Abstract
We compared the prevalence of key pfmdr1 alleles between pretreatment Plasmodium falciparum parasite isolates and parasites that emerged after treatment of uncomplicated malaria in a longitudinal cohort of Ugandan children. The pfmdr1 86N, 184F, and 1246D alleles were selected after treatment with artemether-lumefantrine, but not after artesunate-amodiaquine or amodiaquine-sulfadoxine-pyrimethamine. Remarkably, selection persisted in infections presenting up to about 60 days after treatment with artemether-lumefantrine. Thus, parasites selected for decreased drug sensitivity can appear long after predicted exposure to antimalarial drugs. Continued surveillance of the clinical efficacy and in vitro activity of new combination therapies is warranted.
Artemisinin-based combination therapies (ACTs) are recommended for treating uncomplicated falciparum malaria because of widespread resistance to older drugs. ACTs contain a fast-acting artemisinin and a longer-acting partner drug. Artemisinins rapidly reduce parasite biomass and control malaria symptoms, but treatment with artemisinin monotherapies is often followed by recrudescence. Longer-acting partner drugs increase ACT efficacy by eliminating parasites not cleared during the initial days of treatment. The partner drugs may also protect against selection for artemisinin resistance. However, infections that occur after artemisinins have been cleared, but while long-acting partner drugs continue to circulate, may select for resistance to the partner drugs.
Artemether-lumefantrine (AL) is the most widely used ACT in Africa. Artemether has a plasma half-life of approximately 1 hour. The half-life of lumefantrine has been measured at approximately 3–4 days [1], but it was shorter (33 hours) in children with uncomplicated malaria [2]. These half-lives are considerably shorter than those of the other principal ACT partners amodiaquine, mefloquine, and piperaquine, which have half-lives of approximately 2–4 weeks [1]. AL selects for parasites with genetic polymorphisms that reduce sensitivity to artemether, lumefantrine, and other antimalarial drugs. At various study sites in Africa, new infections that emerged soon after AL therapy had increased prevalence of single-nucleotide polymorphisms (SNPs) in pfcrt and/or pfmdr1, 2 putative drug transporters [3–6]. Of note, AL exerted the opposite effect of chloroquine and amodiaquine on these loci. Specifically, chloroquine and amodiaquine selected for the pfcrt 76T and pfmdr1 86Y, 184Y, and 1246Y alleles, but AL selected for the pfcrt 76K and pfmdr1 86N, 184F, and 1246D alleles [3–6].
The polymorphisms in pfcrt and pfmdr1 selected by prior therapy with antimalarials have been associated with alterations in the in vitro sensitivity of Plasmodium falciparum to various drugs. Considering key ACT components, the pfcrt 76K, pfmdr1 86N, and pfmdr1 1246D alleles (generally considered wild-type based on the reference 3D7 strain [6]) have been linked to increased sensitivity to amodiaquine, but decreased sensitivity to lumefantrine (and its analogue halofantrine), mefloquine, and artemisinins [7–10]. Three polymorphisms primarily seen outside Africa, pfmdr1 1034C, pfmdr1 1042D, and increased copy number of pfmdr1, have been associated with decreased sensitivity of P. falciparum to mefloquine, halofantrine, and artemisinins [4, 9]. Most relevant to this report, polymorphisms 86N and 1246D selected in Africa by prior therapy with AL were associated with decreased sensitivity to both components of this drug [7–10]. Correlations between individual polymorphisms in pfcrt or pfmdr1 and treatment efficacy have not been seen, as AL remains highly efficacious for treating uncomplicated falciparum malaria in Africa [11].
The selective pressure of AL for parasites with diminished drug sensitivity would be predicted to be short-lived, considering the approximately 1 hour and 3–4 day half-lives of its components. To determine the duration of selection by AL and other drugs, we compared the prevalence of key alleles between pretreatment P. falciparum isolates and isolates that emerged over extended periods after treatment with combination regimens in a cohort of children in Kampala, Uganda.
METHODS
We analyzed samples collected between 2004 and 2008 from a previously described clinical trial involving 690 children aged 1–10 years at enrollment [11, 12]. After informed consent, children were assigned to artemether-lumefantrine (AL), artesunate-amodiaquine (AS+AQ), or amodiaquine-sulfadoxine-pyrimethamine (AQ+SP) upon their first episode of malaria. Each child received the assigned treatment regimen for subsequent malaria episodes. They were asked to return for examination on days 1, 2, 3, 7, 14, and 28 after malaria diagnosis and whenever they felt ill. Blood for microscopy was obtained by finger prick, and samples for molecular analyses were stored on filter paper before treatment and on all follow-up days, except day 1. Treatment outcomes were classified following standard criteria, with genotyping based on 6 loci to distinguish recrudescence and new infection after therapy, as previously reported [11, 12].
Molecular analyses were done on 198 randomly selected pretreatment samples and on all 498 parasites that emerged within 120 days after a prior antimalarial treatment. For analysis of polymorphisms of interest and pfmdr1 copy number, DNA was extracted from filter paper with Chelex resin, alleles were identified by nested polymerase chain reaction (PCR) followed by restriction fragment length polymorphism analysis, and copy number was determined by quantitative PCR, all as previously reported [3, 13]. The presence of the pfmdr1 86N/184F/1246D combination was assessed by considering only samples that amplified at all 3 loci. Allele prevalence at various time points was compared by Fisher 2-tailed exact test, considering P < .05 as significant, using GraphPad software version 5.01.
RESULTS AND DISCUSSION
A total of 696 samples were analyzed, but 24 were excluded for failing to amplify after repeated PCR, leaving 672 P. falciparum isolates—195 obtained before the first treatment of study subjects, and 154, 143, and 180 obtained within 120 days after treatment with AL, AS+AQ, and AQ+SP, respectively. Notably, recrudescences were uncommon in the ACT arms of the study, and seen in only 4 of 521 (0.8%) treatments with AL between 2005 and 2008 [12]. Thus, for the AL treatment arm, nearly all of the isolates that caused recurrent disease were from new infections.
A total of 316 isolates (93 randomly selected from the pretreatment samples plus 52, 70, and 101 obtained within 40 days after prior treatment with AL, AS+AQ, and AQ+SP, respectively) were screened for mutations at pfcrt 76, pfmdr1 1034, and pfmdr1 1042. Consistent with recent reports from Uganda, all 305 evaluable isolates (11 did not amplify) had the pfmdr1 1034S and pfmdr1 1042N sequences. For 296 of the 316 samples studied at pfcrt 76 (20 did not amplify), 287 had the pfcrt 76T sequence, 5 76K, and 4 a mixed genotype. Samples with 76K included 1 collected before any treatment, 4 collected after prior treatment with AL, and 4 collected after prior treatment with AQ+SP. Because these loci showed little variability in parasites presenting soon after therapy, when they would be expected to be under the strongest drug pressure, they were not studied further. We also tested for amplification of the pfmdr1 gene in the 52 samples collected within 40 days of prior AL therapy. Consistent with other reports from Africa [3–5], amplification of pfmdr1 was not seen in any of 43 successfully tested samples.
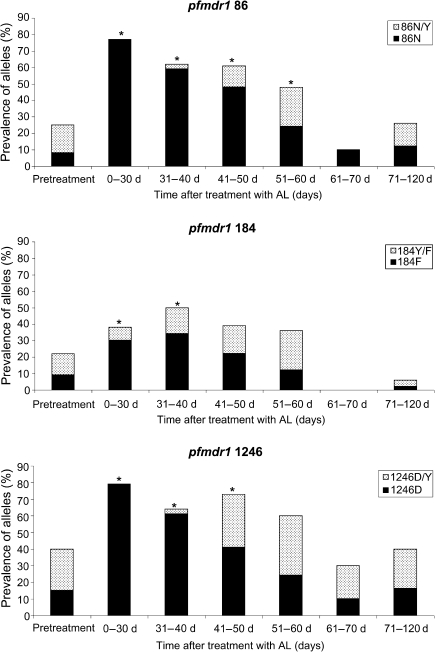
We were particularly interested in the impact of therapy on pfmdr1 N86Y, Y184F, and D1246Y, alleles that are polymorphic in Africa and affected by various antimalarial drugs. For samples collected before the first treatment in our trial, the prevalence of the pfmdr1 86N, 184F, and 1246D alleles was low, with these pure alleles occurring in 8%–15% of samples and mixed genotypes in an additional 13%–25% of samples (Figure 1). In infections that emerged after treatment with AS+AQ or AQ+SP, the prevalences of the pfmdr1 86N, 184F, and 1246D polymorphisms remained similar to those seen in baseline samples, and there was no significant association between time after treatment and prevalence of a particular polymorphism (Table 1). In contrast, as seen previously in a number of African studies [3–5], the prevalences of the 86N, 184F, and 1246D polymorphisms were much higher soon after treatment with AL (Table 1; Figure 1).
Figure 1.
Prevalence of pfmdr1 alleles in pretreatment samples and in samples collected after treatment with artemether-lumefantrine in a cohort of children in Kampala, Uganda. For clarity, prevalences of the pure 86Y, 184Y, and 1246Y alleles are not shown. *Significantly higher than pretreatment prevalence by Fisher exact test (2-tailed; P < .05).
Table 1.
Pfmdr1 Polymorphism Seen in Plasmodium falciparum Isolates From Kampala, Uganda, Following Treatment With Different Drugs
| Time since treatment (d) |
pfmdr1 86N |
pfmdr1 184F |
pfmdr1 1246D |
pfmdr1 86N/184F/1246D |
||||||||
| AL | AS+AQ | AQ+SP | AL | AS+AQ | AQ+SP | AL | AS+AQ | AQ+SP | AL | AS+AQ | AQ+SP | |
| Pretreatment | 16/194 (8%) | 18/194 (9%) | 30/194 (15%) | 15/193 (8%) | ||||||||
| 0–30 | 10/13 (77%)a | 2/46 (4%) | 4/73 (5%) | 4/13 (30%)a | 3/46 (7%) | 8/73 (11%) | 11/14 (79%)a | 5/46 (11%) | 6/68 (9%) | 4/13 (31%)a | 1/46 (2%) | 3/68 (4%) |
| 31–40 | 19/32 (59%)a | 1/17 (6%) | 1/28 (4%) | 11/32 (34%)a | 1/17 (6%) | 2/28 (7%) | 19/31 (61%)a | 3/19 (16%) | 7/26 (27%) | 10/31 (32%)a | 0/17 (0%) | 3/26 (12%) |
| 41–50 | 11/23 (48%)a | 0/10 (0%) | 0/17 (0%) | 5/23 (22%) | 0/10 (0%) | 1/17 (6%) | 9/22 (41%)a | 1/11 (9%) | 3/17 (18%) | 6/23 (26%)a | 0/10 (0%) | 1/17 (6%) |
| 51–60 | 6/25 (24%)a | 0/13 (0%) | 1/13 (8%) | 3/25 (12%) | 0/10 (0%) | 1/13 (8%) | 6/25 (24%) | 2/13 (15%) | 3/13 (23%) | 6/25 (24%)a | 0/13 (0%) | 0/13 (0%) |
| 61–70 | 1/10 (10%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 1/10 (10%) | 1/10 (10%) | 1/10 (10%) | 3/11 (27%) | 2/10 (20%) | 0/10 (0%) | 1/10 (10%) | 0/10 (0%) |
| 71–120 | 6/49 (12%) | 4/40 (10%) | 5/37 (14%) | 1/48 (2%) | 4/39 (10%) | 6/37 (16%) | 8/50 (16%) | 8/41 (20%) | 9/38 (24%) | 0/48 (0%) | 2/39 (5%) | 1/36 (3%) |
For pretreatment samples, 3 of 198 did not amplify for the pfmdr1 gene and 2 additional samples did not amplify for at least 1 allele, leading to the denominators shown. For 86N, P values were < .0001 up to 50 days and .0253 for the 51–60-days time point. For 184F, P values were .0362 for 0–30 days and .0005 for 31–40 days. For 1246D, P values were < .0001 up to 40 days and .007 for 41–50 days. For the combined alleles, P values were .0219 for 0–30 days, .0005 for 31–40 days, .0139 for 41–50 days, and .0204 for 51–60 days. For the individual polymorphisms, only data for the pure alleles are shown. For the combined polymorphisms, data include mixed sequences.
Significantly higher than pretreatment prevalence by Fisher exact test (2-tailed) at P < .05.
The longitudinal design of our trial offered a unique opportunity to determine the duration of selection of alleles of interest after treatment with AL. Remarkably, strong selection of pfmdr1 86N, 184F, and 1246D persisted for about 2 months after treatment with AL (Table 1; Figure 1). Selection was significant for the 86N allele at all studied intervals up to 60 days, for 1246D up to 50 days, and for 184F up to 40 days when considering selection of either only pure or pure and mixed genotypes (except for 184F at 0–30 days, which was significant only for pure genotypes) (Table 1; Figure 1). Moreover, considering all isolates studied, samples containing the 3 polymorphisms (86N, 184F, and 1246D) were more prevalent in parasites from the AL treatment arm (26 of 150, 17%) than in the pretreatment samples (15 of 193, 8%; P = .0075) or in parasites from the AS+AQ (4 of 135, 3%; P < .0001) and AQ+SP (8 of 170, 5%; P = .0004) arms of the study. In vivo, these 3 polymorphisms have been associated with recrudescence after AL treatment in some studies [4, 5]. In vitro, polymorphisms 86N [8] and 1246D [9] have been associated with decreased sensitivity to lumefantrine and artemisinin, respectively. Thus, treatment with AL selected strongly for polymorphisms associated with decreased sensitivity to both components of the combination therapy, and the selective pressure of AL was evident far beyond the half-lives of both components.
Clinically, prolonged selection by AL for particular alleles in pfmdr1 suggests the possibility that resistance to AL may develop as it is increasingly used in Africa. Evidence for this possibility is that the 86N allele has been associated with diminished in vitro sensitivity to artemether, dihydroartemisinin (the active metabolite of artemether) and lumefantrine [7, 8, 10]. Furthermore, the 1246D allele has been associated with diminished sensitivity to halofantrine (an analogue of lumefantrine) [9]. In our study, 17% of samples from children treated at least once with AL had the 86N/184F/1246D SNP combination, which has been associated with AL treatment failure in some studies [4, 5]. Moreover, assessment of children from our cohort with uncomplicated malaria indicated that the current weight-based dosing regimen for AL resulted in a shorter half-life (33 hours) and lower exposure of lumefantrine than in adults [2], potentially also contributing to treatment failures if parasite sensitivity to the drug diminishes over time.
The only other available information on the duration of selection for resistance by antimalarial drugs comes from studies of murine malaria. In mice infected with Plasmodium chabaudi and treated with pyrimethamine, the strength of selection for resistance was highest immediately following drug administration, but, consistent with our results, the selective pressure lasted far longer than would be expected based on the half-life of pyrimethamine [14].
Why does AL, a combination whose components are principally eliminated within days, continue to select for polymorphisms for a few months? First, lumefantrine or an active metabolite may have a longer terminal half-life than is generally appreciated, providing a wider window of selection for resistance than would be expected for a drug with a half-life of 3–4 days [1]. Second, due to genetic variations or other factors, some individuals may exhibit longer artemether or lumefantrine half-lives than others, allowing longer periods of selection. However, in a recent study of Ugandan children with uncomplicated malaria [2], none of the subjects had a lumefantrine half-life >49 hours (S. Parikh, personal communication). Third, the inherent features of malaria may also allow continued selection long after drug levels have decreased to negligible levels. Many new malaria illnesses in semi-immune children may follow extended periods of asymptomatic infection. In this event, the clinical impacts of selection might be seen long after the selection took place.
The strong and prolonged selective pressure of AL for polymorphisms that alter drug sensitivity is concerning. However, there are reasons for reassurance that, for now, AL remains an excellent therapy for uncomplicated malaria in Africa. First, AL has demonstrated outstanding efficacy for the treatment of falciparum malaria in many trials [11, 12]. Second, in vitro drug sensitivities of P. falciparum to artemisinins and lumefantrine have generally remained high [15]. Third, over the duration of our longitudinal trial in Kampala, the prevalence of key polymorphisms did not change noticeably (Table 2). However, AL use was not yet common in Kampala during the course of our study, but it has greatly increased across Africa in the last few years. Of concern are reports of uncommon recrudescences after therapy with AL [4, 5, 11, 12] and occasional African isolates with diminished in vitro response to artemisinins or lumefantrine [8]. Because of difficulties in assigning clinical outcomes and complexities of in vitro drug sensitivity assays, it is uncertain whether early signs of diminished efficacy of AL have been seen in Africa. In any event, as most countries in Africa now rely on AL as the mainstay of malaria therapy, careful surveillance of the clinical efficacy of this drug and of the in vitro activity of both artemisinins and lumefantrine is an urgent priority.
Table 2.
Annual Prevalence of pfmdr1 Polymorphisms in Plasmodium falciparum Isolates From Kampala, Uganda, 2004–2008
| Year | pfmdr 86N | pfmdr 86N/Y | pfmdr 86Y | pfmdr 184F | pfmdr 184Y/F | pfmdr 184Y | pfmdr 1246D | pfmdr 1246D/Y | pfmdr 1246Y |
| 2004 | 3/32 (9%) | 7/32 (22%) | 22/32 (69%) | 4/32 (13%) | 4/32 (13%) | 24/32 (75%) | 5/31 (16%) | 7/31 (23%) | 19/31 (61%) |
| 2005 | 34/280 (12%) | 46/280 (16%) | 200/280 (71%) | 27/279 (10%) | 30/279 (11%) | 222/279 (80%) | 55/280 (20%) | 71/280 (25%) | 154/280 (55%) |
| 2006 | 25/210 (12%) | 32/210 (15%) | 153/210 (73%) | 11/102 (11%) | 20/210 (10%) | 166/210 (79%) | 42/210 (20%) | 35/210 (17%) | 133/210 (63%) |
| 2007 | 20/103 (19%) | 7/103 (7%) | 76/103 (74%) | 11/102 (11%) | 8/102 (8%) | 83/102 (81%) | 25/103 (24%) | 22/103 (21%) | 56/103 (54%) |
| 2008 | 7/35 (20%) | 3/35 (9%) | 25/35 (71%) | 4/35 (12%) | 5/35 (14%) | 26/35 (74%) | 9/35 (26%) | 9/35 (26%) | 17/35 (48%) |
Funding
This work was supported by grants from the National Institutes of Health (AI52142, AI075045, and TW01506 to P. J. R.) and the Doris Duke Charitable Foundation, with which P. J. R. is a Distinguished Clinical Scientist. F. N. B. was supported by a National Institutes of Health T-32/Ruth L. Kirschstein National Research Service Award (5T32AI060537).
Acknowledgments
We thank the participants in the clinical trials from which samples were collected, their parents and guardians, and all members of our clinical study and laboratory teams in Kampala and San Francisco. We thank Bryan Greenhouse for helpful comments on the draft manuscript, and Grant Dorsey, Sunil Parikh, Christian Dokomajilar, Christian Nsanzabana, Danica Helb, and Edwin Ochong for helpful discussions. Control P. falciparum strains were from the Malaria Research and Reference Reagent Resource Center.
References
- 1.Stepniewska K, White NJ. Pharmacokinetic determinants of the window of selection for antimalarial drug resistance. Antimicrob Agents Chemother. 2008;52:1589–96. doi: 10.1128/AAC.00903-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mwesigwa J, Parikh S, McGee B, et al. Pharmacokinetics of artemether-lumefantrine and artesunate-amodiaquine in children in Kampala, Uganda. Antimicrob Agents Chemother. 2010;54:52–9. doi: 10.1128/AAC.00679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ. Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–5. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Happi CT, Gbotosho GO, Folarin OA, et al. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother. 2009;53:888–95. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphreys GS, Merinopoulos I, Ahmed J, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–7. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M. Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 8.Mwai L, Kiara SM, Abdirahman A, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53:5069–73. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–9. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 10.Duraisingh MT, Roper C, Walliker D, Warhurst DC. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol. 2000;36:955–61. doi: 10.1046/j.1365-2958.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- 11.Dorsey G, Staedke S, Clark TD, et al. Combination therapy for uncomplicated falciparum malaria in Ugandan children: a randomized trial. JAMA. 2007;297:2210–9. doi: 10.1001/jama.297.20.2210. [DOI] [PubMed] [Google Scholar]
- 12.Clark TD, Njama-Meya D, Nzarubara B, et al. Incidence of malaria and efficacy of combination antimalarial therapies over 4 years in an urban cohort of Ugandan children. PLoS One. 2010;5:e11759. doi: 10.1371/journal.pone.0011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis. 2001;183:1417–20. doi: 10.1086/319865. [DOI] [PubMed] [Google Scholar]
- 14.Huijben S, Nelson WA, Wargo AR, Sim DG, Drew DR, Read AF. Chemotherapy, within-host ecology and the fitness of drug-resistant malaria parasites. Evolution. 2010;64:2952–68. doi: 10.1111/j.1558-5646.2010.01068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob Agents Chemother. 2010;54:1200–6. doi: 10.1128/AAC.01412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]