complex III defect. In addition, the degree of heteroplasmy and the distribution of wild-type and mutant mtDNAs within the cell and among organelles can further modulate the phenotype. Thus, the degree of heteroplasmy would influence the phenotype of the type A mutations between two extremes, isolated complex III and combined complex I + III defects. By the same token, type B mutations would cause only isolated complex III phenotypes independently of the degree of heteroplasmy. Support for our interpretation comes from the analysis of human patients harboring mutations in nuclear encoded genes that disturb complex III assembly (de Lonlay et al., 2001; Haut et al., 2003). Again, a concomitant reduction in complex I activity was documented when examined (Haut et al., 2003).
We have also confirmed that this structural dependency is not reciprocal, since, as it happens in N. crassa (Marques et al., 2003), the absence of complex I does not impair complex III biogenesis or activity. The latest observation is in agreement with the description of human patients with impaired complex I assembly but unaffected complex III biogenesis or function (Antonicka et al., 2003; Triepels et al., 2001). Therefore, the molecular cause of the induction of a complex III partial deficiency due to mutations in the nuclear encoded NDUFS4 complex I gene (Budde et al., 2000) should be of different nature than that described here and its characterization would require further investigation.
A question that remains to be solved is how the influence of complex III on complex I is exerted. Two main explanations can be envisioned, on one side the natural form of complex I in the cell could be forming superstructures among complexes and if this is not achieved, because complex III is absent, complex I is directed to degradation. Existence of physical association between respiratory complexes has been documented by BNGE (Schägger and Pfeiffer, 2000) and this attractive hypothesis would perfectly accommodate our findings. Alternatively, complex III may participate in a yet undetermined way in the maturation of complex I. In this respect, it has been proposed that core 1 and 2 subunits of complex III may act as a mitochondrial processing peptidase with unknown specificity (Deng et al., 2001). The lack of complex III assembly would impede the expression of the peptidase activity and its potential physiological role. More indirect mechanisms, such as an alteration in the generation of reactive oxygen species (ROS) induced by the physical loss of complex III or by the maintenance of an inactive complex are very unlikely. Thus, we have found no significant difference in the hydrogen peroxide production between control and mutant mouse cells, and, although the treatment of control cells with antimycin A causes an increase in H2O2 production, this does not reduce complex I stability (unpublished data).
In conclusion we have demonstrated the structural dependence between complex I and III and that assembled complex III is required to stabilize complex I in mammalian cells. In this way we have provided a molecular explanation for the variability in the biochemical phenotype induced by mutations in the cytochrome b gene in human patients.
Experimental Procedures
Cell Lines and Media
FBalb/cJ, E9, E14, E18, and FG23-1 cell lines were derived from L929 mouse cells as described elsewhere (Acin-Perez et al., 2003), and were grown in DMEM supplemented with 5% fetal bovine serum (FBS). E9, E14, and E18, called collectively OX6C in Figure 1, are true functional controls for A22 (see comment 2 in Experimental Procedures for a detailed description). FG23-1 harbors an insertion of one C in a track of 6Cs at the ND6 gene, ND613887iC, that introduces a frameshift starting from the 63rd amino acid, and creates a stop codon 51–53 base pairs downstream of the C stretch, resulting in a 79 amino acid-long truncated polypeptide, instead of the 172 amino acid-long full ND6 protein. This mutant was first described by Bai and coworkers (Bai and Attardi, 1998), and independently isolated in our laboratory (Acin-Perez et al., 2003). A22, FA22, and mtDNA-less mouse cells (ρ°929neor) (Tiranti et al., 1998) were grown in DMEM supplemented with 5% fetal bovine serum (FBS), 50 µg/ml of uridine, (ρ° medium) and, in the case of ρ°929neor and FA22, in the presence of 250 µg/ml of Geneticin (G418). The selective medium used for the isolation of mutants was LEIBOVITZ (GIBCO Brl) supplemented with 5% fetal bovine serum (FBS).
The human cell line 4.1 is homoplasmic for the wild-type mtDNA. The human 3.E and 3.B cell lines are homoplasmic for the Δ4-CYT b mutation (Rana et al., 2000). The human cell lines 143B, 4.1, 3.E, and 3.B were grown in DMEM supplemented with 10% FBS, 50 µg/ml of uridine, and 1 mM of sodium pyruvate.
Generation of Galactose Negative/Uridine-Dependent Cell Clones
L929 cells were grown in the presence of 30 µg/ml of TMP (Tri-Methyl Psoralen) for 2 days. Then the cells were irradiated with UV light (4,400*100 µJ/cm2). After irradiation, cells were collected and divided into three groups that were differently treated with EthBr (Ethidium Bromide). Cells of the group A were allowed to recover for 10 days in DMEM supplemented with uridine and pyruvate and then exposed for 11 days to 5 µg/ml of EthBr. Group B cells were exposed for 4 days to 5 µg/ml of EthBr immediately after UV irradiation. Then EthBr was removed for the following 4 days and the EthBr treatment was resumed for 7 more days. Group G cells were allowed to recover for 7 days in DMEM supplemented with uridine and pyruvate and then exposed for 5 days to 15 µg/ml of EthBr and 4 days more to 5 µg/ml of EthBr.
Clones were derived from single cells by harvesting the culture after the EthBr treatment, and plating them at a density of 0.6 to 0.8 cells per well in 96-well plates. In total, 604 clones from TMP/UV treated cells (154 from group A, 176 from group B, and 274 from group G) were isolated. Each of these clones was collected and plated again in replicated 96-well plates. One of the plates was fed with glucose-containing medium and the replica was fed with galactose-containing medium (Leibovitz medium). Those cells that showed no growth or extremely poor growth in galactose (Gal− clones) were then considered as potentially OXPHOS defective, picked up from the glucose-containing plate, and expanded for further analysis. In the experiments described above, Leibovitz medium was chosen for selective conditions because it seemed to provide the more restrictive growing conditions (L929 cells showed the longer doubling time). In addition, all the process was performed without freezing the cells at any stage.
Transfer of Mitochondria from Mutants into ρ°929neor Cell Lines
ρ°929neor cell transformation by cytoplast fusion was carried out as previously described (King and Attardi, 1989). The transmitochondrial cell lines were isolated by growing the cell population in DMEM supplemented with 5% dialyzed fetal bovine serum and 500 µg/ml of Geneticin (G418). The transmitochondrial cell lines obtained were designed with an F letter followed by the original name. An alternative methodology for the generation of transmitochondrial cybrids was employed when A22 cytoplasts were used. ρ°929neor were pretreated with a lethal dose of Rhodamine 6G, a drug that kills the cells unless fresh mitochondria are introduced in their cytoplasm (Moraes et al., 2001; Trounce and Wallace, 1996; Ziegler and Davidson, 1981), and rescued by fusion with A22 derived cytoplasts. Then, geneticin selection was applied to ensure that only cells containing the ρ°929neor nucleus were recovered.
Oxygen Consumption and Enzymatic Activity Measurements
O2 consumption determinations in intact cells or in digitonin-permeabilized cells were carried out in an oxygraph with a Clark electrode (Hansatech) as previously described (Hofhaus et al., 1996; Villani and Attardi, 1997) with small modifications (Acin-Perez et al., 2003). Preparation of mitochondrial fractions and spectrophotometric measurements of the activities of individual complexes were performed as described elsewhere (Birch-Machin and Turnbull, 2001).
Mitocondrial Protein Synthesis Analysis
Labeling of mtDNA encoded proteins was performed with [35S]-methionine in intact cells as described elsewhere (Chomyn, 1996).
DNA Analysis and mtDNA Sequencing
Total DNA was isolated from the different cell lines and sequenced as described elsewhere (Acin-Perez et al., 2003).
The CYT b G15263A mutation was confirmed by RFLP analysis. Thus, a 139 bp fragment containing this site was amplified by PCR with the following primers
Fw: 5′-TTGGCCAACTAGCCTCCATCTC-3′ (positions 15195 to 15216)
Rev: 5′-TTTCAGGTTTACAAGACCAGAG-3′ (positions 15312 to 15333)
The wild-type version generates a recognition site for BbsI while this restriction site is disrupted when the mutation G15263A is present. Fragments generated after the digestion of PCR products with BbsI were visualized by electrophoresis in 2% agarose gels containing 0.5 µg/ml ethidium bromide.
Individual and Assembled Protein Detection by Western Blot
Estimation of the relative level of the assembled respiratory complexes in control and mutant cell lines, as well as in muscle biopsies, was performed by Blue-Native electrophoresis (BNGE) according to Schägger et al. (1996). After electrophoresis, the complexes were electroblotted onto nylon filters and sequentially probed with specific antibodies against complex I (anti-NDUFS3, kindly gifted by Dr. R.A. Capaldi), complex III (anti-core 2, Molecular Probes), complex IV (anti-CO I, Molecular Probes), and complex V (anti β-F1-ATPase, kindly gifted by Dr. J.M. Cuezva). The assembled respiratory complexes in the human cell lines were probed with antibodies against complex I (anti-ND1 subunit), complex II (anti-Fp, 70 kDa subunit, Molecular Probes), and complex IV (anti-subunit II, Molecular Probes). The specific complexes were detected by the “ECL western blotting detection analysis system” from Amersham. The signal obtained with each antibody was quantified on autoradiography films using a laser densitometer.
Estimation of the steady state levels of proteins representative for the different respiratory chain complexes were performed by SDS-PAGE electrophoresis of total cell proteins. Thus, samples were run in a 12.5% acrylamide/bisacrylamide SDS-PAGE and electroblotted onto nylon filters. These filters were probed with specific antibodies: (A) Samples from mouse cells: against complex II (anti-Fp,70 kDa, Molecular Probes), complex III (anti-core 2, Molecular Probes), complex IV (anti-CO I, Molecular Probes) and actin (SIGMA). (B) Samples from human muscle biopsies were probed against complex I (NDUFS3 and NDUFB6, both from Molecular Probes), complex II (anti-Fp, 70 kDa, Molecular Probes), complex III (anti-core 2, Molecular Probes), complex IV (anti-CO I, Molecular Probes), complex V (anti-β-F1-ATPase, kindly gifted by Dr. J.M. Cuezva) and actin (SIGMA). (C) Samples from human cultured cells were sequentially probed with the antibodies against complex I (anti-ND1 subunit), complex II (anti-Fp, 70 kDa subunit, Molecular Probes), complex III (anti-ISP subunit), and complex IV (anti-subunit II, Molecular Probes).
When mitochondrial proteins were isolated and electrophoresed in conditions that keep the respiratory complexes assembled, the β-F1-ATPase antibody detects its targeted protein in a main band that corresponds to the fully assembled FO-F1-ATPase monomer (Figure 3B). In addition, a second band of lower molecular weight could be observed after overexposing the blots, which could correspond to F1-ATPase alone, as proposed elsewhere (Appleby et al., 1999), since it is the only band detected in ρ° cells (not shown). On the other hand, the CO I antibody is able to detect its targeted protein in a main high molecular weight complex that corresponds to the complex IV monomer, but also in two other bands of higher molecular weight, one that would likely contain the complex IV dimers (Schägger, 2001) and the other that may represent the association of complex IV with complex III (Figure 3B). As expected, the assembled complex IV was observed in all cell lines investigated but ρ° cells.
Generation of the Transmitochondrial Cell Line Defective in Complex III (FA22)
Usually, to discriminate between nuclear or mitochondrial origin of the genetic defect causing the OXPHOS impairment, the transmission of the respiration defect along with the mitochondria is investigated by transferring organelles to mtDNA-less cells (ρ° cells). For that purpose we have generated the ρ°929neor cell line (Acin-Perez et al., 2003; Tiranti et al., 1998). Selection in the absence of uridine to avoid the survival of the nonfused ρ°929neor is often applied in the protocols for mitochondrial transference. However, the use of this selection procedure would not allow the establishment of transmitochondrial cell lines carrying, in homoplasmic form, mtDNA mutations that completely abolish complex III or complex IV functionality. Therefore, only if the cause of the complex III defect in A22 were a mutation in a nuclear gene, the new nucleus would be able to compensate for it and the transmitochondrial cells could be selected in the absence of uridine. On the contrary, if the defect were due to a mutation in the mtDNA, the transference of mitochondria to ρ°929neor cells would not allow the generation of transmitochondrial cells by this methodology. In fact, this was the case for A22 cells. Repeated attempts to isolate transmitochondrial cybrids from this cell line were unsuccessful but, on the contrary, we could easily transfer mitochondria from L929 cells and from platelets of different strains of mice: Balb/cJ, C57BL/6J, and CBA/J (Acin-Perez et al., 2003).
To be able to test the mtDNA transmission of the OXPHOS defect together with A22 organelles, an alternative protocol for cybridization without selection against uridine dependence was performed. Thus, ρ°929neor cells were pretreated with a lethal dose of Rhodamine 6G, a drug that kills the cells unless mitochondria from untreated cells are introduced in their cytoplasm (Moraes et al., 2001; Trounce and Wallace, 1996; Ziegler and Davidson, 1981), and rescued by fusion with A22 derived cytoplasts. Then, geneticin selection was applied to ensure that only cells containing the ρ°929neor nucleus were recovered. In that way, transmitochondrial clones from A22 (designed FA22) could be generated.
Interestingly, FA22 cells were unable to grow in galactose, or in glucose-containing medium without uridine. Moreover, the ρ°929neor recovered normal respiration when transformed with mitochondria from the original L929 cell line (FL929) or from platelets of different mouse strains (FBalb/cJ, FC57BL/6J, and FCBA/J; collectively called FOO6C) (Figure 1). On the contrary, FA22 cells showed almost no respiration either coupled or uncoupled, or complex I- and complex III dependent-respiration, whereas complex IV-dependent respiration was normal (Figure 1). Altogether these results show that the respiratory defect observed in A22 cells was transmitted along with their mtDNA.
mtDNA Isogenic Controls for A22 Cells
We discovered after A22 isolation that L929 culture contained three abundant mtDNA haplotypes (Acin-Perez et al., 2003): the COIT6589C (mtDNA haplotype A), the COIT6589C/COIC6063A (mtDNA haplotype B), and the COIT6589C/COIC6063A/ND613887iC (mtDNA haplotype C). Since each of these single-base changes conferred differences in OXPHOS performance, we first identified that the haplotype A was the one selected during the generation of A22. Therefore, several cell lines derived from nonmutagenized L929 and carrying only the mtDNA haplotype A (E9, E14, and E18, called collectively OX6C in Figure 1) were utilized as true functional controls for A22. Thus E9, E14, and E18 were all capable of growing in the absence of uridine and in galactose- containing medium. Moreover, they respired at expected rates (3.4 ± 0.33 fmol/min/cell on average) and uncouplers significantly stimulated their respiration (4.11 ± 0.83 fmol/min/cell on average).
Pharmacological Inhibition of Complex III Activity
Antimycin A binds to the N side of cytochrome b near heme bH and it is believed to inhibit the activity of the complex by blocking electron transfer between bH and bL. It has been shown that antimycin A does not fully impede the reduction of cytochrome b (Matsuno-Yagi and Hatefi, 2001). Therefore, we performed the experiment also with another complex III inhibitor, myxothiazol, that binds to the P side of cytochrome b, between bL and ISP, and also with both inhibitors together, a condition that fully blocks the reduction of cytochrome b (Matsuno-Yagi and Hatefi, 2001). Myxothiazol alone and myxothiazol plus antimycin A treatments immediately render the cells uridine dependent, but in both cases complex I assembly was not significantly affected after 2 weeks of treatment (Figure 4B). Together, these results indicate that it is the physical absence rather than the lack of complex III activity that triggers the catastrophic loss of complex I.
Acknowledgments
We would like to thank Dr. J. Sancho and Dr. A.L. Andreu for their valuable input during the development of this work, Dr. R.A. Capaldi for the kind gift of the anti-NDUFS3 antibody, Dr. J.M. Cuezva for the kind gift of the anti-β-F1-ATPase, and Santiago Morales for technical assistance. Our research was supported by the Spanish Ministry of Education (PM-99-0082) and the Instituto de Salud Carlos III (REDEMETH G03/05) grants to J.A.E., by the Instituto de Salud Carlos III (REDCIEN C03/06-Grupo RC-N34-3) grant to A.P.-M., by the Ramón y Cajal 2001 grant to P.F.S., and by National Institutes of Health (USA) EY10804 and GM55766 grants (to C.T.M.). R.A.-P. and R.M.-L. are recipients of the Spanish F.P.U. and F.P.I. fellowships, respectively.
References
- Acin-Perez R, Bayona-Bafaluy MP, Bueno M, Machicado C, Fernandez-Silva P, Perez-Martos A, Montoya J, Lopez-Perez MJ, Sancho J, Enriquez JA. An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum. Mol. Genet. 2003;12:329–339. doi: 10.1093/hmg/ddg021. [DOI] [PubMed] [Google Scholar]
- Andreu AL, Bruno C, Shanske S, Shtilbans A, Hirano M, Krishna S, Hayward L, Systrom DS, Brown RH, Jr, Di-Mauro S. Missense mutation in the mtDNA cytochrome b gene in a patient with myopathy. Neurology. 1998;51:1444–1447. doi: 10.1212/wnl.51.5.1444. [DOI] [PubMed] [Google Scholar]
- Andreu AL, Hanna MG, Reichmann H, Bruno C, Penn AS, Tanji K, Pallotti F, Iwata S, Bonilla E, Lach B, et al. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N. Engl. J. Med. 1999;341:1037–1044. doi: 10.1056/NEJM199909303411404. [DOI] [PubMed] [Google Scholar]
- Antonicka H, Ogilvie I, Taivassalo T, Anitori RP, Haller RG, Vissing J, Kennaway NG, Shoubridge EA. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 2003;278:43081–43088. doi: 10.1074/jbc.M304998200. [DOI] [PubMed] [Google Scholar]
- Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- Bai Y, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- Bruno C, Santorelli FM, Assereto S, Tonoli E, Tessa A, Traverso M, Scapolan S, Bado M, Tedeschi S, Minetti C. Progressive exercise intolerance associated with a new muscle-restricted nonsense mutation (G142X) in the mitochondrial cytochrome b gene. Muscle Nerve. 2003;28:508–511. doi: 10.1002/mus.10429. [DOI] [PubMed] [Google Scholar]
- Budde SM, van den Heuvel LP, Janssen AJ, Smeets RJ, Buskens CA, DeMeirleir L, Van Coster R, Baethmann M, Voit T, Trijbels JM, et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 2000;275:63–68. doi: 10.1006/bbrc.2000.3257. [DOI] [PubMed] [Google Scholar]
- Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- de Lonlay P, Valnot I, Barrientos A, Gorbatyuk M, Tzagoloff A, Taanman JW, Benayoun E, Chretien D, Kadhom N, Lombes A, et al. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat. Genet. 2001;29:57–60. doi: 10.1038/ng706. [DOI] [PubMed] [Google Scholar]
- Deng K, Shenoy SK, Tso SC, Yu L, Yu CA. Reconstitution of mitochondrial processing peptidase from the core proteins (subunits I and II) of bovine heart mitochondrial cytochrome bc(1) complex. J. Biol. Chem. 2001;276:6499–6505. doi: 10.1074/jbc.M007128200. [DOI] [PubMed] [Google Scholar]
- di Rago JP, Macadre C, Lazowska J, Slonimski PP. The C-terminal domain of yeast cytochrome b is essential for a correct assembly of the mitochondrial cytochrome bc1 complex. FEBS Lett. 1993;328:153–158. doi: 10.1016/0014-5793(93)80984-3. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Haut S, Brivet M, Touati G, Rustin P, Lebon S, Garcia-Cazorla A, Saudubray JM, Boutron A, Legrand A, Slama A. A deletion in the human QP-C gene causes a complex III deficiency resulting in hypoglycaemia and lactic acidosis. Hum. Genet. 2003;113:118–122. doi: 10.1007/s00439-003-0946-0. [DOI] [PubMed] [Google Scholar]
- Hofhaus G, Shakeley RM, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 1996;264:476–483. doi: 10.1016/s0076-6879(96)64043-9. [DOI] [PubMed] [Google Scholar]
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- Keightley JA, Anitori R, Burton MD, Quan F, Buist NR, Kennaway NG. Mitochondrial encephalomyopathy and complex III deficiency associated with a stop-codon mutation in the cytochrome b gene. Am. J. Hum. Genet. 2000;67:1400–1410. doi: 10.1086/316900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: re-population with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Carrara F, Mariotti C, Morandi L, Tiranti V, Zeviani M. A novel nonsense mutation (Q352X) in the mitochondrial cytochrome b gene associated with a combined deficiency of complexes I and III. Neuromuscul. Disord. 2002;12:49–52. doi: 10.1016/s0960-8966(01)00244-9. [DOI] [PubMed] [Google Scholar]
- Marques I, Duarte M, Videira A. The 9.8 kDa subunit of complex I, related to bacterial Na(+)-translocating NADH dehydrogenases, is required for enzyme assembly and function in Neurospora crassa. J. Mol. Biol. 2003;329:283–290. doi: 10.1016/s0022-2836(03)00443-1. [DOI] [PubMed] [Google Scholar]
- Matsuno-Yagi A, Hatefi Y. Ubiquinol:cytochrome c oxidoreductase (complex III). Effect of inhibitors on cytochrome b reduction in submitochondrial particles and the role of ubiquinone in complex III. J. Biol. Chem. 2001;276:19006–19011. doi: 10.1074/jbc.M101446200. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Dey R, Barrientos A. Transmitochondrial technology in animal cells. Methods Cell Biol. 2001;65:397–412. doi: 10.1016/s0091-679x(01)65023-4. [DOI] [PubMed] [Google Scholar]
- Rana M, de Coo I, Diaz F, Smeets H, Moraes CT. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Ann. Neurol. 2000;48:774–781. [PubMed] [Google Scholar]
- Schägger H. Electrophoretic techniques for isolation and quantification of oxidative phosphorylation complexes from human tissues. Methods Enzymol. 1996;264:555–566. doi: 10.1016/s0076-6879(96)64048-8. [DOI] [PubMed] [Google Scholar]
- Schägger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52:119–128. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- Schägger H, Bentlage H, Ruitenbeek W, Pfeiffer K, Rotter S, Rother C, Bottcher-Purkl A, Lodemann E. Electrophoretic separation of multiprotein complexes from blood platelets and cell lines: technique for the analysis of diseases with defects in oxidative phosphorylation. Electrophoresis. 1996;17:709–714. doi: 10.1002/elps.1150170415. [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu A, Beattie DS. Kinetics of assembly of complex III into the yeast mitochondrial membrane. Evidence for a precursor to the iron-sulfur protein. J. Biol. Chem. 1983;258:10649–10656. [PubMed] [Google Scholar]
- Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triepels RH, Hanson BJ, van den Heuvel LP, Sundell L, Marusich MF, Smeitink JA, Capaldi RA. Human complex I defects can be resolved by monoclonal antibody analysis into distinct subunit assembly patterns. J. Biol. Chem. 2001;276:8892–8897. doi: 10.1074/jbc.M009903200. [DOI] [PubMed] [Google Scholar]
- Trounce I, Wallace DC. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somat. Cell Mol. Genet. 1996;22:81–85. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in wild-type and mitochondrial DNA mutation-carrying human cells. Proc. Natl. Acad. Sci. USA. 1997;94:1166–1171. doi: 10.1073/pnas.94.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler ML, Davidson RL. Elimination of mitochondrial elements and improved viability in hybrid cells. Somatic Cell Genet. 1981;7:73–88. doi: 10.1007/BF01544749. [DOI] [PubMed] [Google Scholar]