Abstract
Transcriptional activation of the c-jun gene is a critical event in the differentiation of F9 cells. In our previous studies we characterized an element [differentiation response element (DRE)] in the c-jun promoter that is both necessary and sufficient to confer the capacity for differentiation-dependent up-regulation. This element binds the differentiation regulatory factor (DRF) complex, of which one component is the adenovirus E1A-associated protein p300. We have now identified activation transcription factor-2 (ATF-2) as a DNA-binding subunit of the DRF complex. p300 and ATF-2 interact with each other in vivo and in vitro. The bromodomain and the C/H2 domain of p300 mediate the binding to ATF-2, which in turn requires a proline-rich region between amino acids 112 and 350 for its interaction with p300. The phosphorylation of the serine residue at position 121 of ATF-2 appears to be induced by protein kinase Cα (PKCα) after treatment of cells with retinoic acid (RA) or induction with E1A. In cotransfection assays, wild-type ATF-2 enhanced the transcription of an E2/tk–luciferase construct, in conjunction with p300–E2. However, a mutant form of ATF-2 with a mutation at position 121 (pCMVATF–2Ser121–Ala) did not. These results suggest that ATF-2 and p300 cooperate in the control of transcription by forming a protein complex that is responsive to differentiation-inducing signals, such as RA or E1A, and moreover, that the phosphorylation of ATF-2 by PKCα is probably a signaling event in the pathway that leads to the transactivation of the c-jun gene in F9 cells.
Keywords: p300, ATF-2, PKCα, c-jun, differentiation of F9 cells
Activating transcription factor-2 [ATF-2, also known as CRE-BP-1] is a member of the ATF/CREB family of transcription factors and has a basic region–leucine zipper motif (bZip domain; Hai et al. 1989; Maekawa et al. 1989). This motif is necessary for the binding of heterodimers formed with other members of the ATF family, as well as with members of the Jun/Fos family of factors, in addition to the binding of homodimers, to the cAMP response element (CRE). The CR3 of adenovirus E1A protein interacts with the leucine zipper of ATF-2, which is believed to induce a conformational change that results in exposure of the amino-terminal activation domains (Flint and Jones 1991; Chatton et al. 1993; Liu and Green 1994; Li and Green 1996). Thus, the transactivation domains of ATF-2 can be activated by CR3 of E1A via a mechanism that is independent of the leucine zipper region (Flint and Jones 1991). This type of activation involves the phosphorylation of two threonine residues in the amino-terminal region of ATF-2, namely Thr-69 and Thr-71, by members of the family of stress-activated protein kinases (SAPKs) in cases of serum induction, UV irradiation, exposures to interleukin-1, and genotoxic stress (Derijard et al. 1994; Gupta et al. 1995; Livingston et al. 1995; van Dam et al. 1995). The SAPKs (p46 and p54) that phosphorylate ATF-2 are closely related but not identical to the Jun kinases, JNKs [jun N (amino)-terminal kinases], which also belong to the family of SAPKs and phosphorylate the c-Jun protein in response to cellular stress (Hibi et al. 1993; Kyriakis et al. 1994; Livingston et al. 1995). The E1A product can also activate transcription of the c-jun gene in a CR1-dependent manner (Kitabayashi et al. 1991, 1995; van Dam et al. 1990, 1993). This activation is mediated by two AP-1/ATF-like elements, Jun 1–TRE and Jun 2–TRE, which preferentially bind c-Jun–ATF-2 heterodimers (van Dam et al. 1993, 1995). The CR1-dependent activation was found not to depend strictly on the presence of Thr-69 and Thr-71 at the amino terminus of ATF-2 (Duyndam et al. 1996). Thus, the mechanism of CR-1-dependent activation of the c-jun gene mediated by ATF-2 is still unclear. Recently, ATF-2-deficient mice were generated and exhibited symptoms of chondrodysplasia and neurological abnormalities (Reimond et al. 1996).
p300 and the related CBP (CREB-binding protein; Chrivia et al. 1993; Arany et al 1994, 1995; Eckner et al. 1994; Kwok et al. 1994), referred to below as p300/CBP, are highly homologous proteins that were originally identified as a consequence of the ability to interact with adenovirus E1A proteins and with the transcription factor CREB, respectively. Both p300 and CBP are regulators of transcription that interact with a variety of viral and cellular proteins including components of the basal transcription machinery (Abraham et al. 1993; Kwok et al. 1994; Janknecht and Hunter 1996; Shikawa et al. 1997). In addition, p300/CBP possesses histone acetyltransferase activity that is due either to its intrinsic activity and/or to an associated protein, P/CAF (Bannister et al. 1996; Ogryzko et al. 1996; Yang et al. 1996). The acetylation of histones is thought to be involved in the destabilization and restructuring of nucleosomes, which is probably a crucial event in the control of accessibility of DNA templates to transcription factors.
p300 and CBP are also intimately involved in growth control, progression of the cell cycle, and cell differentiation, as is evident by their interaction with the tumor suppressor p53 (Avantaggiati et al. 1997; Gu et al. 1997; Lill et al. 1997; Somasundaran and El-Deiry 1997) and the viral oncoproteins E1A (Moran 1993) and SV40 large T antigen (Avantaggiati et al. 1996; Eckner et al. 1996). We showed previously that phosphorylation of p300 is associated with retinoic acid (RA)-mediated and E1A-mediated differentiation of F9 cells and identified two factors, designated DRF1 (differentiation regulatory factor 1) and DRF2, that bind specifically to a DRE (differentiation response element), which is necessary and sufficient for RA- and E1A-mediated up-regulation of c-jun expression (Kitabayashi et al. 1992). More recently, we identified p300 as a component of both DRF1 and DRF2 complexes (Kitabayashi et al. 1995). We demonstrate here the molecular association of p300 with ATF-2 in vitro and in vivo ATF-2 and p300 cooperate in the activation of transcription of the c-jun gene, which requires protein kinase Cα (PKCα)-mediated phosphorylation of Ser-121 of ATF-2 within its p300 interaction domain. These findings provide further insight in the regulation of c-jun expression in F9 cells in response to differentiation-inducing signals.
Results
p300 interacts with ATF-2
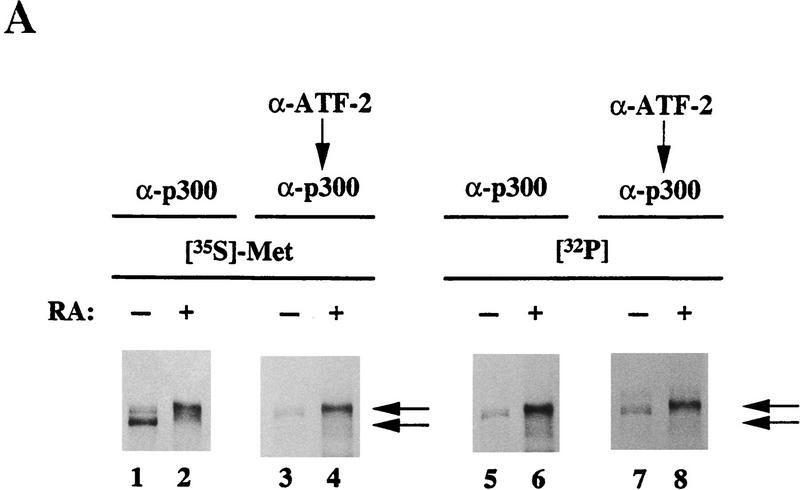
We and others reported previously that the half-site of a DRE inverted repeat sequence of the c-jun promoter overlaps the sequence recognized by Jun 2–TRE, which is sufficient for the activation of the c-jun gene in adenovirus E1-transformed cells (van Dam et al. 1993, 1995; Kitabayashi et al. 1995). Therefore, we performed sequential immunoprecipitations to see the molecular association between ATF-2 and p300 protein (Fig. 1A). The upper band of the doublet or the slowest migrating band of p300 was preferentially radiolabeled upon incubation of undifferentiated F9 cells or RA-induced F9 cells, respectively, with [32P]orthophosphate (Kitabayashi et al. 1995; Fig. 1A, lanes 1,2,5,6). The immunoprecipitates obtained with ATF-2-specific antibodies were boiled in SDS-containing buffer and the released proteins were exposed to p300-specific affinity-purified antibodies. The results revealed the presence of the most slowly migrating p300 proteins, which had been phosphorylated with [32P]orthophosphate, in both undifferentiated and differentiated F9 cells (Fig. 1A, lanes 3,4,7,8). These data indicate that ATF-2 preferentially binds the hyperphosphorylated forms of p300 in both undifferentiated and differentiated F9 cells treated with RA.
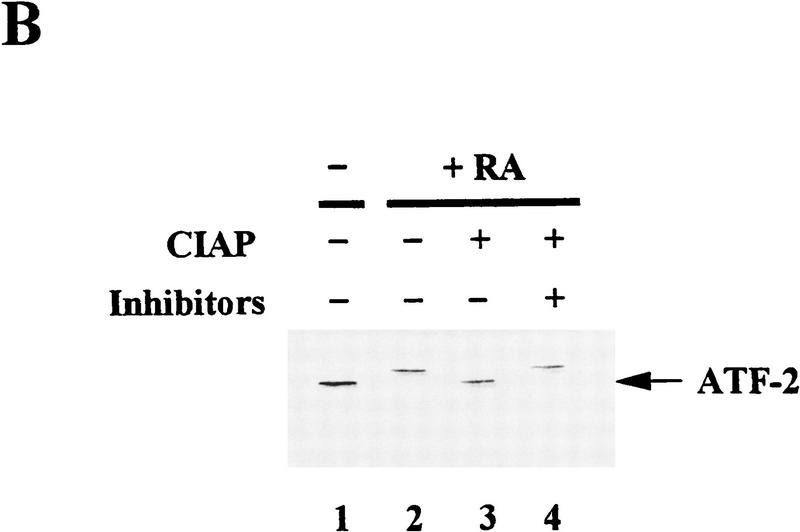
Figure 1.
Selective interaction between p300 and ATF-2 protein species in vivo and the effects of phosphatase in the electrophoretic mobilities of ATF-2. (A) Cell lysates from F9 cells and RA-treated F9 cells were immunoprecipitated directly with antibodies specific for p300 (α-p300; RW128) or were subjected to sequential immunoprecipitations with antibodies against ATF-2 (α-ATF-2; C-19) and antibodies against p300, as described in Materials and Methods. The immunoprecipitated proteins were resolved by SDS-PAGE (5% polyacrylamide). (Lanes 1,3,5,7) Extracts of F9 cells; (lanes 2,4,6,8) p300 from extracts of differentiated F9 cells. (Lanes 1–4) [35S]Methionine-labeled p300; (lanes 5–8) 32P-labeled p300. The positions of both bands of p300 proteins are indicated by arrows. (B) Cell lysates from F9 cells (lane 1) and RA-treated F9 cells (lanes 2–4) were incubated with 50 units of CIAP at 37°C for 30 min in 50 mm Tris-HCl (pH 8.0) in the presence (lane 4) or absence (lane 3) of phosphatase inhibitors (10 mm sodium phosphate, 15 mm sodium pyrophosphate, 5 mm NaF, 0.1 mm Na3VO4). The immunoprecipitated proteins were resolved by SDS-PAGE (10% polyacrylamide) and immunoblotted with antibodies specific for ATF-2 (C-19). The positions of bands of ATF-2 are indicated by arrows.
Similar complementary experiments were performed to determine which forms of ATF-2 protein interacted with p300 protein. The most slowly migrating ATF-2 protein, which had been phosphorylated with [32P]orthophosphate, was detected in the case of both undifferentiated and differentiated F9 cells, on a gel upon the sequential immunoprecipitation of the immunoprecipitate obtained with p300-specific antibodies, with ATF-2-specific monoclonal antibodies (data not shown). The results imply that the phosphorylation of the ATF-2 protein increases during the differentiation of F9 cells (van Dam et al. 1995) and that the hyperphosphorylated form of ATF-2 protein associates to a significant extent with the hyperphosphorylated form of p300 in both undifferentiated F9 cells, RA-induced and E1A-induced differentiated F9 cells in vivo (data not shown). Comparative peptide mapping of ATF-2 immunoprecipitates using Staphylococcus aureus V8 protease suggested that the molecules associated with p300 in uninduced, RA-induced F9 cells, and E1A-transformed F9 cells were the same or biochemically related ATF-2 protein(s) (data not shown).
To determine whether the RA- or E1A-induced changes in mobility of ATF-2 were due to phosphorylation, we prepared 35S-labeled immunoprecipitates of ATF-2 from RA-treated F9 cells and treated them with calf intestine alkaline phosphatase (CIAP). When ATF-2 was treated with CIAP, the slowly migrating form of ATF-2 disappeared, whereas the faster migrating form of ATF-2 was unaffected (Fig. 1B, lane 3). The effect of CIAP was eliminated in the presence of phosphatase inhibitors (lane 4). Upon infection of F9 cells with Ad5, similar changes in ATF-2 protein were observed upon treatment with CIAP in the presence or absence of CIAP inhibitors (data not shown). We conclude from these data that the changes in mobilities of ATF-2 that correlate with the activation of c-jun transcription during the differentiation of F9 cells (Kitabayashi et al. 1992; van Dam et al. 1995; Duyndam et al. 1996) are most likely due to phosphorylation(s).
p300 and ATF-2 are involved in the DRF complexes
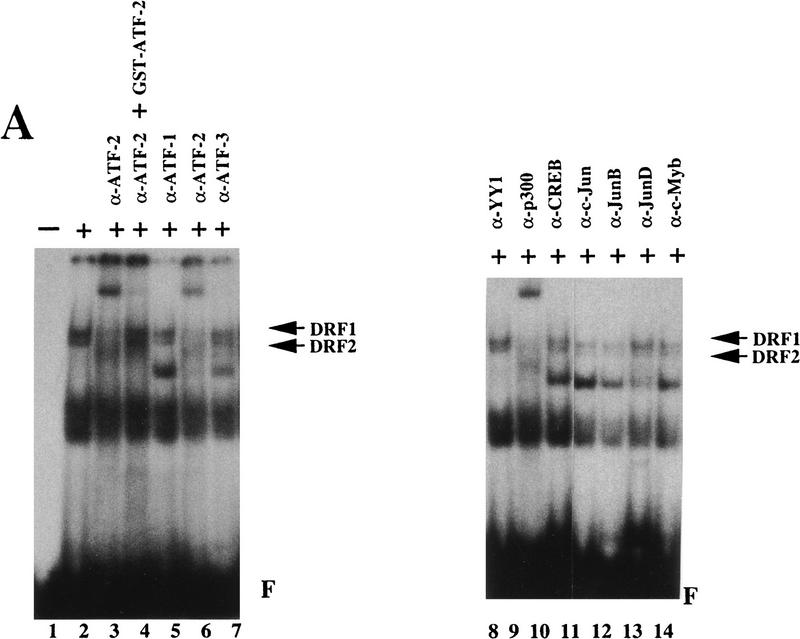
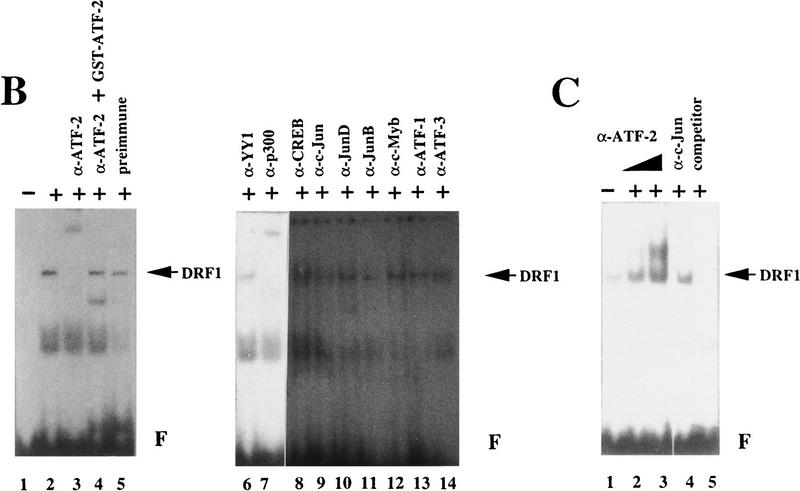
To determine whether ATF-2 is directly associated with DRFs, we performed band-shift assays with DRE as a DNA probe and polyclonal antibodies against ATF-2 (Maekawa et al. 1989; Livingston et al. 1995). As shown in Figure 2A, the retarded bands that corresponded to DRF1 and DRF2 were shifted still further upon addition of the polyclonal antibodies against ATF-2 (lanes 3,6). The supershifting of DRF complexes disappeared after the addition of polyclonal antibodies against ATF-2 that had been absorbed with GST–ATF-2 protein (lane 4). Antibodies against ATF-1, ATF-3, YY-1, CREB, c-Jun, Jun B, Jun D, and c-Myb did not affect the mobilities of the DRF complexes (lanes 5,7,8,10–14). Polyclonal antibodies against p300 did cause supershifting of the complexes of DRF1 and DRF2, as reported previously (lane 9; Kitabayashi et al. 1995). When we used nuclear extracts of RA-treated F9 cells, the band corresponding to DRF1 was shifted still further after the addition of polyclonal antibodies against ATF-2 or p300 (Fig. 2B; lanes 3,7). Again, adsorption of antibodies against ATF-2 with GST–ATF-2 eliminated the supershift (Fig. 2B; lane 4). Other antibodies and control preimmune serum had no effect on the mobilities of the DRF complexes (lanes 5,6, 8–14). The extent of the supershift observed with the polyclonal antibodies against ATF-2 was dose dependent (Fig. 2C; lanes 2,3). A 25-fold molar excess of oligodeoxynucleotides corresponding to the DRE did compete for the binding of DRF (lane 5). Similar results were obtained with extracts of differentiated F9 cells after infection with Ad–E1A but not with dl312 virus (data not shown).
Figure 2.
ATF-2 binds the DRE that responds to p300 in differentiated and undifferentiated F9 cells. (A) Nuclear extracts from F9 cells were incubated on ice for 30 min without (lanes 1,2) or with (lanes 3–14) various antibodies and then analyzed in a band-shift assay with DRE as the probe. (Lane 1) Free DRE probe (F); (lane 2) nuclear extract alone; (lane 3) antibodies against ATF-2 (α-ATF-2; Maekawa et al. 1989); (lane 4) antibodies against ATF-2 that had been preabsorbed with 3 μg of GST–ATF-2; (lane 6) antibodies against ATF-2 (Livingston et al. 1995); (lanes 5,7–14) various antibodies were added to the reactions as indicated (α-n, antibodies against protein n). The positions of bands of DRF1 and DRF2 are indicated by arrows. (B) Nuclear extracts of RA-treated F9 cells were incubated on ice without (lanes 1,2) or with (lane 5) 4 μg of rabbit IgG or with various antibodies (lanes 3,4,6–14) for 30 min and analyzed in a band-shift assay with DRE as the probe. (Lane 1) Free DRE probe (F); (lane 2) nuclear extract alone; (lane 3) antibodies against ATF-2 (Maekawa et al. 1989); (lane 4) antibodies against ATF-2 that had been preabsorbed with 3 μg of GST–ATF-2; (lanes 6–14) various antibodies were added to the reactions as indicated. The positions of bands of DRF1 are indicated by arrows. (C) Nuclear extracts from RA-treated F9 cells were incubated on ice without (lane 1) or with 10−2 dilution (lane 2) or 10−1 dilution (lane 3) of antiserum against ATF-2 (Maekawa et al. 1989; Livingston et al. 1995); with 10−1 dilution of antiserum against c-Jun (lane 4); or with 25-fold molar excess of DRE oligonucleotide as competitor (lane 5; Kitabayashi et al. 1995) for 30 min; then proteins were analyzed in the band-shift assay with DRE as the probe. The positions of bands of DRF1 are indicated by arrows. (F) Free DRE DNA probe.
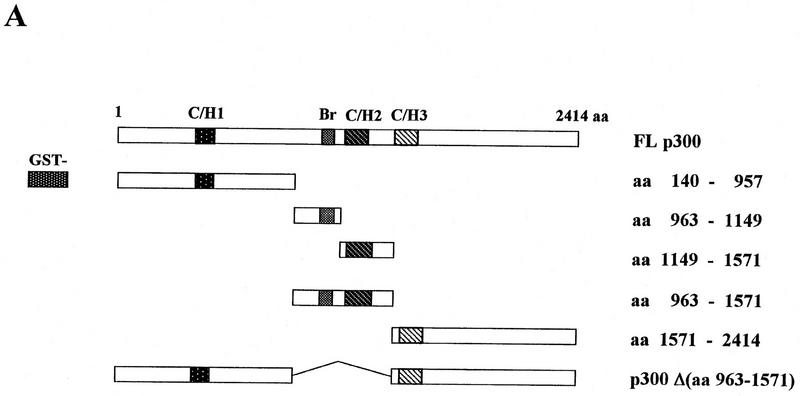
Assignment of domains for the interaction between p300 and ATF-2
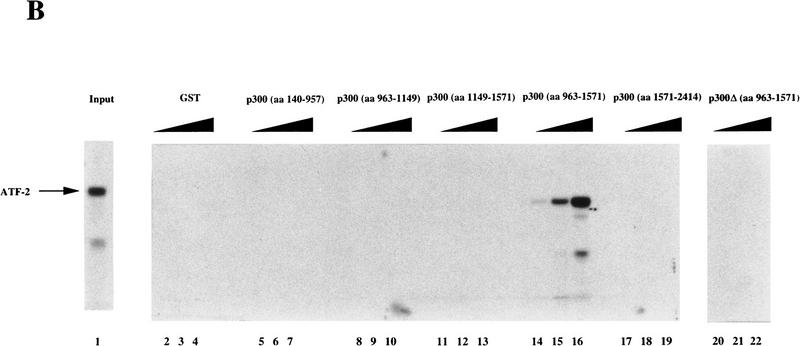
We next performed a GST–affinity matrix-based assay to determine whether or not the p300 protein interacts directly with ATF-2. Various p300 deletion proteins fused to GST were incubated with in vitro-translated [35S]methionine-labeled ATF-2 (Fig. 3). After extensive washing of the matrix, the bound proteins were fractionated by SDS-PAGE. As shown in Figure 3B, in vitro translated [35S]methionine-labeled ATF-2 was captured by GST–p300 (amino acids 963–1571; lanes 14–16) but not by other GST–p300 fusion proteins or by GST alone (lanes 2–13, 17–22). The GST–p300 (amino acids 963–1571) fusion protein contained both the bromodomain and the C/H2 domain. The GST–p300 Δ(amino acids 963–1571) fusion protein, which deleted both regions, did not bind specifically to in vitro-translated ATF-2 protein (lanes 20–22). The GST–p300 fusion proteins, which included each separate domain (amino acids 963–1149 and amino acids 1149–1571), did not bind specifically to in vitro-translated ATF-2 protein (Fig. 3B, lanes 8–13). Thus, association with ATF-2 is required for both regions containing the bromodomain and the C/H2 domain of p300.
Figure 3.
Interaction of in vitro-translated ATF-2 with deletion variants of GST–p300. (A) Schematic representation of the variants of GST–p300. Shown are p300 and deletion derivatives fused to the GST protein. The patterned boxes represent the cysteine/histidine-rich regions C/H1, C/H2, and C/H3, and are labeled; (Br) The bromodomain. The numbers at right indicate the amino acids of p300. (FL p300) Full-length p300 protein; [p300Δ(amino acids 963–1571)], p300 protein lacking amino acids 963–1571; (GST) glutathione S-transferase–truncated protein. (B) In vitro translated [35S]methionine-labeled ATF-2 was incubated with the GST–p300 variants that consisted of the amino-terminal, carboxy-terminal, and central portions of the protein (lanes 5–19) or variants that lacked amino acids 963–1571 (lanes 20–22), or ATF-2 was incubated with GST alone (lanes 2–4). The bound ATF-2 is indicated by an arrow at left. (Lane 1) Input, namely, in vitro-translated [35S]methionine-labeled ATF-2. The input lane contained 1.5% (in terms of cpm) of the radiolabeled protein used in the binding experiments.
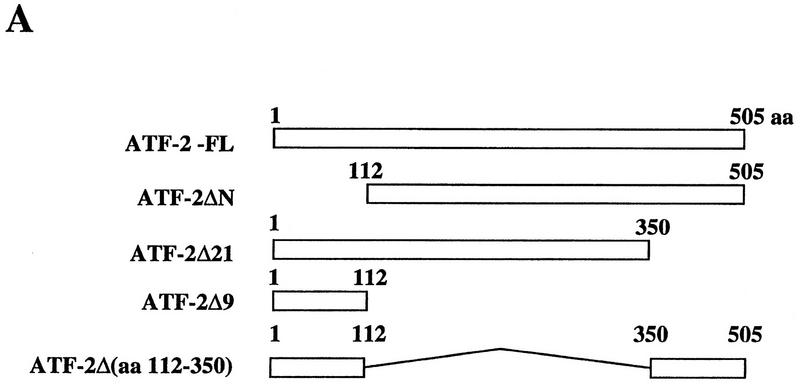
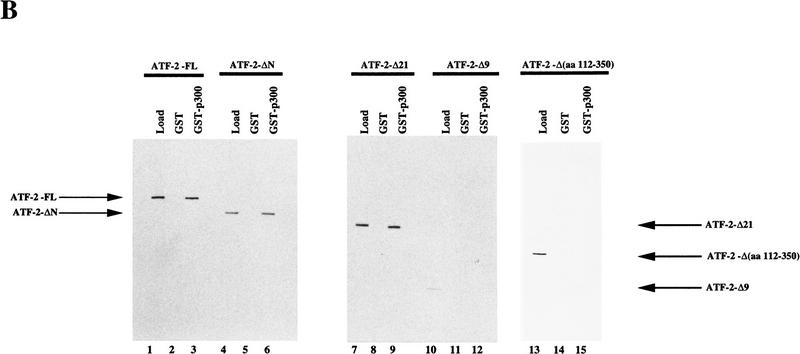
Next we examined the interaction of the GST–p300 (amino acids 963–1571) fusion protein with various mutant derivatives of ATF-2 (Fig. 4A). The GST–p300 (amino acids 963–1571) fusion protein was synthesized in baculovirus-infected insect cells and immobilized on glutathione–Sepharose beads. The beads were incubated with extracts of insect cells that contained overexpressed ATF-2 protein (Patel et al. 1991). The bound proteins were fractionated by SDS-PAGE after extensive washing of the beads, and the presence of ATF-2 was examined by Western blotting with antibodies specific for amino- or carboxy-terminal regions of ATF-2 (Livingston et al. 1995). As shown in Figure 4B, full-length ATF-2 (ATF-2–FL), ATF-2 ΔN (deletion of 111 amino-terminal amino acids), and ATF-2–Δ21 (deletion of 55 carboxy-terminal amino acids) bound to the beads with immobilized GST–p300 (amino acids 963–1571) (lanes 1–9), whereas the derivative of ATF-2 consisting of residues 1–112 (ATF-2–Δ9) did not (lanes 10–12). These results suggest that the amino-terminal region of the ATF-2 plays no significant role in the interaction of p300 and ATF-2. Moreover, ATF-2 with internal deletion of residues 112–350 did not bind to p300 (lanes 13–15). Taken together, the important residues of ATF-2 involved in interactions with p300 represent the proline-rich region between positions 112 and 350.
Figure 4.
Interaction of GST–p300 with deletion variants of ATF-2. (A) Schematic representation of the ATF-2 fusion proteins synthesized in insect cells. (B) Extracts of infected insect cells containing wild-type or mutant ATF-2 proteins were incubated with immobilized GST–p300 fusion protein for 2 hr at 4°C. The beads were washed extensively and bound protein was analyzed by Western blotting analysis with antibodies specific for the amino- or carboxy-terminal-specific region of ATF-2 (lanes 3,6,9,12,15). Beads with bound GST protein (lanes 2,5,8,11,14) were used as a control. Lanes 1,4,7,10, and 13 (Load) were loaded with 0.5% (in terms of cpm) of the input recombinant ATF-2 or the variant proteins used in the binding experiments. The positions of ATF-2–FL, ATF-2–ΔN, ATF-2–Δ21, ATF-2–Δ9 and ATF-2-Δ(amino acids 112–350) are indicated by arrows.
ATF-2 and p300 enhance the DRE-mediated activation of the c-jun promoter
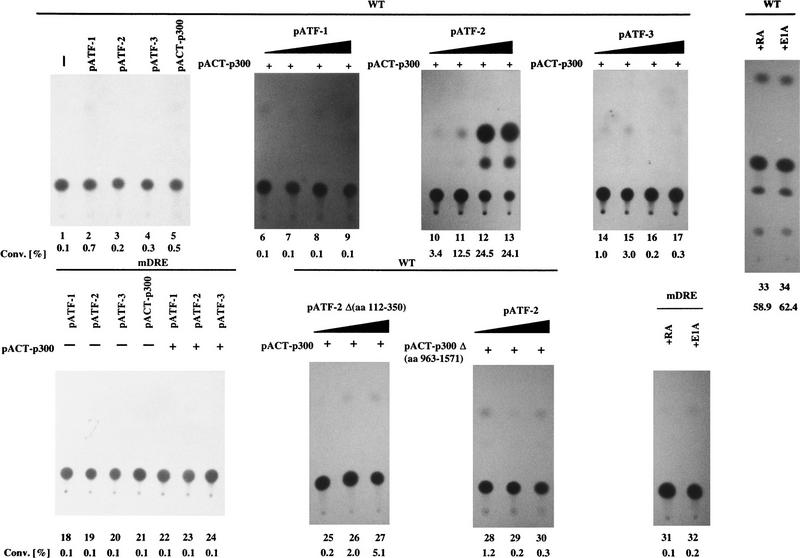
F9 cells that had been stably transformed with c-jun CAT reporter constructs were transfected with either pCMV–ATF-1, pCMV–ATF-2, and pCMV–ATF-3, pACT–p300 expression plasmids, or their combinations. To examine the effect of ATF family and p300 on c-jun expression, only the combination of ATF-2 and pACT–p300 plasmids resulted in a significant transactivation of −730/+874 c-jun CAT in F9 cells in a dose-dependent manner (Fig. 5, lanes 1–17). The mutated c-jun promoter-CAT construct, −730/+874 c-jun CAT–mDRE did not have such activity (lanes 18–24). These results suggest that only ATF-2 among members of the ATF family cooperates with p300 in the DRE-mediated activation of the c-jun promoter in F9 cells. In particular, the mutant ATF-2 encoded by pCMV–ATF-2 Δ(amino acids 112–350), which lacked the putative p300-binding region, was less potent to activate c-jun transcription via the DRE (lanes 25–27). In addition, the mutant of p300, pACT–p300 Δ(amino acids 963–1571), which lacked the region required for association with ATF-2, was unable to activate transcription of the c-jun gene (lanes 28–30). These results indicate that the interaction between p300 and ATF-2 is critical for activation of the transcription of the c-jun gene. However, the CAT activity, a measurement of the extent of the transactivation by ATF-2 and p300 in F9 cells, was still two- or threefold lower than the CAT activity due to the wild-type CAT reporter in F9 cells in the presence of RA and E1A (cf. lanes 10–13 and lanes 33 and 34).
Figure 5.
ATF-2 and p300 cooperate to transactivate the c-jun promoter in F9 cells. F9 cells that had been stably transfected with 5 μg of −730/+874 c-Jun CAT (wild type; lanes 1–17,25–30,33,34) and 5 μg of −730/+874 c-Jun CAT–m182/175 (mDRE; lanes 18–24,31,32) were cotransfected with 4 μg of pCMV–ATF-1 (lanes 6–9), pCMV–ATF-2 (lanes 10–13), pCMV–ATF-2 Δ(amino acids 112–350) (lanes 25–27) or pCMV–ATF-3 (lanes 14–17) in the presence of the pACT–p300 expression plasmid (lanes 6–17,21–27) or pACT–p300 Δ(amino acids 963–1571) (lanes 28–30), or pCMV-E1A (lanes 32,34) plus 2 μg of pRSVLacZ (lanes 1–34), as indicated. The cells were incubated for 72 hr and CAT assays were performed with extracts in which the amount of protein had been normalized by the activity of β-galactosidase. In some cases, F9 cells were incubated with 3 × 10−7 m RA (lanes 31,33). The percent conversion of CAT to its acetylated form is indicated. (Lane 1) Wild type only; (lanes 2–4,18–20) 4 μg of pCMV–ATF-1, pCMV–ATF-2, or pCMV–ATF-3; (lane 5) 4 μg of pACT–p300; (lanes 6–9,10–13,14–17,25–30) 1.0, 4.0, and 8.0 μg of pCMV–ATF-1, pCMV–ATF-2, pCMV–ATF-2 Δ(amino acids 112–350) or pCMV–ATF-3 plasmid, respectively, in the presence of 4 μg of pACT–p300 (lanes 6–17,25–27) or of pACT–p300 Δ(amino acids 965–1571) (lanes 28–30), respectively.
Association of ATF-2 and p300 is critical for activation of DRE-mediated transcription
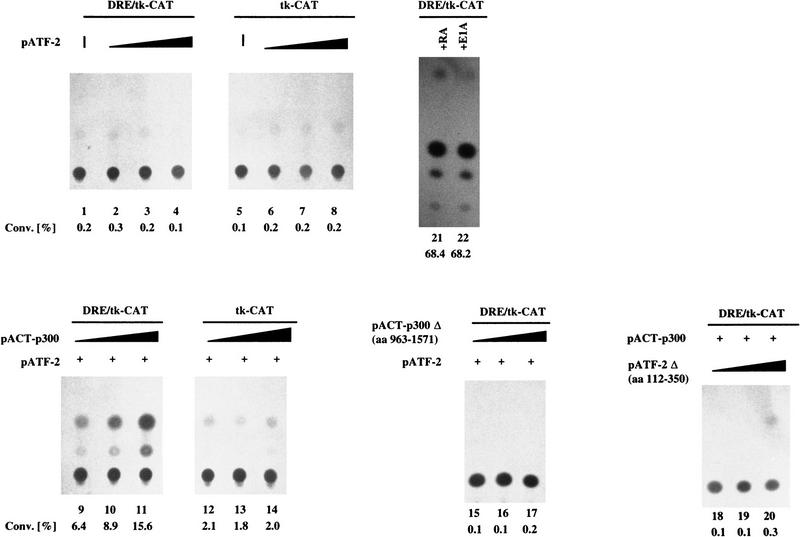
We examined whether the DRE by itself might be sufficient to confer a response to ATF-2 and p300. We observed a large increase of CAT activity of a DRE/tk CAT construct only in the case of the cotransfection with plasmids encoding ATF-2 and p300 (Fig. 6, lanes 1–14). As shown in Figure 6, lanes 15–20, none of the combinations that included either a deleted variant of ATF-2 or a deleted variant of p300-stimulated transcription. The results of treatment with the optimal dose of RA and E1A, like those of Figure 5, yielded CAT activities two- or threefold higher than those observed with the combination of expression vectors for ATF-2 and p300 (cf. lanes 9–11 and lanes 21 and 22). These results strongly suggest an essential role for the DRE in the induction of c-jun by RA and E1A and, moreover, that the activation is due, at least, to the combined action of ATF-2 and p300.
Figure 6.
ATF-2 and p300 cooperate to transactivate DRE-mediated transcription in F9 cells. F9 cells that had been stably transfected with 5 μg of 3× DRE/tk–CAT (lanes 1–4,9–11,15–22) or tk–CAT (lanes 5–8,12–14) were cotransfected with various amounts of pCMV–ATF-2 (lanes 2–4,6–8), pCMV–ATF-2 Δ(amino acids 112–350) (lanes 18–20), pACT–p300 (lanes 9–11,12–14), or pACT–p300 Δ(amino acids 963–1571) (lanes 15–17) and 5 μg (lanes 9–11,15–17), 8 μg (lanes 12–14) of pCMV–ATF-2, or 2 μg of pRSV–LacZ (lanes 1–22). The cells were incubated for 72 hr and CAT assays were performed with extracts in which the amount of protein had been normalized by the activity of β-galactosidase. The percent conversion of CAT to its acetylated forms is indicated. (Lane 1) DRE/tk–CAT only; (lanes 2–4,6–8,18–20) 1.0, 4.0, and 8.0 μg of pCMV–ATF-2 or pCMV–ATF-2 Δ(amino acids 112–350), respectively; (lane 5) tk–CAT only; (lanes 9–17) 1.0, 4.0, and 8.0 μg of pACT–p300 or pACT–p300 Δ(amino acids 963–1571), respectively. (Lane 21) treatment with 3 × 10−7 m RA; (lane 22) transfection with pCMV–E1A (4 μg).
PKCα-mediated phosphorylation of ATF-2
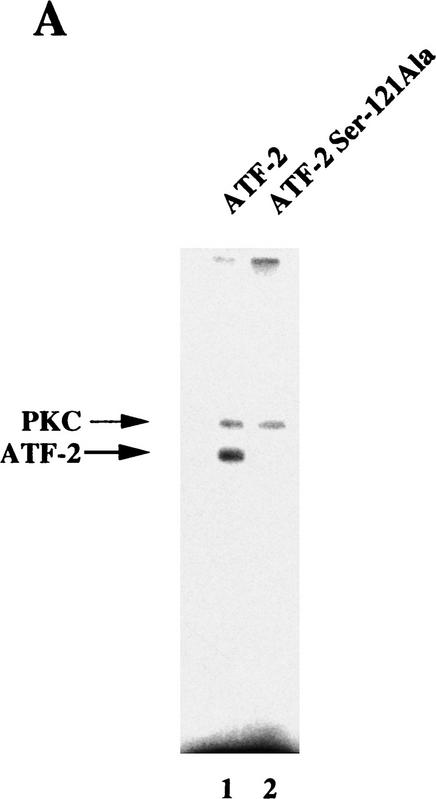
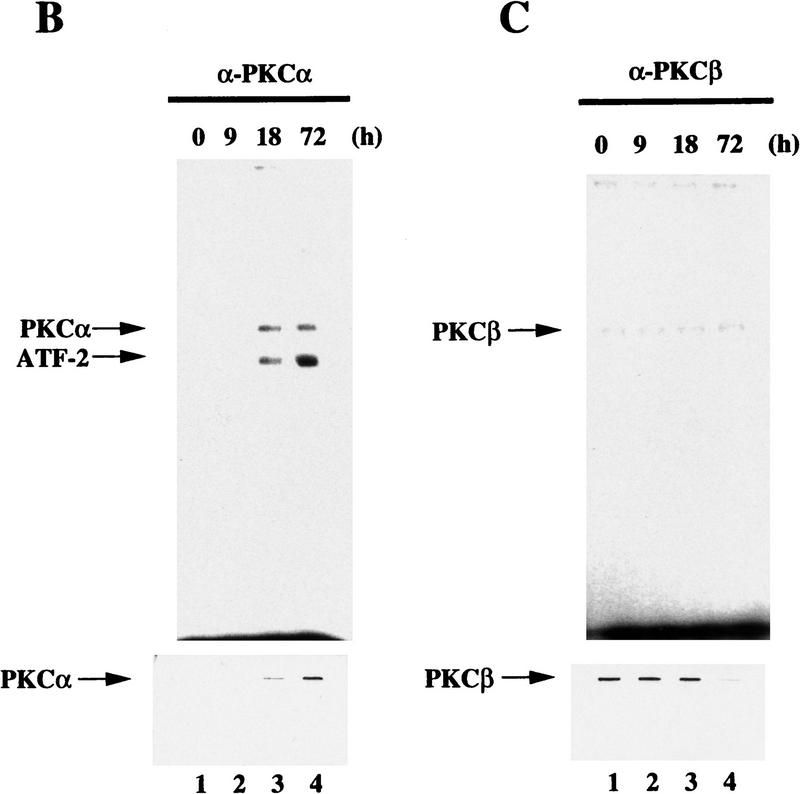
The proline-rich region of ATF-2 that is involved in the interaction with p300 (see Fig. 4) contains putative phosphorylation sites by PKCα (Maekawa et al. 1989). A kinase assay in vitro using recombinant ATF-2 and a mutant protein demonstrated that Ser-121 of ATF-2 was phosphorylated by PKC (Fig. 7A, lanes 1,2). The immunoprecipitation kinase assay showed that ATF-2 protein was phosphorylated by PKCα but not by PKCβ and that the phosphorylated ATF-2 was detectable 18 hr after the start of induction with RA, which is concominant with an increase in PKCα (Fig. 7B,C, lanes 1–4). Thus, RA-induced activation of PKCα might be a key event that leads to the phosphorylation of ATF-2 protein. Similar phosphorylation of Ser-121 of the ATF-2 protein was observed after 1–3 hr of exposure to E1A in F9 cells (data not shown).
Figure 7.
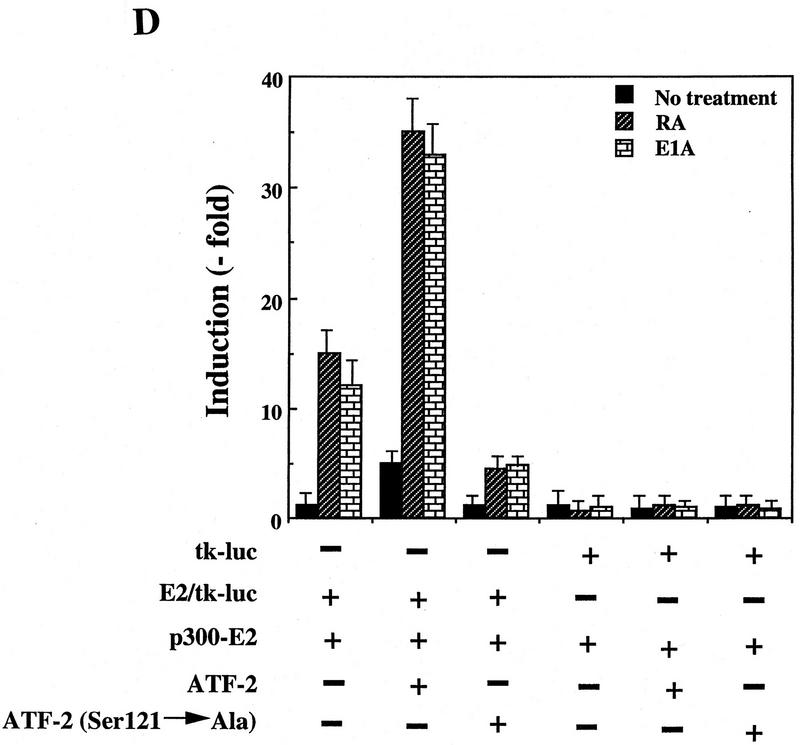
ATF-2 is phosphorylated by PKC and activates a p300–E2 fusion protein. (A) Five micrograms of ATF-2 and mutant ATF-2Ser121–Ala proteins were purified from E. coli and incubated with 25 ng of a mixture of recombinant PKCα, PKCβ, and PKCγ mixture in the appropriate kinase buffer (see Materials and Methods) for 10 min at 30°C. The resultant phosphorylated proteins were analyzed by SDS-PAGE (10% polyacrylamide). (Lane 1) ATF-2; (lane 2) ATF-2Ser121–Ala mutant protein. (B,C) F9 cells were incubated with 3 × 10−7 m RA for the indicated periods of time. Nuclei were isolated and nuclear lysates were subjected to immunoprecipitation with polyclonal antibodies specific for PKCα (B) and PKCβ (C), respectively, and protein A–Sepharose for 2 hr at 4°C. The immune complexes were incubated with 5 μCi of [γ-32P]ATP in kinase buffer for 30 min at 30°C. The resultant phosphorylated protein complexes were subjected to SDS-PAGE (10% polyacrylamide). (Lane 1) F9 cell extract; (lanes 2–4) F9 cells lysate that had been incubated with RA for 9, 18, and 72 hr, respectively. The immunoprecipitates with antibodies specific for PKCα or PKCβ were immunoblotted with the antibodies against the respective PKC (bottom panels). (D) F9 cells that had been stably transfected with 5 μg of tk–luc or 5× E2/tk–luc were cotransfected with 3 μg of pCMV–p300–E2 with or without 4 μg of pCMV–ATF-2 or 4 μg of pCMV–ATF-2Ser121–Ala plus 2 μg of pRSV-LacZ, as indicated. The cells were incubated for 72 hr in the absence or presence of 3 × 10−7 m RA or 2.5 μg of pCMV–E1A. The results are expressed as the extent of induction (x-fold) as compared to the results with the control plasmid (pcDNA3). The results are the means from three independent experiments.
ATF-2 and p300 cooperate in the activation of transcription
To determine whether p300-mediated transcription can be modulated by ATF-2 via the action of PKCα, expression plasmids that encoded wild-type pCMV–ATF-2, the mutant pCMV–ATF-2Ser121–Ala with a mutation at position 121 (Ser → Ala) and p300–E2 were transfected either alone or in combination into RA-treated or E1A-treated F9 cells that had been stably transformed with an E2/tk–luc reporter construct. F9 cells stably transfected with tk–luc served as a control. As shown in Figure 7D, p300–E2 itself activated the transcription of the E2/tk–luc reporter gene to a certain extent after treatment of cells with RA (about sevenfold), as reported previously (Kitabayashi et al. 1995). The introduction of plasmid pCMV–ATF-2 resulted in a significant increase in luciferase activity, in conjunction with p300–E2 (2.5-fold), in the presence of RA (more than sevenfold). The Ser → Ala mutant, pCMV–ATF-2Ser121–Ala, was not associated with any enhancement of luciferase activity but, rather, decreased reporter gene activities. Similar results were obtained in the case of introduction of E1A (Fig. 7D). Taken together, these observations indicate that in F9 cells RA-induced activation of PKCα leads to the phosphorylation of ATF-2, which probably results in the enhancement of transcription of genes, including c-jun by increasing the interaction with p300.
Discussion
ATF-2 and p300 are components of DRF complexes
In the present study, we found that the DRF1 and DRF2 complexes contain the transcription factor ATF-2, which is physically associated with p300 in undifferentiated and RA- or E1A-induced differentiated F9 cells. We characterized this association by examining protein–protein interactions both in vitro and in vivo (Figs. 1, 2, 3, 4) and found that the region that contained the bromodomain and the C/H2 region of p300 was required for the interaction with ATF-2 in vitro (Fig. 3). Further deletion analysis of this region, separating each domain, demonstrated that the bromo- and the C/H2 domains by themselves were unable to participate in the molecular interaction with ATF-2. Thus, both domains of p300 seem to be required for the interaction of p300 with ATF-2 (Fig. 3). ATF-2 is the first protein shown to interact with the bromodomain of p300.
Phosphorylated p300 might be involved in a complex formation with ATF-2 in vivo
Studies in vivo with sequential immunoprecipitations and the results of the immunoprecipitation and Western blotting indicated that the species of p300 protein associated with ATF-2 correponded to be the more slowly migrating species of p300, which is the phosphorylated form (Fig. 1). However, binding studies using GST fusion constructs and deletion mutants demonstrated that p300 protein that had been produced in Escherichia coli was able to bind ATF-2 in vitro, suggesting that the unmodified p300 protein might also have the ability to associate with the ATF-2 protein in vivo (Fig. 3). To fully understand the biological significance of the phosphorylation of p300, the characterization of additional phosphorylation sites within p300 during the differentiation of F9 cells is required.
One also cannot rule out the possibility that the molecule(s) associated with ATF-2 in vivo might be CBP and not the underphosphorylated form of p300. In an earlier study, p300 protein of undifferentiated F9 cells was represented by the more rapidly migrating band of the doublet on gels and, in contrast, the CBP protein was the more slowly migrating protein (Kitabayashi et al. 1995). An immunodepletion experiment using p300-specific monoclonal antibodies, but not one with antibodies against CBP, led to the disappearance of DRF1 and DRF2 complexes in 293 cells (Kitabayashi et al. 1995). In the present study, the immunoprecipitation with antibodies against ATF-2 recovered CBP preferentially, at least in the case of undifferentiated F9 cells. In differentiated F9 cells, the p300 and CBP proteins comigrated, so that a distinction is not possible. The discrepancy between present and earlier results might be due to the use of different cell types such as 293 and F9 cells. Thus, the components, such as p300 or CBP or both, that are coupled to ATF-2 in undifferentiated F9 cells remain to be determined and the changes in their modes of association during the differentiation of F9 cells remain to be clarified. Studies to address this issue using the null mutant mice of either p300 or CBP are now being undertaken.
PKCα phosphorylates Ser-121 of ATF-2 during the differentiation of F9 cells
Adenivirus-transformed cells contain the hyperphosphorylated form of c-Jun, which results from enhanced phosphorylation of Ser-63 and Ser-73 (Hagmeijer et al. 1993), which are also required for CBP-induced stimulation of c-jun gene (Bannister et al. 1995). However, c-Jun is not expressed in F9 cells, which do, however, contain ATF-2. The CR-1- and CR-3-dependent effects of E1A on ATF-2 can occur independently of each other and might even be synergistic. In F9 cells, the 12SE1A protein enhances the transcriptional activity of the amino terminus of ATF-2 fused to GAL4 or GHF 1, whereas the transcriptional activity of the amino terminus of c-Jun fused to GAL4 is not significantly enhanced by this protein (Duyndam et al. 1996). Thus, ATF-2 and not c-Jun appears to be the primary target of 12S E1A in F9 cells. Recently, it was shown that CR3-dependent activation of the amino terminus of ATF-2 required the presence of two threonine residues, Thr-69 and Thr-71, which are potential targets for phosphorylation by SAPKs or JNKs (Gupta et al. 1995; Livingston et al. 1995). However, E1A-dependent activation via CR1 was still significant after these amino acids had been mutated (Duyndam et al. 1996). Moreover, we showed here that the amino-terminal region of ATF-2, including Thr-69 and Thr-71, was not required for interaction with p300 (Fig. 4). Therefore, we can speculate that a different mechanism is involved in the E1A-dependent activation of ATF-2. These observations indicate that a different kinase or inhibition of a phosphatase might be responsible for the hyperphosphorylation of ATF-2, as well as p300, in F9 cells.
Maekawa et al. (1989) reported earlier that a putative site of phosphorylation by PKC exists in the central region (at Ser-121) of ATF-2. The present study indicates that treatment with RA or the introduction of E1A resulted in the specific induction and the increase of the enzymatic activities of PKCα protein in F9 cells (Fig. 7) but not PKCβ and PKCγ (data not shown). These results are consistent with reports by Kindregan et al. (1994) and Khuri et al. (1996). We suggest the RA- or E1A-induced changes in PKCα might be critical as a trigger in the transactivation of the c-jun gene and the differentiation of F9 cells to endoderm-like cells.
Phosphorylation of ATF-2 at Ser-121 is essential for the RA- and E1A-mediated transcription of c-jun in F9 cells
We examined whether members of the ATF-2 family could enhance the DRE-mediated transcription of the c-jun gene via p300. We used a c-jun promoter–CAT construct that included a DRE. Only the combination of expression plasmids for ATF-2 and p300 enhanced the promoter activity of the c-jun gene in undifferentiated F9 cells in the absence of RA and E1A (Fig. 5). However, the observed CAT activity was two- or threefold lower than the CAT activity because of the reporter construct upon stimulation with E1A or treatment with RA (Fig. 5). We confirmed these results by cotransfection and CAT assays with the DRE/tk–CAT construct (Fig. 6). Hagmeijer et al. (1996) reported that the expression of ATF-3 is induced in Ad–E1A-transformed cells, as well as a direct effect of the expression of E1A on the formation of c-Jun/ATF-3. However, we did not find any evidence in support of their results (Figs. 2, 5, and 6).
Ser-121 of the ATF-2 protein was phosphorylated by PKCα during RA- or E1A-induced differentiation of F9 cells (data not shown). Furthermore, the ATF-2 with Ala-121 did not increase transcription of the E2/tk–luc reporter construct in conjunction with p300 after treatment of F9 cells with RA (Fig. 7). We do not know the exact role of the phosphorylation of Ser-121 in ATF-2 during RA- or E1A-induced differentiation. One possible hypothesis is that ATF-2 phosphorylated by PKCα, might increase the histone acetyltransferase (HAT) activity of p300 with a subsequent change in the conformation of the chromatin or that it might stimulate the formation of the DNA-binding complex with p300, to allow transcriptional activation of the c-jun gene.
The RA- or E1A-induced differentiation of F9 cells is a multistage process that involves sequential changes in gene expression (Strickland 1981; Hogan et al. 1983). The commitment to differentiation seems to occur within 18 hr after the start of treatment with RA or 1–3 hr after introduction of E1A into F9 cells (Kitabayashi et al. 1995). We also reported that RA can induce the commitment of F9 cells to irreversible differentiation without protein synthesis de novo (Kitabayashi et al. 1994). In contrast, the transcriptional activation of genes for proteins that are markers of parietal endoderm, such as laminin and type IV collagen—by RA alone or together with dibutyryl cAMP—requires protein synthesis de novo. The expression of these genes is also regulated by PKCα and PKCβ (Kindregan et al. 1994; Khuri et al. 1996). Thus, it is important to determine whether the PKCα gene is directly activated by RA receptors or responds to other intermediates.
It appears that ATF-2 and p300 are, at least, prerequisites for the activation of the c-jun promoter in F9 cells in the absence of any participation of RA or E1A. However, the level of transactivation of the c-jun promoter by p300 and ATF-2 was found to be consistently lower as compared to stimulation by RA- or E1A-dependent c-jun activation (Figs. 5 and 6). UV cross-linking studies with a DRE probe demonstrated that proteins of ∼300, 75–80, and 40–42 kD were present in DRF complexes derived from undifferentiated and differentiated F9 cells (data not shown). The 300- and 75- to-80 kD proteins are most likely p300 and ATF-2, respectively. Thus, another molecule, such as the 40- to 42-kD molecule, identified by UV cross-linking, might represent key regulators that are required for the full activation of the c-jun transcription. This factor is possibly regulated by some unidentified protein kinase(s) or phosphatase(s). The molecular cloning of this 42- to 40-kD protein is now in progress. A precise map of the sites of phosphorylation of p300 combined with further insights into the mechanisms of the PKCα-mediated phosphorylation of ATF-2 as well as phosphorylation or dephoshorylation of other proteins, might ultimately lead to the understanding of the regulation of expression of the c-jun gene and the commitment to differentiation of F9 cells upon exposure to RA or E1A.
Materials and methods
Plasmids
Deletion mutants of the pGST–p300 fusion gene were generated by ligating the respective DNA fragments SpeI–SpeI (amino acids 140–957), EcoRI–SmaI (amino acids 963–1571), SmaI–HindIII (amino acids 1571–2414), PCR-amplified DNA fragments (amino acids 963–1149 and 1149–1571), in-frame, to pGEX2T and its derivatives (Pharmacia Biotech., Uppsala, Sweden). The deletion mutant pGST–p300 Δ(amino acids 963–1571) was generated by ligating the NotI–EcoRI DNA fragment (amino acids 1–957) with the SmaI–HindIII DNA fragment (amino acids 1571–2414) via the pNotI linker. The −730/+874 c-Jun CAT (wild-type), −730/+874 c-Jun CAT–mDRE (mDRE), 3× DRE/tk–CAT, p300–E2, pCMV–VP16–E2, and 5× E2/tk–CAT reporter genes are described elsewhere (Kitabayashi et al. 1992; Eckner et al. 1994). Expression plasmids pCMV–ATF-1 (Liu and Green 1990, 1994), pCMV–ATF-2 (Chatton et al. 1993), and pCMV–ATF-3 (Hai et al. 1989) were generated by ligation of the PCR-amplified DNA fragments of the respective coding sequences in-frame to pcDNA3 (Invitrogene). pGST–ATF-2 was generated by ligating the PCR-amplified DNA fragment that encompassed the coding sequence (amino acids 1–350) for ATF-2 to pGEX2T. pCMV–ATF-2Ser121–Ala, was constructed by ligation of a PCR-amplified DNA fragment, generated with a primer in which the Ala-121-encoding triplet had been substituted for the Ser-121-encoding triplet and an XbaI–ApaI linker. This fragment was subcloned into pcDNA3 or pGEX2T. pCMV–ATF-2 Δ(112–350) was constructed by ligation in-frame of the DNA fragment (amino acids 1–112) with the DNA fragment (amino acids 350–505) via pNotI linker. E2/tk–luc and tk–luc were generated by ligation of a PCR-amplified fragment of luciferase cDNA (Kawasaki et al. 1996) that had a BglII–XhoI linker. pACT–p300, pBLCAT2, pCMV–E1A, and pRSV-lacZ are described elsewhere (Kitabayashi et al. 1995).
Cell culture, transfection, and CAT assay
F9 cells (Bernstine et al. 1973) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS. Stable transfection of F9 cells and CAT assays were performed as described elsewhere (Kitabayashi et al. 1992, 1995), with the exception that the concentration of acetyl-CoA was reduced from 3 to 0.5 mm. The ratio of the activity of CAT to that of β-galactosidase was used for normalization of results.
Assay of luciferase activity
F9 cells were transiently transfected by use of lipofectin (GIBCO BRL, Gaithersburg, MD) in accordance with the manufacturer’s instruction. After incubation for 72 hr, cells were harvested, lysed, and assayed for luciferase activity as described (Kawasaki et al. 1996). Transfection efficiencies were normalized by activity due to cotransfected β-galactosidase-encoding plasmid.
Immunoprecipitation, Western blotting, and immunodepletion
Radiolabeling of F9 cells with [35S]methionine or [32P]orthophosphate, the preparation of cell lysates and the immunoprecipitation with the E1A-specific monoclonal antibody M73 (Harlow et al. 1986), p300-specific polyclonal antibodies (Kitabayashi et al. 1995), p300-specific monoclonal antibodies RW102 or RW128 (Eckner et al. 1994), and CBP-specific monoclonal antibodies AC238 or AC26 (Arany et al. 1995; Eckner et al. 1996) were described elsewhere (Kitabayashi et al. 1992, 1995; Eckner et al. 1994, 1996). After SDS-PAGE, proteins were transferred to filters (Hybond-C Extra; Amersham, Japan) by electroblotting and probed with p300-specific polyclonal antibody (Kitabayashi et al. 1995), ATF-2-specific monoclonal antibody (F2BR-1), polyclonal antibody (C-19; Santa Cruz Biotechnology, Santa Cruz, CA), or ATF-2-specific polyclonal antibodies (Maekawa et al. 1989; Livingston et al. 1995), using the ECL detection system (Amersham Japan) and autoradiography. The complementary immunoprecipitation and Western blotting experiments were carried out by essentially the same procedure. The immunodepletion experiments were performed as described elsewhere (Lassar et al. 1991; Kitabayashi et al. 1995).
Gel mobility shift assays
Preparation of nuclear extracts from F9 cells, the gel mobility shift, and its competition assays was performed as described previously (Kitabayashi et al. 1995). The sequence of DRE is described elsewhere (−190 to −170 of c-jun promoter; Kitabayashi et al. 1995). Antibodies against the following proteins were used: ATF-1 (C41-5.1 and 25G10G; Santa Cruz and UBI, Lake Placid, NY), ATF-2 (UBI), CREB (UBI), c-Jun (UBI), ATF-3 (C-19; Santa Cruz), CREM (Santa Cruz), JunB (210; Santa Cruz), and JunD (329-G; Santa Cruz). Polyclonal affinity-purified antibodies against c-Myb and ATF-2 were kindly provide by T. Maekawa and S. Ishii (RIKEN). Preimmune rabbit serum was used as a control. In some cases, the ATF-2-specific antibodies were absorbed with GST–ATF-2 protein and used for the competition experiment.
Analysis of interactions of ATF-2 and p300 using GST fusion proteins
Synthesis and purification of GST–p300 fusion wild-type protein and mutant derivatives are described elsewhere (Eckner et al. 1994). Lysates of F9 cells that expressed ATF-2 were prepared in 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.5% NP-40, 1 μg/ml each of aprotinin, pepstatin, and leupeptin, 1 mm PMSF, 5 mm NaF, and 10 mm Na2-β-glycerophosphate. ATF-2 was translated in vitro in the presence of [35S]methionine using a TNT kit (Promega, Madison, WI). In vitro-translated products were mixed with 5 μg of protein in a lysate of F9 cells and incubated with glutathione–Sepharose beads (Pharmacia Biotech.) that had been loaded with either GST–p300 or GST in the above-described buffer for 2 hr at 4°C. The beads were then washed extensively with the same buffer, and protein complexes were eluted with sample buffer for SDS-PAGE.
Binding assays with p300
Recombinant baculoviruses encoding ATF-2–FL, ATF-2–Δ21, ATF-2–Δ9, ATF-2–ΔN, and ATF-2–Δ(amino acids 112–350) were constructed in the baculovirus expression system, and lysates of infected cells were prepared as described previously (Patel et al. 1992). GST–p300 (amino acids 963–1571) was amplified by PCR as a KpnI–NotI fragment for baculovirus expression. The resulting fragment was subcloned into a baculovirus transfer vector having the FLAG–tag sequence (Yang et al. 1996). The recombinant viruses were isolated with the BaculoGold system (Pharmingen) according to the manufacturer’s protocol and used for infection of Sf9 cells to express FLAG–GST–p300 (amino acids 963–1571). Recombinant proteins were affinity purified with M2 anti-FLAG antibody–immobilized agarose (Kodak–IBI) according to the manufacturer’s protocol. The binding of GST fusion protein to wild-type ATF-2 and mutant derivatives has been described elsewhere (Livingston et al. 1995). Extracts containing various recombinant ATF-2 proteins were incubated with glutathione–Sepharose beads for 2 hr at 4°C after which time beads were washed extensively with washing buffer (100 mm KCl, 10 mm HEPES at pH 7.9, 5 mm MgCl2, 0.1 mm ZnCl2, 0.1 mm EDTA, 0.1% NP-40, 2 mm DTT). The proteins retained on the beads were eluted and separated by SDS-PAGE. Western blotting analysis was performed using ECL (Amersham Japan) and specific antibodies directed against the amino terminus (amino acids 85–96) and carboxyl terminus (amino acids 490–505) of ATF-2 (Patel et al. 1992). Lysates of cells infected with viruses encoding wild-type and mutant ATF-2 were prepared in the presence of phosphatase inhibitors (10 mm sodium phosphate, 15 mm sodium pyrophosphate, 5 mm NaF, 0.1 mm Na3VO4).
Assay of protein kinase activity in vitro
Assays of PKC activity were carried out with recombinant ATF-2 protein from E. coli as the substrate and mixtures of recombinant PKCα, PKCβ, and PKCγ (Seikagaku Kogyo, Inc., Tokyo, Japan). Phosphorylation was allowed to proceed in the reaction mixture [50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 0.1 mm CaCl2, 1 mm DTT, 0.03% Triton X-100, 0.31 mg/ml of l-phosphatidyl-l-serine, 10–25 ng of PKC, 5 μg of ATF-2] for 10 min at 30°C, and results were analyzed by SDS-PAGE (10% polyacrylamide) and autoradiography.
Immunoprecipitation kinase assay
Nuclear lysates of RA-treated F9 cells (3 × 10−7 m RA) were prepared as described elsewhere (Kitabayashi et al. 1995). The lysates were incubated with polyclonal antibodies raised against PKCα or PKCβ for 2 hr at 4°C. The resultant immunocomplexes were analyzed by assays of their kinase activities in a reaction mixture (50 mm Tris-HCl at pH 7.5, 10 mm MgCl2, 1 mm DTT) that contained 5 μg of ATF-2 in the presence of 2–5 mCi of [γ-32P]ATP (Amersham, Japan) for 30 min at 30°C. The reaction mixtures were then subjected to SDS-PAGE (10% polyacrylamide) and autoradiography.
Acknowledgments
We thank E. Harlow, K. Shiroki, K. Fujinaga, E. Moran, T. Maekawa, S. Ishii, and D.M. Livingston for providing the plasmids and antibodies that allowed us to perform this study. We also thank K. Itakura, G. Gachelin, and C. Geltinger for helpful discussions and H. Gao for technical assistance. This work was supported by a START grant of the Swiss National Science Foundation to R.E. and the Life Science Research Project of RIKEN, the Special Coordination Funds of the Science and Technology Agency, and by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture (Japan) to K.K.Y.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kazunari@rtc.riken.go.jp; FAX 81-298-36-9120.
References
- Abraham SE, Lobo S, Yaciuk P, Wang H-GH, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- Arany Z, Seller WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. A family of transcriptional adapter proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levin AS. The SV40 large T antigen and adenovirus E1a oncoprotein interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser 63/73 are required for CBP-induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- Bernstine EG, Hooper ML, Grandchamp S, Ephrussi B. Alkaline phosphatase activity in mouse teratoma. Proc Natl Acad Sci. 1973;70:3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B, Bocco JL, Gaire M, Hauses C, Reimund B, Goetz J, Kedinger C. Transcriptional activation by the adenovirus larger E1A product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1A. Mol Cell Biol. 1993;13:561–570. doi: 10.1128/mcb.13.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montiminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;265:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu I-H, Barret T, Su B, Deng T, Karin M, Davis R. JNK1: A protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Duyndam MCA, van Dam H, van der Eb AJ, Zantema A. The CR1 and CR3 domains of the adeno-virus type 5 E1A proteins can independently mediate activation of ATF-2. J Virol. 1996;70:5852–5959. doi: 10.1128/jvi.70.9.5852-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adapter. Genes & Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ludlow JW, Lill NL, Oldread E, Arany Z, Modjtuhedi N, DeCaprio JA, Livingston DM, Morgan JA. Association of p300 and CBP with Simian virus 40 large T antigen. Mol Cell Biol. 1996a;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Yao T-P, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes & Dev. 1996b;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Flint KJ, Jones NC. Differential regulation of three members of the ATF/CREB family of DNA-binding proteins. Oncogene. 1991;6:2019–2026. [PubMed] [Google Scholar]
- Gu W, Shi X, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- Gupta S, Campbell D, Derijard B, Davis RJ. Transcription factor ATF-2 regulation by the JNK signal transduction pathway. Science. 1995;276:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- Hagmeijer BM, Konig H, Herr I, Offringa R, Zantema A, van der Eb AJ, Herrlich P, Angel P. Adenovirus E1A negatively and positively modulates transcription of AP-1-dependent genes by dimer-specific regulation of the DNA-binding and transcription activities of Jun. EMBO J. 1993;12:3559–3572. doi: 10.1002/j.1460-2075.1993.tb06030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmeijer BM, Duyndam MCA, Angel P, deGroot RP, Verlaar M, Elfferich P, van der Eb AJ, Zantema A. Altered AP-1/ATF complexes in adenovirus-E1-transformed cells due to E1A-dependent induction of ATF-3. Oncogene. 1996;12:1025–1032. [PubMed] [Google Scholar]
- Hai T, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: An extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes & Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Harlow E, Whyte P, Franze BR, Jr, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-jun activation domain. Genes & Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Barlow DP, Tilley R. F9 teratocarcinoma cells as a model system for the differentiation of parietal endoderm and visceral endoderm in the mouse embryo. Cancer Surv. 1983;2:115–140. [Google Scholar]
- Janknecht R, Hunter T. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Ohkawa J, Tanishige N, Yoshinari K, Murata T, Yokoyama KK, Taira K. Selection of the best target site for ribozyme-mediated cleavage within a fusion gene for adenovirus E1A-associated 300-kD protein (p300) and luciferase. Nucleic Acids Res. 1996;24:3010–3016. doi: 10.1093/nar/24.15.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuri FR, Cho Y, Talmage DA. Retinoic acid-induced transition from protein kinase Cβ to protein kinase Cα in differentiated F9 cells: Correlation with altered regulation of proto-oncogene expression by phorbol esters. Cell Growth Differ. 1996;7:595–602. [PubMed] [Google Scholar]
- Kindregan HC, Rosenbaum SE, Ohno S, Niles RM. Characterization of conventional protein kinase C (PKC) isotype expression during F9 teratocarcinoma differentiation. J Biol Chem. 1994;269:27756–27761. [PubMed] [Google Scholar]
- Kitabayashi I, Chiu R, Gachelin G, Yokoyama K. E1A-dependent up-regulation of c-jun/Ap-1 activity. Nucleic Acids Res. 1991;19:649–655. doi: 10.1093/nar/19.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I, Kawakami Z, Chiu R, Ozawa K, Matsuoka T, Toyoshima S, Umezono K, Evans R M, Gachelin G, Yokoyama K. Transcriptional regulation of the c-jun gene by retinoic acid and E1A during differentiation of F9 cells. EMBO J. 1992;11:167–175. doi: 10.1002/j.1460-2075.1992.tb05039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I, Chiu R, Umesono K, Evans RM, Gachelin G, Yokoyama K. A novel pathway for retinoic acid-induced differentiation of F9 cells that is distinct from receptor mediated trans-activation. In Vitro Cell Dev Biol. 1994;30A:761–768. doi: 10.1007/BF02631299. [DOI] [PubMed] [Google Scholar]
- Kitabayashi I, Eckner R, Arany Z, Chiu R, Gachelin G, Livingston DM, Yokoyama K. Phosphorylation of the adenovirus E1A-associated 300 kDa protein in response to retinoic acid and E1A during differentiation of F9 cells. EMBO J. 1995;14:3496–3509. doi: 10.1002/j.1460-2075.1995.tb07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RPS, Lundblad JR, Chirivia JC, Richards A, Bachinger HP, Brennan RG, Roberts SGE, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Banerjee P, Nikolaki E, Dai T, Rubie A, Ahmad MF, Avruch J, Woodgett JR. The stress-activated protein kinase subfamily of c-jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vitro. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lee J-S, Zhang X, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated E1A-associated protein p300. Genes & Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- Li X, Green MR. Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes & Dev. 1996;10:517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Liu F, Green MR. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1A protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- ————— Promoter targeting by adenovirus E1A through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- Livingston C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Sakura H, Kanei-Ishii C, Sudo T, Yoshimura T, Fujisawa J, Yoshida M, Ishii S. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP-response element in brain. EMBO J. 1989;8:2023–2028. doi: 10.1002/j.1460-2075.1989.tb03610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran E. DNA tumor viruses and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Patel G, Nasmyth K, Jones N. A new method for the isolation of recombinant baculovirus. Nucleic Acids Res. 1992;20:97–104. doi: 10.1093/nar/20.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Grusby MJ, Kosaras B, Fries JWV, Mori R, Maniwa S, Clauss IM, Collins T, Sidman RL, Glimcher MJ, Glimcher LH. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature. 1996;379:262–265. doi: 10.1038/379262a0. [DOI] [PubMed] [Google Scholar]
- Shikawa N, Lyon J, LaThangue NB. The p300/CBP family: Integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- Somasundaram K, El-Deiry WS. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- Strickland S. Mouse teratocarcinoma cells: Prospects for the study of embryogenesis and neoplasia. Cell. 1981;24:277–278. doi: 10.1016/0092-8674(81)90313-5. [DOI] [PubMed] [Google Scholar]
- van Dam H, Offringa R, Meijer I, Stein B, Smits AM, Herrich P, Bos JL, van der Eb AJ. Differential effects of the adenovirus E1A oncogene on members of the AP-1 transcription factor family. Mol Cell Biol. 1990;10:5857–5864. doi: 10.1128/mcb.10.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Duyndam MCA, Ropttier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb AJ. Heterodimer formation of c-Jun and ATF-2 is responsible for induction of c-Jun by 243-amino-acid adenovirus E1A protein. EMBO J. 1993;12:479–487. doi: 10.1002/j.1460-2075.1993.tb05680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-J, Ogryzko VV, Nishikawa J-I, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]