Abstract
Background
Aberrant activation of the hedgehog (Hh) signaling pathway is implicated widely in both pediatric and adult malignancies. Inactivation of the Hh regulator PTCH is responsible for the Gorlin cancer predisposition syndrome. The spectrum of tumors found in Gorlin Syndrome includes basal cell carcinoma, medulloblastoma, and rarely, rhabdomyosarcoma (RMS). A previous report utilizing in situ hybridization has provided initial evidence for the expression of Hh targets GLI1 and PTCH in RMS tumors.
Procedure
To investigate the role of Hh pathway signaling in pediatric rhabdomyosarcoma (RMS) and undifferentiated sarcoma (US) tumors, the expression of Hh pathway targets GLI1 and PTCH was measured. RNA was extracted from archival human tumor specimens collected from pediatric patients enrolled on Intergroup Rhabdomyosarcoma Study III and IV, and subjected to quantitative reverse transcriptase-polymerase chain reaction.
Results
Expression of GLI1 with or without PTCH was detected in substantial subsets of embryonal RMS (ERMS) and US tumors but only rarely in alveolar RMS tumors. Neither PTCH mutations nor activating SMO mutations were detected in ERMS tumors with high GLI1 expression. Microarray analysis demonstrated relative overexpression of downstream Hh targets in ERMS tumors with high or intermediate GLI1 expression. Unlike a recent report, Hh pathway activity in ERMS tumors did not correlate with a unique clinical phenotype.
Conclusions
Our findings support a role for Hh pathway activation in the genesis of a subset of ERMS and US tumors. Hh signaling may represent a novel therapeutic target in affected tumors.
Keywords: Rhabdomyosarcoma, Undifferentiated sarcoma, Gorlin’s syndrome, GLI, Hedgehog
INTRODUCTION
Signaling of the evolutionarily conserved Hedgehog (Hh) pathway is a critical component of embryonic development [1]. The vertebrate hedgehog homologues, sonic (Shh), Indian (Ihh), and desert (Dhh), initiate pathway signaling by binding to the 12 pass transmembrane PTCH receptor protein. In the unbound state, PTCH inhibits the G-protein coupled phosphoprotein receptor smoothened (SMO). In the presence of Hh, the Hh-PTCH complex is internalized and the repression of PTCH on SMO is relieved. SMO signaling is linked through the activity of Suppressor of fused (SUFU) to the expression of Hh pathway effectors GLI1, GLI2, and GLI3. While GLI3 functions as a transcriptional repressor, GLI2 is primarily a transcriptional activator. The zinc finger transcription factor GLI1, a potent oncogene and a key effector of normal and aberrant SHH pathway signaling, functions solely as a transcriptional activator [2]. While hedgehog signaling is generally absent and not essential in adult organisms, aberrant post-embryonic Hh signaling has been detected in a variety of pathological conditions including many pediatric and adult malignancies such as basal cell carcinoma (BCC), medulloblastoma, gastrointestinal cancers, and prostate carcinoma [3-6].
Signaling of the Hh pathway is also implicated in the genesis of a subset of the childhood tumor rhabdomyosarcoma (RMS). Heterozygous Ptch +/− mice on the CD1 genetic background develop RMS tumors with a histology consistent with the embryonal (ERMS) subtype at a frequency of 10%, suggesting a role for aberrant Hh signaling in ERMS tumorigenesis [3]. Likewise, several case reports describe RMS tumors in children with Nevoid Basal Cell Carcinoma Syndrome (NBCCS or Gorlin Syndrome), an autosomal dominant cancer predisposition syndrome linked to germline mutations of the tumor suppressor gene PTCH [4,5]. Of note, genomic amplification of chromosomal region 12q13-15 containing GLI1 has been identified in a subset of alveolar RMS (ARMS) tumors [6].
A study utilizing in situ hybridization (ISH) has detected GLI1 and PTCH mRNA expression in formalin-fixed samples of paraffin-embedded RMS tumors, establishing a role for Hh signaling in the genesis of human RMS tumors [7]. A more recent study found that Hh genes PTCH, GLI1, and GLI3 were more highly expressed in ERMS and PAX3- and PAX7-FOXO1 fusion negative ARMS tumors than fusion positive ARMS [8]. To further explore Hh pathway activity in RMS tumors, we measured the mRNA expression of the Hh pathway members GLI1 and PTCH in frozen archival pediatric RMS and undifferentiated sarcoma (US) specimens. In tumors with evidence of marked Hh pathway activity, molecular mechanisms of pathway activation were sought. The presence of Hh pathway activation was also correlated with clinical features.
METHODS
RMS cell lines and gene expression by ribonuclease protection assay (RPA)
Established ERMS cell lines (Birch, SMS-CTR, TTC-442, and RD) and ARMS cell lines (RH5, RH18, RH28, RH30, RHRKMP4, and CW9019) were maintained in Dulbecco modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The human myoblast cell line HMP8 was cultured in Ham’s F10 media supplemented with 20% FBS, 0.5% chick embryo extract, and 0.12 M CaCl2. RNA was extracted from actively dividing cells by guanidinium isothiocyanate cell lysis using RNA-STAT (Tel-Test B, Friendswood, TX) followed by phenol/chloroform extraction and ethanol precipitation. To investigate the role of the Hh pathway in RMS cell lines, antisense riboprobes were constructed to measure the expression of various Hh pathway genes in ARMS and ERMS tumor-derived cell lines. To construct a PTCH riboprobe, a 410 base pair portion of PTCH cDNA corresponding to exons 12 to 14 was subcloned into the pSP72 vector. A 430 base pair portion of human GLI1 cDNA corresponding to exons 3 to 6 was subcloned into pSP72 to create a GLI1 riboprobe. In vitro transcription using either SP6 (PTCH) or T7 (GLI1) RNA polymerase (Maxiscript, Ambion, Austin, TX) and 32P-labeled UTP produced radiolabeled antisense probes. Total RNA was hybridized to the antisense probe at 42°C for 18 to 20 hours. Samples were then treated with RNases A1 and T1 at 37°C for 30 minutes, degrading any non-hybridized RNA. Hybridized samples were fractionated by denaturing polyacrylamide gel electrophoresis for 4.5 hours. Autoradiography and phosphorimaging were used to visualize the bands. In both the GLI1 and PTCH assays, an internal GAPDH probe was used to control for loading variation and RNA integrity.
Analysis of human tumor specimens
Human RMS specimens were derived from Intergroup Rhabdomyosarcoma Study (IRS)-III (1984 to 1991) and IV (1991 to 1997) patients. Total RNA was extracted from frozen tissue by guanidinium isothiocyanate cell lysis using RNA-STAT (Tel-Test), followed by phenol/chloroform extraction and ethanol precipitation. RNA was assayed by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) (ABI Prism 7700, Applied Biosystems, Foster City, CA). Each experimental reaction was duplexed with an 18S rRNA internal control assay (Applied Biosystems) to account for sample loading error and RNA degradation. The qRT-PCR reactions used 200 ng of DNase-treated RNA. Reagents obtained from Applied Biosystems were used at the following concentrations: TaqMan buffer (1x), magnesium chloride (5.5 mM), nucleotides including dUTP (300 μM), AmpliTaq Gold DNA polymerase (0.025 units/ml), Multiscribe reverse transcriptase (0.25 units/mL), and RNase inhibitor (0.4 units/mL). The GLI1 forward and reverse primer concentrations were 0.25 μM, and the GLI1 probe concentration was 0.1 μM. The PTCH primer concentrations were 0.4 μM and probe concentrations was 0.1 μM. The 18S internal control primer concentration was 0.07 μM and probe concentration was 0.1 μM. After determining the cycle number at which the fluorescent level reaches a designated threshold level, the relative levels of RNA transcripts were extrapolated from standard curves generated during the run. Standard curves were generated by serial dilutions of RNA from RMS cell lines RH30-A and RH30-P for GLI1 and PTCH assays, respectively. All experiments were run in triplicate. Multiplexed reactions measured the experimental gene and 18S control simultaneously.
Measurement of GLI1 copy number in RMS tumors
Amplification of GLI1 was measured in genomic DNA derived from RMS tumors by quantitative PCR (qPCR). The GLI1 gene copy number was compared to that of the MGP gene, an unrelated gene on the short arm of chromosome 12. Various dilutions of lymphocyte (diploid GLI1 copy number) and RH30 (approximately 10-fold GLI1 amplification) DNA were used as controls and to construct standard curves. PCR primer sequences are listed in Supplemental Appendix I. The relative GLI1 to MGP ratio was then calculated based on the threshold cycle identified for a particular DNA sample. Southern hybridization of selected ARMS tumors with known GLI1 amplification was used to validate the qPCR methodology. The distribution of ERMS, ARMS, and US tumors among high, intermediate, and low GLI1 expression groups was compared using the likelihood ratio chi-square test (JMP software, SAS, Cary, NC).
Sequence analysis of PTCH and SMO
To identify mutations of PTCH, tumor RNA was reverse-transcribed and the entire open reading frame was amplified with RNA LA PCR kit version 1.1 (Takara Bio, Madison, WI) and then gel purified (Qiagen, Valencia, CA). To avoid contamination from non-specific PCR products, a second nested PCR reaction was performed followed by gel purification. The amplification products underwent automated direct sequencing. PTCH was screened for mutations in 6 tumors that demonstrated Hh pathway activation as evidenced by GLI1 and PTCH expression. DNA sequencing was performed at the University of Pennsylvania sequencing core with ABI 377 and 373A Stretch Sequencers (Applied Biosystems).
Two recurrent SMO mutations in exons 9 and 10, detected in BCC, were screened by restriction enzyme digestion of amplified DNA sequences. Exon 9 was amplified by PCR and digested with BstN1 as described previously [9]. The recurrent G→T transversion (W535L) removes the BstN1 site and can be detected by electrophoretic analysis of PCR products digested with BstN1. A similar method using Alw1 was used to screen for a recurrent mutation in exon 10.
Analysis of SHH expression
Tumor and cell line RNA samples were reverse transcribed with 1 mM dNTP, 5 mM MgCl2, 25 μM random hexamers, and reverse transcriptase at 2.5 units/μL in PCR buffer II (Applied Biosystems). Amplification of the cDNA utilized 1.25 mM MgCl2, PCR buffer II, and SHH specific primers spanning exons 1 and 2 of the gene. (See supplemental materials for PCR primer sequences). PCR parameters included 35 cycles of 94°C for 60 seconds, 65°C for 60 seconds, and 72°C for 120 seconds. PCR products were visualized by ethidium bromide staining on 2% agarose gels. The FOXO1 gene was used as an internal control to confirm the integrity of the RNA. To further increase sensitivity, RT-PCR products were hybridized to a SHH-specific oligonucleotide end-labeled with 32P-dATP and visualized by autoradiography. The A2243 synovial sarcoma cell line and HEK293 human embryonic kidney cell line, identified from a panel of available cell lines, were used as positive controls.
Clinical correlation with Hh activity
Tumors with GLI1 expression greater than 0.1 of that expressed by RH30-A were considered high expressers. We compared patients with high GLI1 expression to those with intermediate or low expression. Previously collected clinical data from all evaluated IRS-III and IV cases was analyzed. The clinical characteristics considered in tumors with and without SHH activation included: age at diagnosis, gender, primary tumor site, tumor stage, IRS clinical group, local invasiveness, and presence of nodal or distant metastasis. Overall and event-free survival were determined by the method of Kaplan and Meier [10]. Survival curves were compared by the log-rank test [11]. The comparisons of the distribution of GLI1/18S ratio by patient characteristics were made using Fisher’s exact test for contingency tables.
Microarray analysis
Microarray analysis
The Pediatric Cooperative Human Tissue Network tumor bank (Columbus, OH) and the Children’s Hospital Los Angeles institutional tumor bank provided frozen RMS tissue for analysis. Data management and analysis were conducted using the Genetrix suite of tools for microarray analysis (Epicenter Software, Pasadena, CA). Probe set modeling and data preprocessing were derived using the ProbeProfiler algorithm (Corimbia, Berkeley, CA). The Affymetrix (Santa Clara, CA) U133A GeneChip tumor microrray data set, sample clinical covariates, and microarray protocols can be found on the National Cancer Institute Director’s Challenge1 and University of Southern California/Children’s Hospital Los Angeles Genome Core2 Web sites. ANOVA was also used for statistical analysis of differential gene expression between GLI1 expression groups.
Hh pathway analysis
Data was analyzed through the use of Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA). A data set containing gene identifiers and corresponding expression values was uploaded into the application. Each identifier was mapped to its corresponding object in Ingenuity’s Knowledge Base. A p-value cutoff of 0.05 was set to identify molecules whose expression was significantly differentially regulated. These molecules, called network eligible molecules, were overlaid onto a global molecular network developed from information contained in Ingenuity’s Knowledge Base.
RESULTS
Hh activity in human RMS cell lines
SHH, GLI1, SMO, and PTCH mRNA expression in RMS cell lines were measured by RPA, comparing the expression of each gene to a GAPDH internal control. We studied 3 subclones of RH30, an ARMS cell line with the characteristic PAX3-FOXO1 fusion. The RH30 subclones have comparable amplification of GLI1 (8 to 12-fold). ‘RH30-D’ is an early passage cell line obtained directly from St. Jude Children’s Research Hospital. ‘RH30-P’ is a subclone of RH30-D, and ‘RH30-A’ was purchased from American Type Culture Collection (ATCC, Manassas, VA). RH30A and RH30P were found to have elevated levels of GLI1 mRNA expression. Four other PAX3-FOXO1-positive ARMS cell lines (RH5, RH18, RH28, and RHKMP4), an ARMS cell line with the variant PAX7-FOXO1 fusion (CW9019), four ERMS cell lines (RD, SMS-CTR, TTC-442, and Birch) and a human myoblast cell line (HMP8) expressed relatively low or undetectable levels of GLI1 and PTCH (Figure 1). RH30A demonstrated a 13-fold higher expression of GLI1 than other ARMS tumor cell lines. Some cell lines had essentially no GLI1 expression (RHKMP4 and the human myoblast HMP8), while others had detectable, but significantly lower expression compared to RH30-A (RH5 and RH18). GLI1 expression in the four ERMS cell lines was similar to non-amplified ARMS cell lines.
Figure 1. GLI1 and PTCH expression in human RMS cell lines.
The levels of GLI1 (A) and PTCH (B) mRNA were measured by RNase protection assay (RPA) using GAPDH as an internal control. The relative mRNA levels were extrapolated from a standard curve generated during the run. RPA expression of GLI1 was validated by qRT-PCR (C).
Varying degrees of PTCH expression were detected in the 3 subclones derived from RH30 (Figure 1). While RH30-D expressed relatively low levels of GLI1 and PTCH, RH30-A expressed high GLI1, but low PTCH. Finally, RH30-P was found to express higher levels of GLI1 and PTCH. Other ARMS cell lines as well as ERMS cell lines expressed low PTCH mRNA levels.
Hh activity in human RMS and US tumors
Quantitative RT-PCR (qRT-PCR) was used to measure GLI1 and PTCH mRNA levels in 70 RMS tumor specimens. GLI1 expression in RH30-A (10-fold GLI1 amplification and high level of expression) and RH5 (normal GLI1 copy and low detectable expression) were used as positive controls. We confirmed that GLI1 expression in these cell lines differs by an order of magnitude. For these studies, we selected 0.1 of the GLI1/18S ratio of RH30-A as the lower cut off for high GLI1 expression and 0.01 as the upper cut off for low GLI1 expression. These values were based upon the calculation of 5 and 2 standard deviations, respectively, above the mean GLI1 level in non-amplified RMS cell lines. Levels between 0.01 and 0.1 were designated intermediate.
A spectrum of GLI1 mRNA levels was detected among ERMS tumors (Figure 2). We classified ERMS tumors into high, intermediate, and low GLI1 expressing groups. Fifteen of seventy (21%) ERMS tumors expressed high levels of GLI1, which was defined as greater than 0.1 or 10% of the GLI1/18S ratio expressed in RH30-A. Twenty-seven tumors (39%) were classified as intermediate GLI1 expressers, producing between 0.01 and 0.1 of the level in RH30-A. The remaining tumors (40%) expressed low GLI1 levels, which was defined as less than 0.01 of the level of RH30-A GLI1/18S ratio. To assess the significance of high GLI1 mRNA levels in ERMS tumors, we measured the expression of PTCH, a downstream target of the Hh pathway via autoregulatory feedback. While ERMS tumors with intermediate or low GLI1 mRNA levels expressed mean PTCH PTCH/18S ratio of 0.016 of RH30-P, tumors with high GLI1 levels expressed significantly higher PTCH/18S ratios (mean = 0.096, p = 0.011). The finding of up-regulation of GLI1 with or without upregulation of PTCH provides the potential for downstream activation of the Hh pathway and suggests a role for the Hh pathway in the development and pathogenesis of a subset of ERMS tumors.
Figure 2. GLI1 and PTCH expression in ERMS tumors.
GLI1 and PTCH RNA levels were measured in triplicate by qRT-PCR with an 18S rRNA internal control. The expression of GLI1/18S and PTCH/18S RNA in ERMS tumors is depicted in box plots. The boxes represent the 25th and 75th percentile of GLI1 or PTCH expression. The median GLI1/18S ratio represented by the bars within the boxes was 0.013, 0.03, and 0.0035 for high, intermediate, and low GLI1 expressing tumors, respectively. For the same categories of GLI1 expression, PTCH/18S expression was 0.035, 0.0092, and 0.0054. Standard deviation error bars are shown. Dots represent the distribution of GLI1 and PTCH expression within each ERMS subset.
Significant GLI1 or PTCH expression was considerably less frequent in ARMS tumors than in ERMS tumors. None of the ARMS tumors expressed high GLI1/18S ratios corresponding to greater than 0.1 of the ratio in RH30-A. Only 6 of 20 (30%) ARMS tumors expressed intermediate GLI1 expression, and the remaining 14 (70%) demonstrated low GLI1 expression. A significant difference was found in the distribution of ARMS and ERMS among high, intermediate, and low GLI1 expression subsets, p < 0.005. ARMS tumors with and without GLI1 gene amplification were assayed, demonstrating that involvement of GLI1 in the chromosome 12q13-15 amplicon found in some ARMS tumors does not usually lead to increased GLI1 gene expression.
GLI1 and PTCH expression was also detected in a subset of US tumors collected during the IRS-IV study (Figure 3). Of 13 tumors, 4 tumors demonstrated GLI1/18S expression over 0.1 of that expressed by RH30-A. Again, mean PTCH expression was higher in the group of tumors that expressed more abundant GLI1 mRNA (0.82 versus 0.01, p = 0.049). The expression of GLI1 among ARMS, ERMS, and US tumors is depicted Figure 4.
Figure 3. GLI1 and PTCH expression in US tumors.
GLI1 and PTCH RNA levels were measured in triplicate by qRT-PCR with an 18S rRNA internal control.
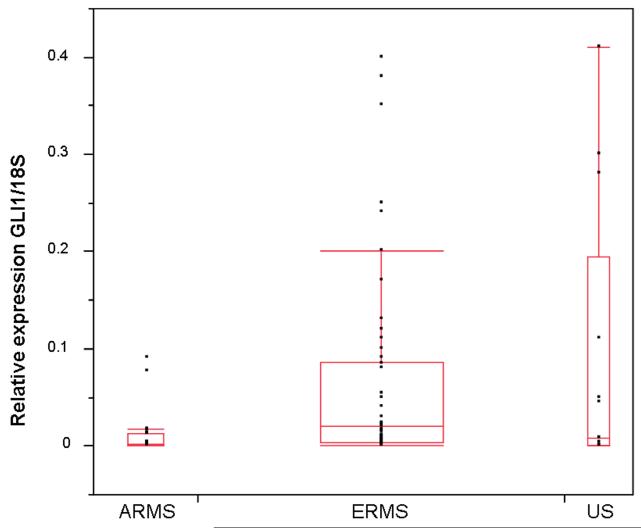
Figure 4. Comparison of GLI1/18S ratios in ARMS, ERMS, and US tumors.
The ratios are depicted in box plots. The median GLI1/18S ratios found are depicted by bars within the squares: ARMS 0.0014, ERMS 0.02, and US 0.0075. The boxes represent the 25th and 75th percentile of GLI1 expression. Standard deviation error bars are shown. Dots represent the distribution of GLI1 tumor expression within each tumor type.
Analysis of mechanisms of Hh pathway signaling
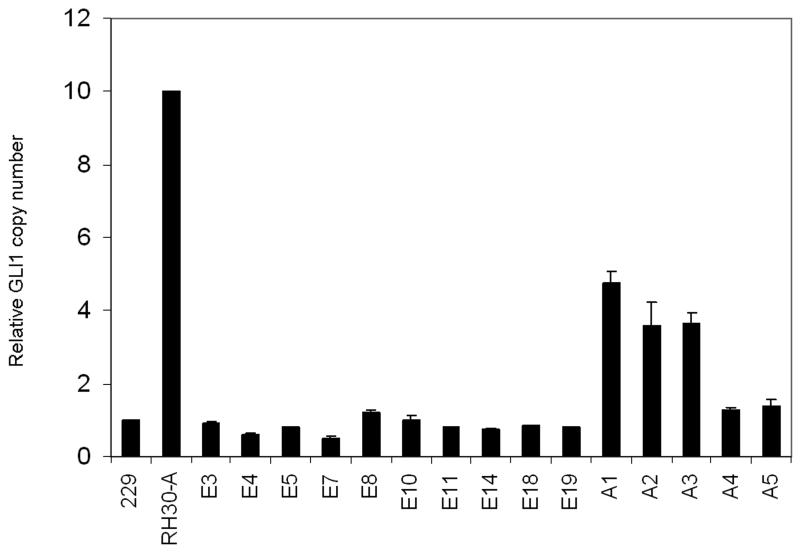
ERMS tumors with high GLI1 and PTCH expression were evaluated for potential molecular mechanisms of Hh pathway activation. Analysis of GLI1 genomic copy number was undertaken by qPCR. Our results confirmed the previous finding of GLI1 amplification by Southern blot in a subset of ARMS tumors. However, none of the ERMS tumors tested, including tumors with high (n=8) and intermediate (n=2) GLI1 expression, demonstrated GLI1 amplification (Figure 5).
Figure 5. GLI1 copy number in ERMS and ARMS tumors.
Using qPCR, the relative GLI1 copy number was determined in comparison to MGP, an unrelated gene on the opposite arm of chromosome 12. The methodology was validated by comparison to results obtained by Southern methodology. Standard curves were generated from analysis of DNA from normal lymphocyte (229) and the RH30-A cell line.
As a negative regulator of Hh signaling, PTCH was evaluated by sequence analysis of cDNA derived from reverse transcribed tumor RNA. Since the ERMS tumors of interest expressed high levels of PTCH, tumor RNA was utilized in the sequencing of the PTCH gene. This method decreased the risk of contamination from surrounding normal tissue or stroma since the majority of PTCH RNA would likely originate from tumors cells with high PTCH expression. Six ERMS tumors with high GLI1 and PTCH expression (#2, 3, 4, 5, 7, and 10) were evaluated. No point mutations of PTCH were detected, but a number of polymorphisms were found (Table I). Four of six tumors demonstrated heterozygous sequence variation, confirming biallelic expression of PTCH. This finding suggests that loss of PTCH function through large deletions or gene silencing was not required for Hh pathway activation in this cohort of tumors.
Table I.
PTCH polymorphisms
| Tumor | Sequence variation | Consequence |
|---|---|---|
| E2 | None detected | |
| E3 | 3158 T>C (heterozygous) | Asparagine>Asparagine |
| E4 | 2430 A>G (heterozygous) | Asparagine>Serine |
| E5 | 2910 T>C (heterozygous) 3932 C>T (heterozygous) |
Serine>Serine Proline>leucine |
| E7 | None detected | |
| E10 | 1641 C>T (homozygous) | Serine>serine |
To analyze recurrent activating SMO mutations, PCR products of exons 9 and 10 of selected ERMS tumors were digested by BstN1 and Alw1 restriction enzymes. This methodology allows the detection of a G-to-T transversion of exon 9 at base pair 1604, which changes codon 535 from tryptophan to leucine, and G-to-A transition of exon 10 at base pair 1685 resulting in a change of codon 562 from arginine to glutamine. Gel electrophoresis of digested PCR products revealed wild-type sequences of the SMO mutation hot spots in ERMS tumors with high (E2, E3, E4, E7, E8, E10, and E11) and intermediate (E16, and E18) expression of GLI1.
The expression of the PTCH ligand SHH was also analyzed in RMS specimens. Using conventional RT-PCR followed by hybridization to a radiolabeled probe to maximize sensitivity, total tumor RNA was measured for SHH expression. SHH mRNA was not detected in human ERMS tumors with GLI1 and PTCH expression, likely ruling out autocrine SHH production. In addition, SHH expression was not detected in the panel of RMS cell lines including RH30, an ARMS cell line with high GLI1 and PTCH expression, supporting a mechanism of GLI1 upregulation independent of autocrine SHH expression.
Microarray analysis of ERMS tumors
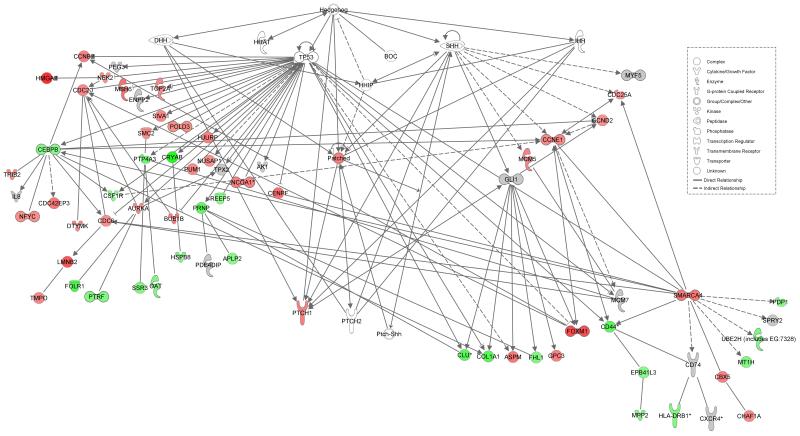
To compare the gene expression profiles of tumors with varying degrees of GLI1 expression, oligonucleotide microarray analysis of frozen RMS tumors was performed. Microarray data was available for 9 high, 6 intermediate, and 12 low GLI1 expressing ERMS tumors. When comparing the expression profiles of high + intermediate tumors versus low GLI1 expressing tumors, 170 differentially expressed genes were identified (p < 0.05). Analysis of the differentially expressed genes by Ingenuity software demonstrated that 56 (33%) of the differentially expressed genes were in pathways downstream of Hh (Figure 6). Among these downstream genes are those that are associated with p53-inducible pathways [12]. The gene list is included in Supplemental Appendix II.
Figure 6. Differentially expressed genes found to be downstream of Hedgehog.
Ingenuity Pathways Analysis was used to identify genes downstream of Hh that were differentially expressed between high + intermediate versus low GLI1. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation. Genes depicted in gray were not differentially expressed in a statistically significant manner (p > 0.05). Nodes are displayed using various shapes that represent the functional class of the gene product.
Clinical Correlation
The level of GLI1 expression of ERMS tumors was correlated with patient clinical characteristics and survival outcome. Of the 70 tumors evaluated, clinical data was available for all but 2 of the tumors, both of which were low GLI1-expressing tumors. The GLI1/18S ratio was not significantly associated with any patient or disease characteristic (Table II). The median follow–up was 5.8 years with a range of 0 to 10.3 years; one patient had no follow-up information. While patients with high GLI1 expression had superior survival outcome, neither failure-free nor overall survival was statistically significant (Figure 7).
Table II.
Clinical characteristics of patients with high, intermediate, and low GLI1 expression
| GLI1/18S ratio | High (n=15) |
Intermediatez (n=27) |
Low (n=28) |
|
|---|---|---|---|---|
| Age (years) (p=0.64) | <5 5 to <10 10 to <15 > or =15 |
5 (16%) 6 (23%) 3 (50%) 1 (25%) |
15 (47%) 10 (38%) 1 (17%) 1 (25%) |
12 (38%) 10 (38%) 2 (33%) 2 (50%) |
| Gender (p=0.48) | Male Female |
9 (20%) 6 (27%) |
17 (37%) 10 (45%) |
20 (43%) 6 (27%) |
| Race (p=0.73) | White Non-white |
11 (26%) 4 (16%) |
16 (37%) 11 (44%) |
16 (37%) 10 (40%) |
|
IRSG tumor stage (p=0.83) |
1 2 3 4 |
4 (22%) 2 (18%) 8 (25%) 1 (14%) |
6 (33%) 3 (27%) 14 (44%) 4 (57%) |
8 (44%) 6 (55%) 10 (31%) 2 (29%) |
| Clinical group (p=0.39) | I II III IV |
3 (19%) 1 (13%) 10 (27%) 1 (14%) |
6 (38%) 1 (13%) 16 (43%) 4 (57%) |
7 (44%) 6 (75%) 11 (30%) 2 (29%) |
|
Primary tumor site (p=1.0 favorable vs. *unfavorable) |
Orbit Head/Neck *Parameningeal GU (non-bladder or prostate) *Bladder/Prostate *Extremity Other |
1 3 3 1 2 1 4 |
- 4 10 4 5 1 3 |
2 - 9 6 4 1 4 |
| Tumor size (p=0.86) | <5cm >5cm Unknown |
5 (17%) 8 (23%) 2 |
13 (43%) 13 (37%) 1 |
12 (40%) 14 (40%) - |
Figure 7. GLI1 expression correlated with patient outcome.
Failure-free survival and GLI1/18S expression (top). Overall Survival and GLI1/18S expression (bottom).
DISCUSSION
Our findings demonstrate that a substantial subset of pediatric ERMS and US tumors possess Hh pathway activity, as evidenced by the expression of GLI1 and PTCH RNA transcripts. In contrast, we found that ARMS tumors did not express GLI1 and/or PTCH over the same predetermined threshold. The finding of Hh pathway activation in ERMS tumors sheds further light on the biology underlying RMS tumorigenesis and may point to novel therapeutic approaches to ERMS tumors. The essential role of Hh signaling in embryogenesis highlights a critical link between development and tumorigenesis of childhood tumors.
Utilizing qRT-PCR methodology, our finding of Hh activation in ERMS tumors is comparable to that of the ISH analysis of RMS tumors described by Tostar et al [7]. While the previous study only quantified GLI1 and PTCH expression as negative, positive (1+), or very strong (2+), our methodology allowed us to further discriminate the relative magnitude of GLI1 and PTCH expression between ARMS and ERMS tumors. More recently, Zibat et al. using microarray and qRT-PCR analysis has shown that Hh activation measured by GLI1, GLI3, PTCH, and MYF5 was most closely associated with ERMS and fusion-negative ARMS tumors [8].
Activation of the Hh pathway is linked to the genesis of a wide variety of human cancers such as BCC [13], medulloblastoma (MB) [14], and digestive tract tumors [15], among other tumor types. While a number of genetic alterations have been linked to aberrant Hh pathway activation in cancer, the unifying effect of these genetic changes is upregulation of the GLI transcription factors. The role of GLI1 as an oncogene is well characterized. Activation of GLI1 induces a transcription program (including feedback upregulation of PTCH) that promotes several aspects of tumorigenesis [16]. Expression of GLI1 has been demonstrated to transform cells and promote local invasion [2].
Previous studies have suggested that a 9q22 tumor suppressor locus may be involved in the pathogenesis of RMS. The study by Tostar et al. utilizing ISH to study Hh pathway activity in RMS tumors detected GLI1 and PTCH mRNA expression in a majority of 23 ERMS and 6 ARMS tumors [7]. Four of nine informative rhabdomyomas and RMS tumors demonstrated allelic loss of the PTCH locus at 9q22. Previous comparative genomic hybridization studies of ERMS tumors have also identified frequent genomic losses involving the 9q22 locus that includes PTCH [6,17]. Despite this previous evidence implicating a 9q22 locus in ERMS, we did not identify PTCH mutations in the 6 analyzed tumors with the high GLI1 and PTCH mRNA levels in the present study. Four of the six tested tumors revealed heterozygous polymorphisms, providing support of biallelic PTCH expression and thereby ruling out allelic loss and epigenetic mechanisms for inactivating one PTCH locus. The other 2 tumors could conceivably be affected by allelic loss, mitotic recombination, epigenetic silencing, or promoter mutation of PTCH. We also acknowledge that the methodology used in our studies did not assess mutations within the PTCH introns, which may affect splicing patterns.
The finding of high GLI1 and PTCH expression is suggestive of Hh pathway activation in a subset of ERMS tumors. While PTCH expression was statistically higher among ERMS tumors demonstrating high GLI1 expression, we note that not all tumors in this category expressed high PTCH levels. We speculate that those tumors with high GLI1 and low PTCH may represent a subset of tumors with silencing of PTCH either through gene deletion or promoter methylation among other causes. Without further inhibition by PTCH, the high GLI1/low PTCH subset may potentially exert more active Hh signaling. Additional findings also support the concept that higher GLI1 expressing tumors are associated with Hh pathway activation. Analysis of oligonucleotide microarray expression data demonstrated that genes differentially expressed between high and intermediate versus low GLI1 expression include an abundance of Hh target genes (Figure 6). Together, the statistically increased expression of PTCH and association of downstream Hh targets support the concept that high GLI1 expression is linked to Hh pathway activity in ERMS tumors.
Our findings suggest that the expression of GLI1 +/− PTCH in these ERMS tumors may be due to a novel mechanism. Though we have ruled out autocrine synthesis of SHH, one possible mechanism that deserves further exploration in ERMS is paracrine or endocrine release of SHH, such that it would not be detectable in the tumor RNA [18]. Likewise, the present study did not analyze potential loss of SUFU function [19]. Likewise, a more detailed analysis of SMO may have identified mutations or other genetic alterations responsible for Hh pathway activation. Another novel mechanism for PTCH and SMO independent GLI1 expression has been implicated in the Ewing Sarcoma Family of Tumors (ESFT) [20]. Expression of the t(11;22)-associated ESFT fusion oncoprotein EWS-FLI was demonstrated to enhance the expression of GLI1, contributing to the transforming properties of EWS-FLI in NIH3T3 fibroblasts. These findings provide evidence that GLI1 expression may result from mechanisms independent of classic Hh pathway mutations.
Comparative genomic hybridization studies have identified a subset of ARMS tumors with an amplicon involving chromosome 12q13-15, which includes GLI1 and other cancer-related genes [17]. We assayed several ARMS tumors with genomic amplification of GLI1, yet evidence of GLI1 overexpression was not detected. These findings suggest that genes included in the 12q13 amplicon other than GLI1 such as CDK4 or MDM2 may be the more critical, amplified components of that chromosomal region in affected ARMS tumors. This finding may be relevant to the often studied ARMS cell line RH30 which has a 10-fold amplification of GLI1 and correspondingly high expression of GLI1 RNA levels. This finding appears to represent an acquired event during the establishment of the tumor cell line rather than a characteristic typical of ARMS tumors.
In a limited number of US tumors, we also found evidence of Hh signaling among this ill-defined group of tumors. US tumors are a heterogeneous group of mesencyhmal malignancies with no immunohistochemical evidence of differentiation or cytogenetic findings characteristic of other soft-tissue sarcomas such as RMS [21]. Our findings suggest a common mechanism of tumorigenesis between a subset of ERMS and a subset of US tumors. Alternatively, affected US tumors may represent unrecognized, poorly differentiated, ERMS tumors.
The expression of GLI1 mRNA transcripts in ERMS tumors did not correlate with survival or other clinical characteristics such as age, tumor stage, group, or primary anatomic site. Despite activity in various types of malignancies, the Hh pathway has not commonly been utilized for risk stratification. For instance, in the case of the desmoplastic variant of the embryonal tumor MB, Hh activity is not associated with a clinical phenotype distinct from other MB tumors. A recent report found that PTCH expression correlates with a poor clinical outcome in ERMS and fusion-negative ARMS patients [8]. Analysis of PTCH expression in the present study did not identify such a clinical correlation, though we acknowledge that this study focused on a convenience sample and not a truly randomized cohort. To further determine the clinical significance of these molecular events, a systematic prospective analysis of Hh signaling in RMS tumors would be a worthwhile undertaking in future RMS trials. A larger cohort of patients, such as may be available from a single large trial, may potentially identify unique clinical characteristics associated with Hh signaling in ERMS.
Patients with ERMS and US tumors demonstrating Hh pathway activity may be candidates for targeted therapies that inhibit Hh signaling. While the majority of ERMS patients experience favorable outcomes, treatment failure and toxicity remain substantial [22]. Small molecule inhibitors of SMO are being studied in various Hh related malignancies [23-25]. Such agents would be most appealing in patients with tumors that possess activating mutations of PTCH or SMO but likely not alterations more distal in the pathway [26]. Without identifying a consistent mechanism of Hh pathway activation, inhibitors of GLI1 or more distal effectors of the pathway may ultimately be necessary to effectively inhibit signaling in affected ERMS tumors [27].
Supplementary Material
Acknowledgements
We wish to thank Charles Emerson Ph.D. for providing the human myoblast cell line HMP8, Timothy Cripe M.D., Ph.D. for providing the ARMS cell line RHRKMP4, and the IRSG pathologists for supplying the RMS tumor specimens.
Funding:
This work was supported by U10 CA98543, U10 CA98413, U10 CA24507 from the National Cancer Institute, National Institutes of Health, Bethesda, MD, USA. Additional support provided by the Group Chair’s Translational Research Developmental Fund Award from the Children’s Cancer Group (FGB), Children’s Oncology Group Young Investigator’s Award (JGP), and Ladies Auxiliary to the VFW Cancer Research Fellowship (JGP).
Footnotes
Conflicts of interest: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Ruppert JM, Vogelstein B, Kinzler KW. The zinc finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol Cell Biol. 1991;11(3):1724–1728. doi: 10.1128/mcb.11.3.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahn HW, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcoma and radiation hypersensitivity in a mouse model of Gorlin Syndrome. Nature Medicine. 1998;4(5):619–622. doi: 10.1038/nm0598-619. L. [DOI] [PubMed] [Google Scholar]
- 4.Gorlin R. Nevoid Basal-Cell Carcinoma Syndrome. Medicine. 1987;66(2):98–113. doi: 10.1097/00005792-198703000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Beddis IM, Bullimore J. Case Report: Nasopharyngeal Rhabdomyosarcoma and Gorlin’s Naevoid Basal Cell Carcinoma Syndrome. Medical And Pediatric Oncology. 1983;11:178–179. doi: 10.1002/mpo.2950110309. MG. [DOI] [PubMed] [Google Scholar]
- 6.Bridge JA, Liu J, Qualman SJ, et al. Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer. 2002;33(3):310–321. doi: 10.1002/gcc.10026. [DOI] [PubMed] [Google Scholar]
- 7.Tostar U, Malm CJ, Meis-Kindblom JM, et al. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208(1):17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 8.Zibat A, Missiaglia E, Rosenberger A, et al. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 29(48):6323–6330. doi: 10.1038/onc.2010.368. [DOI] [PubMed] [Google Scholar]
- 9.Lam CW, Xie J, To KF, et al. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene. 1999;18(3):833–836. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan E, Meier P. Nonparametric estmation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 11.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50(3):163–170. [PubMed] [Google Scholar]
- 12.Po A, Ferretti E, Miele E, et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 29(15):2646–2658. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bale AE, Gailani MR, Leffell DJ. The Gorlin syndrome gene: a tumor suppressor active in basal cell carcinogenesis and embryonic development. Proc Assoc Am Physicians. 1995;107(2):253–257. [PubMed] [Google Scholar]
- 14.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 15.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425(6960):846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 16.Yoon JW, Kita Y, Frank DJ, et al. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J Biol Chem. 2002;277(7):5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- 17.Bridge JA, Liu J, Weibolt V, et al. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes Chromosomes Cancer. 2000;27(4):337–344. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Theunissen JW, de Sauvage FJ. Paracrine Hedgehog signaling in cancer. Cancer Res. 2009;69(15):6007–6010. doi: 10.1158/0008-5472.CAN-09-0756. [DOI] [PubMed] [Google Scholar]
- 19.Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31(3):306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 20.Zwerner JP, Joo J, Warner KL, et al. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27(23):3282–3291. doi: 10.1038/sj.onc.1210991. [DOI] [PubMed] [Google Scholar]
- 21.Pawel BR, Hamoudi AB, Asmar L, et al. Undifferentiated sarcomas of children: pathology and clinical behavior--an Intergroup Rhabdomyosarcoma study. Med Pediatr Oncol. 1997;29(3):170–180. doi: 10.1002/(sici)1096-911x(199709)29:3<170::aid-mpo3>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 22.Crist WM, Anderson JR, Meza JL, et al. Intergroup rhabdomyosarcoma study-IV: results for patients with nonmetastatic disease. J Clin Oncol. 2001;19(12):3091–3102. doi: 10.1200/JCO.2001.19.12.3091. [DOI] [PubMed] [Google Scholar]
- 23.Dierks C. GDC-0449--targeting the hedgehog signaling pathway. Recent Results Cancer Res. 184:235–238. doi: 10.1007/978-3-642-01222-8_17. [DOI] [PubMed] [Google Scholar]
- 24.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 26.Yauch RL, Dijkgraaf GJ, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauth M, Bergstrom A, Shimokawa T, et al. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104(20):8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.