Abstract
We have identified a gene, DRhoGEF2, which encodes a putative guanine nucleotide exchange factor belonging to the Dbl family of oncogenes. DRhoGEF2 function is essential for the coordination of cell shape changes during gastrulation. In the absence of maternal DRhoGEF2 gene activity, mesodermal and endodermal primordia fail to invaginate. The phenotype seen in DRhoGEF2 mutants is more severe than the defects associated with mutations in two previously identified gastrulation genes, folded gastrulation and concertina, suggesting that DRhoGEF2 acts in a signaling pathway independent of these genes. Expression of dominant-negative DRhoA during gastrulation results in phenocopies of the DRhoGEF2 mutant, suggesting that a signaling cascade involving DRhoGEF2 and the small GTPase DRhoA is responsible for the regulation of cell shape changes during early Drosophila morphogenesis.
Keywords: Drosophila, RhoGEF, GTPase, Rho, gastrulation
The first outwardly visible sign of differentiation in all animals is the formation of the germ layers during gastrulation. In Drosophila, gastrulation is associated with a major morphological rearrangement of the embryo and begins with the invagination of the mesodermal primordium on the ventral side. Immediately after cellularization, the previously rounded apical surfaces of cells in an ∼20-cell-wide stripe flatten and cells move into close contact with each other. Slightly later, a shallow groove, the ventral furrow, forms along the ventral midline. The furrow deepens as more lateral cells become incorporated in it and eventually closes to complete invagination of the mesoderm. A few minutes after the onset of ventral furrow formation, cells of the posterior midgut primordium start to invaginate. The mechanism underlying these invaginations is based on a series of coordinated cell shape changes. By constricting apically, and displacing cell nuclei from the apical to the basal end, cells change their shapes from columnar to trapezoidal, thus forcing the cell sheet to bend inward. Subsequently, this process is enhanced by a shortening of the cells along their apical–basal axes resulting in complete internalization of the epithelium. The force driving these cell shape changes is thought to be generated by the actin/myosin network underlying the cell surface, and myosin has been shown to accumulate specifically in the apices of cells undergoing constriction (for review, see Leptin 1995; Sweeton et al. 1991).
Genetic analyses have identified a number of genes that are required for gastrulation to proceed normally. Formation of the ventral furrow has been shown to be under the control of the dorsal–ventral maternal system and its zygotic target genes twist (twi) and snail (sna) (Leptin and Grunewald 1990). In twi and sna mutant embryos, specific but distinct aspects of the cell shape changes observed in wild-type embryos do not occur. Consequently, both mutants form only a temporary ventral furrow and the mesoderm is not internalized. In twi, sna double mutant embryos almost no cell shape changes can be observed, and for this reason both genes have been suggested to control ventral furrow formation in a combinatorial fashion. Invagination of the posterior midgut (PMG) is controlled in a similar manner by the zygotic target genes of the terminal maternal system huckebein (hkb) and tailless (tll). In hkb,tll double mutant embryos, no posterior midgut invagination is formed.
All of the genes mentioned above do not only affect cell shape changes in invaginating tissues but are essential for the determination of mesodermal and endodermal cell fates, respectively (Leptin 1995). Two genes, folded gastrulation (fog) and concertina (cta), have been identified that are specifically involved in gastrulation. fog is a transcriptional target of twi, sna, hkb, and tll in the respective tissues and encodes a zygotically acting putative secreted protein that is expressed locally in the presumptive invaginating tissues (Costa et al. 1994). In contrast to fog, cta, which encodes a putative G-protein α-subunit, acts strictly maternally and cta mRNAs are distributed ubiquitously throughout the embryo (Parks and Wieschaus 1991). Mutations in both genes result in virtually identical phenotypes, which are most severe in the endoderm where invagination of the posterior midgut is blocked. In the mesoderm, the phenotype of fog and cta mutants is significantly less severe compared to twi or sna mutants. Although the induction of cell shape changes is delayed and coordination of the invagination process is disrupted, the mesoderm is eventually internalized.
The close resemblance of the phenotypes caused by fog and cta mutations, and the fact that both genes encode putative signaling molecules, has led to a model tentatively placing both genes in a common signal transduction pathway. Because of the nonessential function of fog and cta in the mesoderm, however, this pathway has been implicated in the coordination, rather than the control of cell shape changes during gastrulation (Costa et al. 1994).
We have identified a gene, named DRhoGEF2, which is essential for the invagination of mesodermal and endodermal primordia during gastrulation. Mutations in DRhoGEF2 disrupt the coordinated induction of cell shape changes. DRhoGEF2 encodes a putative guanine nucleotide exchange factor belonging to the Dbl family of oncoproteins, suggesting that a signaling cascade involving Rho family GTPases regulates cell shape changes during early Drosophila morphogenesis.
Results
Identification of DRhoGEF2 mutations
DRhoGEF2 was isolated from a large screen designed to characterize the maternal effects of zygotic lethal mutations (Perrimon et al. 1996). Examination of cuticle phenotypes of mutant lines and stainings of embryos derived from germ-line clones using tissue-specific markers led to the identification of a P-element insertion, l(2)04291 (Spradling et al. 1995) (referred to as DRhoGEF2l(2)04291), and three chemically induced alleles (see Materials and Methods) falling into the same complementation group. Embryos homozygous for DRhoGEF2l(2)04291 die during late embryogenesis or early larval stages with no obvious phenotypes. However, embryos derived from mothers homozygous mutant for DRhoGEF2l(2)04291 in the germ line (henceforth referred to as DRhoGEF2l(2)04291 embryos) develop with ventral open cuticles. Examination of these embryos revealed deep transverse folds forming during germ-band elongation (see Fig. 1L). This phenotype is reminiscent of mutants that fail to invaginate the endodermal germ layer and suggests that DRhoGEF2l(2)04291 embryos develop defects during gastrulation. Because all embryos show identical defects the observed phenotype depends solely on maternal contribution and cannot be rescued zygotically by a paternal wild-type chromosome. The zygotic lethality of the DRhoGEF2l(2)04291 mutation is most likely caused by reutilization of the corresponding gene during late embryonic and early larval development. The lethality and the maternal effect phenotype can be reverted by excision of the P element, suggesting that no other zygotic lethal mutations reside on the chromosome and the P-element insertion is responsible for the observed phenotype.
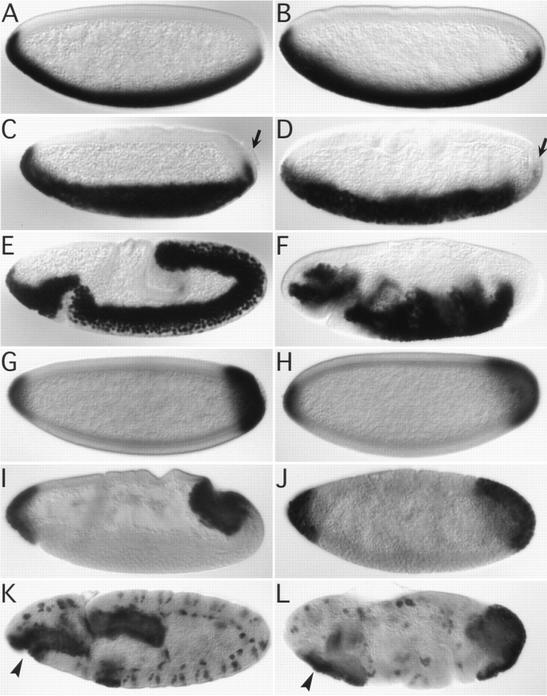
Figure 1.
Invagination of mesodermal and endodermal primordia in wild-type and DRhoGEF2 mutant embryos. All embryos are stained immunohistochemically for the transcription factors Twi (A–F) or Fkh (G–L). Expression of Twi is initiated in the wild-type (A) and DRhoGEF2l(2)04291 (B) embryos during syncytial blastoderm. At stage 6 the ventral furrow has invaginated in the wild type. The PMG primordium has flattened and moved to a dorsally shifted position (C). The pole cells are attached to the PMG primordium (arrow in C). In DRhoGEF2l(2)04291 embryos no ventral furrow is formed (embryo shown is at a slightly later stage than in C). The mesoderm is not extending to the posterior pole as in the wild type. The PMG primordium is not moving dorsally and the pole cells are deeply embedded into the epithelium (arrow in D). At stage 9 the mesoderm, with the exception of the most lateral cells of the primordium, has completely invaginated in the wild type. The germ band is extending around the posterior pole to the dorsal side of the embryo (E). In DRhoGEF2l(2)04291 embryos at the same stage the mesoderm remains on the surface of the embryo. The germ band is not moving around the posterior pole (F). Expression of Fkh is established in the PMG primordium at the late syncytial blastoderm stage in wild type (G) and DRhoGEF2l(2)04291 embryos (H). At stage seven the posterior midgut has started to invaginate in the wild type (I). In DRhoGEF2l(2)04291 embryos no invagination is formed at this stage. Note the folds forming in the dorsal epithelium indicating that the germ band has started to elongate (J). At stage 10 the PMG and the AMG (arrowhead) are fully internalized and have started to elongate in the wild type (K). In DRhoGEF2l(2)04291 embryos PMG and AMG remain uninvaginated (arrowhead). The germ band forms deep transverse folds (L).
DRhoGEF2 is essential for the invagination of mesodermal and endodermal primordia
To characterize the effect of the DRhoGEF2 mutation during gastrulation we stained DRhoGEF2l(2)04291 embryos using antibodies against the transcription factors twi (Thisse et al. 1988) and forkhead (fkh) (Weigel et al. 1989). twi determines mesodermal cell fates and is expressed in all cells undergoing ventral furrow invagination throughout extension of the germ band (Leptin and Grunewald 1990). fkh expression is initiated by hkb (Brönner et al. 1994) and tll (Pignoni et al. 1990) at the late syncytial blastoderm in the gut primordia and is maintained in these tissues throughout embryonic development. Expression of twi and fkh is initiated normally in DRhoGEF2l(2)04291 embryos (Fig. 1, cf. A with B and G with H), suggesting that mesodermal and endodermal cell fates are established correctly in the absence of DRhoGEF2 gene activity.
DRhoGEF2l(2)04291 embryos develop defects during gastrulation. On the ventral side of the embryo no ventral furrow is formed (Fig. 1D). As a consequence mesodermal cells remain on the surface of the embryo even after germ-band extension is completed (Fig. 1F). At the posterior pole, DRhoGEF2l(2)04291 embryos do not form the amnioproctodeal invagination. Although the PMG primordium is initially pushed into a slightly dorsally shifted position (Fig. 1J), probably by the force of the extending germ band, it never moves around the posterior end onto the dorsal side (Fig. 1L). At the anterior end, DRhoGEF2l(2)04291 embryos fail to invaginate the anterior midgut (AMG) (Fig. 1L, arrowhead). These observations suggest that cell shape changes necessary to internalize endodermal and mesodermal germlayers are not initiated in DRhoGEF2l(2)04291 embryos.
DRhoGEF2 regulates cell shape changes during gastrulation
Defects in the ability of DRhoGEF2l(2)04291 embryos to induce cell shape changes during gastrulation were analyzed by scanning electron microscopy (SEM). In wild-type embryos the first sign that gastrulation has begun is a flattening of cell surfaces in an ∼20-cell-wide stripe along the ventral midline (Fig. 2A) (Sweeton et al. 1991). This step is thought to be independent of the actin cytoskeleton and appears to occur in DRhoGEF2l(2)04291 embryos (Fig. 2E). Subsequently apical constriction of cells is induced in the wild type at the ventral midline resulting in formation of a shallow groove (Fig. 2A). This process is severely disrupted in the DRhoGEF2 mutant. Although some cells undergo apical constriction in a spatially disorganized fashion, others expand their apices and remain at the surface (Fig. 2E). As the germ band starts to elongate and the ventral furrow has closed in the wild type (Figure 2B), temporary clefts induced by randomly constricting cells form across the mesoderm in DRhoGEF2l(2)04291 embryos, and no ventral furrow is formed (Fig. 2F).
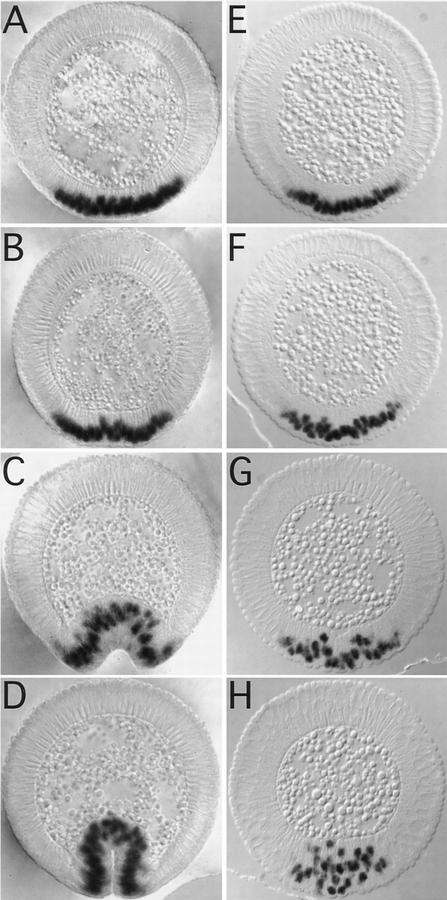
Figure 2.
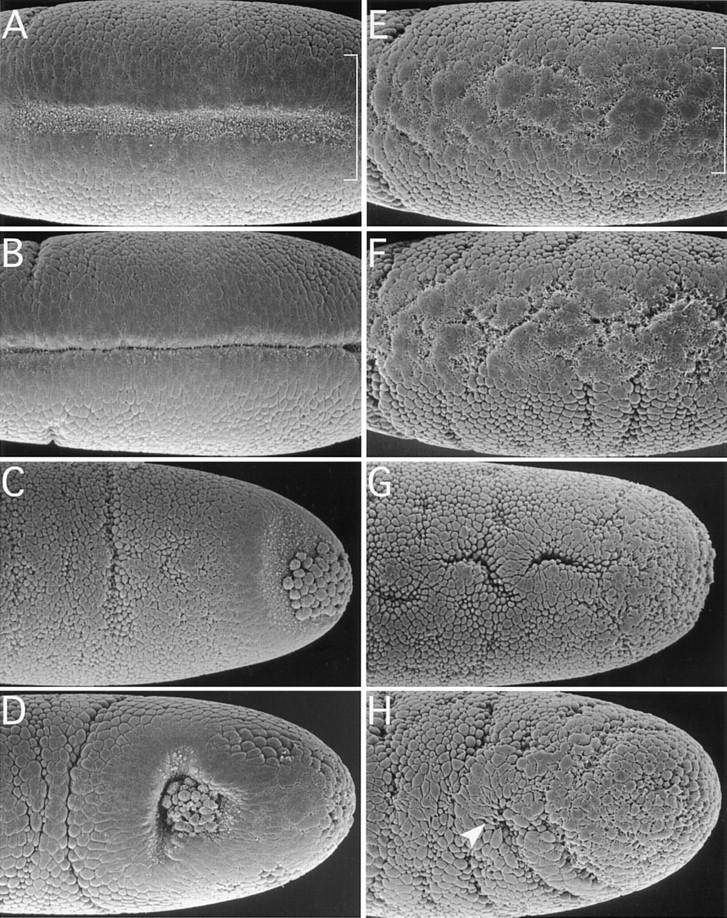
Cell shape changes in wild type (A–D) and DRhoGEF2 mutants (E–H) analyzed by SEM. Following cellularization, cells in an ∼20-cell-wide stripe on the ventral surface of the embryo (indicated by brackets in A and E) flatten their surfaces and move into close contact with each other. Subsequently apical constriction of cells near the midline (notice the membrane blebs indicative of this process) forces the epithelium to bend inward and form a shallow groove (A). As more lateral cells constrict, the furrow deepens and closes over to invaginate the mesoderm (B). In DRhoGEF2l(2)04291 embryos the initial flattening of the cell surfaces appears to occur. However, only very few cells undergo apical constriction in a spatially disorganized fashion (E). As the germ band starts to elongate (indicated by the formation of transverse folds laterally) temporary clefts form at random in the ventral mesoderm (F). No ventral furrow is formed. The PMG primordium is invaginated by a very similar mechanism as the mesoderm. After the initial flattening, cells located dorsal to the pole cells constrict first, as seen by the occurrence of membrane blebs in this region (C). Subsequently a cup shaped invagination is formed harboring the pole cells (D). In DRhoGEF2l(2)04291 embryos, cells in the dorsal polar region do not undergo any specific shape changes and no invagination is formed. The pole cells are deeply embedded into the epithelium. On the dorsal surface of the embryo, groups of cells undergo apical flattening or apical constriction in random regional patterns (G). As the germ band starts to elongate some dorsal cells form cytoplasmic protrusions (arrowhead in H). Altogether cell shape changes appear randomized (H). A, B, E, and F are ventral views; C, D, G, and H are dorsal views. All embryos are oriented with anterior to the left.
In the wild-type PMG primordium, cell shape changes are first induced dorsally to the pole cells (Fig. 2C). In DRhoGEF2l(2)04291 embryos no coordinated changes in cell shape are seen in this region. Most cells remain unconstricted with rounded surfaces (Fig. 2G). Shortly after the onset of germ-band elongation a deep invagination harboring the pole cells has formed in the wild type (Fig. 2D). In DRhoGEF2l(2)04291 embryos cell shape changes appear to occur at random and no invagination is formed (Fig. 2H). Interestingly, not only cells in invaginating tissues but those throughout the embryo are affected. On the dorsal side, groups of cells with flattened surfaces can frequently be observed to form around constricted cells embedded in temporary clefts. The pole cells are deeply embedded into the epithelium and indistinguishable from neighboring cells (Fig. 2G; cf. Figure 1D, arrow). Later, some cells form cytoplasmic protrusions (Fig. 2H, arrow). These observations suggest that cells do not lose the ability to change shape in the DRhoGEF2l(2)04291 mutant but, rather, undergo random shape changes because of loss of an instructive signal.
The behavior of cells in the ventral mesoderm at the onset of gastrulation was examined in further detail in cross sections of embryos stained with anti-Twi antibodies. Ventral furrow formation begins in the wild type immediately following cellularization (Fig. 3A) when cell nuclei start to be displaced from the apical to the basal end in cells at the ventral midline (Fig. 3B). By constricting apically, cells change from a columnar to a trapezoidal shape forcing the epithelium to bend inward (Fig. 3C). This process is subsequently supported by a shortening of the cells along their apical–basal axis (Fig. 3D). Although cellularization appears to be unaffected in DRhoGEF2l(2)04291 embryos (Fig. 3E), the basally directed movement of cell nuclei occurs at random positions across the mesoderm. (Fig. 3F). Only very few cells constrict their apices in a spatially uncoordinated fashion (Fig. 3G). The shortening of cells along the apical–basal axis is not observed in DRhoGEF2l(2)04291 embryos. Instead, cells lose their epithelial character and pile up in multiple layers failing to form a ventral furrow (Fig. 3H).
Figure 3.
Analysis of cell shape changes during ventral furrow formation in wild-type (A–D) and in DRhoGEF2 (E–H) mutants in cross sections immunohistochemically stained for Twi. Shortly after cellularization is completed on the ventral side of the embryo (A), nuclei of cells at the ventral midline start to move basally (B). Apical constriction of midventral cells forces the epithelial sheet to bend inward (B,C). Shortening of cells along their apical–basal axis supports this process and results in internalization of the mesoderm (D). DRhoGEF2l(2)04291 embryos appear normal during cellularization. Cell nuclei are lined up along the apical end of cells (E). At the onset of gastrulation, nuclei are seen to move basally in a random manner (F). Very few cells undergo apical constriction (G). Cells do not shorten along their apical basal axes; they lose their epithelial character and pile up in several layers. No ventral furrow is formed (H).
DRhoGEF2 encodes a putative guanine nucleotide exchange factor of the Dbl family
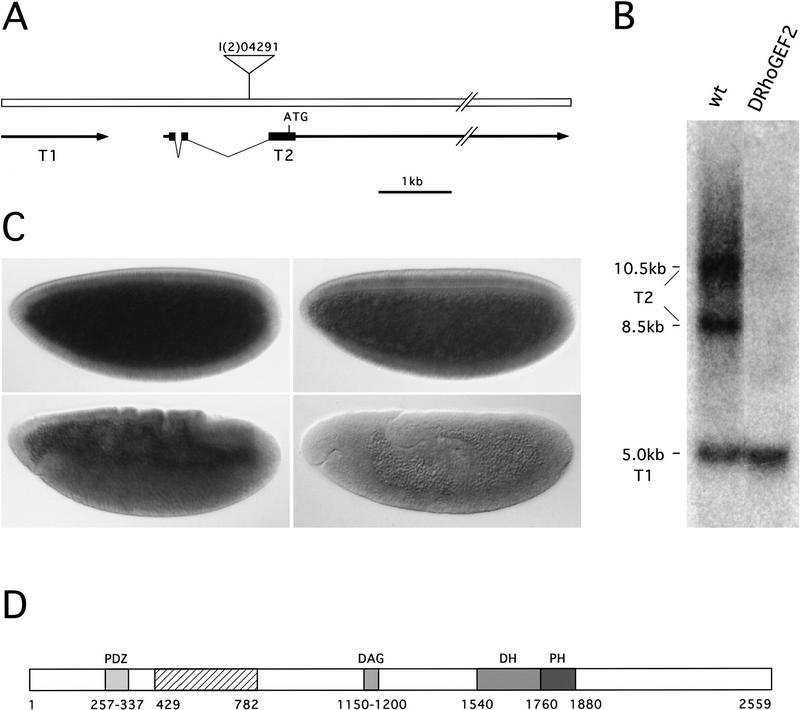
To molecularly characterize the gene affected by the DRhoGEF2 mutation, we isolated genomic DNA adjacent to the insertion site of P-element l(2)04291 by plasmid rescue (Cooley et al. 1988). The plasmid rescue DNA fragment was used to screen a genomic DNA library, and 24 kb of DNA encompassing the genomic region flanking the P element was obtained. A screen for cDNAs located in this region led to the identification of two transcription units, T1 and T2. T1 codes for a 5-kb transcript terminating 1.9 kb short of the P-element insertion point (Fig. 4A). Expression of this transcript was found to be unaltered in DRhoGEF2l(2)04291 embryos, suggesting that T1 is unaffected by the insertion (Fig. 4B). Transcript T2 is located downstream of T1 and oriented in tandem. The 5′ end of T2 was mapped 1.1 kb upstream of the P element, leaving a short intergenic region of 0.8 kb between T1 and T2 and placing the P element inside the T2 transcript. Northern blot analyses showed that two mRNAs, 8.5 and 10.5 kb in length, are derived from the T2 locus. Both of these transcripts are absent from DRhoGEF2l(2)04291 embryos (Fig. 4B). This observation, together with the insertion of the P element inside the T2 transcript, provides compelling evidence that T2 identifies the DRhoGEF2 gene. Furthermore the absence of T2 transcripts, from the 0- to 12-hr embryonic poly(A)+ RNA preparation used in the Northern blot analysis indicates that there is no significant zygotic contribution to the expression of DRhoGEF2 during the first 12 hr of embryogenesis. This is consistent with the lack of paternal rescue in the DRhoGEF2 mutant and with the ubiquitous early expression of DRhoGEF2 observed by in situ hybridization to whole mount embryos (Fig. 4C).
Figure 4.
Molecular cloning, Northern analysis, and expression of the DRhoGEF2 gene. (A) Genomic map of the region surrounding the P-element insertion l(2)04291. Two transcription units, T1 and T2, were identified near the P-element insertion point. The P element was found to be inserted into an intron in the untranslated leader region of the T2 transcript. The 3′ end of T2 was mapped ∼20 kb from the P-element insertion. Solid boxes indicate exons; arrows denote transcribed regions. (B) Northern blot analysis of T1 and T2 on poly(A)+ RNA derived from 0- to 12-hr wild type (left lane) and DRhoGEF2l(2)04291 (right lane) embryos. A single T1 transcript, 5.0 kb in length, is detected by the T1 cDNA in RNA prepared from wild-type and DRhoGEF2l(2)04291 embryos, suggesting that the P insertion is not interfering with the expression of this gene. In wild-type RNA cDNAs of T2 detect two transcripts, 8.5 and 10.5 kb in length. Both transcripts are absent in RNA derived from DRhoGEF2l(2)04291 embryos indicating that expression of T2 is abolished by the P element insertion. (C) In situ hybridization of a genomic DNA fragment, encompassing part of the T2 transcription unit, to wild-type embryos. DRhoGEF2 transcripts are abun dant at the syncytial blastoderm stage (top left). DRhoGEF2 mRNAs are distributed evenly throughout the embryo during gastrulation (top right and bottom left). DRhoGEF2 transcripts are no longer detectable after extension of the germ band (bottom right). (D) Schematic representation of the DRhoGEF2 protein. The DRhoGEF2 cDNA contains an ORF for a protein of 2559 amino acids with several regions of homology to sequences present in the databases. A PDZ domain is located near the amino terminus (PDZ). In the central region DRhoGEF2 contains a cysteine-rich diacylglycerol-binding motif (DAG). A DH and a PH domain are located in tandem in the carboxy-terminal half of the protein and identify DRhoGEF2 as a putative GEF for small GTPases of the Rho family. The hatched box in the amino-terminal third of the protein represents the region not encoded in the shorter version of the cDNA (see Materials and Methods). Numbers represent amino acid positions.
Sequencing of a DRhoGEF2 cDNA revealed an ORF of 7677 bp coding for a putative hydrophilic protein of 281 kD, which contains several regions with similarity to sequences present in the databases (Fig. 4D). A region of ∼80 amino acids near the amino terminus of DRhoGEF2 encodes a PDZ domain (for review, see Fanning and Anderson 1996), which shows the highest homology to a motif found in Rhophilin (Watanabe et al. 1996), a mouse protein implicated in G-protein-coupled signaling events (Figs. 4D and 5A). Interestingly the GLGF motif, which is thought to comprise the ligand binding pocket of PDZ domains, is changed to GYGM in DRhoGEF2. In the central region of DRhoGEF2 a cysteine-rich zinc butterfly motif is found, which is also present in isoforms of protein kinase C (PKC) and the mouse Dbl family oncoprotein Lfc (Figs. 4D and 5B) (Kazanietz et al. 1994; Whitehead et al. 1995). The carboxy-terminal region of DRhoGEF2 contains an extensive region of homology to two separate protein motifs characteristic of the Dbl family of oncoproteins (Fig. 4D and 5C,D) (Cerione and Zheng 1996). The first motif, termed Dbl homology domain (DH domain), was found to promote the exchange of guanine nucleotides on Rho family GTPases (Hart et al. 1994). The second domain, located carboxy-terminally juxtaposed to the DH domain, is a Pleckstrin homology domain (PH domain) (Haslam et al. 1993; Mayer et al. 1993). Both domains are invariantly present in all members of the Dbl family and identify DRhoGEF2 as a putative guanine nucleotide exchange factor (RhoGEF) for Rho family GTPases.
Figure 5.
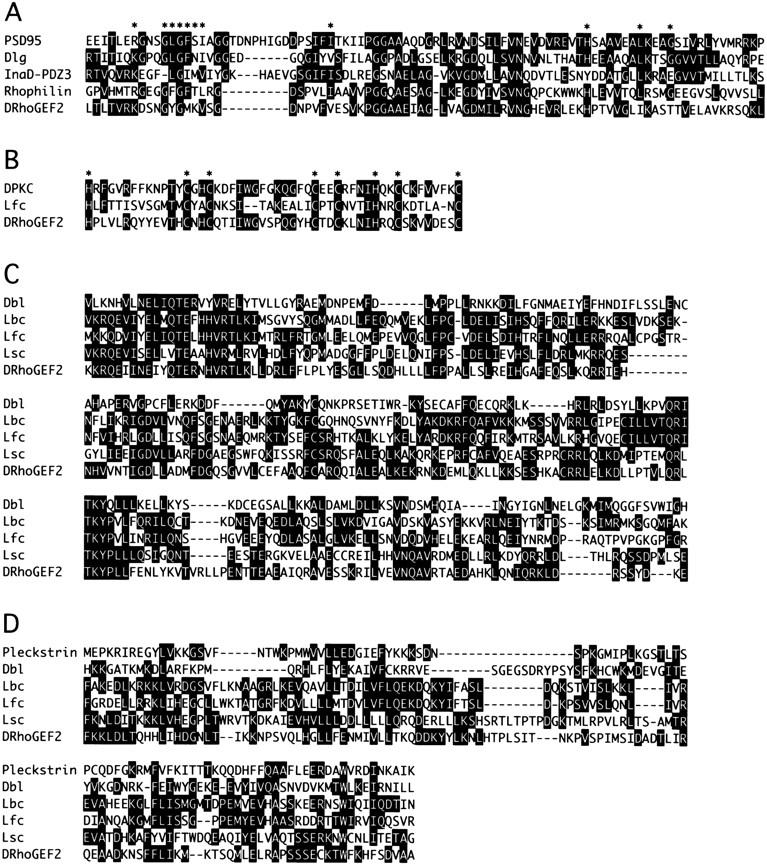
Conserved sequence motifs found in DRhoGEF2. (A) Sequence alignment of the PDZ domains of PSD95, Discs large (Dlg), the third PDZ domain of InaD, Rhophilin, and DRhoGEF2. Stars above the sequence indicate residues implicated in substrate binding. (B) Sequence alignment of the DAG-binding motifs of PKC (Drosophila eye isoform), the mouse Dbl family oncogene Lfc, and DRhoGEF2. Stars indicate conserved histidine and cysteine residues. (C) Sequence alignment of the DH domains of the human oncoproteins Dbl and Lbc, the mouse oncoproteins Lfc, and Lsc, and DRhoGEF2. (D) Sequence alignment of the PH domains of Pleckstrin and the Dbl-family genes Dbl, Lbc, Lfc, Lsc, and DRhoGEF2. Sequences where aligned by the Clustal method using DNASTAR software. Solid boxes indicate residues identical to a consensus sequence.
DRhoA is involved in early morphogenesis
The identification of DRhoGEF2 as a RhoGEF suggests a role for Rho family GTPases during Drosophila gastrulation. To test this hypothesis we expressed dominant-negative forms of three Rho family members isolated in Drosophila, DRhoAN19 (Strutt et al. 1997), DRacN17, and DCdc42N17 (Luo et al. 1994) in the mesoderm using the GAL4–UAS system (Brand and Perrimon 1993). When expression was driven by a twi–GAL4 driver line (Greig and Akam 1993), all embryos developed severe cuticle defects (not shown) indicating that the dominant-negative GTPases were expressed and biologically active. When embryos carrying twiGAL4,UASDRhoAN19 were stained with anti-Twi antibodies, patches of mesodermal cells that failed to invaginate were seen frequently along the germ band (Fig. 6B). In addition, staining of the same embryos with anti-Fkh antibodies revealed that invagination of the AMG was blocked (Fig. 6C, arrowhead), a phenotype also observed in DRhoGEF2l(2)04291 embryos (cf. Figure 1L, arrowhead). The defects seen in twiGAL4,UASDRhoAN19 embryos coincide with the onset of expression of twiGAL4 in the mesoderm at stage seven (Greig and Akam 1993) and thus are likely to be specific to the loss of DRhoA activity. No similar defects were seen in invaginating tissues in either twiGAL4,UASDRacN17 or twiGAL4,UASDCdc42N17 embryos, indicating that DRhoA activity is specifically required for the invagination of mesodermal and endodermal cells during gastrulation in a manner distinct from DRac and DCdc42.
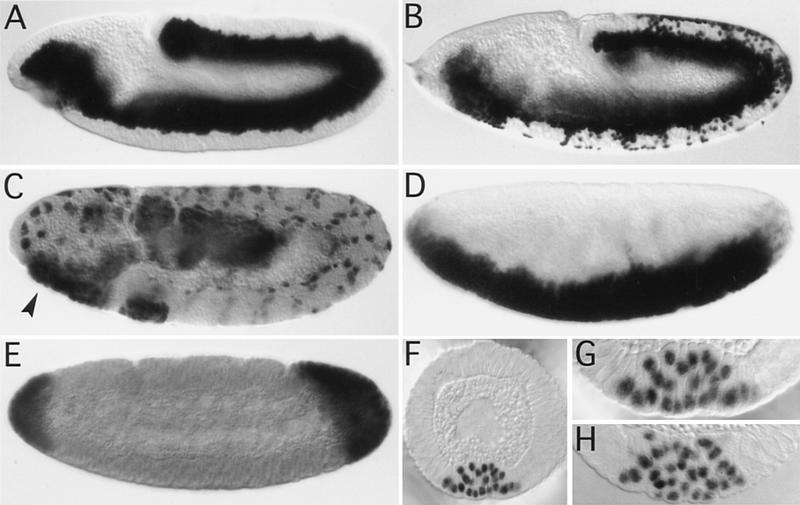
Figure 6.
Expression of dominant-negative forms of Rho family GTPases during gastrulation. Embryos are stained immunohistochemically for either Twi (A,B,D,F–H) or Fkh (C,E). In wild-type embryos all mesodermal cells have been internalized at stage 10 (A). In twiGAL4/UASDRhoAN19 embryos at the same stage patches of mesodermal cells along the germ band have failed to invaginate (B). Invagination of the AMG is blocked (arrowhead in C, cf. Fig. 1L). (D–G) matα4-GAL–VP16/UASDRhoAN19 embryos: The phenotype caused by the maternal expression of dominant-negative DRhoA strongly resembles that seen in DRhoGEF2l(2)04291 embryos. No ventral furrow is formed (D; cf. Fig. 1D) and no PMG invagination is seen in the posterior dorsal region (E, cf. Fig. 1J). Cells fail to undergo shape changes during ventral furrow formation (F,G) in a way very similar to DRhoGEF2l(2)04291 embryos (H).
To examine an earlier role of DRhoA during gastrulation, we expressed DRhoAN19 using a maternal GAL4 driver. When DRhoAN19 was expressed using matα4-GAL–VP16 (a gift from Daniel St Johnston, Wellcome/Cancer Research Campaign Institute, Cambridge, UK), ventral furrow formation as well as PMG invagination were blocked resulting in a phenocopy of DRhoGEF2l(2)04291 embryos (Fig. 6D,E). Examination of matα4-GAL–VP16,UASDRhoAN19 embryos in cross sections revealed that cell shape changes are disrupted in a manner very similar to DRhoGEF2l(2)04291 embryos (Fig. 6, cf. F and G to H). Our observations suggest that DRhoA is required for invagination of mesodermal and endodermal primordia during gastrulation and its activity is likely to be regulated by DRhoGEF2.
Discussion
We have identified a gene, DRhoGEF2, which encodes a putative guanine nucleotide exchange factor belonging to the Dbl family of oncogenes. Mutations in DRhoGEF2 have a maternal effect phenotype disrupting the coordination of cell shape changes during gastrulation. Unlike previously identified gastrulation genes, DRhoGEF2 is essential for the invagination of mesodermal and endodermal primordia. In addition, we have obtained evidence that the small GTPase DRhoA plays a role during gastrulation very similar to DRhoGEF2, suggesting that a signal transduction pathway involving DRhoGEF2 and DRhoA regulates cell shape changes during early Drosophila morphogenesis.
DRhoGEF2 regulates the induction of cell shape changes
Embryos mutant for DRhoGEF2 appear normal during cellularization. The first developmental defects coincide with the onset of gastrulation suggesting that DRhoGEF2 is specifically required during this process. The failure of cell shape changes, necessary to invaginate mesodermal and endodermal primordia to occur in DRhoGEF2 mutants, is reminiscent to the phenotype associated with the gastrulation mutants fog and cta. Both genes encode putative signaling molecules and have been tentatively placed in a common signal transduction pathway. It has been proposed that a localized signal generated by Fog is received by an unknown receptor and transduced via the putative G protein Cta to the cytoskeleton. The fact that two transcriptional activators of fog, twi, and fkh are expressed normally in DRhoGEF2 mutants and the identification of DRhoGEF2 as a putative RhoGef suggest that DRhoGEF2 may be required to transduce the signal elicited by Fog to the cytoskeleton. However, there is a significant difference between the mutant phenotypes of fog and cta on one hand and DRhoGEF2 on the other: In contrast to DRhoGEF2, fog and cta are not essential for ventral furrow formation. For this reason DRhoGEF2 must be activated in a Fog/Cta-independent way (Fig. 7). In fact, because of the nonessential function of fog and cta in the mesoderm, a second pathway instructing cells to undergo shape changes has been postulated (Costa et al. 1994). We propose that DRhoGEF2 identifies this pathway as a G-protein-coupled signaling cascade involving the GTPase DRhoA. Whether this pathway is also required to transduce the Fog signal is presently unclear and this question will require further attention in the future.
Figure 7.
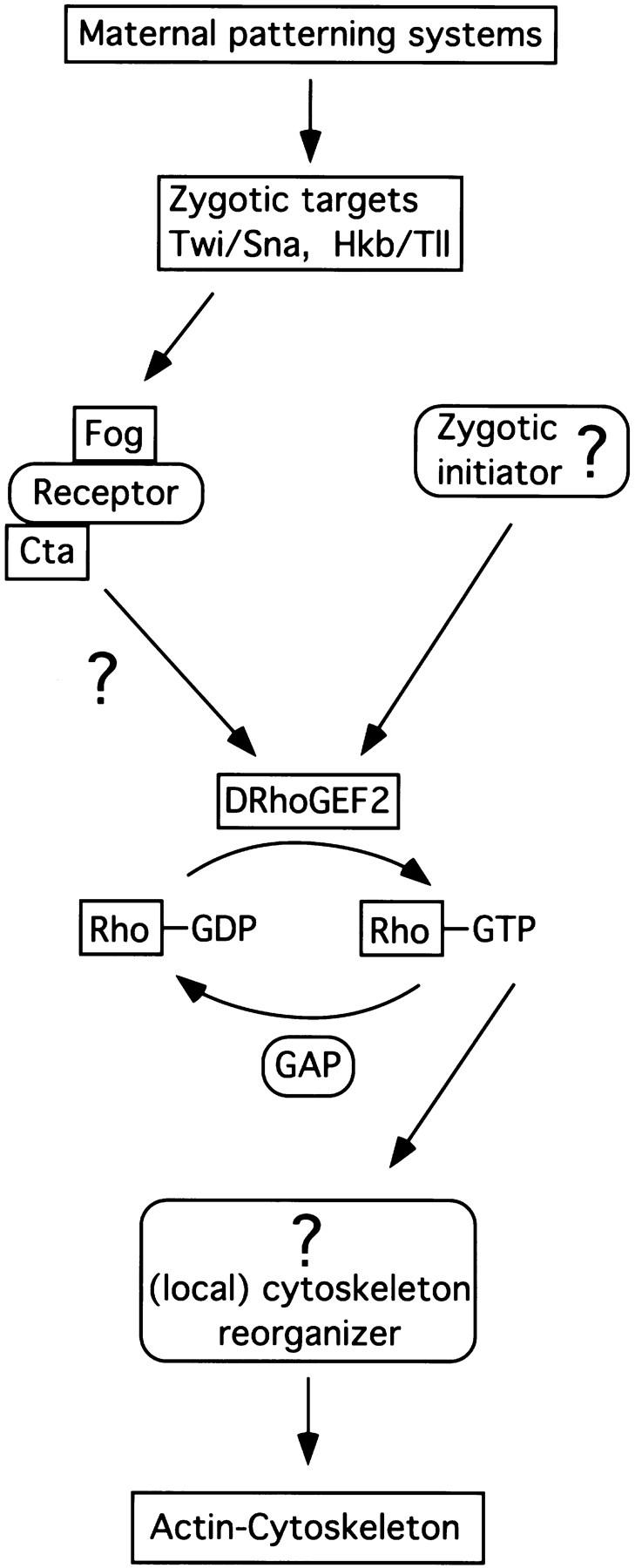
Model of the signaling events regulating cell shape changes during gastrulation. Boxed components have been identified in Drosophila. Boxes with rounded corners represent postulated components. (see text for details).
The phenotype of DRhoGEF2 mutants and the broad expression of DRhoGEF2 transcripts show that DRhoGEF2 is not only required for the generation of cell shape changes in invaginating tissues but that it plays an important role in the control of cell shape throughout the early embryo (Fig. 2). This raises the question: How are cells in different regions of the embryo induced to respond differently to DRhoGEF2 activity during gastrulation? One can envision several mechanisms to regulate cell shape in a localized fashion. Specificity could be generated through differential subcellular activation of DRhoGEF2 to different levels in response to localized, tissue-specific upstream factors. Fog could be one of these factors contributing to a synergistic activation of DRhoGEF2 in invaginating tissues; however, as ventral furrow formation does still occur in fog mutants, at least one additional activator must exist. Alternatively, fog could be involved in a different aspect of cytoskeletal regulation and cell shape changes in the mesoderm could depend solely on the localized activity of an unidentified “zygotic initiator.”
In an alternative model DRhoGEF2 could be constitutively active in all cells to maintain cell shape throughout the epithelium. Mesodermal cells could be induced to change their shape by the spatially and temporally regulated expression of specific cytoskeleton-associated proteins linking the signaling pathway to the actin cytoskeleton in a Rho-dependent manner. Although the isolation of factors conferring specificity during gastrulation has proven to be difficult in the past, we believe that the identification of DRhoGEF2 will open the path for a new approach to this problem.
The Dbl family of oncogenes
The Dbl family includes a growing number of oncoproteins that are characterized by the presence of two closely juxtaposed regions of homology (Cerione and Zheng 1996). The first motif, the DH domain, spans ∼220 amino acids and has been demonstrated to promote the exchange of guanine nucleotides on small GTPases of the Rho family resulting in the activation of these molecules (Hart et al. 1994). Deletions within this domain result in a loss of the transforming activity in the prototype gene of the family, the oncogene Dbl (Ron et al. 1991). The second region of homology is a widespread protein motif that was first described in the cytoskeleton-associated protein Pleckstrin (Tyers et al. 1988). Although poorly conserved at the sequence level this motif seems to adopt a common three-dimensional structure as determined by nuclear magnetic resonance and x-ray crystallography. The PH domain is most likely involved in signal-dependent membrane localization (for review, see Shaw 1996) and may direct proteins to specific cytoskeletal locations to activate Rho family GTPases.
In vertebrates the Dbl family presently consists of ∼15 genes, of which 2, FGD1 and vav, have been shown to be involved in embryonic development. The human FGD1 gene was determined to be the locus responsible for faciogenital dysplasia (Aarskog–Scott syndrome), a multisystemic developmental disease affecting skeletal and urogenital systems (Pasteris et al. 1994). In the mouse, a knockout of the vav oncogene has been found to result in embryonic lethality (Zmuidzinas et al. 1995). While the function of these vertebrate genes has been inferred from their molecular identity, the Cdc24 gene, a member of the Dbl family isolated from yeast, has been shown by biochemical and genetic means to function as the physiological activator of the small GTPase Cdc42 (Sloat et al. 1981; Zheng et al. 1994).
In Drosophila two members of the Dbl family have been identified previously. One of these genes, named still life, has been shown to play a role during differention of synaptic terminals (Sone et al. 1997). The second Drosophila RhoGEF, termed DrtGEF or DRhoGEF1, is expressed during oogenesis and throughout embryogenesis in morphogenetically active tissues (Werner and Manseau 1997), suggesting a role for another RhoGEF during gastrulation.
In addition to the DH and PH domains, DRhoGEF2 contains an amino-terminal PDZ domain. The PDZ domain is a widespread modular protein motif and has been implicated in the assembly of signaling molecules to the plasma membrane by binding to the carboxyl terminus of transmembrane receptors or by forming heterodimers with other PDZ domain-containing proteins (for review, see Fanning and Anderson 1996). Interestingly the family typical GLGF motif that is thought to specifically interact with a (S/T)XV motif commonly found at the carboxyl terminus of transmembrane receptors is not conserved in DRhoGEF2. Consequently, the DRhoGEF2–PDZ motif might have a different binding specificity or may exclusively form heterodimers. Consistent with this idea, the PDZ adaptor protein InaD, which has been implicated in the assembly of a signaling complex during G-protein-mediated photoreceptor signaling in the eye, contains five PDZ domains with modified substrate-binding motifs (Tsunoda et al. 1997). The DRhoGEF2–PDZ domain might link the RhoGEF to a similar adaptor protein present in invaginating tissues during gastrulation.
In its central region DRhoGEF2 contains a cysteine-rich zinc butterfly motif that is also found in another member of the Dbl family, the mouse oncogene lfc (Whitehead et al. 1995) and in isoforms of PKC (Kazanietz et al. 1994). In PKC this domain has been shown to bind the second messenger diacylglycerol, which is generated following signal-dependent hydrolysis of membrane lipids (Ahmed et al. 1991; Bell and Burns 1991; Hubbard et al. 1991; Quest et al. 1992). Similar to the PH and the PDZ domains, this motif could promote membrane association of DRhoGEF2 and activation in response to specific signals. The presence of three different regulatory domains might provide a mechanism to activate DRhoGEF2 to different levels in response to distinct cellular signals; for example, the second messenger diacylglycerol could activate DRhoGEF2 throughout the epithelium to maintain cell shape while the PDZ domain mediates specific signals present locally in the invaginating tissues.
A role for Rho during gastrulation
The members of the Rho family, Rho, Rac, and Cdc42, belong to the Ras superfamily of GTP-binding proteins and cycle between a GDP and a GTP bound form. The level of activity of these molecules is regulated by guanine nucleotide exchange factors (GEFs) like DRhoGEF2, which convert the GTPase into its active GTP-bound form, and GTPase-activating proteins (GAPs) which stimulate the hydrolysis of GTP (Fig. 7). The alternation between an active and an inactive form allows small GTPases to act as bimodal switches in various intracellular signal transduction pathways (for review, see Ridley 1996).
In Drosophila, homologs of several members of Rho family GTPases have been cloned. In studies using dominant-negative as well as activated forms of these molecules DRho, DRac, and DCdc42 have been shown to play a role in regulation of cell shape changes during dorsal closure and head involution as well as in morphogenesis during axon outgrowth muscle development and oogenesis (Luo et al. 1994; Harden et al. 1995; Murphy and Montell 1996). In addition, mutations in DRhoA have been shown recently to disrupt tissue polarity during imaginal development (Strutt et al. 1997).
DRho, DRac, and DCdc42 are expressed uniformely throughout development and are most likely supplied maternally to the embryo (Hariharan et al. 1995). An important question arising from this observation is: How is specificity generated among these very similar molecules? It has been demonstrated that RhoL, DRac, and DCdc42 fulfill specific but distinct functions during oogenesis (Murphy and Montell 1996). Similarly, expression of dominant-negative forms of DRac and DCdc42 in neurons during axon outgrowth and during muscle development results in distinct phenotypes. Our studies indicate that the function of DRac and DCdc42, if any, during gastrulation is clearly distinguishable from DRhoA in that only DRhoAN19, but not DRacN17, or DCdc42N17, interferes with the invagination of mesodermal and endodermal precursors. Furthermore, maternal expression of DRhoAN19 resulted in the generation of phenocopies of DRhoGEF2 mutant embryos. Based on these observations, we propose that DRhoGEF2 acts as a DRhoA-specific RhoGEF and that specificity in G-protein-coupled signaling pathways acting simultaneously in the same tissues is generated at least in part by the use of GTPase-specific GEFs.
Materials and methods
Drosophila stocks
UASDRhoAN19/Cyo was obtained from M. Mlodzik (Strutt et al. 1997). UASDRacN17 and UASDCdc42N17 were provided by Y.N. Jan (Luo et al. 1994) and the twiGAL4–driver line was from M. Akam (Greig and Akam 1993). The matα4-GAL–VP16 line contains a construct consisting of the DNA-binding domain (amino acids 1–144) of GAL4 fused to the VP16 transcriptional activation domain expressed from the α4–tubulin promoter and was a gift of D. St Johnston.
Genetics of DRhoGEF2
A single P[LacZ, ry+] element insertion, l(2)04291, was identified in a screen for maternal effects of zygotic lethal mutations. Association of the observed phenotype with the P-element insertion was confirmed by mobilization of the P element using a y w; Δ2-3, Sb/TM6 strain (Robertson et al. 1988). Of 108 excision lines, 83 were homozygous viable. From a parallel screen, using chemically induced mutagenesis, three further alleles, DRhoGEF2B27.8, DRhoGEF23W18, and DRhoGEF25K19, were identified by complementation analysis. Embryos derived from germ-line clones of all these alleles show phenotypes identical to DRhoGEF2l(2)04291 null embryos during gastrulation. Females with germ-line clones were generated using the autosomal FLP–DFS technique (Chou and Perrimon 1996).
Cloning of DRhoGEF2
A 5-kb genomic DNA fragment flanking the DRhoGEF2 P-element insertion was obtained by plasmid rescue (Cooley et al. 1988). This fragment was subsequently used to screen a genomic DNA library (Stratagene) to obtain clones covering 24 kb of genomic DNA. Using subfragments covering this entire region, 36 cDNAs were isolated from poly(dT)-primed (Brown and Kafatos 1988) and random-primed (Clontech) cDNA libraries. Two of the cDNAs correspond to the T1 transcript (see Fig. 4B) and are 3.5 and 5.0 kb in length, respectively. All other cDNAs were between 1.2 and 5.5 kb in length and encode different subregions of the T2 transcipt. Restriction analysis and DNA sequencing allowed the reconstitution of two distinct transcripts from these cDNAs, 8607 and 7545 bp in length. Both transcripts are represented by multiple independent cDNAs and contain all the regions of homology to other proteins found in the T2 sequence. In the shorter transcript the region of the ORF from amino acid 429 to 782 (DAPTVK....GGQATG) is not present (see Fig. 4D). We presume that the two alternative proteins are encoded by the two respective transcripts seen on Northern blots. The discrepancy in transcript sizes observed between the Northern blot analysis and the T2 cDNAs is most likely due to gel migration artifacts caused by secondary structure formation in the RNA.
DNAs were sequenced using the dideoxy chain termination method (Sanger et al. 1977). The precise point of P-element insertion was determined by sequencing the P-element rescue DNA fragment using a primer derived from the P-element LTR as well as genomic DNA fragments. The P element is inserted into an intron in the untranslated leader region of the transcript (as determined by comparison of genomic and cDNA sequences) 260 bp upstream of the 3′ intron boundary. The intron is 1.1 kb in length.
Northern Blot analysis
Northern blots were carried out by standard procedures (Sambrook et al. 1989) using 5 μg poly(A)+—RNA per lane. Transcript sizes of T1 and T2 were first determined in separate experiments (not shown) using the 5-kb T1 cDNA or the 5.5-kb T2 cDNA (extending from the 5′ end and encompassing both transcripts) as probes. Northern blots using both probes simultaneously were carried out on poly(A)+–RNA prepared from 0- to 12-hr collections of wild-type or DRhoGEF2l(2)04291 embryos. The absence of both T2 transcripts from DRhoGEF2l(2)04291 embryos suggests that DRhoGEF2l(2)04291 is a null allele and that there is no significant zygotic expression of DRhoGEF2 during the first 12 hr of embryogenesis.
Immunohistochemistry
Embryos were fixed by shaking in 4 ml of PEM (100 mm PIPES, 1 mm EGTA, 20 mm MgSO4), 4% formaldehyde, and 5 ml of heptane for 20 min and devitellinized by addition of 10 ml of methanol and vigorous shaking for 30 sec. Stainings were carried out by standard procedures. The polyclonal rabbit anti-Twi antibody (gift of Siegfried Roth, Max-Planck-Institut für Entwicklungsbiologie, Tübingen, Germany) was used at a dilution of 1/5000. The polyclonal Fkh antiserum (gift of Pilar Carrera, CSIC, Barcelona, Spain) was diluted 1/2000.
In situ hybridization to whole mount embryos
In situ hybridization was carried out by standard protocols using digoxigenin (Boehringer)-labeled double-stranded DNA probes. The stainings shown in Figure 4C were reproducible with several genomic DNA fragments encompassing different regions of the T2 transcript. Control fragments yielded no staining.
SEM
Embryos were collected at timed intervals, dechorionated, fixed in 2.5% glutaraldehyde for 2 hr, and devitellinized with methanol. Embryos were then washed twice in 0.1 m sodium cacodylate (pH 7.4), postfixed in 1% osmiumtetroxide and 0.1 m sodium cacodylate (pH7.4) for 1 hr, and dehydrated in an ethanol series. After exchange of ethanol against carbon dioxide in a critical point dryer, embryos were mounted on stubs, gold–palladium coated in a sputter coater, and photographed in the SEM.
Sectioning of embryos
Embryos stained with polyclonal antiserum against Twi were dehydrated in an ethanol series and then transferred into ethanol/xylene (1:1) and finally into xylene. After mounting in araldite (Serva) on a slide, embryos were staged under the light microscope and selectively embedded (in araldite) for sectioning. After polymerization of the araldite overnight at 80°C, 5 μm sections were cut on a Leitz microtome, mounted in araldite, and photographed under Nomarski optics on a Zeiss Axiophot microscope.
Acknowledgments
We thank D. Strutt and M. Mlodzik for sending us the UASDRhoAN19 line, Y.N. Jan for the UASDRacN17 and UASDCdc42N17 lines, A. Michelson for providing the twi–GAL4 line, and D. St Johnston for providing us with the matα4-GAL–VP16 line prior to publication. We extend our special gratitude to S. Roth for the anti-Twi antibody and to P. Carrera who made the anti-Fkh antibody. We also thank E. Seling for his expertise with the SEM and K. Barrett and J. Settleman for discussing results prior to publication. We are grateful to members of the Perrimon laboratory, and in particular to B. Mathey-Prevot for comments on the manuscript. This work was supported by a Human Frontier Science Program Organization long-term fellowship to U.H. and the Howard Hughes Medical Institute which N.P. is an Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
Initially the gene described in this article was named akkordeon (akk); however, we subsequently became aware that the same gene had been independently isolated and named shar pei by K. Barrett and J. Settleman (pers. comm.). To avoid confusion and to reflect its putative function, we mutually agreed to rename the gene DRhoGEF2.
The DNA and protein sequences of DRhoGEF2 have been submitted to GenBank and have been assigned accession number AF031930.
Footnotes
E-MAIL perrimon@rascal.med.harvard.edu; FAX (617) 432-7688.
References
- Ahmed S, Kozma R, Lee J, Monfries C, Harden N, Lim L. The cysteine-rich domain of human proteins, neuronal chimaerin, protein kinase C and diacylglycerol kinase binds zinc. Evidence for the involvement of a zinc-dependent structure in phorbol ester binding. Biochem J. 1991;280:233–241. doi: 10.1042/bj2800233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RM, Burns DJ. Lipid activation of protein kinase C. J Biol Chem. 1991;266:4661–4664. [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brönner G, Chu-LaGraff Q, Doe CQ, Cohen B, Weigel D, Taubert H, Jäckle H. Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature. 1994;369:664–668. doi: 10.1038/369664a0. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Chou T-B, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melangaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76:1075–1089. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM. Protein-protein interactions: PDZ domain networks. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- Greig S, Akam M. Homeotic genes autonomously specify one aspect of pattern in the Drosophila mesoderm. Nature. 1993;362:630–632. doi: 10.1038/362630a0. [DOI] [PubMed] [Google Scholar]
- Harden N, Loh HY, Chia W, Lim L. A dominant inhibitory version of the small GTP-binding protein Rac disrupts cytoskeletal structures and inhibits developmental cell shape changes in Drosophila. Development. 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Hu KQ, Asha H, Quintanilla A, Ezzell RM, Settleman J. Characterization of rho GTPase family homologues in Drosophila melanogaster: Overexpressing Rho1 in retinal cells causes a late developmental defect. EMBO J. 1995;14:292–302. doi: 10.1002/j.1460-2075.1995.tb07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Bishop WR, Kirschmeier P, George SJ, Cramer SP, Hendrickson WA. Identification and characterization of zinc binding sites in protein kinase C. Science. 1991;254:1776–1779. doi: 10.1126/science.1763327. [DOI] [PubMed] [Google Scholar]
- Kazanietz MG, Bustelo XR, Barbacid M, Kolch W, Mischak H, Wong G, Pettit GR, Bruns JD, Blumberg PM. Zinc finger domains and phorbol ester pharmacophore. Analysis of binding to mutated form of protein kinase C zeta and the vav and c-raf proto-oncogene products. J Biol Chem. 1994;269:11590–11594. [PubMed] [Google Scholar]
- Leptin M. Drosophila gastrulation: From pattern formation to morphogenesis. Annu Rev Cell Dev Biol. 1995;11:189–212. doi: 10.1146/annurev.cb.11.110195.001201. [DOI] [PubMed] [Google Scholar]
- Leptin M, Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110:73–84. doi: 10.1242/dev.110.1.73. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes & Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Ren R, Clark KL, Baltimore D. A putative modular domain present in diverse signaling proteins. Cell. 1993;73:629–630. doi: 10.1016/0092-8674(93)90244-k. [DOI] [PubMed] [Google Scholar]
- Murphy AM, Montell DJ. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J Cell Biol. 1996;133:617–630. doi: 10.1083/jcb.133.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks S, Wieschaus E. The Drosophila gastrulation gene concertina encodes a G α-like protein. Cell. 1991;64:447–458. doi: 10.1016/0092-8674(91)90652-f. [DOI] [PubMed] [Google Scholar]
- Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL. Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: A putative Rho/Rac guanine nucleotide exchange factor. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- Quest AF, Bloomenthal J, Bardes ES, Bell RM. The regulatory domain of protein kinase C coordinates four atoms of zinc. J Biol Chem. 1992;267:10193–10197. [PubMed] [Google Scholar]
- Ridley AJ. Rho: Theme and variations. Curr Biol. 1996;6:1256–1264. doi: 10.1016/s0960-9822(02)70711-2. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Zannini M, Lewis M, Wickner RB, Hunt LT, Graziani G, Tronick SR, Aaronson SA, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G. The pleckstrin homology domain: An intriguing multifunctional protein module. BioEssays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- Sloat BF, Adams A, Pringle JR. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y, Hama C. Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin G. Gene disruption using P transposable elements: An integral component of the Drosophila geneome project. Proc Natl Acad Sci. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt DI, Weber U, Mlodzik M. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 1997;387:292–295. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–789. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J. 1988;7:2175–2183. doi: 10.1002/j.1460-2075.1988.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G- protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- Tyers M, Rachubinski RA, Stewart MI, Varrichio AM, Shorr RG, Haslam RJ, Harley CB. Molecular cloning and expression of the major protein kinase C substrate of platelets. Nature. 1988;333:470–473. doi: 10.1038/333470a0. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jürgens G, Kuttner F, Seifert E, Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Werner LA, Manseau LJ. A Drosophila gene with predicted RhoGEF, pleckstrin homology and SH3 domains is highly expressed in morphogenic tissues. Gene. 1997;187:107–114. doi: 10.1016/s0378-1119(96)00732-9. [DOI] [PubMed] [Google Scholar]
- Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- Zmuidzinas A, Fischer KD, Lira SA, Forrester L, Bryant S, Bernstein A, Barbacid M. The vav proto-oncogene is required early in embryogenesis but not for hematopoietic development in vitro. EMBO J. 1995;14:1–11. doi: 10.1002/j.1460-2075.1995.tb06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]