Abstract
Genetic analysis in Arabidopsis has led to the identification of several genes that are required for auxin response. One of these genes, AXR1, encodes a protein related to yeast Aos1p, a protein that functions to activate the ubiquitin-related protein Smt3p. Here we report the identification of a new gene called TRANSPORT INHIBITOR RESPONSE 1 (TIR1). The tir1 mutants are deficient in a variety of auxin-regulated growth processes including hypocotyl elongation and lateral root formation. These results indicate that TIR1 is also required for normal response to auxin. Further, mutations in TIR1 display a synergistic interaction with mutations in AXR1, suggesting that the two genes function in overlapping pathways. The TIR1 protein contains a series of leucine-rich repeats and a recently identified motif called an F box. Sequence comparisons indicate that TIR1 is related to the yeast protein Grr1p and the human protein SKP2. Because Grr1p and other F-box proteins have been implicated in ubiquitin-mediated processes, we speculate that auxin response depends on the modification of a key regulatory protein(s) by ubiquitin or a ubiquitin-related protein.
Keywords: F-box protein, Arabidopsis, auxin response, TIR1, human SKP2, yeast Grr1p
Physiological studies have implicated the plant hormone indole-3-acetic acid (IAA or auxin) in the regulation of diverse developmental processes including stem elongation, apical dominance, photo- and gravitropism, and lateral root initiation (Klee and Estelle 1991). At the cellular level, auxin acts to regulate these processes through changes in cell division and cell expansion (Evans 1984). Although the mechanism or mechanisms of auxin action are largely unknown, the existing evidence suggests that the hormone acts both at the plasma membrane and within the cell (Vesper and Kuss 1990; Venis and Napier 1995). Auxin treatment results in rapid hyperpolarization of the plasma membrane and induction of specific gene expression (Barbier-Brygoo 1995; Abel and Theologis 1996).
In Arabidopsis, genetic studies have resulted in the identification of a number of genes that are required for normal auxin response (Hobbie and Estelle 1994, Leyser 1997). For three of the genes, AXR1, AXR4, and AUX1, recessive mutations result in reduced auxin response as well as an array of auxin-related growth defects (Hobbie and Estelle 1994). By genetic criteria, AXR1 and AUX1 appear to act in distinct pathways. AUX1 is a membrane protein with similarity to amino acid permeases from plants and fungi (Bennett et al. 1996). Because IAA is an indolic compound structurally related to tryptophan, AUX1 may function in cellular auxin uptake and not response per se. In contrast, the axr1 and axr4 mutations display a synergistic interaction, suggesting that the wild-type genes function in the same or overlapping pathways (Hobbie and Estelle 1995). Genetic experiments indicate that another gene, called SAR1, can also be placed in this group. The sar1 mutants were isolated as suppressors of the axr1 mutations and have a phenotype that is distinct from that of either axr1 or wild type (Cernac et al. 1997). In double-mutant combinations, sar1 is epistatic to axr1. This combination of suppression and epistasis suggests that SAR1 functions after AXR1. Thus, AXR1, AXR4, and SAR1 form a genetically related group of genes that appear to function together to mediate auxin response.
The AXR1 gene encodes a protein related to the ubiquitin-activating enzyme (E1), the first enzyme in the ubiquitin conjugation pathway (Leyser et al. 1993). The function of E1 is to form a thiol–ester linkage between the carboxyl terminus of ubiquitin and an internal cysteine. The ubiquitin moiety is subsequently transferred to a ubiquitin-conjugating enzyme (E2). Finally, ubiquitin is attached to the target protein by an isopeptide linkage between a lysine and the carboxyl terminus of ubiquitin. This last step is catalyzed by a protein or protein complex called a ubiquitin–protein ligase (E3). Proteins related to AXR1 have now been identified in humans (Chow et al. 1996) and fungi (Johnson et al. 1997; Shayeghi et al. 1997). These proteins all share extensive sequence similarity with the amino terminus of E1, but lack the active-site cysteine required for E1 activity. Recent studies in Saccharomyces cerevisiae indicate that at least one member of this family, called Aos1p, functions as a dimer with a second protein called Uba2p (Johnson et al. 1997). Uba2p is similar to the carboxyl terminus of E1 and includes the region containing the active-site cysteine (Dohmen et al. 1995). The Aos1p/Uba2p dimer forms a thiol–ester linkage with a ubiquitin-related protein called Smt3p. Thus, it is possible that AXR1 and other members of the AXR1 protein family each interact with a second protein to form a functional enzyme. The substrate for each heterodimer-type E1, ubiquitin and/or a ubiquitin-related protein, remains to be determined.
To identify additional genes involved in auxin physiology, we have screened for Arabidopsis mutants that are resistant to the growth-inhibiting properties of the auxin-transport inhibitors naphthylphthalamic acid (NPA) and 2-carboxyphenyl-3-phenylpropane-1,2-dione (CPD) (Ruegger et al. 1997). This screen resulted in the recovery of mutants that are altered in auxin transport as well as auxin response. In this report, we show that the transport inhibitor response 1 (tir1) mutants are deficient in auxin response. Molecular characterization indicates that the TIR1 protein is, like AXR1, a member of a family of proteins implicated in ubiquitin-mediated processes.
Results
Genetic characterization of the tir1 mutants
Previously, we reported the isolation of 16 tir mutants by screening for resistance to auxin-transport inhibitors (Ruegger et al. 1997). Genetic analysis indicated that five of these mutations were independent alleles at a locus we called TIR1. To determine the genetic basis for resistance, we examined segregation of CPD resistance in F1 and F2 populations. In our initial studies, seedlings that were heterozygous for tir1 alleles were often significantly less affected by CPD than wild-type seedlings, suggesting that the mutations are at least partially dominant. When F2 populations were analyzed, a graded distribution of phenotypes was observed. An example of a population scored on 5 μm CPD is shown in Table 1. The genotype of individual F2 plants was determined by examining the response of F3 progeny to CPD. This analysis revealed that homozygous tir1 plants are clearly resistant to the compound and heterozygous plants tended to be somewhat resistant compared with homozygous wild-type plants. To confirm that tir1 mutations are semidominant, we directly compared populations of wild-type, tir1/+ and tir1/tir1 seedlings on medium containing CPD. The results are shown in Table 2. For each of the three alleles tested, heterozygous plants were weakly resistant. The five mutants differ only slightly with respect to CPD response. The tir1-2 allele is slightly weaker, whereas tir1-1 and tir1-3 are slightly stronger than tir1-6 and tir1-7. All of the phenotypic analysis described below was performed with the tir1-1 mutant.
Table 1.
Segregation of CPD resistance in a tir1-2 × wildtype F2 population
| F2 root length (mm)
|
F2 genotype1
|
||
|---|---|---|---|
|
tir1/tir12
|
tir1/+2
|
+/+2
|
|
| 17 | 2 | ||
| 16 | 5 | ||
| 15 | 1 | ||
| 14 | |||
| 13 | |||
| 12 | 1 | ||
| 11 | |||
| 10 | 6 | ||
| 9 | 6 | ||
| 8 | 7 | 6 | |
| 7 | 3 | 4 | |
| 6 | 2 | 4 | |
| 5 | |||
| 4 | 1 | ||
| 3 | 1 | ||
| Totals | 8 | 27 | 14 |
Genotype determined by analyzing CPD resistance in F3 plants.
Number of seedlings.
Table 2.
CPD resistance in tir homozygous and heterozygous plants
| Line
|
Root length (mm)
|
±s.e.
|
No.
|
|---|---|---|---|
| +/+ | 7.8 | 0.1 | 32 |
| tir1-1/tir1-1 | 15.7 | 0.3 | 33 |
| tir1-3/tir1-3 | 15.8 | 0.4 | 34 |
| tir1-7/tir1-7 | 13.0 | 0.5 | 33 |
| tir1-1/+ | 10.9 | 0.3 | 15 |
| tir1-3/+ | 12.5 | 0.3 | 17 |
| tir1-7/+ | 12.6 | 0.2 | 17 |
The tir1 mutants are affected in auxin response
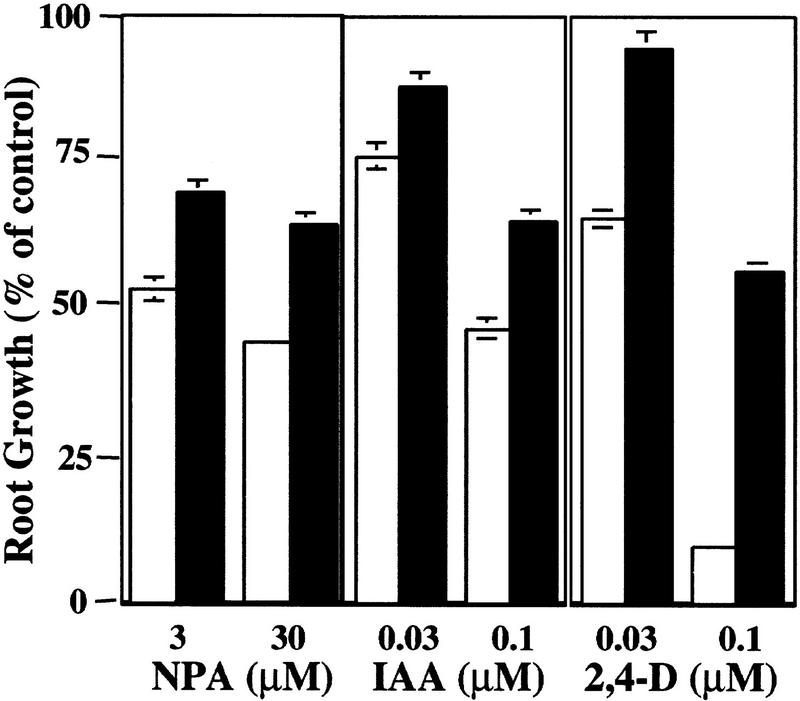
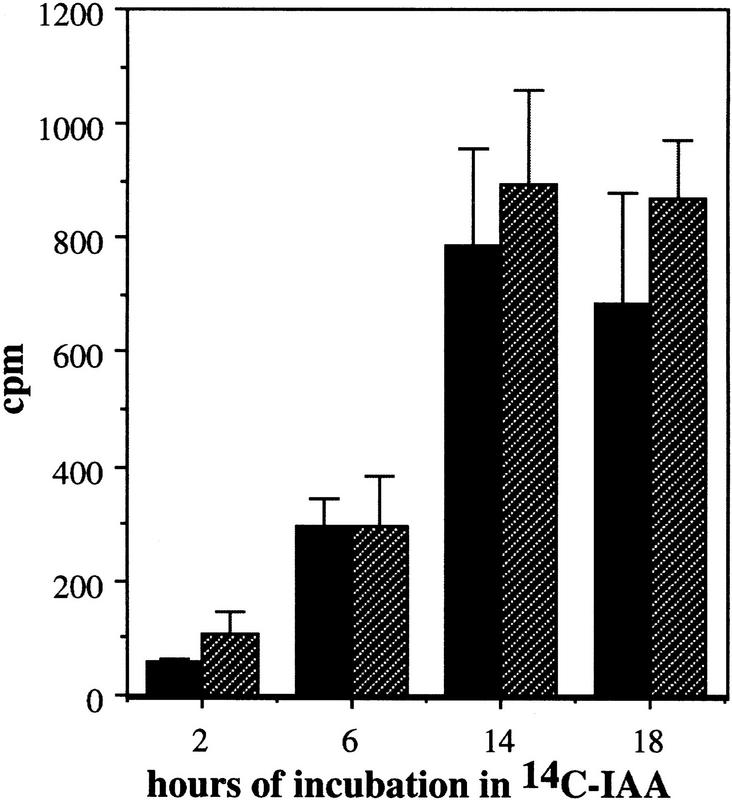
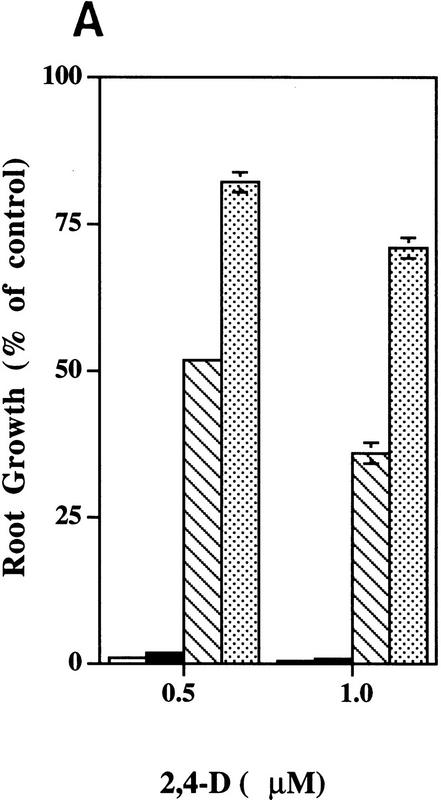
Treatment of Arabidopsis seedlings with auxin-transport inhibitors such as CPD or NPA inhibits root elongation and promotes swelling of the root tip (Ruegger et al. 1997). Both of these effects are thought to be caused by accumulation of auxin in this region as a result of the inhibition of auxin transport away from the root tip (Muday and Haworth 1994; Muday et al. 1995; Ruegger et al. 1997). Because auxin-transport inhibitors affect root growth by increasing auxin levels in the root tip, resistance could result from a change in auxin transport or a reduction in auxin response. To distinguish between these possibilities, we examined the growth of tir1-1 seedlings on media containing NPA, IAA, or the synthetic auxin, 2,4-dichlorophenoxyacetic acid (2,4-D). The results are shown in Figure 1. As expected, tir1-1 seedlings display increased root growth on NPA compared with wild type. Mutant seedlings, however, were also less sensitive to the growth-inhibiting effects of the auxins, suggesting that the primary effect of the mutation is on auxin response and not auxin transport. Consistent with this conclusion, polar auxin transport in tir1-1 stem segments is similar to wild type (Fig. 2). This contrasts with the behavior of the tir3 mutant, which is resistant to NPA but not auxin and has a clear defect in auxin transport (Ruegger et al. 1997). In addition, the response of tir-1 roots to abscisic acid and the cytokinin benzyladenine was similar to wild type.
Figure 1.
The roots of wild-type (open bars) and tir1-1 (solid bars) seedlings are resistant to the growth-inhibiting properties of NPA, IAA, and 2,4-D. Seeds were germinated on nutrient medium. After 3 days, seedlings were transferred to media containing the indicated compound. Five days later, new root growth was measured and plotted as a percentage of root growth on medium without compound. Bars represent standard errors. Absence of bar indicates error less than thickness of line (n = 12).
Figure 2.
Polar auxin transport is similar in wild-type (solid bars) and tir1-1 (hatched bars) inflorescence stems. Two and one-half centimeters of stem was excised, and the apical end was placed in a nutrient solution containing 1 μm [14C]IAA for the times indicated. The amount of radioactive IAA transported to the basal end of the stem was assayed by liquid scintillation. Each column represents the mean of three replicates; the bar represents the standard error of the mean.
Cell proliferation in response to auxin-transport inhibitors is reduced in tir1 seedlings
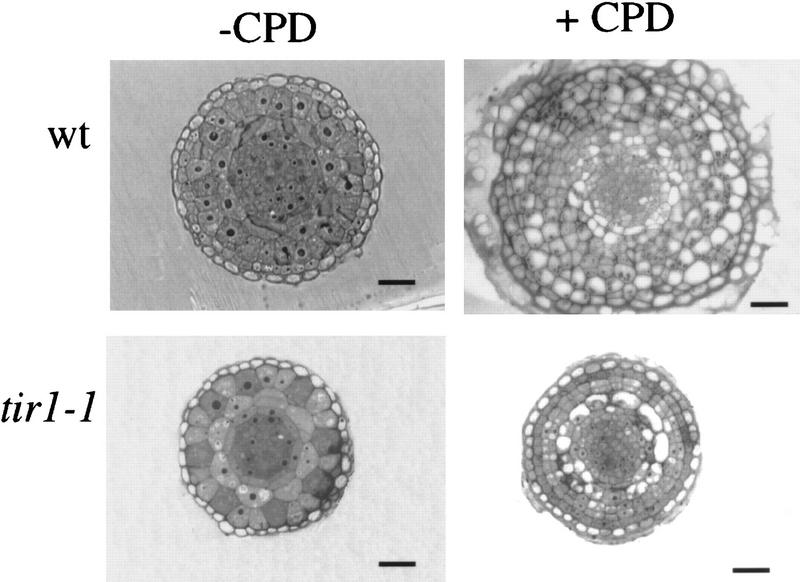
CPD treatment results in pronounced swelling of the root tip and expansion of the zone of cell division away from the root tip (Ruegger et al. 1997). Root tip swelling is the result of extra periclinal cell divisions in the meristem. To determine whether these events occur in tir1 roots, we examined cross sections of wild-type and tir1-1 roots grown on media with or without CPD. The sections shown in Figure 3 were cut 300 μm back from the root tip at a position where the different cell layers of the mature root are clearly defined. The organization of Arabidopsis roots is highly regular and consists of the epidermis, cortex, endodermis, pericycle, and vascular tissue. The appearance of untreated tir1-1 roots was similar to that of untreated wild-type roots. When grown on CPD, wild-type roots exhibited a dramatic increase in cell number as expected (Ruegger et al. 1997). In contrast, tir1-1 roots grown on CPD displayed modest changes in cell number and organization, indicating that CPD-induced cell proliferation is reduced in tir1-1 seedlings.
Figure 3.
Cell proliferation in response to CPD is reduced in the roots of tir1-1 seedlings. Seedlings were grown for 7 days on nutrient medium plus or minus 5 μm CPD, fixed, and embedded in Spurrs (Ruegger et al. 1997). Root cross sections 300 μm from the root tip are shown. Bars, 20 μm.
The TIR1 gene is involved in lateral root formation
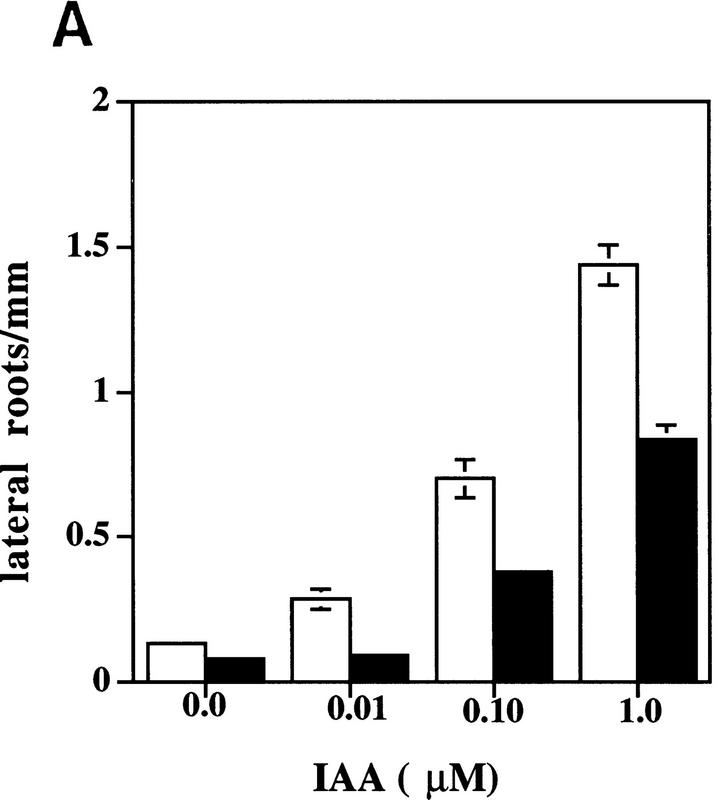
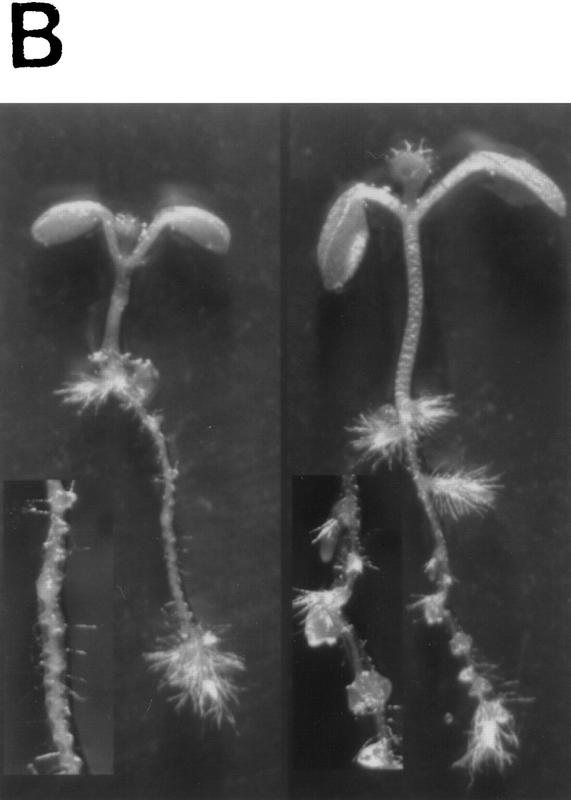
Physiological and genetic studies have shown that auxin is required for lateral root initiation and growth (Wightman and Thimann 1980; Celenza et al. 1995; Hobbie and Estelle 1995). Formation of a lateral root primordium starts with a local increase in anticlinal divisions in the xylem-radius pericycle cells. This is followed by radial expansion of these pericycle cells and several periclinal divisions to form a 4-cell-layer dome-shaped primordium (Malamy and Benfey 1997). When seedlings are treated with auxin, these early events occur in a synchronized fashion along the entire length of the root (Laskowski et al. 1995). Examination of seedlings grown on agar medium indicated that tir1 plants are deficient in lateral root formation. After 10 days of growth, wild-type seedlings had 6.3 ± 0.4 lateral roots (n = 10), whereas tir1-1 seedlings had 1.3 ± 0.4 (n = 10). When tir1-1 seedlings were examined by confocal microscopy, no additional lateral root primordia were observed, suggesting that mutant seedlings are deficient in an early step in formation of a lateral root (data not shown). To determine whether the TIR1 gene functions in auxin-induced lateral root formation, we exposed excised root segments from wild-type and tir1-1 seedlings to various concentrations of IAA or 2,4-D. The results for IAA are displayed in Figure 4A. Both compounds induced lateral root formation in wild-type and tir1-1 root segments. The number of new lateral roots on tir1-1 segments, however, was reduced compared with wild type. This effect can also be observed in Figure 4B. When grown on medium containing 0.5 μm 2,4-D, a continuous row of lateral roots was initiated along the length of the wild-type root. In contrast, initiation of lateral roots on tir1-1 seedlings is sporadic. These results indicate that the TIR1 gene functions in auxin induction of lateral root formation.
Figure 4.
The tir1-1 mutant is deficient in IAA-induction of lateral roots. (A) Eight-millimeter root segments were excised from 5-day-old wild-type (open bar) and tir1-1 (solid bar) seedlings grown on nutrient medium and transferred to medium with IAA. Lateral roots were counted after 5 days by use of a dissecting microscope. Bars represent standard errors. Absence of bar indicates error less than thickness of line (n = 10). (B) Ten-day-old wild-type (left) and tir1-1 (right) seedlings grown on 0.5 μm 2,4-D. (Insets) Higher magnification images of roots.
The tir1 mutants are also deficient in a cell-elongation response
Growth of Arabidopsis seedlings at 28°C under moderate light conditions results in a dramatic increase in hypocotyl elongation compared with seedlings grown at 20°C (Fig. 5; W.M. Gray and M. Estelle, unpubl.). After 9 days of growth, the hypocotyls of wild-type seedlings are 4.5-fold longer at 28°C compared with seedlings grown at 20°C. This increase is primarily caused by increased cell elongation because epidermal cells are 3.7-fold longer at the higher temperature. We have shown that this response is absent in the axr1-12 mutant and reduced in plants that are transgenic for the iaaLys gene and, consequently, have reduced IAA levels (W.M. Gray and M. Estelle, unpubl.). In contrast, mutants deficient in gibberellin biosynthesis or ethylene response are unaffected. These results indicate that increased hypocotyl elongation at elevated temperature is auxin dependent. To determine whether the TIR1 gene functions in this response, we grew wild-type and tir1-1 seedlings at the two temperatures. The results shown in Figure 5 indicate that mutant seedlings are deficient in hypocotyl elongation under these conditions. Mutant hypocotyls are 2.6-fold longer at the higher temperature (compared with 4.5 for wild type). Similarly the epidermal cells of tir1-1 hypocotyls are 2.2-fold longer at 28°C compared with 20°C (wild-type cells are 3.7-fold longer).
Figure 5.
Hypocotyl elongation in response to elevated temperature is reduced in tir1-1 seedlings compared with wild-type seedlings. Seedlings were grown on vertically oriented agar medium for 9 days at the two temperatures.
The tir1 and axr1 mutants display a synergistic interaction
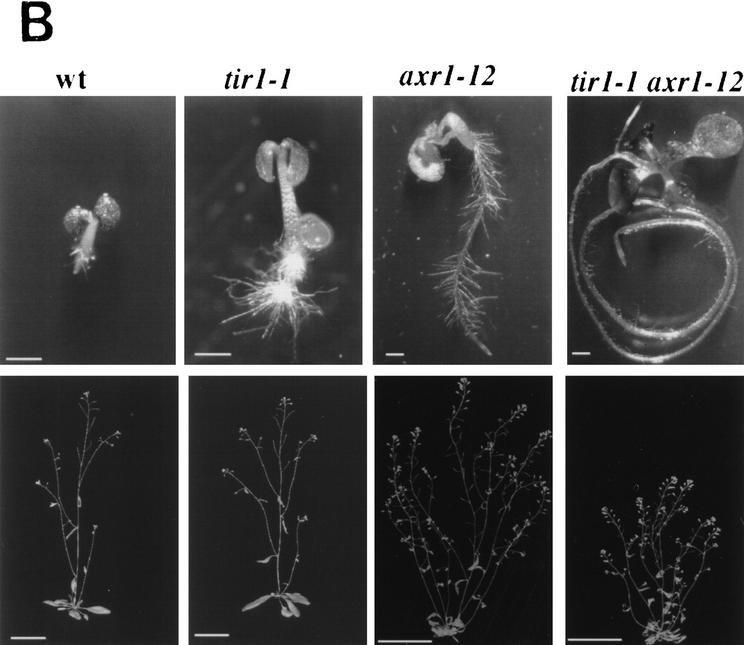
The AXR1 gene is also required for auxin response (Lincoln et al. 1990; Timpte et al. 1995). To explore possible interactions between AXR1 and TIR1, we generated plants that were homozygous for both the axr1-12 and tir1-1 mutations and examined their phenotype. When grown on medium containing 0.1 or 0.5 μm 2,4-D, axr1-12 tir1-1 seedlings are more resistant than axr1-12 seedlings (Fig. 6A). Because this effect occurs in a concentration range where tir1-1 does not confer resistance by itself, the interaction between the two mutations is synergistic. A similar conclusion was reached when the morphology of double-mutant plants was examined. In most respects, the tir1-1 rosette and inflorescence are identical in appearance to wild type (Fig. 6B). There was a slight increase in the number of secondary inflorescences growing from the rosette (wild type 1.5 ± 0.2 compared with tir1 2.3 ± 0.3; n = 10), suggesting an effect on apical dominance. In an axr1-12 background, the tir1-1 mutation caused a further reduction in stature (Fig. 6B). This result indicates that, despite the lack of aerial phenotype, the TIR1 gene functions in aerial structures. Further, it suggests that TIR1 is involved in AXR1-mediated processes.
Figure 6.
tir1-1 and axr1-12 have synergistic effects on auxin response and plant morphology. (A) The tir1-1 mutation decreased auxin response in an axr1-12 background at a concentration at which tir1-1 has no effect by itself. Seedlings were grown and treated as described for Fig. 1. (Open bars) Wild type; (solid bars) tir1; (hatched bars) axr1; (stippled bars) axr1 tir1. Bars represent standard error and absence of bar indicates error less than thickness of line. (B) Seedlings (top row) were photographed after 12 days of growth on 1 μm 2,4-D. Bar, 2.5 mm. Mature plants (bottom row) were photographed 35 days after germination in soil. Bar, 5 cm.
Isolation of the TIR1 gene
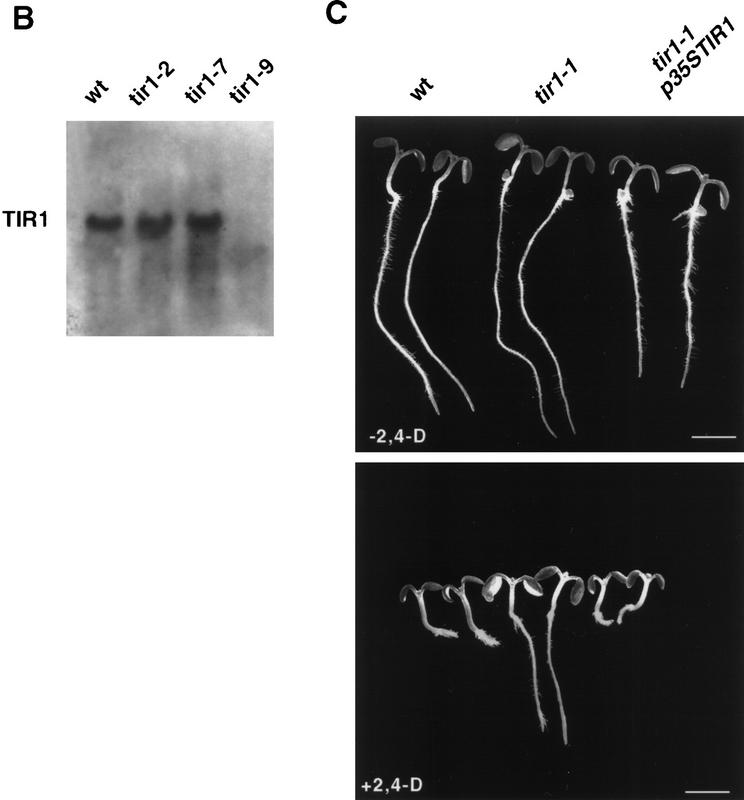
The tir1-9 mutant was isolated from a population of T-DNA transformed plants (Ruegger et al. 1997) generated by K. Feldmann and colleagues (Feldmann 1991). Genetic analysis indicated that the tir1-9 line carried a single T-DNA insert that cosegregated with the tir1-9 mutation. The left end of the T-DNA insert and flanking Arabidopsis sequences were cloned by plasmid rescue. Southern analysis revealed that a 5.5-kb EcoRI–SalI restriction fragment recovered in this fashion detected a polymorphism between wild type (WS) and the tir1-9 mutant, confirming that this Arabidopsis DNA lies adjacent to the T-DNA insert (data not shown). The EcoRI–SalI restriction fragment was used to isolate cDNA clones. The longest cDNA recovered was 2.2 kb in length. DNA sequence analysis of the insert revealed an open reading frame (ORF) encoding a predicted protein of 594 amino acids (Fig. 7A). To confirm that this ORF corresponds to the TIR1 gene, two tir1 alleles were sequenced by RT-PCR. The tir1-1 mutation is a glycine to aspartate substitution at position 147 and the tir1-2 mutation is a glycine to aspartate change at position 441. Sequence analysis of a genomic clone spanning the cDNA indicated that the gene contains two introns. The tir1-9 allele has a T-DNA insert in the first intron. RNA blot experiments with RNA isolated from wild-type and mutant tissues detected a transcript of ∼2.2 kb in wild-type tissues. This transcript was absent in RNA from the tir1-9 mutant (Fig. 7B). The TIR1 transcript was detected in all tissues examined including roots, rosettes, stems, and flowers (data not shown). The TIR1 cDNA was placed adjacent to the CaMV 35S promoter and introduced into tir1-1 plants. Of five independent transgenic lines examined, auxin response was restored to the wild-type level in four, providing final proof that the candidate cDNA was TIR1 (Fig. 7C).
Figure 7.
The characterization of three mutant alleles and transformation rescue of the tir1-1 mutant with a candidate cDNA identify the TIR1 gene. (A) Predicted amino acid sequence of the TIR1 protein. The position of the tir1-1 and tir1-2 mutations, both (G → D), are indicated with an asterisk above the affected residue. The GenBank accession nos. for the cDNA and genomic sequences are AF005048 and AF005047, respectively. (B) RNA blot analysis of TIR1 transcript in tir1 mutants. Twenty-five micrograms of total RNA isolated from 13-day-old seedlings was loaded in each lane. (C) Transformation rescue of the tir1-1 mutant. The p35S:TIR1 plasmid was introduced into tir1-1 plants by vacuum infiltration. Transgenic lines were tested for auxin resistance by plating on medium containing 0.085 μm 2,4-D.
To determine the map position of the TIR1 gene, a restriction fragment length polymorphism (RFLP) was identified between the Columbia and Landsberg ecotypes with the EcoRI–SalI fragment containing the TIR1 gene as a probe. The polymorphism was mapped by use of 44 recombinant inbred lines (Lister and Dean 1993) to position 128 on chromosome 3, close to nga128.
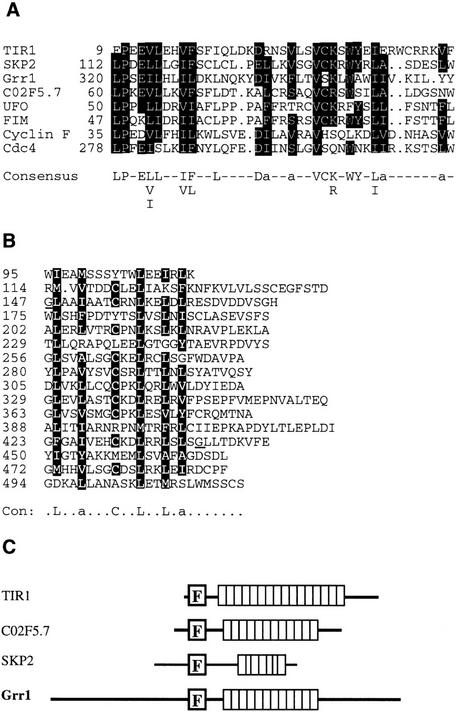
The TIR1 protein contains an F-box domain and leucine rich repeats
Examination of the TIR1 amino acid sequence revealed several features of interest. First, TIR1 contains an F-box domain (Fig. 8A; Bai et al. 1996). This motif is present in a variety of regulatory proteins in mammals and yeast including the S-phase kinase-associated protein (SKP2) and Cyclin F proteins from humans and the Cdc4 and Grr1 proteins from yeast (Bai et al. 1996). Each of these proteins binds a second protein called SKP1 in an interaction that requires the F-box (Bai et al. 1996; Li and Johnston 1997). Recent evidence has also shown that the UFO protein from Arabidopsis contains an F box and binds Arabidopsis orthologs of SKP1 called Ask1 and Ask2 (A. Samach, S. Kohalmi, G. Haughn, and W. Crosby, pers. comm.). Second, TIR1 has 16 degenerate leucine-rich repeats (LRRs) (Fig. 8B). Similar repeats have been identified in a wide variety of proteins although the repeat consensus sequence, length, and level of redundancy vary widely among different proteins (Kobe and Deisenhofer 1995a,b). Data base searches indicate that the TIR1 repeats are most similar to those found in SKP2, Grr1p, and an uncharacterized ORF in Caenorhabditis elegans called C02F5.7, a protein that also has an F box.
Figure 8.
The TIR1 protein has an F box and 16 LRRs. (A) Alignment of F-box motifs from diverse proteins. Identical residues are boxed. (B) Alignment of LRRs in TIR1. The affected residues in the tir1-1 and tir1-2 mutants are underlined. In consensus sequences listed in A and B, aliphatic residues are indicated with an a. (C) Comparison of TIR1, SKP2, Grr1, and C02F5.7. Boxes represent the LRRs.
The presence of an F box and the similarity of the LRRs in TIR1, Grr1p, SKP2, and C02F5.7 suggests that these proteins are related. A comparison of the four proteins is shown in Figure 7C. Although the biochemical function of the F-box proteins and their presumed binding partner SKP1 is uncertain, recent studies indicate that these proteins may function together as a ubiquitin–protein ligase complex called an SCF (Skp1–Cdc53–F-box protein) complex (Bai et al. 1996; Feldman et al. 1997; Li and Johnston 1997; Skowyra et al. 1997).
TIR1 is a member of a family of leucine-rich repeat F-box proteins in Arabidopsis
In addition to the animal and fungal proteins mentioned above, data base searches revealed the existence of a number of TIR1-related genes in Arabidopsis and rice. We have called these genes LRF for LEUCINE-RICH REPEAT F-BOX. Two of the sequences (LRF1 and LRF2) were identified as ESTs. We obtained these clones from the Arabidopsis Biological Resource Center (ABRC) and screened cDNA libraries in an attempt to obtain full-length clones. Sequence analysis indicated that for each gene, the longest cDNA we recovered did not contain the entire coding sequence. Because the LRF1 and LRF2 transcripts are similar in size to TIR1 (data not shown), we estimate that ∼50 nucleotides of coding sequence is missing from the 5′ end of the two cDNAs. The sequence of the third gene, LRF3, was determined by the Arabidopsis Genome Initiative. LRF1 and LRF2 are 69% and 60% identical with TIR1 respectively, whereas LRF3 is 34% identical to TIR1. There is also one highly related rice EST present in the dBest database (accession no. D22807).
Discussion
TIR1 functions in auxin-dependent cell division and cell elongation
The tir1 mutants are deficient in several auxin-mediated responses including auxin-dependent hypocotyl elongation, auxin inhibition of root elongation, CPD-mediated stimulation of cell proliferation in the root tip, and auxin stimulation of lateral root formation. In addition, tir1 seedlings have reduced numbers of lateral roots when they are grown on medium without auxin. These defects strongly suggest that TIR1 functions in auxin response and not auxin transport. Because both cell elongation (hypocotyl elongation) and cell division (cell proliferation at the root tip) are affected, we conclude that TIR1 has a general role in auxin response.
The nature of the lesion in the tir1-9 allele, and the absence of detectable full-length TIR1 RNA in this mutant, strongly suggests that homozygous tir1-9 seedlings completely lack TIR1 protein. Because this mutant still responds to auxin, the TIR1 protein is not essential for auxin response. Somewhat paradoxically, the semidominant nature of the tir1 mutations indicates that Arabidopsis plants are sensitive to TIR1 gene dose. One wild-type gene is not sufficient to confer a wild-type phenotype, but complete loss of TIR1 function results in only a modest increase in the severity of the phenotype. One possible explanation for this behavior is that the closely related LRF proteins are functionally redundant with TIR1. In the absence of TIR1, the LRF proteins may provide sufficient function to maintain near normal growth and development.
The TIR1 protein may function in ubiquitin–protein conjugation
Among animal and fungal proteins, TIR1 is most closely related to the SKP2 protein from human cells, the Grr1 protein from S. cerevisiae, and an uncharacterized ORF in Caenorhabditis elegans called C02F5.7. Each of these proteins has a motif called an F box as well as a series of LRRs. The F box was first identified by Bai et al. (1996), and shown to be required for binding to a conserved protein called SKP1. This interaction has now been shown for the F-box proteins cyclin F and SKP2 from mammals, Cdc4p and Grr1p from yeast, and the UFO protein from Arabidopsis (Zhang et al. 1995; Bai et al. 1996; Li and Johnston 1997; A. Samach, S. Kohalmi, G. Haughn, and W. Crosby, pers. comm.). The function of SKP1 and the F-box proteins is uncertain. Several lines of evidence, however, suggest that they act as components of one or more ubiquitin–protein ligases (E3) to facilitate transfer of ubiquitin from an E2 enzyme to a target protein. In yeast cells, Skp1p binds the F-box proteins Cdc4p and Grr1p. These two proteins are essential for ubiquitin-mediated degradation of the Clb–CDK inhibitor Sic1p, and the G1 cyclins (Cln1p and Cln2p), respectively (Bai et al. 1996; Li and Johnston 1997). In skp1 mutants, both Sic1p and the G1 cyclins are stabilized. Taken together, these results suggest that Skp1p works in conjunction with an F-box protein to target the ubiquitin conjugation machinery to specific proteins. In two recent studies, Feldman et al. (1997) and Skowyra et al. (1997) shown that Skp1p, Cdc53p, and one of several F-box proteins form ubiquitin-ligase complexes called SCFs. The F-box protein is apparently involved in conferring specificity to the complex. There are at least two SKP1 homologs in Arabidopsis (Nadeau et al. 1996; A. Samach, S. Kohalmi, G. Haughn, and W. Crosby, pers. comm.) and at least one CDC53 homolog (Genbank accession no. AC002330). TIR1 may interact with these proteins or similar proteins and promote ubiquitin modification of one or more regulatory proteins required for auxin response.
AXR1 and TIR1 function together to mediate auxin response
We have determined previously that the AXR1 protein is similar to the amino-terminal half of the ubiquitin-activating enzyme E1 (Leyser et al. 1993). Although AXR1 is missing key residues known to be essential for E1 activity, recent studies in yeast indicate that AXR1 and related proteins probably function to activate either ubiquitin or another small ubiquitin-like protein. In S. cerevisiae, two AXR1 homologs (Aos1p and Enr2p) have been investigated. Aos1p functions together with another E1-related protein called Uba2p to activate a ubiquitin-like protein called Smt3p (Johnson et al. 1997). Enr2p is required for conjugation of a different ubiquitin-related protein, called Rub1p, to the cell cycle protein Cdc53p (D. Lammer, N. Mathias, J. Laplaza, Y. Liu, J. Callis, M. Goebl, and M. Estelle, unpubl.). These observations suggest that AXR1 forms a dimer with a second protein that carries the active-site cysteine, similar to the situation with yeast Aos1p and Uba2p. This heterodimer would then activate ubiquitin or a ubiquitin-related protein.
The results of our genetic experiments suggest that the AXR1 and TIR1 genes function in the same or overlapping pathways. This is consistent with our current understanding of the AXR1 and TIR1 proteins. AXR1 may participate in the activation of either ubiquitin or a ubiquitin-related protein and TIR1 could function later in the pathway as part of an E3 complex. The nature of the targeted protein is unknown. We hypothesize that auxin response depends on ubiquitin (or ubiquitin-related protein) modification of one or more regulatory proteins. Ubiquitination may then result in degradation by the proteosome (Hochstrasser 1996). This is consistent with a model proposed by Theologis and colleagues in which transcription of auxin-regulated genes is normally repressed by the action of short-lived repressor proteins (Ballas et al. 1995; Abel and Theologis 1996). Auxin may relieve this repression by stimulating degradation of the repressors in an AXR1–TIR1 dependent manner. Alternatively, the modification may alter the activity or cellular localization of a regulatory protein. For example, ligand-dependent ubiquitin modification of the α-factor receptor in yeast results in endocytosis of the receptor (Hicke and Riezman 1996). In mammalian cells, localization of the protein RanGAP1 to the nuclear pore complex depends on modification by the ubiquitin-related protein SUMO1 (Mahajan et al. 1997). Clearly, we will need to identify the substrates of the AXR1–TIR1 pathway(s) to establish a mechanism. One potential substrate is the product of the SAR1 gene. By genetic criteria, SAR1 has a negative role in auxin response, and acts downstream of AXR1 (Cernac et al. 1997).
How auxin affects the pathway is also uncertain, particularly because neither the auxin receptor nor other signal transduction components have been identified. One attractive possibility is suggested by experiments of Li and Johnston (1997) on Grr1p. As mentioned above, Grr1p is required for ubiquitin-mediated degradation of the G1 cyclins Cln1p and Cln2p in a process that appears to involve an SCF complex that contains Grr1p (Feldman et al. 1997; Li and Johnston 1997; Skowyra et al. 1997). Grr1p also plays a central role in glucose repression and glucose induction of genes encoding glucose transporters, however, a function that also requires Skp1p. When wild-type cells are grown in glucose, there is a substantial increase in the amount of Grr1p–Skp1p complex that is immunoprecipitated relative to cells grown in glycerol even though the levels of Grr1p and Sk1p are similar in the two extracts. Li and Johnston (1997) suggest that this difference may reflect a mechanism by which the cell links nutrient availability to cell cycle regulation. Because TIR1 may bind a SKP1 homolog in Arabidopsis, it is possible that auxin affects this interaction in some way, thereby altering the level of ubiquitin modification of one or more targets. We are currently working to identify TIR1-interacting proteins to test this model.
Summary
Genetic studies in Arabidopsis have led to the identification of eight genes that are required for normal auxin response (Hobbie and Estelle 1994). To date, three of these genes have been isolated. The structures of the proteins encoded by two of the genes, AXR1 (Leyser et al. 1993) and TIR1 (this study), suggest that they function in a ubiquitin–protein conjugation pathway. Thus, our results provide strong support for a role for ubiquitin (or related protein) modification in auxin response. Recently, D. Xie and J. Turner (University of East Anglia, Norwich, UK) have independently shown that the LRF3 gene described here is identical to a previously identified gene called COI1 (J. Turner, pers. comm.). The recessive coi1 mutants are completely insensitive to jasmonic acid, suggesting that the gene plays a key role in jasmonate signaling or response (Feys et al. 1994). It appears likely that the TIR1/COI1 genes represent the first members of a novel class of signaling proteins in plants.
Materials and methods
Plant materials and growth conditions
All mutant lines were from the Columbia ecotype. Plants were grown at 21–23°C under continuous fluorescent illumination (100–150 μE/m2 per sec) in 13-cm clay pots containing Metro-Mix (W.R. Grace & Co.) or an equivalent soilless mixture. During the first 3 weeks of growth, a mineral nutrient solution containing 5 mm KNO3, 2.5 mm KPO4 (adjusted to pH 5.8), 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm Fe–EDTA, 70 μm H3BO3, 14 μm MnCl2, 0.5 μm CuSO4, 1 μm ZnSO4, 0.2 μm Na2MoO4, 10 μm NaCl, and 10 nm CoCl2 was supplied to the plants. For many experiments, plants were grown under sterile conditions in petri plates containing the above nutrient solution plus 0.7% agar, 1.0% sucrose (wt/vol), and various hormones or inhibitors as indicated in the text. Before plating, seeds were surface-sterilized by agitation for 10–20 min in 20% commercial bleach and 0.02% Triton X-100, rinsed several times with sterile water, and held at 4°C for 3–4 days to enhance germination. The seeds were dispersed on the growth media with sterile water. The plates were oriented vertically and held in an incubator containing fluorescent lighting (30–50 μE/m2 per sec; 16 hr photoperiod) and a temperature of 20–21°C. In all experiments where plants are grown, day 0 is considered to be the time when seeds are first placed in the growth conditions described above. For hypocotyl and cell elongation studies, seedlings were grown under 24 hr light at 85 μE/m2 per sec.
Genetic analysis
For genetic analyses, resistance to inhibition of root growth by CPD (a gift from G.F. Katekar, Commonwealth Scientific and Industrial Research Organization, Canberra City, Australia) was assayed by the following procedure. Surface-sterilized, cold-treated seeds were plated on nutrient medium, as described above, supplemented with 5 μm CPD. On day 7 (unless otherwise stated) the seedlings were straightened with forceps and the root length (distance from the root/hypocotyl junction to the root tip) was measured. Resistance to 0.2 μm 2,4-D was assayed in a similar manner except that roots were scored visually as either 2,4-D-resistant or wild type.
To generate axr1-12 tir1-1 double mutant plants, crosses between the single mutant lines axr1-12 and tir1-1 were performed. The genotypes of individual F2 plants from these crosses were determined by test crosses to both of the parental lines and tests for CPD or 2,4-D resistance as described above.
Linkage analysis was performed with a 9-kb EcoRI–SalI fragment of Arabidopsis genomic DNA containing the coding sequences of the TIR1 gene. This fragment detected a RFLP between EcoRV-digested Columbia and Landsberg erecta (Ler) genomic DNA. Mapmaker I (S. Lincoln, M. Daly, J. Abrahamson, A. Barlow, and L. Newburg; Massachusetts Institute of Technology, Cambridge, MA) was used to compile the results of the DNA blot analysis of 44 RI lines (Lister and Dean 1993) with this fragment as a probe.
Auxin-transport assays
Auxin-transport assays were performed according to the method of Okada et al. (1991). Stem segments (2.5 cm) of primary inflorescence were incubated in a 1.5-ml Eppendorf centrifuge tube containing 30 μl of nutrient solution with 1 μm [14C]IAA (1.74 nCi/ml). The segments were incubated with their apical end in the solution for various times from 2 to 18 hr. After the incubation, a 5-mm section from the basal end of the segment was excised and added to 3 ml of liquid scintillation cocktail (Bio-Safe II; RPI, Mount Pleasant, IL). The samples were shaken at 100 rpm for at least 2 hr and left overnight at room temperature before scintillation counting in a scintillation counter (model LS 6500; Beckman Instruments). The experiment involved three stem segments for each time point. Wild-type and tir1-1 plants were grown in 24 hr of light (105 μE/m2 per sec) for 36 days.The experiment was repeated three times with similar results. Stems incubated with 15 μm NPA or with the basal end of the stem segment in the solution transported low levels of [14C]IAA.
Induction of lateral root formation by IAA
Seedlings were grown on nutrient medium for 5 days. Eight-millimeter root segments were excised with a razor blade from mature root and transferred to nutrient medium supplemented with various concentrations of IAA. The number of lateral roots was determined by use of a dissecting microscope and expressed as the number of lateral roots per millimeter of primary root.
Isolation of sequences flanking the T-DNA insert in tir1-9
Plasmid rescue of T-DNA sequences was done following the procedure of Behringer and Medford (1992). Genomic DNA, prepared from rosette leaves of the tir1-9 mutant line, was digested with either SalI or EcoRI to isolate DNA flanking the T-DNA left border or right border, respectively. The digested DNA was ligated with T4 DNA ligase, and electroporated into JS4 E. coli (Bio-Rad). Four colonies derived from the SalI-digested DNA (L1–L4) and >100 colonies derived from the EcoRI-digested DNA were recovered. Plasmid DNA from L1–L4 and from 14 of the EcoRI-derived colonies (R1–R14) was prepared and analyzed by restriction analysis to eliminate clones derived entirely from the T-DNA insert (Behringer and Medford 1992). Two left border isolates (L3 and L4) had identical restriction digest patterns and potentially contained Arabidopsis DNA. When used as a probe in DNA blot analysis, a 5.5-kb EcoRI–SalI fragment from L3 identified RFLPs between tir1-9 and wild-type WS genomic DNA.
Isolation and characterization of genomic and cDNA clones
The L3 5.5-kb probe was used to isolate six genomic clones (D1–D6; library, a gift from R. Davis, Stanford University, CA) and four cDNA clones (E1–E4; library, a gift from J. Ecker, University of Pennsylvania, Philadelphia). E4, which contained the largest insert, was subcloned for sequencing (Sequenase kit, U.S. Biochemical). A 9-kb EcoRI–SalI fragment from D1 (D109ES), which hybridized to E4, was subcloned into pBluescript (Stratagene). A 1.5-kb HincII fragment, a 2.1-kb SacI ClaI fragment, and a 1.5-kb HindIII fragment, which together encompassed a 3.8-kb region of D109ES containing the TIR1 coding region, were subcloned into pBluescript and sequenced (Sequitherm, Epicentre Technologies).
Characterization of tir1 mutant alleles
RT-PCR (Stratagene) was used to amplify the expressed sequences of two tir1 alleles from total RNA prepared from seedlings grown in liquid culture. The PCR products were separated in a low-melt agarose gel and sequenced directly by use of a modified Sequenase protocol (B. Robertson, University of Colorado). The position of the T-DNA insertion of tir1-9 was determined by sequencing a subcloned fragment of the L3 plasmid that hybridized to probes made from the TIR1 cDNA and from the T-DNA left border (plasmid pBSH10).
RNA blot analysis
Total RNA was isolated from 13-day-old seedlings according to Newman et al. (1993) with modifications by Timpte et al. (1995), and blotted as described in Timpte et al. (1995).
Generation of transgenic plants
The 2.2-kb TIR1 cDNA was cloned into the pBI121 vector (replacing the GUS gene) to generate the p35S:TIR1 plasmid. This plasmid was introduced into the Agrobacterium strain GV3101 by electroporation. tir1-1 plants were inoculated with this strain by vacuum infiltration according to Bechtold et al. (1993). Transformed seedlings were identified by selection on 50 μg/ml kanamycin. Transgenic lines were tested for TIR1 function by plating on medium that contained 0.085 mm 2,4-D.
Acknowledgments
We are grateful to members of the Estelle laboratory and the Indiana University Arabidopsis group for stimulating discussion throughout the course of this work, to Roger Innes for careful reading of the manuscript, to the ABRC for cDNAs, and to L. Washington for assistance with DNA sequencing. This research was supported in part by the National Science Foundation (postdoctoral fellowship MCB-9008316 to L.H., IBN-9307134 to M.E.) and National Institutes of Health (U.S. Public Health Service grant-GM43644 to M.E.) and an Eli Lilly/Indiana Institute for Molecular Biology predoctoral fellowship to M.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mestelle@bio.indiana.edu; FAX (812) 855-6705.
References
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Ballas N, Wong L-M, Ke M, Theologis A. Two auxin-responsive domains interact positively to induce expression of the early indolacetic acid-inducible gene PS-IAA4/5. Proc Natl Acac Sci. 1995;92:3483–3487. doi: 10.1073/pnas.92.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H. Tracking auxin receptors using functional approaches. Crit Rev Plant Sci. 1995;14:1–25. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Behringer FJ, Medford JI. A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol Biol Rep. 1992;10:190–198. [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes & Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Cernac A, Lincoln C, Lammer D, Estelle M. The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development. 1997;124:1583–1591. doi: 10.1242/dev.124.8.1583. [DOI] [PubMed] [Google Scholar]
- Chow NW, Korenberg JR, Chen XN, Neve RL. APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein. J Biol Chem. 1996;271:11339–11346. doi: 10.1074/jbc.271.19.11339. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Evans ML. Functions of hormones at the cellular level of organization. In: Scott TK, editor. Hormonal regulation of development II, encyclopedia of plant physiology. Vol. 10. Berlin, Germany: Springer-Verlag; 1984. pp. 23–79. [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: Mutation spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hobbie L, Estelle MA. Genetic approaches to auxin action. Plant Cell Environ. 1994;17:525–540. doi: 10.1111/j.1365-3040.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- ————— The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 1995;7:211–220. doi: 10.1046/j.1365-313x.1995.7020211.x. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteosomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H, Estelle M. Molecular genetic approaches to the biology of plant hormones. Annu Rev Plant Physiol Mol Biol. 1991;42:529–551. [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995a;374:183–185. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- ————— Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995b;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln C, Timpte C, Lammer D, Turner J, Estelle M. The auxin-resistance gene AXR1 of Arabidopsis encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Leyser O. Auxin: Lessons from a mutant weed. Physiol Plantarum. 1997;100:407–414. [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis pathway through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Muday GK, Haworth P. Tomato root growth, gravitropism and lateral root development: Correlations with auxin transport. Plant Physiol & Biochem. 1994;32:193–203. [PubMed] [Google Scholar]
- Muday GK, Lomax TL, Rayle DL. Characterization of the growth and auxin physiology of roots of the tomato mutant, diageotropica. Planta. 1995;195:548–553. doi: 10.1007/BF00195714. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Zhang XS, Li J, O’Neill SD. Ovule development: Identification of stage-specific and tissue-specific cDNAs. Plant Cell. 1996;8:213–239. doi: 10.1105/tpc.8.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TC, Ohem-takagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M. Reduced NPA-binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell. 1997;9:745–757. doi: 10.1105/tpc.9.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayeghi M, Doe CL, Tavassoli M, Watts FZ. Characterization of a Schizosaccharomyces pombe RAD31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res. 1997;25:1162–1169. doi: 10.1093/nar/25.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Venis MA, Napier RM. Auxin receptors and auxin binding proteins. Crit Rev Plant Sci. 1995;14:27–47. [Google Scholar]
- Vesper MJ, Kuss KL. Physiological evidence that the primary site of auxin action in maize coleoptiles is an intracellular site. Planta. 1990;182:486–491. doi: 10.1007/BF02341022. [DOI] [PubMed] [Google Scholar]
- Wightman F, Thimann KV. Hormonal factors controlling the initiation and development of lateral roots. Physiol Plant. 1980;49:13–20. [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]