Abstract
Objective
Variants of two genes, CYP2C9 and VKORC1, explain approximately 30% of variability in warfarin maintenance dose requirements. However, the clinical utility of using this information in addition to clinical and demographic data (‘pharmacogenomic-guidance’) is unclear, as few comparative clinical trials have been conducted to date. The objective of this study was to explore the incremental effect of pharmacogenomic-guided warfarin dosing under various conditions using clinical trial simulation.
Methods
We utilized an existing PK/PD model to perform clinical trial simulations of pharmacogenomic-guided versus standard of care warfarin therapy. The primary outcome was the percentage of patient time spent in therapeutic range over the first month of therapy. We assessed the influence of the frequency of INR monitoring, and the use of a loading dose and dose increase delay in patients with CYP2C9 variants.
Results
Pharmacogenomic guidance resulted in a 3-4% absolute increase in time spent in therapeutic range over the first month of therapy compared to standard of care. The improvement in time in range was greater when the frequency of INR monitoring in both arms was assumed to be lower. The absolute difference increased to 6-8% with use of a loading dose and dose increase delay in patients with a CYP2C9 variant.
Conclusion
Our initial results imply that pharmacogenomic-guided warfarin dosing may be more useful in settings with less intensive patient follow-up, and when adjustments are made for slower therapeutic response in patients with a CYP2C9 variant. Further PK/PD model development may be useful for warfarin pharmacogenomic trial design.
Keywords: warfarin, trial simulation, pharmacogenomic, pharmacokinetic, pharmacodynamic
Introduction
Warfarin is effective in reducing the risk of thromboembolic events (TEs), but is also one of the most common causes of serious adverse drug events.1 Variants of CYP2C9 and VKORC1 have been shown to explain approximately 35% of the variability in warfarin dose requirements, and CYP2C9 variants have been associated with a higher risk of serious bleeding events.2, 3 While there is strong evidence that genetics influence warfarin dose, there are limited data on the clinical impact of using this information in addition to clinical and demographic data to guide warfarin therapy. Anderson et al reported no difference in time in therapeutic range (TTR) between pharmacogenomic-guided and standard dose initiation in an intensively monitored population, while Caraco et al found a decrease in time to first therapeutic INR.4, 5 The optimal care setting and implementation strategies for pharmacogenomic (PGx)-guided dosing are not yet clear.6
Pharmacokinetic-pharmacodynamic (PK/PD) models that incorporate the respective effects of CYP2C9 and VKORC1 variants on drug serum levels and INR have been developed for dosing of warfarin.7, 8 The implementation of such models within a dynamic dosing and monitoring framework affords a means of assessing the potential impact of PGx-guided dosing upon various measures of clinical outcome. We conducted a set of clinical trial simulations with the following objectives: 1) to estimate the magnitude of the effect size for PGx-guided versus standard care warfarin dose initiation, 2) to explore the influence of INR monitoring frequency on the effect size, and 3) to assess the potential benefits of dosing adjustments that account for the slower therapeutic response in patients with a CYP2C9 variant.
Methods
A warfarin clinical trial simulation tool was developed as an R interface that calls upon the nonlinear mixed effects modeling software NONMEM.9, 10 The simulator was designed with the capability of adjusting dosing and follow-up appointment scheduling based on a simulated subject's INR at each clinical appointment. We used a recently published population model of S-warfarin pharmacokinetics and pharmacodynamics developed by Hamberg and colleagues to simulate warfarin concentration and effect, as well as variability, in a hypothetical clinical trial population.8 Subject demographics, including baseline INR, age, and CYP2C9 and VKORC1 genotype, were simulated based on the median and range for the continuous variables or fractions for the categorical distribution reported by Hamberg and colleagues. Each subject's dosing history, appointment schedule and INR were tracked during the simulated protocol. We did not assess the effects of parameter uncertainty in our simulations; in part because parameter uncertainty was low (generally < 10% RSE) compared to variability (26-99% CV), so the expected effect on predictions was limited11 and in part due to lack of access to the full parameter estimate covariance matrix from the Hamberg analysis.
Simulation outcome
The primary outcome was the average percentage of time patients spent in the therapeutic range (INR 2-3) over the first month of therapy. The percentages of time spent above and below therapeutic range were also assessed. The amount of time patients spent above/in/below INR range was interpolated from INRs at individual clinic visits using the technique of Rosendaal.12, 13 This method provides a more informative measure of patient exposure to over- and under-anticoagulation than the proportion of INR values above and below range, respectively, at specific clinic visits.
Warfarin initiation
We compared the following warfarin initiation protocols. In the “standard” initiation, all hypothetical subjects received 5 mg/day for days 1-4 if under age 75 years, and 2.5 mg/day otherwise. In “pharmacogenomic” (PGx) initiation, day 1-4 doses were specified based on CYP2C9 and VKORC1 genotype as well as age of each hypothetical subject (Table 1).14 While the initiation equation specified dosing based on patient weight, the Hamberg PK/PD model did not identify weight as an important covariate in warfarin PK-PD, and so we assumed a weight of 150 lb for all subjects.8
Table 1.
Pharmacogenomics-based dose initiation algorithm.14
| Factor | Dose adjustment |
|---|---|
| Base Dose | 5.67 mg/day |
| AGE (yrs) | − 0.05*(AGE-61) |
| WT (lbs) | + 0.01*(WT-191) |
| CYP2C9 (each *2) | − 1.04*(no. of *2) |
| CYP2C9 (each *3) | − 1.14*(no. of *3) |
| VKORC1 (GG) | + 1.85 |
| VKORC1 (AA) | − 2.33 |
CYP2C9 adjustments as compared to *1/*1
VKORC1 adjustments as compared to AG
Minimum dose 0.5 mg/day
Warfarin maintenance
We adapted an algorithm developed by Wilson and colleagues (Table 2), which features moderately frequent INR monitoring and is reflective of a maintenance protocol for patients in an anticoagulation clinic setting.15 We simulated dosing and INR over the first month of therapy. For comparison purpose, we implemented two parallel algorithms, which we refer to as “more frequent” and “less frequent” monitoring – corresponding to the lower and upper values of the follow-up range in the Wilson algorithm. All subjects received follow-up appointments on days 4 and 8 to determine doses 4-7 and from 8-30, respectively. After day 8, all appointments were individually specified based on INR.
Table 2.
Dose adjustment algorithm adapted from Wilson et al.15
| INR | Suggested Dose Adjustment (for maintenance of INR in [2,3]) |
INR Recheck (days) Frequency: |
|
|---|---|---|---|
| More | Less | ||
| ≤1.3 | Increase dose by 50% | 5 | 7 |
| 1.4 | Increase dose by 33% | 5 | 7 |
| 1.5-1.8 | Increase dose by 25% | 5 | 7 |
| 1.9 | Increase dose by 10% | 7 | 14 |
| 2.0-2.8 | No Change | 7* | 14* |
| 2.9-3.1 | Reduce dose by 10% | 7 | 14 |
| 3.2-3.5 | Reduce dose by 25% | 5 | 7 |
| 3.6-3.7 | Reduce dose by 33% | 5 | 7 |
| 3.8-4.4 | Skip 1 day, Reduce dose by 33% | 3 | 7 |
| 4.5-5.0 | Skip 2 days, Reduce dose by 33% | 3 | 7 |
| 5.1-6.0 | Skip 3 days, Reduce dose by 33% | 3 | 7 |
| ≥6.1 | Determined by physician** | – | – |
was 14-28 days in the Wilson algorithm.15
not implemented, treated as for INR 5.1-6.0.
Trial simulation
For each protocol, Monte Carlo simulation was performed for 2000 subjects and the percent of time subjects were above/in/below INR range (INR ‘in-range’ defined as 2 - 3) was assessed. Results were stratified based on patient's genotype: a) subjects with CYP2C9 variants and any VKORC1 genotype; b) subjects with VKORC1 variants and CYP2C9 wild type; c) wild type for both CYP2C9 and VKORC1; and d) all subjects. These groups are not mutually exclusive, and were defined to explore the relative effects of CYP2C9 variants, which affect warfarin clearance and half-life, and VKORC1 variants, which do not.
To explore the effect of the trial size, we performed multiple trial simulation of 200, 500, 1000 and 2000 subject trials. Due to lengthy run times, we simulated 1000, 400, 200, and 100 trials of each size, respectively. The standard deviations of percent above/in/below range time results (or standard error of the mean) were computed for each trial size. The standard deviations provide estimates of between-trial variability for trials of each sample size. Comparable results were independently obtained with a less computationally intensive approach based on bootstrap runs from a simulated pool of 2,000 subjects (results not shown).
Simulation Strategy
The warfarin protocol simulation routine was written utilizing a modular architecture as a function call in the R programming language with system calls to NONMEM for Monte Carlo pharmacokinetic/pharmacodynamic simulation and a system call to SED for changing the random seed in the NONMEM control file. After initial dosing and prescheduled appointments, each simulated subject receives individual appointment scheduling and dose adjustment, based on the specified protocol, for the duration of the simulation.
The simulation function allows the user to set simple inputs such as the number of subjects to simulate, the number of days to simulate the protocol, random seeds for population simulation from both demographic and pharmacokinetic/dynamic variability, and prescheduled appointments that all study subjects receive (e.g. days 4 and 8 to adjust doses 4-7 and 8-until the next individually scheduled appointment; scheduling is based on simulated INR prior to the appointment). More complex inputs to the simulation function include a list of dosing algorithms (functions) that collectively specify the current protocol (e.g. initiation, transition and maintenance routines) and a function specifying when to switch to the next dosing algorithm (either for all subjects based on protocol day or individually based on current INR or time in range).
The initial modules of the simulation create the NONMEM “dataset”, which specifies for each subject the dose and observation schedule as well as demographic covariates (such as age, baseline INR, and CYP2C9 and VKORC1 variant) which are sampled from specified covariate distributions. The initial doses are specified based on the initiation algorithm and a default dose (to be changed as the simulation progresses) is specified for all later dose times. The dose amounts in this initial dataset are updated as the simulation progresses and doses are adjusted. Individual appointment schedules are specified and tracked via an array in R.
Once the initial dataset is constructed, the individual inputs to the pharmacokinetic and pharmacodynamic models are fully specified. NONMEM is then called to choose individual pharmacokinetic and pharmacodynamic model parameters (from within the specified distributions) and run the time-dependent simulation. The output includes daily INR values for each subject. For each day of the simulation, each subject with a scheduled appointment receives adjustment to future doses (via the appropriate dose selection algorithm) and a scheduled future appointment. The new doses are then updated in the dataset and the simulation is then run forward. Each subject with an appointment on the next simulation day then receives adjustment to future doses and a scheduled future appointment. For the multiple trial simulations, the trial simulation function is wrapped in a loop within which the appropriate random seeds are changed for each trial (thus simulating a new study population) and the running mean and standard deviation of results (time above/in/below range) are computed.
Protocols simulated
The following protocols were simulated:
Base-case protocol: standard dose initiation vs. PGx-guided dose initiation, as described above, with more- and less-frequent monitoring.
A protocol similar to (1), except CYP2C9 variants under the PGx initiation strategy received a loading dose 2× the predicted maintenance dose on days 1-2 only, and no dose increase for the first 18 days (dose decrease was allowed).
As a test of the effect of dose timing compliance, dosing times were allowed to vary randomly (normally distributed) with a standard deviation of 4 hours.
As a test of the effect of missed doses, 10% of doses in the simulated population were skipped at random.
To explore the impact of within-subject variability (drift) due, for example, to dietary variation (e.g., vitamin K intake), we allowed each subject's prescribed dose to wander in a random walk, starting at a dose multiplier of 1 (i.e. no change) with step chosen uniformly from the interval [-0.025, 0.025] and with ultimate dose multiplier bounds of [0.9, 1.1].
Results
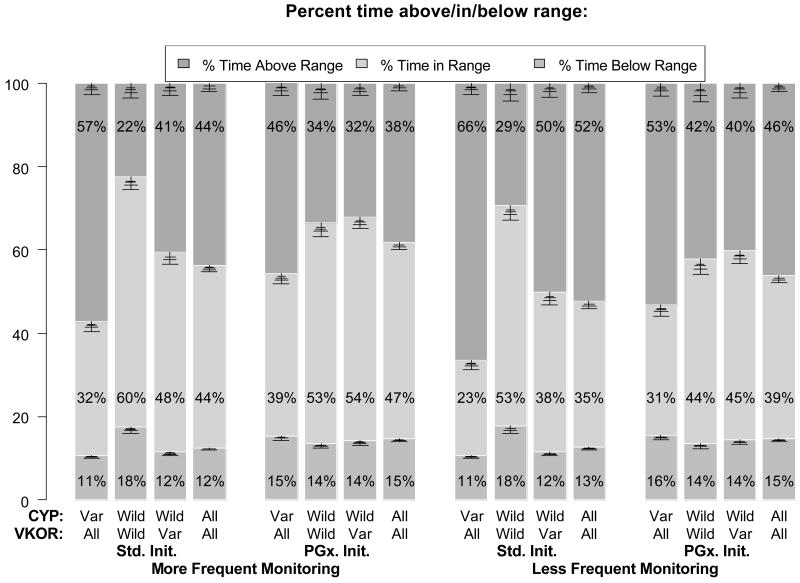
The primary results are shown in Figure 1: standard (Std) versus pharmacogenomic initiation (PGx), each with more frequent or less frequent monitoring. The results for each protocol are stratified by genotype. The vertical bars represent the percent time over the first month of therapy spent above, in, and below therapeutic range.
Figure 1.
Trial simulation results from standard vs. pharmacogenomic-based (PGx-based) dose initiation under 2 monitoring scenarios, over the first month of therapy. Each column shows time in, above and below range for all 2000 simulated subjects, as well as three subgroups, based on CYP2C9 and VKORC1 genetic polymorphisms. Each pair of clusters shows results for more and less frequent monitoring, respectively. Both more or less frequent monitoring trials are each initiated by either standard or PGx-based initiation of therapy. Error bars visualize standard deviation for population sizes of 200, 500, 1000 and 2000 (corresponding to decreasing error bars). See text for further details.
The key findings that can be extracted from this figure are as follow. Firstly, under the more frequent monitoring scenario, the percentage of time in therapeutic range (TTR) for all patients was 44% for standard initiation vs. 47% in the PGx initiation arm (4th and 8th columns, respectively). In contrast, under the less frequent monitoring scenario, the TTR for both the Std and PGx arms were lower (35% and 39%, respectively). Thus the absolute increase in TTR due to PGx initiation was slightly larger in the less frequent monitoring scenario compared to the more frequent monitoring scenario (4Δ% vs. 3Δ%, Δ% = absolute change in %TTR). Secondly, the TTR improved for VKORC1 variants by 6-7Δ% with PGx initiation. On the other hand, TTR decreased for CYP2C9/VKORC1 wild type/wild type patients by 7-9Δ% with PGx initiation, despite higher starting doses of 6.0 – 8.4 mg. Thirdly, the % time above range for CYP2C9 variants was lower with PGx versus Std initiation by 11Δ% (46 vs. 57%) and 13Δ% (53 vs. 66%) in the more and less frequent monitoring protocols, respectively. Furthermore, in the Std vs. PGx initiation protocols, 3.5% vs. 1.0% of patient time was above an INR of 5 (more frequent monitoring) and 8.4% vs. 3.8% (less frequent monitoring). Lastly, the average number of clinic visits was 5.4 vs. 5.2 (more frequent monitoring) and 3.8 vs. 3.7 (less frequent monitoring) for Std and PGx initiation, respectively.
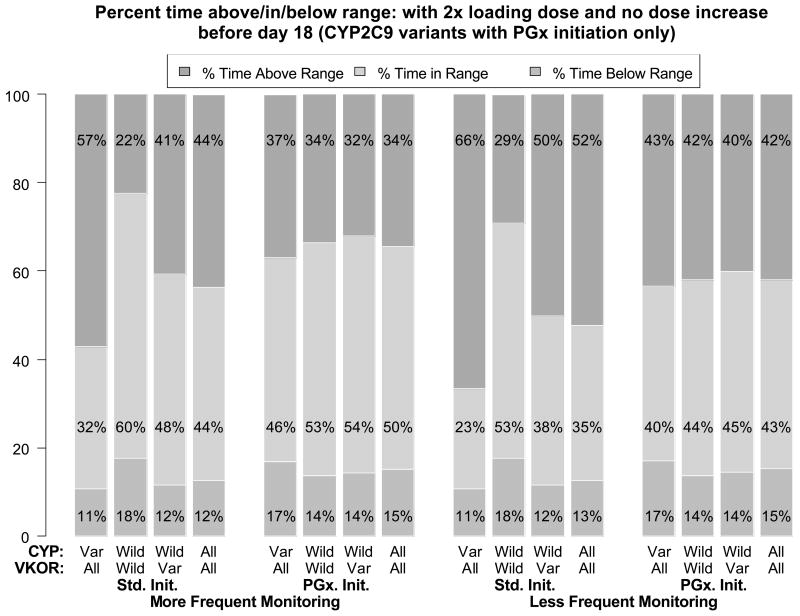
The results of protocol (2) in which patients with a CYP2C9 variant were administered a 2× loading dose and did not have any dose increases for the first 18 days are shown in Figure 2. The TTR for all patients in the PGx initiation simulations was 50% and 43% for more and less frequent monitoring, respectively, as compared to 47% (± 0.6% for 2000-subject or ± 1.7% for 200-subject trials) and 39% (± 0.7% for 2000-subject or ± 1.8% for 200-subject trials) for the base-case simulation. Compared to the base-case simulation (Figure 1), this represents a doubling of the difference between PGx and Std. initiation strategies (i.e., from 3-4Δ% to 6-8Δ%). These differences are within the simulation variability for the small (N=200) trials, but not for the larger (N=2,000) trial. The improvement was primarily the result of a decrease in the % time above range for CYP2C9 variants. However, the approximately 20% absolute decrease in time above range for patients with CYP2C9 variants is offset to some degree by an approximately 6% absolute increase in the time below range – thus there is potentially a slight increase in thromboembolic risk.
Figure 2.
Trial simulation results similar to those in Fig. 1, except CYP2C9 variants in the PGx-based initiation of therapy arm received double the predicted maintenance dose days 1-2 and no dose adjustment for the first 18 days. See text and Fig. 1 for further details.
In protocol (3) in which dose times were allowed to vary randomly, the results were nearly unchanged from the base-case simulation. When 10% of doses were skipped at random (protocol 4), the %TTR increased by 1-3% as a result of a small decrease in the % time above range for patients across genotype categories. Lastly, in protocol (5) in which each subject's prescribed dose was allowed to wander in a random walk, the %TTR across all patients was slightly decreased (e.g., <1%).
Discussion
Comparison to observed data
The percent of time in therapeutic range (TTR) over the first month of therapy simulated in this study for standard initiation, 35-44%, is somewhat lower than what has been observed over the first month of therapy both in our outpatient academic medical center anticoagulation clinic (50%) and in a first-month analysis of the COUMAGEN trial (48%).5, 14 The difference between the simulated and clinical data are primarily driven by a greater % time above range for CYP2C9 variants in the simulations, 57% - 66%, versus 25-37% observed. These results imply that clinician specialists may be managing patients with CYP2C9 variants more effectively than strict adherence to a simulated maintenance protocol. Overall, in the COUMAGEN randomized controlled trial, genotyping did not improve TTR, with 49.8% in the PGx group and 51.9% in the standard of care group, p=0.54. These results again suggest that clinicians may be able to adapt to the impact of CYP2C9 variants on warfarin dose and clearance in a way that is not reflected by the algorithm simulated in this study. Of note, however, 80% of patients in the COUMAGEN trial were inpatients and had INR monitored daily during initiation. Comparison to results of Caraco et al is difficult because %TTR was reported between day 9 and “stable” anticoagulation, rather than over a fixed time period.4
Limitations
There are several important limitations of our study worth noting. There likely were systematic differences in the nature of the simulated individuals compared to patients in the actual clinical studies. The simulated individuals were reflective of those in the Hamberg study, whereas observed data were from anticoagulation patients with potentially different clinical and demographic characteristics. The lack of reported correlation of the covariates could potentially cause systematic overestimation of demographic variability in our simulated subjects.
The PK/PD model upon which we based our analysis was developed using 150 patients, and likely did not have sufficient sampling of all patient subgroups.8 For instance, the model suggests that *2/*2 patients have a greater pharmacodynamic response (INR) to warfarin than *2/*3 patients, which is in contrast to mechanistic and observed data.7, 16 In addition, weight was not found to be a predictive covariate, although dose prediction algorithms and PK/PD models have recognized it to be significant.2, 17
We found simulation of the dynamic, individualized doses and dose scheduling inherent with warfarin therapy to be challenging. In the early stage of our simulation work, several maintenance dosing and monitoring protocols were evaluated.5, 18 We found that implementation of protocols with less frequent (e.g., 7-10 days) INR monitoring for patients with an INR in or near therapeutic range during the first 1-2 weeks of therapy led to poor INR control over the first month of therapy. However, continual simulation of periods beyond 1 month led to extremely tight INR control, which also does not match clinical experience or literature data. A more realistic model, possibly incorporating variables such as intra-subject PK variability due to diet, etc., may be ultimately needed for realistic simulations beyond a 1-month time period. Given these limitations, the findings of this study should be considered exploratory in nature.
We allowed dose time to vary with a SD of 4 hours to assess dose-time variation, but we did not account for decreased adherence in terms of proportion of dosing actually taken. Investigation of such effects is warranted, but beyond the scope of the current study. Furthermore, although not evaluated in this study, an additional strategy to improve anticoagulation outcomes would be the use of a PK/PD model to individualize dosing, or algorithms that incorporate initial INR measurements with genotype information.8, 19
Implications
Our findings suggest that the effect of PGx initiation may be relatively small, but potentially clinically relevant in certain populations, such as those monitored less frequently than in an anticoagulation clinic. The clinical implications can be estimated based on epidemiologic studies of INR and the risk of clinical events. For example, in a recent study, van Walraven and colleagues assessed 6,400 patient years of anticoagulant exposure.20 Using their estimates for the risk of bleeding and thromboembolic events above, within, and below INR range, we estimate that pharmacogenomic-guided dosing in 1,000 patients would avert approximately 1 bleed, with little difference in clotting events.
The results also imply that utilizing information about the slower therapeutic response in patients with a CYP2C9 variant may improve the effectiveness of the intervention. Furthermore, given the ability to identify patients that require a lower dose, and in addition the subset of those that will be slow to respond to warfarin, the benefit-risk tradeoff of using a loading dose may be improved.18
Our findings may have implications for trial design. For instance, using standard statistical power calculations (at 80% power), a trial with %TTR in the first month of therapy as the primary outcome, and assuming a %TTR of 39% in the standard care arm and 43% in the PGx arm based on the trial simulations in the current study, would require 4,800 patients. The simulation results utilizing a 2× loading dose and dose increase delay (39% and 47% %TTR) would imply a trial of 1,200 patients.
Conclusion
The use of quantitative modeling approaches to explore trial design issues for pharmacogenomic interventions, involving dosage and monitoring refinements, offer significant promise. Further investment in PK/PD models and trial simulation approaches is warranted.
Acknowledgments
This study was supported in part by Centers for Disease Control (CDC) Seed Funding for Public Health Genomics Research (DLV), Grant# 1U18 GD000005-01 (CDC Office of Genomics and Disease Prevention), and the University of Washington Drug Metabolism, Transport, and Pharmacogenomic Research (DMTPR) Program (supported by unrestricted gifts from Abbott, Allergan, Amgen, Bend Research, Bristol-Myers Squibb, Eli Lilly, Hoffman La Roche, Johnson & Johnson and Pfizer).
References
- 1.Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis. 2008;25:45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 2.IWPC, Consortium TIWP. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28:1084–97. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–70. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 5.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, Kahn SF, May HT, Samuelson KM, Muhlestein JB, Carlquist JF. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 6.Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 7.Linder MW, Looney S, Adams JE, 3rd, Johnson N, Antonino-Green D, Lacefield N, Bukaveckas BL, Valdes R., Jr Warfarin dose adjustments based on CYP2C9 genetic polymorphisms. J Thromb Thrombolysis. 2002;14:227–32. doi: 10.1023/a:1025052827305. [DOI] [PubMed] [Google Scholar]
- 8.Hamberg AK, Dahl ML, Barban M, Scordo MG, Wadelius M, Pengo V, Padrini R, Jonsson EN. A PK-PD model for predicting the impact of age, CYP2C9, and VKORC1 genotype on individualization of warfarin therapy. Clin Pharmacol Ther. 2007;81:529–38. doi: 10.1038/sj.clpt.6100084. [DOI] [PubMed] [Google Scholar]
- 9.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: 2007. [Google Scholar]
- 10.Beal SL, Sheiner LB, Boeckmann AJ. NONMEM Users Guides. Icon Development Solutions; 1989-2006. [Google Scholar]
- 11.Samtani MN, Perez-Ruixo JJ, Brown KH, Cerneus D, Molloy CJ. Pharmacokinetic and pharmacodynamic modeling of pegylated thrombopoietin mimetic peptide (PEG-TPOm) after single intravenous dose administration in healthy subjects. J Clin Pharmacol. 2009;49:336–50. doi: 10.1177/0091270008329559. [DOI] [PubMed] [Google Scholar]
- 12.Rosendaal FR, Cannegieter SC, van der Meer FJ, Briet E. A method to determine the optimal intensity of oral anticoagulant therapy. Thromb Haemost. 1993;69:236–9. [PubMed] [Google Scholar]
- 13.Torn M, Cannegieter SC, Bollen WL, van der Meer FJ, van der Wall EE, Rosendaal FR. Optimal level of oral anticoagulant therapy for the prevention of arterial thrombosis in patients with mechanical heart valve prostheses, atrial fibrillation, or myocardial infarction: a prospective study of 4202 patients. Arch Intern Med. 2009;169:1203–9. doi: 10.1001/archinternmed.2009.176. [DOI] [PubMed] [Google Scholar]
- 14.Meckley LM, Wittkowsky AK, Rieder MJ, Rettie AE, Veenstra DL. An analysis of the relative effects of VKORC1 and CYP2C9 variants on anticoagulation related outcomes in warfarin-treated patients. Thromb Haemost. 2008;100:229–39. [PubMed] [Google Scholar]
- 15.Wilson SE, Costantini L, Crowther MA. Paper-based dosing algorithms for maintenance of warfarin anticoagulation. J Thromb Thrombolysis. 2007;23:195–8. doi: 10.1007/s11239-006-9025-4. [DOI] [PubMed] [Google Scholar]
- 16.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. Jama. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 17.Tham LS, Goh BC, Nafziger A, Guo JY, Wang LZ, Soong R, Lee SC. A warfarin-dosing model in Asians that uses single-nucleotide polymorphisms in vitamin K epoxide reductase complex and cytochrome P450 2C9. Clin Pharmacol Ther. 2006;80:346–55. doi: 10.1016/j.clpt.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs MJ, Rodger M, Anderson DR, Morrow B, Kells G, Kovacs J, Boyle E, Wells PS. Comparison of 10-mg and 5-mg warfarin initiation nomograms together with low-molecular-weight heparin for outpatient treatment of acute venous thromboembolism. A randomized, double-blind, controlled trial. Ann Intern Med. 2003;138:714–9. doi: 10.7326/0003-4819-138-9-200305060-00007. [DOI] [PubMed] [Google Scholar]
- 19.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, Milligan PE, Grice G, Lenzini P, Rettie AE, Aquilante CL, Grosso L, Marsh S, Langaee T, Farnett LE, Voora D, Veenstra DL, Glynn RJ, Barrett A, McLeod HL. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Walraven C, Oake N, Wells PS, Forster AJ. Burden of potentially avoidable anticoagulant-associated hemorrhagic and thromboembolic events in the elderly. Chest. 2007;131:1508–15. doi: 10.1378/chest.06-2628. [DOI] [PubMed] [Google Scholar]