Abstract
The homeobox gene extradenticle (exd) acts as a cofactor of Hox function both in Drosophila and vertebrates. It has been shown that the distribution of the Exd protein is developmentally regulated at the post-translational level; in the regions where exd is not functional Exd is present only in the cell cytoplasm, whereas it accumulates in the nuclei of cells requiring exd function. We show that the subcellular localization of Exd is regulated by the BX-C genes and that each BX-C gene can prevent or reduce nuclear translocation of Exd to different extents. In spite of this negative regulation, two BX-C genes, Ultrabithorax and abdominal-A, require exd activity for their maintenance and function. We propose that mutual interactions between Exd and BX-C proteins ensure the correct amounts of interacting molecules. As the Hoxd10 gene has the same properties as Drosophila BX-C genes, we suggest that the control mechanism of subcellular distribution of Exd found in Drosophila probably operates in other organisms as well.
Keywords: extradenticle, Hox genes, nuclear translocation, post-translational control
The homeotic genes determine the characteristic development (identity) of Drosophila body segments (Lawrence and Morata 1994; Lawrence and Struhl 1996). They encode proteins that contain a DNA-binding domain known as a homeodomain and regulate the transcription of target genes by binding to their regulatory sequences. Although the various homeotic genes determine different segment identities in vivo, the homeotic proteins show similar binding specificity in vitro (Hoey and Levine 1988) raising the problem of how the in vivo specificity is achieved.
One factor that contributes to the specificity of the homeotic function is the gene extradenticle (exd). Mutations in exd alter the identity of some body segments without altering the expression pattern of the homeotic genes (Peifer and Wieschaus 1990; Rauskolb et al. 1993). The Exd protein contains a homeodomain and acts as a cofactor of the homeotic proteins; in vitro binding experiments have also shown that Exd can cooperatively bind to DNA with the Ultrabithorax (Ubx) and Abdominal-A (Abd-A) proteins, increasing their affinity for some target sites (Chan et al. 1994; van Djik and Murre 1994). Given the high degree of conservation of the Exd product (Rauskolb et al. 1993), it is likely that exd vertebrate homologs, the Pbx genes, also act as cofactors. In fact, there is evidence that the Pbx proteins interact with Hox proteins to mediate their binding specificity (Kamps et al. 1990; Nourse et al. 1990; Monica et al. 1991; Flegel et al. 1993; Chang et al. 1994; Phelan et al. 1994; Lu et al. 1995).
Besides the evidence for molecular interactions with homeotic products, there are some results indicating distinct levels of regulation of exd function. In wild-type embryos the exd RNA is distributed uniformly throughout the body segments in early development, but at the extended germ-band stage there are fewer exd transcripts in the posterior abdomen than in the thorax (Rauskolb et al. 1993). As embryos deficient for the Bithorax Complex (BX-C) genes show high and uniform levels of transcripts in thorax and abdomen at this stage (Rauskolb et al. 1993), it suggests a negative regulation of exd transcription by the BX-C genes. In addition, there is another level of regulation: The Exd product accumulates in early embryos only in the cell cytoplasm and is later translocated to the nucleus (Mann and Abu-Shaar 1996; Aspland and White 1997). The nuclear accumulation is also spatially regulated; there is more nuclear Exd in the cells of the thoracic segments than in those of the abdominal segments, both in the epidermis and the CNS. In the imaginal discs the nuclear localization of Exd is restricted to the proximal regions of the legs, wings, halteres, and antennae and is cytoplasmic in the distal regions (Mann and Abu-Shaar 1996; Aspland and White 1997). Functional tests (Gonzalez-Crespo and Morata 1995; Rauskolb et al. 1995) indicate that exd is developmentally active only in the regions where its product is nuclear.
Mosaic analyses have also shown a general requirement for exd in various adult patterns (Gonzalez-Crespo and Morata 1995; Rauskolb et al. 1995). The elimination of exd activity in clones of imaginal cells give rise to a large variety of homeotic phenotypes in the different body segments. One particularly intriguing phenotype is the antenna to leg transformation, characteristic of the dominant Antp mutations and of experiments forcing gain of Antennapedia (Antp) function (Schneuwly et al. 1987; Gibson and Gehring 1988; Kaufman et al. 1990). A similar Antp-like transformation is also produced when the BX-C genes Ubx, abd-A and Abd-B are expressed ectopically in the antenna (Mann and Hogness 1990; Kuziora 1993; Casares et al. 1996). The transformation induced by abd-A and Abd-B is surprising because these genes normally specify the development of abdominal segments (Sanchez-Herrero et al. 1985) that do not possess legs. Moreover, in other body regions the phenotype of exd mutant clones resembles the loss of function of some homeotic genes. For example, exd− clones in the metanotum produce a transformation into mesonotum, a typical Ubx− phenotype (Morata and Garcia-Bellido 1976), or a thoracic transformation of the head capsule that resembles a labial− (lab−) phenotype (GonzalezCrespo and Morata 1995).
These observations suggest the existence of various interactions between exd and the homeotic genes, some of which are described in this paper. We show that the Drosophila proteins Ubx, Abd-A, and Abd-B and the mouse Hoxd10 protein can reduce or eliminate exd function by preventing nuclear translocation of Exd, suggesting that in normal development they modulate exd activity. This effect on exd provides an explanation for the unexpected phenotypes caused by ectopic expression of abd-A and Abd-B. Conversely, nuclear localization of Exd is necessary for the normal expression and function of Ubx and abd-A. We propose that there is a mechanism to ensure the appropriate relative amounts of interacting Hox/Exd products and that the BX-C genes contribute to this mechanism.
Results
Distribution and regulation of the Exd product
The distribution of exd transcripts during embryogenesis has been described (Rauskolb et al. 1993). exd RNA has both a maternal and a zygotic component; the maternal component is uniform and persists until embryonic stage 9. The expression of the zygotic component is modulated (Rauskolb et al. 1993), and as a result the overall pattern of exd transcripts is nonuniform, being higher in the thorax than in the abdomen. In the embryo Exd protein is detected at cellular blastoderm stage, but then it is exclusively cytoplasmic (Mann and Abu-Shaar 1996; Aspland and White 1997). After the extended germ-band stage accumulates predominantly in the nuclei of the thoracic segments in both the epidermis and the CNS, whereas there is a gradation of nuclear expression in the abdomen that diminishes toward the posterior end, where the Exd protein is detected only in the cytoplasm.
We have studied and compared the distribution of exd RNA and protein in embryos containing only either the maternal (mat+ zyg−) or the zygotic (mat− zyg+) component of exd. Maternal exd RNA lasts for a few hours (Fig. 1A–C), and by stage 9 of embryogenesis it is undetectable, in agreement with previous work (Rauskolb et al. 1993). Protein produced by maternal RNA is stable; in mat+ zyg− embryos it can be detected until late stages of embryogenesis although at lower levels than the protein produced by zygotic expression. Most importantly, maternally derived protein translocates into nuclei (Fig. 1D) and the pattern of cells that do so is similar or identical to the wild type. This suggests that the protein translated from maternal RNA is functional, a result that is consistent with experiments showing that the presence of extra copies of the exd gene during oogenesis can rescue the exd zygotic phenotype (Peifer and Wieschaus 1990). It also indicates that regulation of protein distribution is not dependent on transcriptional control.
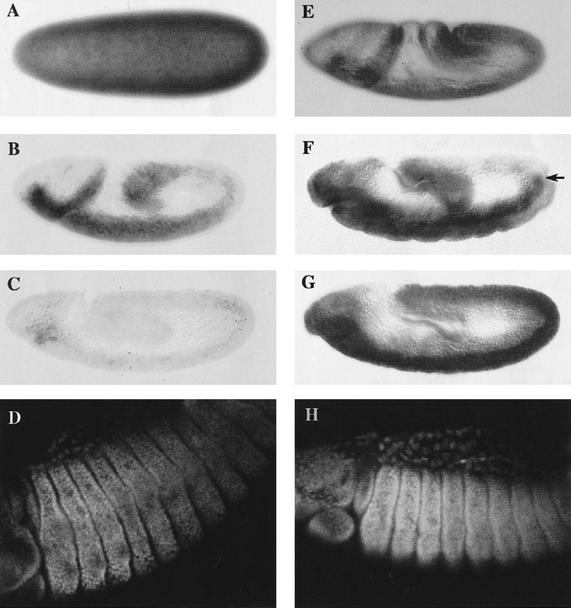
Figure 1.
Distribution of exd RNA and protein in maternal and zygotic exd mutants. All panels show embryos with anterior to the left and dorsal up. (A–D) Embryos of exdY012 genotype and therefore, have Exd product exclusively from maternal origin (mat+ zyg−). (E–H) Embryos containing only Exd product of zygotic origin (mat− zyg+). (A) mat+ zyg− embryo at the blastoderm stage stained for exd RNA. The maternal product is ubiquitously distributed throughout the embryo. (B) mat+ zyg− embryo at stage 8–9 of embryogenesis. Maternal exd RNA is still present, but at lower levels. (C) Stage 10 embryo of the same genotype in which the maternal RNA is almost undetectable. (D) mat+ zyg− embryo after germ-band retraction stained for Exd protein. Although no maternal RNA is detectable in the embryo from stage 11 onwards, the protein is still present in later stages of embryogenesis. Note the nuclear localization of Exd in the cells of thoracic segments. (E) Stage 8 mat− zyg+ embryo lacking the maternal contribution stained for exd mRNA. The zygotic product is present throughout the body. (F) Stage 11 embryo showing a modulation in the levels of zygotic RNA along the A/P axis. In the ectoderm there is very little expression in the more posterior segments, but note that there is mesodermal exd expression (arrow) that is not in register with the ectoderm. (G) Embryo of approximately the same age as in F stained with the anti-Exd antibody. The Exd zygotic protein is still present in the posterior segments of the embryo, where the mRNA is not detected anymore. (H) Stage 14 mat− zyg+ embryo stained with anti-Exd antibody. The zygotic protein shows high levels of nuclear distribution in the thoracic segments and lower levels in the abdominal segments. The distribution of the zygotic protein is the same as the maternal one, but the levels of the maternal protein are lower (cf. D and H).
Zygotic exd RNA is first synthesized around stage 5 of embryogenesis. In mat− zyg+ embryos, it is originally uniform but is later modulated (Fig. 1E,F). Transcripts cannot be detected in the posterior segments of the body after the extended germ-band stage, although there are differences between the ectoderm and mesoderm (Fig. 1F). Protein derived from zygotic RNA (Fig. 1G) can be detected shortly after initiation of transcription and translocates into nuclei beginning at stage 8–9. The pattern of cells with nuclear Exd is similar to wild type (Fig. 1H).
The finding that Exd is subjected to a subcellular regulation may provide an explanation for previous results concerning exd ectopic expression (Rauskolb et al. 1995). Heat shock-driven exd expression does not have a phenotypic effect, but it is able to rescue the zygotic phenotype of exd mutants (Rauskolb et al. 1995). The excess of product in the regions where it is not translocated to the nuclei may remain in the cytoplasm and therefore, not be functional. We have checked this hypothesis by making use of the GAL4/UAS method (Brand and Perrimon 1993) to generate embryos containing greater amounts of Exd in body regions where it is normally cytoplasmic. The Gal4-444 and patched (ptc)–Gal4 (Hinz et al. 1994) lines drive expression of the UAS–exd gene (Gonzalez-Crespo and Morata 1996) all over the body or in the anterior compartments respectively. We did not detect any morphological alteration in the cuticle of Gal4-444/UAS–exd or in ptc–Gal4/UAS–exd first instar larvae. Despite their normal phenotype, ptc–Gal4/UAS–exd embryos show high levels of Exd in the anterior part of every thoracic and abdominal segment (Fig. 2A) at the germ-band elongation stage. By the time these embryos retract the germ band, the protein is translocated normally into the nucleus in the thoracic segments, but it is mostly cytoplasmic in the posterior abdominal segments (Fig. 2B). We also observed the normal low levels corresponding to this region, suggesting that the ectopic cytoplasmic Exd protein is unstable (Fig. 2B). These observations suggest that the nuclear transport of Exd is tightly regulated during embryonic development. There is a negative control in the posterior body segments that can cope with high levels of Exd product.
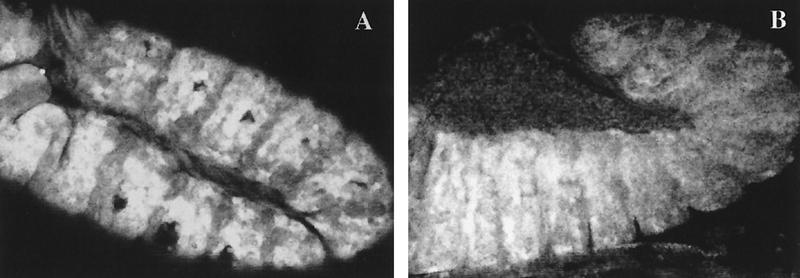
Figure 2.
Ectopic expression of exd. (A) Stage 11 embryo with induced exd expression in the ptc domain stained for Exd protein. The levels of ectopic Exd are high in every anterior compartment. (B) Embryo with the same genotype as A shortly before completion of germ band retraction, close to stage 12, stained as in A. The high levels of Exd are not observed in the posterior end of the embryo where Exd is clearly cytoplasmic.
The Drosophila BX-C genes and the mouse Hoxd10 gene regulate the subcellular localization of Exd at the post-translational level
Because both exd transcripts and nuclear distribution of Exd protein changes along the A/P axis, it seems plausible that the BX-C genes are involved in regulating exd function. We therefore analyzed how the different BX-C genes affect Exd protein distribution.
We find that embryos homozygous for the deficiency Df(3R)P9, lacking the entire Bithorax complex (Lewis 1978), contain Exd at high levels in the nuclei of epidermal cells of both thoracic and abdominal segments (Fig. 3B). Only in the Keilins’ organs of each segment does Exd remain cytoplasmic (Fig. 3A,B), as expected from previous results (Mann and Abu-Shaar 1996; Aspland and White 1997). This rise in the level of nuclear Exd in the abdominal segments (in comparison with the wild-type distribution) already indicates an involvement of the BX-C genes with the subcellular distribution of the product. In contrast, the thorax-determining genes Sex combs reduced (Scr) and Antp do not appear to affect Exd localization, as Scr− Antp− homozygous embryos exhibit normal distribution of Exd (not shown). Furthermore, high levels of Antp protein in ptc–Gal4/UAS–Antp embryos or of Deformed (Dfd) protein in hsp70–Dfd embryos fail to produce any significant effect on Exd subcellular localization.
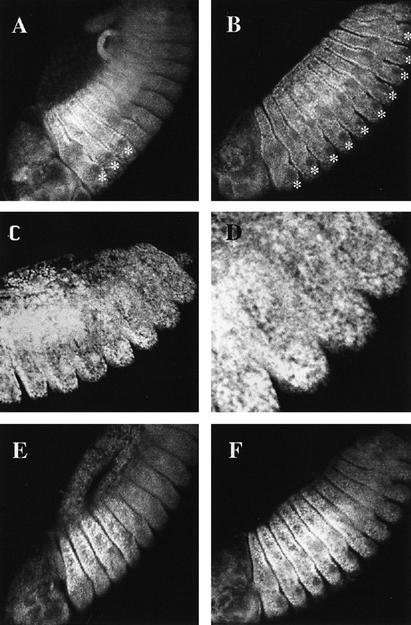
Figure 3.
Effect of BX-C mutations on the subcellular localization of Exd. (A) Wild-type distribution (see text for details). Note the predominant nuclear localization in the three thoracic segments, except in the precursors of the Keilin’s organs (*). (B) Exd distribution in homozygous Df(3R)P9 embryos lacking the BX-C genes. Exd is nuclear in all the thoracic and abdominal segments. Because of the homeotic transformation, all thoracic and abdominal segments develop Keilin’s organs whose precursor cells (*) have cytoplasmic Exd. (C) Exd localization in a mat+ zyg− embryo also homozygous for Df(3R)P9. The product is nuclear and evenly localized in thoracic and abdominal segments. (D) Magnification of the posterior segments of the embryo in C. (E) Embryo homozygous for the UbxPlacZ mutation. The region of high nuclear expression of Exd extends to the A1 segment (cf. A). (F) Embryo homozygous for the UbxMX9 abd-AMI chromosome and, therefore, lacking the Ubx and abd-A functions. The region of high nuclear Exd expression now extends to segment A4 (cf. with A and E).
To demonstrate that the effect of the BX-C genes on Exd nuclear translocation is not mediated by transcription, we studied Exd localization in exdY012; Df(3R)P9 embryos only containing Exd of maternal origin. These embryos show lower levels of Exd protein than the wild type, but the protein is observed in the nuclei of all the segments, including the more posterior ones (Fig. 3C,D), and is uniformly distributed. This observation demonstrates that regulation of Exd subcellular localization by the homeotic genes occurs at the post-translational level and is independent of exd transcription.
To discriminate the roles of individual BX-C genes in the nuclear translocation of Exd, we looked at the distribution of Exd protein in two more BX-C mutant combinations: Ubx− abd-A+ Abd-B+ and Ubx− abd-A− abd-B+. As shown in Figure 3E, embryos homozygous for the first combination, defective only for Ubx function, show an increased level of nuclear Exd in the first abdominal segment as compared to wild type. In more posterior abdominal segments the levels and distributions of Exd are normal (Fig. 3E). In the second combination, lacking Ubx and abd-A functions, Exd is detected at high levels in the nuclei of the abdominal segments A1 to A4. Because in Df(3R)P9 (Ubx− abd-A− Abd-B−) embryos Exd nuclear localization extends to A8, these results indicate that each BX-C gene is capable of preventing or reducing the nuclear translocation of Exd.
We have explored this property further by making use of the UAS/Gal4 system (Brand and Perrimon 1993) to induce ectopic expression of each of the three BX-C genes during the embryonic period. We describe the results for Ubx, although similar results were obtained for abd-A and Abd-B. The ptc–Gal4 line was used to drive a UAS–Ubx construct encoding the major form (Ia) of the Ubx protein (O’Connor et al. 1988; Kornfeld et al. 1989); in ptc–Gal4/UAS–Ubx embryos Gal4-induced Ubx protein is present in the anterior compartment of each segment in addition to the normal Ubx protein. The effects on exd RNA and protein localization are illustrated in Figure 4. The distribution of exd RNA is not altered by ectopic or high levels of Ubx product. As shown in Figure 4, G–H, for the thoracic segments, the presence of Ubx protein in the anterior compartments does not affect exd transcription. We do not obvserve any intrasegmental modulation that would be expected if Ubx expression in the ptc domain were transcriptionaly repressing exd. This normal exd RNA distribution observed cannot be the consequence of perdurance of the maternal RNA because at this stage the maternal exd RNA has already disappeared (Fig. 1C). In contrast, ectopic or high levels of Ubx protein clearly affect the subcellular localization of Exd. As shown in Figure 4, A–F, the level of nuclear Exd in the anterior compartments of the three thoracic segments is reduced, whereas it remains high in the posterior compartments. As the same result is observed after similar experiments with UAS–abd-A and UAS–Abd-B (data not shown), we conclude that each BX-C gene can inhibit nuclear translocation of Exd in the thorax, the region of maximal exd expression.
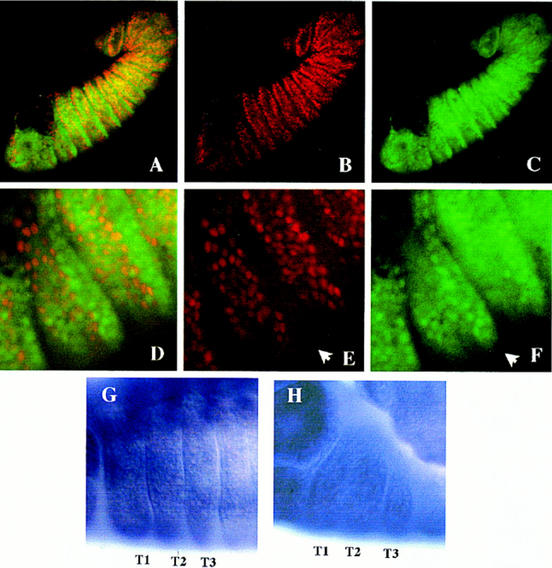
Figure 4.
Ectopic expression of Ubx in embryos. (A–F) Exd antigen is in green, and Ubx is in red. (A) Embryo with Ubx ectopic expression in the ptc domain doubly stained for Exd and Ubx proteins. The domains of high nuclear expression of both proteins do not overlap. (B) Same embryo as A, showing ectopic Ubx protein in the anterior compartments. (C) exd expression in the same embryo as A. Nuclear Exd localization in the thoracic segments is restricted to the posterior compartment, where no ectopic Ubx is induced. (D) Higher magnification of the thoracic segments of the embryo in A, showing that Ubx and Exd are mutually exclusive. (E) Ubx expression induced in the thorax by the ptc–Gal4 driver. The posterior compartments do not show ectopic Ubx (arrow). (F) Expression of exd in the same region as in E. Nuclear protein is mostly confined to the posterior compartments (arrow). (G, H) exd RNA expression in the thoracic segments (T1, T2, and T3) of wild-type (G) and in ptc–Gal4/UAS–Ubx embryo (H), showing no significant differences.
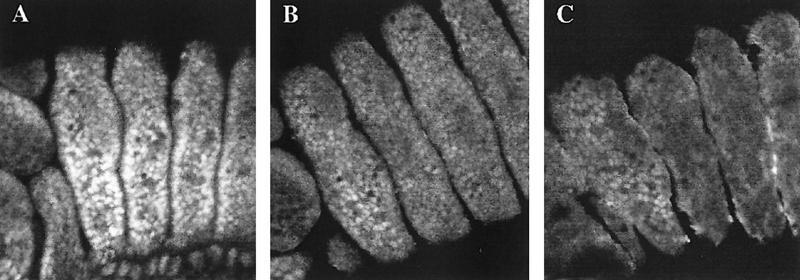
Although the experiments above indicate that any BX-C gene has the potential to inhibit Exd nuclear translocation, wild-type embryos show distinct levels of nuclear Exd along the abdominal segments that gradually decrease toward the most posterior segments, where Exd is almost entirely cytoplasmic. This indicates that at normal physiological levels the different BX-C genes prevent Exd nuclear transport to different extents. We have tested this possibility by comparing the effect of Ubx and Abd–Bm proteins when under the control of the same Antp–Gal4 driver. This driver will induce ectopic Ubx or Abd–Bm expression in the Antp domain, which extends from the posterior compartment of the first to the third thoracic segment (Wirz et al. 1986). As shown in Figure 5 Antp–Gal4/UAS–Ubx embryos, compared to the wild type, reared at 17°C show a reduction but not elimination of nuclear Exd in the second and third thoracic segments. In comparison, Antp–Gal4/UAS–Abd–Bm embryos, reared under the same conditions, show no detectable nuclear Exd, suggesting that the Abd–Bm product is more effective in preventing Exd nuclear transport.
Figure 5.
Ectopic expression of Ubx and Abd-B in embryos shows their different ability in preventing the nuclear translocation of Exd. (A) Stage 13 wild-type embryo showing the nuclear distribution of Exd in the three thoracic segments, aligned left to right. (B) Stage 13 embryo of genotype Antp–Gal4/UAS–Ubx reared at 17°C stained for Exd protein. The Antp domain occupies the posterior part of the first thoracic to the end of the third thoracic segment (see text). The moderate excess of Ubx protein in the second and third thoracic segments reduce, but do not eliminate, the nuclear transport of Exd. Note higher levels of nuclear Exd in the anterior region of the first thoracic segments, outside the Antp domain. (B) Stage 13 embryo of the genotype Gal4–Antp/UAS–Abd-B, also reared at 17°C. The presence of the Abd–Bm protein abolishes Exd nuclear expression, except in the anterior first thoracic segment.
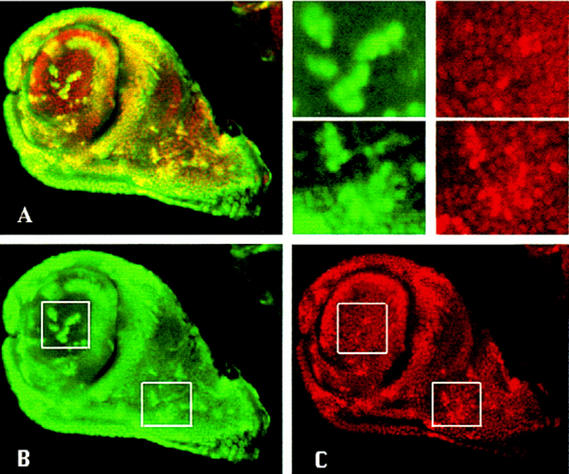
The ability of BX-C genes to inhibit Exd nuclear transport can also be demonstrated in imaginal cells. In the following experiments we tested the effect of Ubx and the mouse Hoxd10 gene, and Abd-B ortholog, for we have previously observed that the expression of Hoxd10 in the fly produces morphological alterations suggestive of exd inactivation (see below). We generated clones of cells containing Ubx+ or Hoxd10+ activity and examined the subcellular localization of Exd protein in the cells of the antennal disc. Ubx-expressing cells were detected with anti-Ubx antibody and Hoxd10-expressing clones were monitored by in situ hybridization with a Hoxd10 probe. We find that expression of either Ubx or Hoxd10 protein in antennal cells prevents nuclear translocation of Exd; in all of the Ubx+ clones found in the region where Exd is normally nuclear, it disappears from the nucleus (Fig. 6D–F). We find a similar result with Hoxd10+ clones (Fig. 6A–C), although in this case we cannot demonstrate the coincidental expression of the Hoxd10 and the Exd products. However, patches of cytoplasmic exd expression appeared with the same frequency as those labeled by in situ hybridization in sibling discs, and they never appear in control non-heat-shocked larvae, which suggests strongly that the loss of nuclear Exd in the clones is due to Hoxd10 activity. Furthermore, we tested the possibility of an effect of Hoxd10-expressing cells on exd transcription by in situ hybridization of the antennal discs with an exd RNA probe. We failed to observe any patch of cells lacking exd RNA (data not shown).
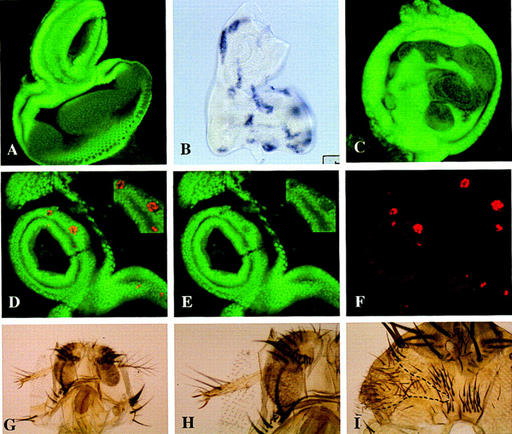
Figure 6.
Ectopic expression of Hoxd10 or Ubx prevents the nuclear translocation of Exd in the antennal cells. (A) Confocal image with the expression of Exd in a wild-type eye–antennal disc. Exd is nuclear in the proximal and cytoplasmic in the more distal regions. (B) Eye–antennal disc dissected from a larvae of FLP; Arm>y>Gal4/UAS–Hoxd10 genotype heat shocked at 37°C and stained for Hoxd10 mRNA. Clones of Hoxd10-expressing cells can be detected all over the disc. (C) Confocal image of an eye–antennal disc from a larva of the same genotype and treated as in B, stained with anti-exd antibody. The patches of cytoplasmic Exd are likely to be clones of Hoxd10-expressing cells in which the nuclear translocation of Exd is prevented. (D–F) Clones of Ubx-expressing cells (red) affecting Exd (green) localization. (D) Confocal image of an eye–antennal disc with several clones containing ectopic Ubx activity stained for Exd and Ubx. No overlapping of the nuclear expression of both proteins is observed. (E) Confocal image of the same disc as D, stained for Exd and showing cytoplasmic expression. (F) The same portion of the disc as in E showing the clones containing Ubx expression. (G) Adult head with a clone of Hoxd10-expressing cells marked with y and showing a leg transformation. (E) Higher magnification of the clone shown in G. Note the presence of two claws. (F) Clone of Hoxd10+ cells (encircled) in the head capsule exhibiting a transformation toward thorax.
The inhibition of Exd nuclear transport by BX-C genes and Hoxd10 causes exd-like phenotypes
The experiments described above indicate that high levels of the BX-C products interfere with the translocation of Exd into the nucleus and presumably cause a functional inactivation of exd. This effect should be reflected in the appearance of exd-like mutant phenotypes. We have tested this by inducing ectopic Ubx expression with several Gal4 lines during embryonic development and examining the segmental transformations produced. In the presence of normal exd function, Ubx specifies the pattern of the first abdominal (Al) segment. In contrast, in embryos lacking exd function, Ubx specifies a pattern resembling a more posterior segment, of A3–A5 type (Peifer and Wieschaus 1990).
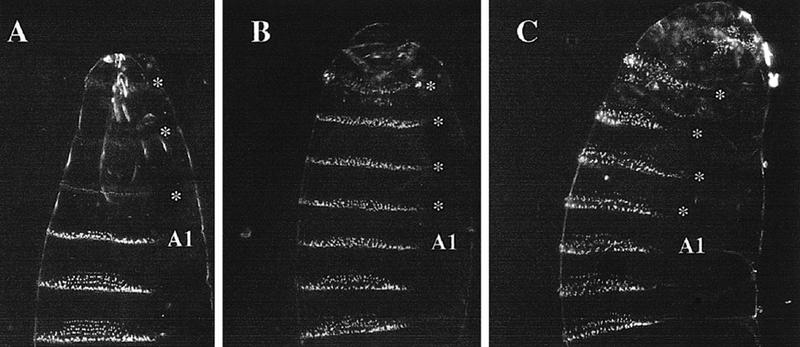
The results obtained with the Gal4-444 line, which induces high to moderate levels in the thoracic and abdominal segments, are shown in Figure 7. When grown at 17°C (the temperature at which the Gal4 product is less active) the thoracic and some head segments of Gal4-444/UAS–Ubx first instar larvae display A1 identity. This transformation (Fig. 7B) is expected for ectopic Ubx activity, in agreement with previous work (Gonzalez-Reyes and Morata 1990; Mann and Hogness 1990). However, when grown at 29°C, a temperature at which levels of Gal4 activity are higher, larvae of the same genotype develop all segments anterior to A2 with an A3–A5 pattern (Fig. 7C). This segment pattern resembles closely that found in the A1 segment of zygotic exd− larvae and is the same overall pattern observed after heat shock-inducing Ubx expression in zygotic exd− embryos (Peifer and Wieschaus 1990). These observations indicate that high levels of Ubx protein are able to produce an exd-like phenotype, in good agreement with the observed negative effect of BX-C genes on the nuclear translocation of Exd. We have also observed that the transformation into an A3–A5 pattern in Gal4-444/UAS–Ubx larvae can be partially reverted by adding an additional dose of exd+. In an experiment crossing females homozygous for the first chromosome Gal4-444 driver to UAS–Ubx homozygous males, all the progeny contain both the Gal4-444 and the UAS–Ubx chromosomes, and at 29°C the majority of the embryos (65 out of 72) develop the thoracic and A1 segments with an A3–A5 pattern. In comparison, in a similar cross in which parental males carry an additional dose of the exd gene in the Y chromosome, 33 out of 73 embryos develop thoracic segments of A1 type, whereas 40 develop an A3–A5 pattern. This suggests strongly that the presence of the extra dose of exd in the male embryos reduces the exd-like phenotype induced by the excess of Ubx protein.
Figure 7.
Different amounts of Ubx protein cause different transformations. (A) Ventral aspect of a wild-type larva showing head, thorax, and the first three abdominal segments. The first abdominal segment (A1) has a characteristic thin belt of denticles, whereas those of more posterior abdominal segments are wider and with a trapezoidal shape. (B) Larva of genotype Gal4-444; UAS–Ubx, grown at 17°C, in which Ubx has been overexpressed during embryogenesis (see text). At this temperature the three thoracic and one head segments (*) are transformed toward A1, and the A1 segment remains unaltered. (C) Larva of the same genotype as B grown at 28°C. The greater amount of Ubx protein produces a transformation of A1, the three thoracic segments, and one head segment (*) into an A3–A5 segment type.
Induction of high levels of Ubx, Abd-A and Abd-B products in the antennal cells is known to produce transformation into leg (this paper; Mann and Hogness 1990; Kuziora 1993; Casares et al. 1996). Because an antenna-to-leg transformation is also produced in exd− clones (Gonzalez-Crespo and Morata 1995; Rauskolb et al. 1995), it is possible that any of the BX-C products may produce an inactivation of Exd in the antennal cells. The lack of specificity of this phenotype is further emphasized by previous work showing that the mouse Hoxb9 gene (an Abd-B homolog) can induce the antenna-to-leg transformation (Malicki et al. 1993) and by our finding that a similar transformation is observed in Distal-less Dll–Gal4/UAS–Hoxd10 flies in which the mouse Hoxd10 gene is expressed in the Dll domain of the antenna. Hoxd10 also interferes with head and thorax development when expressed with other Gal4 lines (N. Azpiazu and G. Morata, unpubl.). To discriminate better the local effect of the Hoxd10 gene, we generated flip-out clones (see Materials and Methods) of Hoxd10-expressing cells labeled with the cuticle marker y. These clones autonomously differentiate leg structures in the antenna (Fig. 6G–H). Moreover, in the head capsule they differentiate thoracic structures (Fig. 6I) resembling those produced by lab− clones, whereas in the thorax they alter the bristle arrangement. All of these phenotypic aspects are similar to those described for exd− clones (Gonzalez-Crespo and Morata 1995; Rauskolb et al. 1995).
exd function is necessary for maintenance of Ubx expression in the metathorax
The preceding results indicate that the BX-C genes negatively regulate exd function. However, previous work also indicates that the lack of exd function can produce phenotypes resembling loss of function of homeotic genes. Notably, exd− clones in the metanotum (Gonzalez-Crespo and Morata 1995) produce a transformation into mesonotum similar to that reported for the loss of Ubx function (Morata and Garcia-Bellido 1976). This suggests that in this region the normal function of Ubx requires exd activity.
Using an antibody directed against the Ubx protein we examined Ubx expression in haltere imaginal discs containing exd− clones induced at different times during larval development. We find that in exd− clones induced at 48–60 hr after egg laying (AEL), Ubx expression is abolished or greatly reduced (Fig. 8A–C) in the region of the disc that will develop into metanotum. In exd− clones induced later (72–84 hr AEL), Ubx expression is variable; some cells retain the Ubx antigen, whereas it is undetectable in others (Fig. 8D–F). As expected, exd− clones in the region of the disc giving rise to the haltere pouch, where Exd is cytoplamic, have no effect on Ubx expression. This observation indicates that exd is involved in the maintenance of Ubx activity in the trunk region of the segment (metanotum), but not in the appendage.
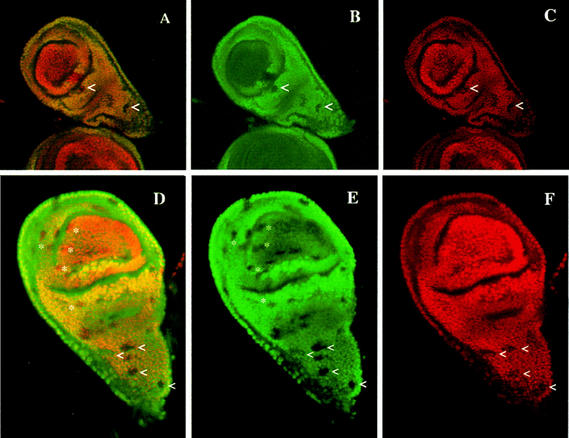
Figure 8.
exd function is necessary to maintain Ubx expression in the haltere imaginal disc. (A–F) Exd protein is in green; Ubx is in red. Overlapping regions of expression appear yellow. (A–C) Confocal image of a haltere disc with exd− clones induced at 48–72 hr, doubly stained for Exd and Ubx. exd− clones are detected by the lack of Exd protein. The upper pointer (<) indicates an exd− clone crossing the border of nucleo/cytoplasmic exd expression. The portion of the clone in the appendage part contains Ubx protein, but in the proximal part of the clone Ubx protein is not detected. Other exd− clones showing loss of Ubx antigen are also visible in the thoracic part of the haltere (<). (B, C) The same clones in the green (Exd) and red (Ubx) channels. (D–F) Confocal image of another haltere disc with several small exd− clones induced at 72–96 hr of development and double-stained as in A–C. exd− clones in the haltere pouch (*) have no effect on Ubx expression, whereas many of those in the metanotum (<) reduces Ubx expression.
The involvement of exd in Ubx expression is supported by the next experiment in which we induced clones in the haltere disc, using the armFRT<y+<FRT/Gal4 system, that express high levels of exd activity. In the metanotum, where cells normally possess exd function, the excess of Exd results in an increase of Ubx protein that can be readily detected (Fig. 9). We did not expect an effect of exd+ clones on the haltere appendage, but we found, first, that the excess of Exd protein results in its appearance in the nucleus (Fig. 9B), and second, that the presence of nuclear Exd in the appendage cells gives rise in many of those cells, although not in all of them, to an increase of Ubx protein above the normal levels (Fig. 9C). It appears that unlike embryonic cells, an increase in the amount of gene product in imaginal cells may result in the breakdown of the control mechanism of subcellular localization of Exd. A similar observation has been made recently in the leg disc (Gonzalez-Crespo and Morata 1996).
Figure 9.
Effect of increasing levels of Exd on Ubx expression in the haltere imaginal disc. Confocal images of a third instar disc containing clones of excess of exd expression double-stained with anti-Exd (green) and anti-Ubx (red) antibody. (A) Double staining of the disc, where cells with both proteins appear yellow. Clones can be seen in the proximal and distal part of the disc and are detected by higher levels of the two proteins. (B) The same disc only showing Exd protein. (C) Ubx protein in the same disc to show the general correspondence between the increase of Exd and Ubx proteins. Insets in B and C are amplified in the upper right panel for a better assessment of the protein levels in the clones. Note that the excess of Exd protein in the clones in the pouch gives rise to nuclear Exd localization and that the increase of nuclear Exd produces an increase of Ubx protein, although the correspondence is sometimes not strictly cell by cell.
As it is known that Ubx negatively autoregulates its own expression, an excess of Ubx protein reduces Ubx transcription (Irvine et al. 1993; Casares et al. 1997), we searched for Ubx loss-of-function phenotypes associated with the increase of Ubx product observed in the exd+ clones in the haltere. A Ubx-like phenotype would have consisted of an easily scorable wing transformation marked with y. However, we failed to observe any y− clone showing wing transformation. Unfortunately, y− clones that do not produce a transformation into wing or mesonotum cannot be detected, but as we find many y− clones in other parts of the fly, we assume that they are also present in the haltere, but do not produce a transformation.
Discussion
Regulation of exd expression and function by Hox genes
The exd gene has a maternal and a zygotic component, both of which contribute to exd function during embryogenesis. Our results show that the two components have the same pattern of distribution in the organism and in cells.
Examination of the distribution of the exd RNA and protein products in wild-type embryos reveals two levels of regulation. In the first place, there is transcriptional control, the number of transcripts in the posterior abdominal segments is lower than in the thorax, and this down-regulation depends on the presence of the BX-C genes (Rauskolb et al. 1993). This suggests that the BX-C genes are negative transcriptional regulators of exd. Obviously, transcriptional regulation during embryogenesis can affect only the zygotic component. In the second place, there is post-translational control, Exd protein is translocated to the cell nucleus in some body regions, whereas it remains in the cytoplasm in others (Mann and Abu-Shaar 1996; Aspland and White 1997). This process operates in embryonic as well as in imaginal cells and appears to be a control mechanism of exd function, as only the nuclear protein is functional (Gonzalez-Crespo and Morata 1995; Rauskolb et al. 1995). During embryogenesis, as indicated by our ptc–Gal4/UAS–exd experiment (Fig. 2), this mechanism can keep excess Exd protein out of the nucleus, thereby impeding a developmental effect. In the experiments reported by Rauskolb et al. (1995), the heat shock–driven product can rescue exd− phenotype in the thoracic segments because the nuclear translocation mechanism is operating in those segments and, therefore, the exogenous Exd product can be translocated to the nucleus. However, the heat shock has no effect in the abdominal region where Exd nuclear translocation is prevented.
Our results show that each of the three Drosophila BX-C genes and the mouse Hoxd10 gene can prevent Exd nuclear transport and consequently reduce or eliminate exd function; thus, the BX-C genes are involved in the control of Exd subcellular localization. This control takes place at the post-translational level because the BX-C genes exert this role even in mat+ zyg− embryos (Fig. 3) in which transcriptional control is not possible. The BX-C (and likely Hoxd10) genes do not act directly on exd, but trigger a control mechanism presumably activating some target genes responsible for the process. The lack of effect of GAL4 or heat shock–induced excess of Exd in the abdomen is due to the presence of the abd-A and/or Abd-B products that prevent Exd nuclear transport.
This regulation by the BX-C genes is also reflected in the pattern of subcellular localization of Exd in wild-type embryos (see Fig. 3A). In the thoracic segments Exd is predominantly nuclear, but becomes gradually more cytoplasmic toward the abdomen, and in the most posterior segment (A8) it is only cytoplasmic. This segment is part of the Abd-B domain and contains almost exclusively Abd-B product (Sanchez-Herrero et al. 1985). This suggests that the physiological levels of Abd-B in A8 are sufficient to prevent the nuclear translocation of Exd. More anterior segments contain low amounts of nuclear Exd coincidental with Ubx and abd-A proteins, indicating that the physiological levels of Ubx and abd-A products in those segments reduce but do not eliminate the nuclear translocation of Exd. This observation is consistent with the finding that Ubx and abd-A product possess the hexapeptide motif and interact with Exd (Johnson et al. 1995) and, as discussed below, the normal function of Ubx and abd-A requires exd activity, that is, Exd nuclear localization. In the case of Ubx there is a correlation between protein levels and nuclear translocation of Exd. In the T3 and A1 segments where Ubx is the only BX-C gene product present, the low amounts of Ubx protein in T3 correlate with the higher level of nuclear Exd, whereas the higher Ubx levels in A1 are coincidental with lower levels of nuclear Exd.
All together these results indicate that each BX-C protein has a different ability to suppress Exd nuclear translocation and, hence, exd function. It is higher in Abd-B, more moderate in abd-A, and lower in Ubx. It also suggests that this property is used in normal development to modulate the amount of Exd in the nuclei of cells of different segments.
This negative control can be visualized at the phenotypic level. Both in embryos (Fig. 6) and in adult structures (Fig. 5, see also Fig. 1B–D in Casares et al. 1996), the Ubx, abd-A, Abd-B, or Hoxd10 products acting in ectopic places or in higher than normal expression levels can produce exd−-like phenotypes. In fact, this property can readily account for previously unexplained results such as the antenna-to-leg transformation produced by inappropriate expression of abdominal genes like abd-A or Abd-B (Kuziora 1993; Casares et al. 1996). We propose that these transformations are produced by functional inactivation of exd.
Thus, considering all of the results, the BX-C genes appear to affect negatively both exd transcription (Rauskolb et al. 1993) and Exd nuclear transport mechanism. However, we believe that the effect on transcription may be a secondary consequence of the inhibition of Exd nuclear transport. This is based on the finding that ectopic expression of any BX-C gene affects the subcellular localization of Exd at a time when it has no detectable expression on zygotic transcription. We presume that exd may have an autoregulatory mechanism to maintain its own expression that requires nuclear localization of the product. This process is prevented by the BX-C genes and, as a consequence exd transcription, is abolished or diminished.
The negative control of exd function by the Hox genes, especially for Ubx, is intriguing, for there is evidence that Exd and Ubx proteins interact and Exd is a necessary cofactor for normal Ubx function. For example, the patterning function of Ubx in T3 and A1 segments depends on the presence of the sufficient amount of active Exd (Peifer and Wieschaus 1990), and the control of dpp expression by Ubx in the visceral mesoderm requires normal exd function (Chan et al. 1994; Rauskolb and Wieschaus 1994). In imaginal cells the elimination of exd function produces a Ubx phenotype in the metanotum, which we show is due to the exd requirement for Ubx maintenance. In the adult A1 segment the loss of exd function also alters the normal patterning role of Ubx (Gonzalez-Crespo and Morata 1995). In the A2–A4 segments, where the predominant role is played by abd-A (Sanchez-Herrero et al. 1985), exd mutant clones give rise to abnormal patterns (Gonzalez-Crespo and Morata 1995), indicating that normal abd-A function also requires exd activity.
Thus, on one hand, normal function of Ubx and abd-A is shown to require exd activity, and on the other, excess of Ubx and abd-A products is shown to repress exd activity. These two observations together suggest that Exd and Hox products have to be in each segment in the appropriate relative amounts. The current view about Exd/Hox interaction (Chan and Mann 1996) is that Exd and the different Hox proteins form heterodimers and the binding specificity and affinity to target DNA depend on the heterodimer itself. Increasing amounts of the homeotic product will probably result in some free homeotic product in addition to the heterodimer. The free homeotic product, which by itself has little binding specificity (Hoey and Levine 1988), is able to bind DNA and may interfere with the activity of the heterodimer. Furthermore, it has been proposed recently (Pinsonneault et al. 1997) that by themselves the exd and Hox genes act as repressors while the Exd/Hox complexes act as activators. In such a manner of function any free Exd or Hox product would interfere with the normal function of the Exd/Hox complex. This situation calls for a mechanism to regulate precisely the relative number of interacting molecules.
exd and the maintenance of Ubx expression
Our results also indicate a hitherto unknown aspect of the maintenance of Ubx expression, that is, its dependence on exd function; the lack of exd activity results in the loss of Ubx product in the metanotum and, conversely, increasing amounts of Exd product anywhere in the disc produce a corresponding increase in the amount of Ubx product. Thus, Exd has the property of promoting Ubx expression. It may simply indicate that during imaginal development exd acts as transcriptional activator of Ubx. Another possibility is that Exd partakes as a cofactor with the Ubx protein in the process of Ubx maintenance, a role that it plays in autoregulatory circuits of other homeotic genes (Chan and Mann 1996; Ponsonneault et al. 1997). At this point we do not have evidence to favor any of these alternatives. It is of interest, however, that the regulation of Ubx expression appears to be different in the trunk region of the disc, where it requires exd, and in the appendage, where it does not. There is another aspect of the homeotic function that also discriminates clearly between the trunk region and the appendage: Ectopic expression of any of the three BX-C genes (Casares et al. 1996), and also of Hoxd11 (N. Azpiazu and G. Morata, unpubl.), produce a nonspecific wing-to-haltere transformation, whereas the same BX-C genes produce specific transformations in the trunk region of the wing segment. Where the regions show specificity or lack of it coincides with the presence or absence of nuclear Exd.
The interactions between Ubx and exd may provide an explanation for the phenomenon of negative autoregulation of some homeobox genes like Ubx and others (Chouinard and Kaufman 1991; Heemskerk et al. 1991; Irvine et al. 1993; Casares et al. 1996). Negative autoregulation for Ubx has been described in embryos and in the haltere disc; the observation is that an excess of Ubx protein induced via heat shock (Irvine et al. 1993) or Gal4 (Casares et al. 1997) results in loss or reduction in the transcription of the endogenous Ubx gene. As we have observed that the excess of Ubx produces a loss of exd function and the latter is in turn necessary for Ubx maintenance in the metanotum, we propose that negative autoregulation may simply be the result of the negative effect that Ubx has on exd function. This hypothesis does not explain negative autoregulation in the appendage, where exd is not functional. The explanation that we suggest is based on the recent model by Pinsonneault et al. (1997), proposing that the Hox genes by themselves act as repressors. If we assume that in the appendage Ubx transcription is maintained by an interaction of Ubx with another cofactor, the increase in the amount of Ubx product may produce free Ubx protein, which inhibits transcription. It is noteworthy that in our experiments the increase of Ubx product observed in the exd+ clones in the haltere appendage does not produce loss of Ubx function, as these clones do not show indication of Ubx− phenotype. We speculate that in this case there is an increase of both Exd and Ubx product, which prevents the repressing function of Ubx alone.
Conservation of Exd nuclear transport
The conservation in the entire animal kingdom of the general organization of the Hox complex (Duboule and Dollé 1989; Graham et al. 1989) and of some properties of Hox genes, such as spatial colinearity (Gaunt et al. 1988; Duboule and Dollé 1989; Graham et al. 1989) or phenotypic suppression/posterior prevalence (Gonzalez-Reyes and Morata 1990; Bachiller et al. 1994), suggests that the general aspects of Hox function are also conserved. The Exd gene product is a general cofactor in Drosophila and is also highly conserved, not only in the homeodomain, but also in most of the protein sequence (Rauskolb et al. 1993). Moreover, similar cooperative interactions for DNA binding have been described at the molecular level for Exd and homeotic products in Drosophila and for the mouse Hoxb7 and Hoxb8 and Pbx1 and Pbx2 (vertebrate homologs of exd) gene products (van Dijk et al. 1995). Thus, the exd/Hox interaction appears to be conserved. To date, however, there is no indication that subcellular control of Exd localization occurs in organisms other than Drosophila. We have demonstrated that in Drosophila some Hox genes are involved in the nuclear translocation of Exd and consequently regulate exd function. We have also shown that one Abd-B mouse homolog, the Hoxd10 gene, is able to prevent nuclear translocation of Exd when expressed in Drosophila imaginal cells. It also produces homeotic transformations in the Drosophila antenna, head capusle, and thorax, similar to those produced by the loss of exd function in those regions. This ability of Hoxd10 may be indicative that the Pbx genes may also be subjected to a similar control by vertebrate Hox genes. It is possible that the Hox proteins modulate the intracellular location of Pbx proteins. The Hoxd10 gene and perhaps other vertebrate Hox genes, such as Hoxb9, which also produces exd−-like phenotypes in Drosophila (Malicki et al. 1993), may activate or repress the set of genes that control Pbx translocation into the nucleus. The existence of such mechanism in vertebrates may be of importance in understanding the overall role of Pbx proteins. In humans the fusion protein E2A–Pbx1 is responsible for the pre-B acute lymphoblastoid leukemia, and it would be of interest to know whether a nucleo/cytoplasmic control mechanism is compromised in cases like this.
Materials and methods
Drosophila mutant strains
The following homeotic and exd mutant lines were used in this study: UbxMX9 abdAM1, Df(3R)P9, exdY012/FM7eve–lacZ, exdY012/FM7eve–lacZ; Df(3R)P9/Mc. The exdY012 allele was provided by Cordelia Rauskolb (Princeton University, NJ). Germ-line clones homozygous for exdY012 were generated by using the yeast recombinase-based FLP–DTS system (Chou and Perrimon 1992). Recombination was induced in females carrying two copies of a FRT insertion in the X chromosome, and a single copy of the dominant female sterile mutation ovoD2, in trans to the chromosome carrying the exdY012 mutation. The FLP recombinase was provided by a 1.5-hr heat shock treatment of 24–48 hr larvae. In experiments combining mutant combinations for BX-C genes and exd, homozygous homeotic mutant embryos were identified using the appropriate antibodies to recognize the lack of homeotic gene product, and the zygotic exd mutant embryos were identified by using a FM7/eve lacZ balancer.
UAS/GAL4 strains
The Gal4/UAS system (Brand and Perrimon 1993) was used to drive the expression of the different BX-C proteins. The UAS–Ubx and UAS–abd-A lines drive the expression of the Ia form of Ubx (O’Connor et al. 1988; Kornfeld et al. 1989) and abd-A, respectively, and have been described previously (Mitchelson 1994; Casares et al. 1996). The UAS–Abd–Bm line drives the expression of the Abd–Bm form (Casanova et al. 1986; Zavortink and Sakonju 1989) and has also been described (Castelli-Gair et al. 1995). All of these lines were provided by Ernesto Sanchez-Herrero (CSIC–UAM, Madrid, Spain). The UAS–Antp was a gift from Eduardo Moreno (CSIC–UAM, Madrid, Spain). The UAS–Hoxd10 line was generated in the laboratory using the 1.6-kb BamHI cDNA fragment of the Hoxd10 provided by D. Duboule (Zappavigna et al. 1991).
The following driver lines were utilized: GAL4-444 (general expression throughout the embryo, provided by Fernando Jimenez, CSIC–UAM, Madrid, Spain), or ptc-GAL4 [expression in the ptc domain, a gift from Campos-Ortega (see Hinz et al. 1994)]. Embryos from UAS/GAL4 crosses were collected at 29°C when used for antibody or in situ staining. To prepare larval cuticles the embryos were grown either at 29°C or 17°C.
To generate clones of ectopic expression of the Hoxd10 gene we used the FLP; Armadillo [Arm–FRT>y>FRT–Gal4/UAS–Hoxd10] genotype (Sanson et al. 1996). For clones of ectopic expression of Ubx we used the FLP/actin-FRT>y>FRT–Gal4; UAS–Ubx genotype. Clones were induced by giving a heat shock at 37°C at convenient times during larval development and discs were dissected from mature third instar larvae. The arm–FRT>y+>FRT–Gal4 transgenic flies were provided by P. Simpson (Sanson et al. 1996), and the heat shock–flipase stock (FLP122) by G. Struhl (Struhl and Basler 1993). Transgenic actin–FRT>y>FRT–Gal4 flies were provided by L. Zipursky (Pignoni and Zipursky 1997).
Preparation of larval cuticles
Eggs were collected on agar plates over 12-hr intervals and allowed to develop to larvae at the desired temperature (either 29°C or 17°C). They were dechorionated in bleach, fixed in methanol–heptane (1:1) for 40 sec, washed in methanol, rinsed in 0.1% Triton, mounted in a 1:1 mixture of Hoyer’s mountant and lactic acid, and incubated at 60°C for 12 hr.
Antibody staining of embryos
Embryos were fixed for 20 min in a 4% paraformaldehyde–heptane solution (1:1) at room temperature and then washed in methanol. They were blocked in 10% BSA for 1 hr and incubated with the first antibody in PBT (PBS, 0.1% Tween 20) overnight at 4°C. Washes were performed in PBT, and the secondary antibody incubated for 2 hr in PBT at room temperature. The secondary antibodies were biotinylated and an avidin/biotin/horseradish peroxidase (HRP) complex was used for the detection (Vectastain from Vector Laboratories). The HRP substrate was diaminobenzidine (DAB) in PBS. Antibodies were provided by Sergio Gonzalez-Crespo (Centro de Investigacíon y Desarollo–CSIC, Barcelona, Spain; anti-Exd), R. White (University of Cambridge, UK; anti-Ubx). β-Gal antibody (rabbit) was purchased from Cappel. After staining, the embryos were dehydrated and mounted in Permount. Photographs were taken on Zeiss Axiophot with Nomarski optics and Fujichrome 64T film.
In situ hybridization
The in situ hybridization with DIG-labeled probe to embryos was done as described in Tautz and Pfeiffle (1989), with slight modifications. The exd probe used was a 2.2-kb ClaI fragment from the exd cDNA. To stain discs, they were dissected from third instar larvae in PBS, kept on ice for 30 min, and fixed in 4% paraformaldehyde during 20 min at room temperature. They were washed three times in PBS, incubated for 2 min in 0.25% glutaraldehyde on ice, washed in PBS, and fixed in 4% formaldehyde, 0.1% Triton X-100, 0.1% sodium deoxycholate for 20 min at room temperature. After a short wash in PBS, discs were rinsed in PBT, incubated with proteinase K (50 μg/ml) for 3 min, postfixed in 4% paraformaldehyde during 20 min at room temperature, washed in PBT, and prehybridized at 48°C in hybridization solution for 1 hr. The following steps were done as for embryos. The Hoxd10 probe used was a 1.6-kb BamHI fragment from the Hoxd10 cDNA.
Immunofluorescence staining
Imaginal discs were dissected from third instar larvae, fixed in 4% paraformaldehyde in PBS for 20 min at room temperature, postfixed in 4% paraformaldehyde, 0.1% Triton X-100, and 0.1% sodium deoxycholate in PBS for 20 min, washed in PBS for 5 min, and blocked in 1% BSA and 0.3% Triton X-100 in PBS for 1 hr. Discs were incubated in the first antibody (anti-Exd diluted 1/300) overnight at 4°C. Washes were performed with PBS, 1% BSA, and 0.3% Triton X-100, and the discs were incubated for 2 hr in the secondary biotinylated anti-rat antibody (Amersham). After washing the secondary antibody, a FITC–streptavidin complex (Jackson) was added for 2 hr and the discs were washed and mounted in PBS, 4% propylgalate. For embryos, the fixation was done as described previously (antibody staining for embryos), and the rest of the procedure was performed as for imaginal discs. Photographs were taken in a laser scan Zeiss microscope and subsequently processed using Adobe Photoshop.
Preparation of adult cuticle
The adult flies with appropriate phenotypes were dissected in water and cut into pieces. They were subsequently treated with 10% KOH at 95°C for 3–5 min to digest the internal tissues, washed with water, rinsed in ethanol, and mounted in Euparal. The preparations were photographed on a Nikon AFX-II microscope using Fujichrome 64T film.
Acknowledgments
We thank E. Sanchez-Herrero, S. Gonzalez-Crespo, F. Jimenez, J.A. Campos-Ortega, P. Simpson, G. Struhl, and L. Zipursky for the UAS and Gal4 lines; Thomas Kornberg, Peter Lawrence, Manfred Frasch, Ernesto Sanchez-Herrero, Isabel Guerrero, and Sergio Gonzalez-Crespo for their comments on the manuscript; and Rosa Gonzalez for technical assistance. The work was supported by grants from the Human Science Frontier Scientific Programme and the Dirección General de Investigatión Científica y Técnica. N.A. was a recipient of an European Molecular Biology Organization long-term fellowship and is now a postdoctoral fellow from the Spanish Ministerio de Educación y Ciencia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gmorata@trasto.cbm.uam.es; FAX 34 1 397 4799.
References
- Aspland SE, White RAH. Nucleocytoplsmic localisation of extradenticle protein is spatially regulated throughout development in Drosophila. Development. 1997;124:741–747. doi: 10.1242/dev.124.3.741. [DOI] [PubMed] [Google Scholar]
- Bachiller D, Macias A, Duboule D, Morata G. Conservation of a functional hierarchy between mammalian and insect Hox/HOM genes. EMBO J. 1994;13:1930–1941. doi: 10.1002/j.1460-2075.1994.tb06462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Casanova J, Sanchez-Herrero E, Morata G. Identification and characterisation of a parasegment specific regulatory element of the Abdominal-B gene of Drosophila. Cell. 1986;47:627–636. doi: 10.1016/0092-8674(86)90627-6. [DOI] [PubMed] [Google Scholar]
- Casares F, Calleja M, Sanchez-Herrero E. Functional similarity in appendage specification by the Ultrabithorax and abdominal-A Drosophila Hox genes. EMBOJ. 1996;15:3934–3942. [PMC free article] [PubMed] [Google Scholar]
- Casares F, Bender W, Merriam J, Sanchez-Herrero E. Interactions of Drosophila Ultrabithorax regulatory regions with native and foreign promoters. Genetics. 1997;145:123–137. doi: 10.1093/genetics/145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli-Gair J, Greig S, Micklem G, Akam M. Dissecting the temporal requirements for homeotic gene function. Development. 1995;120:1983–1995. doi: 10.1242/dev.120.7.1983. [DOI] [PubMed] [Google Scholar]
- Chan S-K, Mann R. A structural model for a homeotic protein-extradenticle-DNA complex accounts for thechoice of Hox protein in the heterodimer. Proc Natl Acad Sci. 1996;93:5223–5228. doi: 10.1073/pnas.93.11.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S-K, Jaffe L, Capovilla M, Botas J, Mann RS. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with Extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chang C-P, Shen W-F, Rosenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. EMBO J. 1994;13:5967–5976. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Chou T-B, Perrimon N. Use of a yeast site specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard S, Kaufman TC. Control of expression of the homeotic labial (lab) locus of Drosophila melanogaster. Evidence for both positive and negative autogenous regulation. Development. 1991;113:1267–1280. doi: 10.1242/dev.113.4.1267. [DOI] [PubMed] [Google Scholar]
- Duboule D, Dollé P. The structural and functional organisation of the murine Hox gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel WA, Singson AW, Margolis JS, Bang AG, Posakony JW, Murre C. Dpbx, a new homeobox gene closely relates to the human proto-oncogene pbx1 molecular structure and developmental expression. Mech Dev. 1993;41:155–161. doi: 10.1016/0925-4773(93)90045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, S.J., P.T. Sharpe, and D. Duboule. 1988. Spatially restricted domains of homeo-gene transcripts in mouse embryos: relation to a segmented body plan. Development 104 (Suppl): 71–82.
- Gibson G, Gehring W. Head and thoracic transformations caused by ectopic expression of Antennapedia during Drosophila development. Development. 1988;102:657–675. [Google Scholar]
- Gonzalez-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- ————— Genetic subdivision of the arthropod limb into coxopodite and telopodite. Development. 1996;122:3921–3928. doi: 10.1242/dev.122.12.3921. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Morata G. The development effect of overexpressing a Ubx product in Drosophila embryos is dependent on its interactions with other homeotic products. Cell. 1990;61:512–522. doi: 10.1016/0092-8674(90)90533-k. [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R. The murine and Dropsophila homeobox gene complexes have common features of organisation and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S, Kostriken R, O’Farrell P. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–412. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz V, Giebel B, Campos-Ortega JA. The basic helix-loop-helix of Drosophila lethal of scute protein is sufficient for pro-neural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Hoey T, Levine M. Divergent homeobox proteins recognise similar DNA sequences in Drosophila. Nature. 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Botas J, Jha S, Mann RS, Hogness DS. Negative autoregulation by Ultrabithorax controls the level and pattern of its expression. Development. 1993;117:387–399. doi: 10.1242/dev.117.1.387. [DOI] [PubMed] [Google Scholar]
- Johnson FB, Parker E, Krasnow M. Extradenticle protein is a selective cofactor for the Drosophila homeotics: Role of the homeodomain and YPWM amino acid motif in the interaction. Proc Natl Acad Sci. 1995;92:739–743. doi: 10.1073/pnas.92.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps MP, Murre C, Sun X-H, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organisation in the Antennapedia gene complex of Drosophila melanogaster. Adv Genet. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- Kornfeld K, Saint RB, Beachy PA, Harte PJ, Peattie DA, Hogness DS. Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation. Genes & Dev. 1989;3:243–258. doi: 10.1101/gad.3.2.243. [DOI] [PubMed] [Google Scholar]
- Kuziora MA. Abdominal-B protein isoforms exhibit distinct cuticular transformations and regulatory activities when ectopically expressed in Drosophila embryos. Mech Dev. 1993;42:125–137. doi: 10.1016/0925-4773(93)90002-f. [DOI] [PubMed] [Google Scholar]
- Lawrence P, Morata G. Homeobox genes: Their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- Lawrence P, Struhl G. Morphogens, compartments and patterns: Lessons from Drosophila? Cell. 1996;85:951–961. doi: 10.1016/s0092-8674(00)81297-0. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lu Q, Knoepfler P, Scheele J, Wright D, Kamps M. Both Pbx1 and E2A-Pbx1 bind the DNA motif ATCAATCAA cooperatively with the products of multiple murine Hox genes, some of which are themselves oncogenes. Mol Cell Biol. 1995;15:3786–3795. doi: 10.1128/mcb.15.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J, Bogarad LD, Martin MM, Ruddle FH, McGinnis W. Functional analysis of the mouse homeobox gene HoxB9 in Drosophila development. Mech Dev. 1993;42:139–150. doi: 10.1016/0925-4773(93)90003-g. [DOI] [PubMed] [Google Scholar]
- Mann R, Abu-Shaar M. Nuclear import of the homeodomain protein Extradenticle in response to Wg and Dpp signaling. Nature. 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]
- Mann R, Hogness D. Functional dissection of Ultrabithorax proteins in Drosophila melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- Michelson AM. Muscle pattern diversification in Drosophila is determined by the autonomous function of homeotic genes in the embryonic mesoderm. Development. 1994;120:755–768. doi: 10.1242/dev.120.4.755. [DOI] [PubMed] [Google Scholar]
- Monica K, Galila N, Nourse J, Saltman D. Pbx2 and Pbx3, new homeobox genes with extensive homology to the human proto-oncogene Pbx1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Garcia-Bellido A. Developmental analysis of some mutants of the bithorax system of Drosophila. Wilhem Roux’s Arch. 1976;179:125–143. doi: 10.1007/BF00848298. [DOI] [PubMed] [Google Scholar]
- Nourse J, Mellentin JD, Galili N, Wilkinson J, Stanbridge E, Smith SD, Cleary ML. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- O’Connor MB, Binari R, Perkins LA, Bender W. Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J. 1988;7:435–445. doi: 10.1002/j.1460-2075.1988.tb02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Wieschaus W. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes & Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Sadoul R, Featherstone MS. Cooperative interactions between Hox and Pbx proteins mediated by a conserved peptide motif. Mol Cell Biol. 1994;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Pinsonneault J, Florence B, Vaesin H, McGinnis W. A model for extradenticle function as a switch that changes Hox proteins from repressors to activators. EMBO J. 1997;16:2032–2043. doi: 10.1093/emboj/16.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Wieschaus E. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Peifer M, Wieschaus E. extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx-1. Cell. 1993;74:1101–1112. doi: 10.1016/0092-8674(93)90731-5. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Smith KM, Peifer M, Weischaus E. extradenticle determines segmental identities throughout Drosophila development. Development. 1995;121:3663–3673. doi: 10.1242/dev.121.11.3663. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organisation of Drosophila Bithorax Complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent J-P. Uncoupling cadherin-based adhesion from wingless signaling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homeotic gene Antennapedia. Nature. 1987;325:816–828. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organising activity of the wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeiffle C. A non-radioactive in situ hybridisation method for the localisation of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- van Dijk MA, Murre C. extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- van Dijk MA, Peltenburg LTC, Murre C. Hox gene products modulate the DNA binding activity of Pbx1 and Pbx2. Mech Dev. 1995;52:99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]
- Wirz J, Fessler LI, Gehring WJ. Localization of the Antennapedia protein in Drosophila embryos and imaginal discs. EMBO J. 1986;5:327–334. doi: 10.1002/j.1460-2075.1986.tb04647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappavigna V, Renucci A, Izpisúa-Belmonte J-C, Urier C, Peschle C, Duboule D. Hox 4 genes encode transcription factors with potential auto- and cross-regulatory capacities. EMBO J. 1991;10:4177–4187. doi: 10.1002/j.1460-2075.1991.tb04996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavortink M, Sakonju S. The morphogenetic and regulatory functions of the Drosophila Abdominal-B gene are encoded in overlapping RNAs transcribed from separate promoters. Genes & Dev. 1989;3:1969–1981. doi: 10.1101/gad.3.12a.1969. [DOI] [PubMed] [Google Scholar]