The transition between the closed and open conformations of the protein translocation channel controls the efficiency of protein translocation and the fidelity of signal sequence recognition. Mutations in Sec61 that delay or accelerate this structural transition have antagonistic effects on translocation efficiency and fidelity.
Abstract
The transition between the closed and open conformations of the Sec61 complex permits nascent protein insertion into the translocation channel. A critical event in this structural transition is the opening of the lateral translocon gate that is formed by four transmembrane (TM) spans (TM2, TM3, TM7, and TM8 in Sec61p) to expose the signal sequence–binding site. To gain mechanistic insight into lateral gate opening, mutations were introduced into a lumenal loop (L7) that connects TM7 and TM8. The sec61 L7 mutants were found to have defects in both the posttranslational and cotranslational translocation pathways due to a kinetic delay in channel gating. The translocation defect caused by L7 mutations could be suppressed by the prl class of sec61 alleles, which reduce the fidelity of signal sequence recognition. The prl mutants are proposed to act by destabilizing the closed conformation of the translocation channel. Our results indicate that the equilibrium between the open and closed conformations of the protein translocation channel maintains a balance between translocation activity and signal sequence recognition fidelity.
INTRODUCTION
An evolutionarily conserved heterotrimeric protein-conducting channel (SecYEG in eubacteria, SecYEβ in archae, Sec61 complex in eukaryotes) mediates the translocation of secreted proteins and integration of membrane proteins (recent reviews in Driessen and Nouwen, 2008; Mandon et al., 2009). Protein translocation channels are composed of a large subunit (SecY, Sec61, or Ssh1p) that has 10 transmembrane (TM) spans plus smaller β (SecG, Secβ, Sec61β, Sbh1p/Sbh2p) and γ (SecE, Sec61γ, Sss1p) subunits. Protein photo-cross-linking experiments have shown that the signal sequence and mature regions of secretory proteins are in continuous contact with Sec61 during protein translocation (Mothes et al., 1994), indicating that Sec61 (or SecY) forms the transport pore through which nascent polypeptides pass. In budding yeast, the Sec61 complex can also assemble with the Sec62/Sec63 complex to form a heptameric Sec complex involved in posttranslational translocation (Deshaies et al., 1991; Panzner et al., 1995).
Protein translocation across the yeast endoplasmic reticulum can occur by cotranslational or posttranslational pathways that depend on recognition of the signal sequence by the signal recognition particle (SRP) for cotranslational translocation or by the heptameric Sec complex for posttranslational translocation. Targeting of the ribosome-nascent chain (RNC) complex to the rough endoplasmic reticulum via the interaction between the SRP and the SRP receptor leads to the GTP-dependent transfer of the RNC to the Sec61 complex or the closely related Ssh1 complex (Finke et al., 1996; Jiang et al., 2008). The Ssh1 complex, which is nonessential in yeast, is exclusively involved in cotranslational translocation (Wittke et al., 2002). Recent structures of the RNC-Sec61, RNC-Ssh1, ribosome-SecY, and ribosome-Sec61 complexes that have been obtained by cryo–electron microscopy show that a single Sec61, Ssh1, or SecY heterotrimer serves as a protein-conducting channel (Menetret et al., 2007, 2008; Becker et al., 2009). Cytosolic loops 6 and 8 of Sec61p (or Ssh1p) interact with the RNC at the polypeptide exit site on the large ribosomal subunit (Cheng et al., 2005; Becker et al., 2009), making contacts that are critical for cotranslational translocation (Cheng et al., 2005). The molecular mechanism that promotes signal sequence insertion into the translocation pore upon binding of the RNC to the cytosolic face of Sec61 has not been elucidated. The mechanism of nascent chain insertion into the Sec complex (Sec61 complex plus the Sec62/Sec63 complex) is likewise poorly understood, as structures of the Sec62/Sec63 complex have yet to be obtained.
The high-resolution structure of the Methanocaldococcus jannaschii SecYEβ complex provides a detailed model for the closed conformation of a protein translocation channel (Van den Berg et al., 2004). TM spans 1–5 and TM spans 6–10 of SecY form two sides of an hourglass-shaped transporter that can open lumenally to allow translocation of soluble proteins and laterally to the membrane bilayer to permit integration of membrane proteins. The first structural insight into how a signal sequence could insert into the signal sequence–binding (SSB) site of Sec61 was provided by the structures of the SecYEG–SecA complex (Zimmer et al., 2008) and a SecYEG–Fab complex (Tsukazaki et al., 2008). The lateral gate of the translocation channel, which includes the SSB site, is formed by extensive side-chain contacts between residues in TM2, TM3, TM7, and TM8 (Van den Berg et al., 2004; Mandon et al., 2009). Binding of the ATP-bound conformation of SecA to SecYEG induces a partial separation of the lateral translocon gate so that an α-helical segment of the signal sequence could insert adjacent to TM2 and TM7 (Zimmer et al., 2008). Photo-cross-linking experiments showed that signal sequences can be cross-linked to TM2 and TM7, and hence this portion of the lateral gate serves as the SSB site (Plath et al., 1998). Covalently linking TM2 to TM7 via a disulfide between cysteine residues introduced into the lateral gate of Escherichia coli SecY blocks translocation of secretory protein substrates, thereby demonstrating that lateral gate separation is an essential event in protein translocation (du Plessis et al., 2009). TM7 and TM8, which form the C-terminal half of the lateral gate, are connected by a long lumenal loop (L7). The importance of L7 in translocon function is suggested by the observation that the sec61-3 allele corresponds to a G341E mutation (Wilkinson et al., 1997) at a highly conserved residue in L7.
A class of particularly informative SecY and Sec61 mutants cause the prl phenotype, which corresponds to enhanced translocation of proteins with defective signal sequences (Bankaitis and Bassford, 1985; Junne et al., 2007). It has been proposed that prl mutations promote translocation of signal-defective precursors by stabilizing the open conformation or destabilizing the closed conformation of the protein translocation channel (Van den Berg et al., 2004; Smith et al., 2005). This hypothesis is based on the observation that most mutations that cause the prl phenotype map to the pore ring, lateral gate, and plug domain of SecY. Biochemical evidence to support the conclusion that prl mutations destabilize the closed conformation of the translocation channel is scant, particularly in the case of the eukaryotic translocation channel. SecY prl mutants show enhanced interactions with the SecA ATPase and a reduced dependence on proton motive force for precursor transport (van der Wolk et al., 1998; de Keyzer et al., 2002).
Here we tested whether structure-perturbing mutations in L7 of Sec61p cause defects in cotranslational and posttranslational translocation. Mutations in L7 had a more severe impact on translocation of posttranslational substrates than cotranslational substrates. Defects in cotranslational translocation correlate with a delay in translocon gating, suggesting that L7 mutations interfere with the concerted movement of TM7 and TM8 during channel gating. The translocation defect of the sec61 L7 mutants could be suppressed by a panel of second-site mutations in Sec61p that cause the prl phenotype, thereby restoring the normal balance between the open and closed conformations of the protein translocation channel.
RESULTS
Mutations in L7 cause translocation defects
To screen for functionally important segments of L7, the hemagglutinin (HA)-epitope peptide (YPYDVPDYA) was inserted into the yeast Sec61 sequence directly after one of four residues (P315, I320, S340 or E354) to perturb the structure of L7 at surface exposed sites (Figure 1A). A plasmid shuffle procedure was used to replace wild-type Sec61 with the sec61 insertion mutants in a haploid yeast strain that lacks the nonessential Ssh1 translocon. The resulting strains were viable and do not display marked growth rate defects relative to the parental ssh1Δ strain. We chose to analyze newly constructed Sec61 mutants in an ssh1Δ background because Ssh1p acts as a bypass suppressor for sec61 alleles that cause defects in the cotranslational translocation pathway (Cheng et al., 2005).
FIGURE 1:
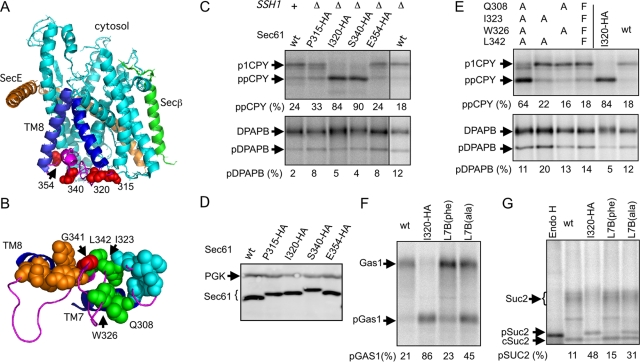
Mutations in loop 7 cause translocation defects. (A) HA-tag insertion sites (red spheres) map to exposed sites on the lumenal surface of yeast Sec61. TM7 and TM8 are blue; L7 is magenta; other segments of SecY are cyan. (B) Residues in group A (cyan; L310, L319, I320, and L322), B (green; Q308, I323, W326, and L342), and C (orange; Y344, Y345, D358, and K361) are shown as spheres on the ribbon diagram of residues 267–322 of M. jannaschii SecY. The sec61-3 (G341E) mutation is shown as a red sphere. (C) CPY and DPAPB were immunoprecipitated from pulse-labeled yeast cultures. The glycosylated ER forms of CPY (p1CPY) and DPAPB were resolved from nontranslocated precursors (ppCPY and pDPAPB) by SDS-PAGE. (D) Protein immunoblots of total-cell extracts using antisera for Sec61 and the cytosolic protein phosphoglycerate kinase (PGK), which served as a gel loading control. CPY (E), DPAPB (E), Gas1(F), and Suc2 (G) were immunoprecipitated from pulse-labeled cultures of wild-type and sec61 group B mutant yeast. All strains in E–G are ssh1Δ. Nontranslocated precursors (pGas1 and pSuc2) were resolved from cytoplasmic invertase (cSuc2) and glycosylated Gas1 and Suc2 by PAGE in SDS. Quantified values are the average of two or more experiments, one of which is shown here. A and B were created using MacPyMOL software (DeLano Scientific, Palo Alto, CA) using the M. jannaschii SecYEβ structure (PDB 1RHZ).
The sec61 L7 mutants were tested for defects in translocation of carboxypeptidase Y (CPY) and dipeptidylaminopeptidase B (DPAPB) by pulse labeling (Figure 1C). Posttranslational translocation of CPY through the heptameric Sec complex is detected by the N-glycosylation–induced gel mobility difference between the untranslocated precursor (prepro-CPY [ppCPY]) and the endoplasmic reticulum (ER) form of proCPY (p1CPY). The p1 form of CPY migrates as a doublet due to the presence of proCPY glycoforms that have three or four N-linked oligosaccharides. Defects in CPY translocation for the mutants ranged between undetectable (sec61 E354-HA) and severe (sec61 I320-HA and sec61 S340-HA) (Figure 1C). Pulse-chase experiments showed that the primary fate for ppCPY in the sec61 I320-HA cells was degradation rather than delayed translocation.
Cotranslational integration of DPAPB (Ng et al., 1996), which is mediated by Sec61 or Ssh1 heterotrimers, is detected by the acquisition of seven or eight N-linked oligosaccharides. Although deletion of Ssh1p causes a slight reduction in DPAPB integration as observed previously (Cheng et al., 2005), the HA-tag insertions into Sec61 do not cause an additional reduction in DPAPB integration (Figure 1C). The defect in posttranslational translocation of CPY is not explained by reduced expression of Sec61p by the L7 HA-tag insertion mutants (Figure 1D). As expected, the insertion of the HA tag reduced the SDS gel mobility of Sec61. Native immunoprecipitation experiments did not reveal any reduction in the assembly of Sec61 into the heptameric Sec complex that is responsible for translocation of CPY (Supplemental Figure S1).
To determine which structural features of L7 were perturbed by the HA-tag insertions, point mutations were introduced at selected residues in the vicinity of I320 or S340 (Figure 1B). A cluster of four aliphatic residues (group A, cyan spheres) in the vicinity of the I320 insertion site was replaced with alanine or phenylalanine. Four residues (group B, green spheres) that structurally link the L7 minihelix to TM7 were replaced with alanine (sec61 L7B(ala)) or phenylalanine (sec61 L7B(phe)). Four polar residues (group C, orange spheres) that link the L7 minihelix to TM8 via a hydrogen bond network were replaced with alanine. The sec61-3 mutation (G341E, red sphere) is adjacent to the group B cluster and the S340 insertion site.
Replacing the four group B residues with alanine (A), but not phenylalanine (F), caused a CPY translocation defect that was almost as severe as the I320-HA insertion (Figure 1E). Two interactions (W326 with Q308, and I323 with L342) in the group B cluster link TM7 to the L7 minihelix (Figure 1B). Alanine substitutions that eliminated only one of these contacts did not cause a translocation defect (Figure 1E). Likewise, mutagenesis of group A or C residues did not cause significant protein translocation defects (Supplemental Figure S2). We noticed that the reduction in CPY translocation caused by the sec61 L7B(ala) mutant was accompanied by an increase in the percentage of the hypoglycosylated variant of p1CPY. The reduction in CPY glycosylation may be a secondary consequence of the protein translocation defect that affects glycosylation of CPY but not DPAPB.
Additional substrates were then analyzed to determine whether the sec61 L7 mutants only affect the posttranslational translocation pathway. Posttranslational translocation of Gas1p (Ng et al., 1996) is accompanied by N-linked glycosylation, which causes a reduction in gel mobility relative to the untranslocated precursor (pGas1). The Gas1p precursor was the major form synthesized by the sec61 I320-HA mutant and was elevated in the sec61 L7B(ala) mutant (Figure 1F).
The secreted protein invertase (Suc2p) was selected as a second example of a protein that is translocated by a cotranslational pathway (Johnsson and Varshavsky, 1994). Suc2 translocation is less sensitive to depletion of SRP54 or SRα than DPAPB integration (Hann and Walter, 1991; Ogg et al., 1992). This is likely explained by redirection of Suc2 into a posttranslational translocation pathway when the SRP-targeting pathway is compromised (Mason et al., 2000). In ssh1Δ cells, cytoplasmic invertase (cSuc2) and core-glycosylated secretory invertase (Suc2) are the predominant forms of Suc2p detected after a brief pulse-labeling period. The nontranslocated precursor of secretory invertase (pSuc2) was detected in both the sec61 I320-HA and sec61 L7B(ala) mutants (Figure 1G).
Although the sec61 L7 mutations have a more severe impact on translocation of posttranslational substrates (CPY and Gas1p) than a cotranslational substrate (Suc2p), both translocation pathways are clearly affected. The observation that DPAPB translocation was apparently normal in the sec61 L7 mutants was interesting but not entirely unexpected, based on previous pulse-labeling experiments conducted using the sec61-2 and sec61-3 mutants at the permissive and restrictive temperatures (Rothblatt et al., 1989; Stirling et al., 1992). For both sec61-2 and sec61-3, DPAPB translocation defects are only manifested at the restrictive temperature and are less pronounced than translocation defects for CPY and prepro–α-factor. In contrast to the sec61-3 mutation (Stirling et al., 1992), the sec61 L7B(ala) mutation does not cause a restrictive growth defect at 18 or 37°C, nor does it cause an obvious reduction in Sec61p expression.
Intragenic suppressors of the sec61 L7B(ala) mutant
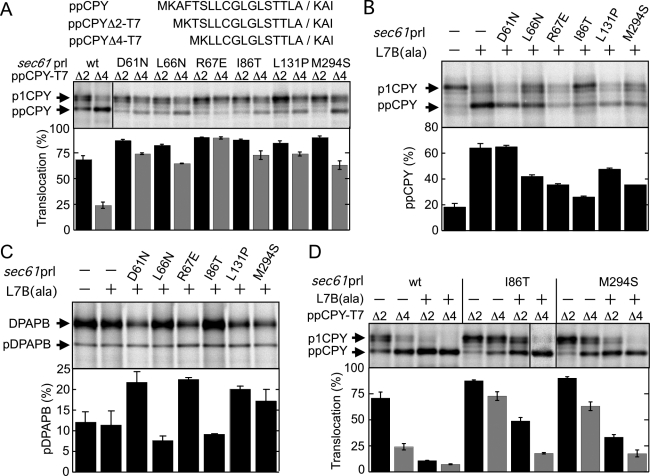
On the basis of the hypothesis that SecY prl alleles increase the open probability of the protein translocation channel (Smith et al., 2005), we decided to test whether sec61 prl alleles could act an intragenic suppressors of the sec61 L7B(ala) mutation. The sec61 prl alleles we tested as potential intragenic suppressors of the sec61 L7B(ala) mutation correspond to nonconservative substitutions in the pore ring (I86T, M294S), the lateral gate (L131P), or the plug domain (D61N, L66N, and R67E) of Sec61p (Junne et al., 2007). Except for sec61 M294S, these sec61 prl alleles were characterized previously using a CPY precursor that lacks three residues in the signal sequence (Junne et al., 2007). Here the sec61 prl alleles were analyzed using CPY derivatives that lack either two (ppCPYΔ2-T7) or four (ppCPYΔ4-T7) signal sequence residues as reporters (Figure 2A). The four-residue deletion reduces both the length and overall hydrophobicity of the signal sequence (Supplemental Figure S3A). As expected, ppCPYΔ4-T7 is a poor substrate for translocation through the wild-type Sec complex (Figure 2A). In contrast, translocation of CPYΔ4 ranged between 60 and 90% for the sec61 prl mutants (Figure 2A). CPYΔ2 serves as a sensitive reporter for the posttranslational translocation pathway; despite the two-residue deletion from the signal sequence, the majority of ppCPYΔ2 is translocated by wild-type Sec61p and all six sec61 prl mutants.
FIGURE 2:
Intragenic suppression of sec61 L7B(ala) by prl mutations. All strains are ssh1Δ. (A) The prl phenotype of sec61 mutants was assayed by pulse labeling using the ppCPYΔ2-T7 and ppCPYΔ4-T7 reporters. CPY (B) and DPAPB (C) were immunoprecipitated from pulse-labeled yeast strains and resolved by PAGE in SDS. (D) Translocation of ppCPYΔ2-T7 and ppCPYΔ4-T7 was assayed by pulse labeling. Percentage translocation (A, D) or percentage precursor (B, C) is the average of two or more determinations; error bars designate SD or the individual data points.
Double mutants (e.g., sec61 D61N L7B(ala)) were then constructed and tested for growth rate defects in the ssh1Δ background. As reported previously (Junne et al., 2007), most sec61 prl mutations do not cause a growth rate defect at 30 or 37°C (Supplemental Figure S4A). The sec61 R67E mutant grows more slowly than the parental strain at 37°C, but this growth defect was not aggravated by the presence of the L7B(ala) mutation. Three of the six tested sec61 prl L7B(ala) double mutants (sec61 D61N L7B(ala), L131P L7B(ala), and M294S L7B(ala)) grow more slowly than the parental strain (SEC61 ssh1Δ) at 30 and 37°C (Supplemental Figure S4A). Protein immunoblot analysis indicated that Sec61 levels were elevated severalfold in three of the double mutants (sec61 L7B(ala) combined with D61N, R67E, or L131P) and reduced in several others (sec61 L7B(ala) combined with L66N or I86T). Although the presence of higher levels of Sec61p in the slow-growing strains was unexpected, this result indicates that a reduction in Sec61p expression is not responsible for the growth defects.
The majority of the sec61 prl alleles reduced the CPY translocation defect caused by the sec61 L7B(ala) mutation (Figure 2B), with the strongest suppression shown by the two pore-ring substitutions (I86T and M294S). The slow-growing double mutants had slightly elevated levels (5–10%) of the DPAPB precursor (Figure 2C). The elevated levels of DPAPB precursor suggest that the reduction in growth rate is explained by a general defect in protein translocation. The reduced incorporation of radiolabel into proteins by the slow-growing strains (e.g., sec61 D61N L7B(ala)) is expected, as ribosome biosynthesis in yeast is coordinately regulated by protein flux through the secretory pathway (Mizuta and Warner, 1994).
We next asked whether the prl phenotype was retained by the sec61 L7B(ala) I86T and sec61 L7B(ala) M294S mutants (Figure 2D). In both cases, the double mutants showed reduced translocation of ppCPYΔ2 and ppCPYΔ4 relative to the wild-type strain or to the single sec61 prl mutants (Figure 2A), indicating that the prl phenotype was completely suppressed by the L7B(ala) mutation.
Comparison of the sec61-3, sec61 R406E, and sec61 L7B(ala) mutants
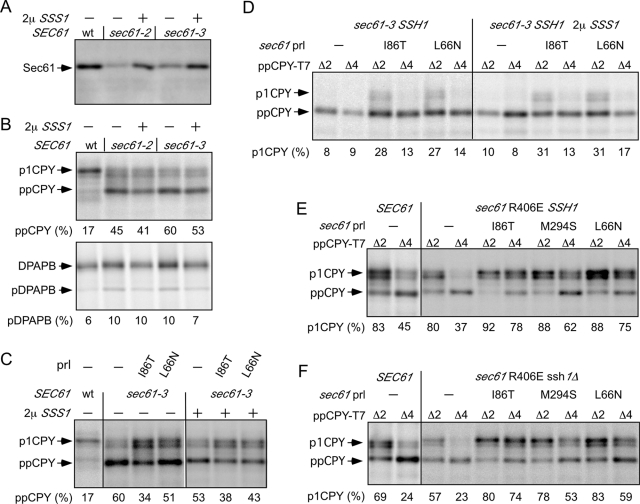
The translocation defects and temperature sensitivities of the sec61-2 and sec61-3 mutants are believed to be explained by degradation of Sec61p, particularly at the restrictive temperature (Sommer and Jentsch, 1993; Biederer et al., 1996). The sec61-2 (G213D; Nishikawa et al., 2001) and sec61-3 (G341E; Wilkinson et al., 1997) alleles were tested in the SSH1 background. Overexpression of the Sss1p subunit of the yeast Sec61 complex suppresses the temperature-sensitive lethality of the sec61-2 and sec61-3 mutants by enhancing the stability of Sec61p (Esnault et al., 1993, 1994). The lower cellular content of Sec61p in the sec61-2 and sec61-3 mutants was largely corrected by overexpression of Sss1p (Figure 3A). Pulse-labeling experiments conducted at 30°C revealed significant defects in CPY translocation for both sec61-2 and sec61-3 but essentially normal translocation of DPAPB (Figure 3B). Overexpression of Sss1p did not correct the defect in CPY translocation (Figure 3B), indicating that the G213D (sec61-2) and G341E (sec61-3) mutations reduce both the activity and stability of the Sec complex.
FIGURE 3:
Intragenic suppression of sec61-3 by prl mutations. Wild-type and sec61 mutants in the SSH1 (A–E) or ssh1Δ (F) backgrounds were transformed with a high-copy SSS1 plasmid as indicated and grown at 30°C. (A) Protein immunoblots of total-cell extracts using antisera for Sec61p. CPY (B, C) and DPAPB (B) were immunoprecipitated from pulse-labeled yeast strains and resolved by PAGE in SDS. (D–F) Translocation of ppCPYΔ2-T7 and ppCPYΔ4-T7 was assayed by pulse labeling. Percentage precursor (B, C) or percentage translocation (D–F) is the average of two or more determinations, one of which is shown here.
Two representative prl alleles (L66N and I86T) were combined with the sec61-3 mutant and tested as intragenic suppressors of the CPY translocation defect (Figure 3C). The pore-ring mutation (I86T) was a more effective suppressor than the plug-domain mutation (L66N). The combination of a prl mutation and Sss1p overexpression caused at best an additive improvement in CPY translocation. The sec61-3, sec61-3 I86T, and sec61-3 L66N mutants are severely defective in translocation of the prl reporters (Figure 3D) relative to the SEC61SSH1 control strain.
As a control for these experiments, we tested whether the prl phenotype is suppressed by a mutation (sec61 R406E) that interferes with ribosome binding to Sec61, thereby causing a defect in the cotranslational translocation pathway (Cheng et al., 2005). The sec61 R406E mutation was combined with prl alleles (L66N, I86T, or M294S) in both the SSH1 and ssh1Δ backgrounds. In the presence of the Ssh1p translocon, the sec61 R406E strain grows normally and lacks a detectable translocation defect (Cheng et al., 2005). All three double mutants (e.g., sec61 R406E I86T SSH1) showed more efficient translocation of ppCPYΔ4 than the wild-type strain (Figure 3E). When the sec61 R406E ssh1Δ strain is shifted from nutrient-poor media (synthetic ethanol glycerol, using ethanol and glycerol as carbon sources) into media containing dextrose as a carbon source (synthetic defined [SD] media with dextrose), the stain displays a transient yet severe defect in translocation of both DPAPB and CPY (Cheng et al., 2005). After undergoing an adaptation process (Mutka and Walter, 2001), which includes a fourfold reduction in growth rate, the sec61 R406E ssh1Δ strain displays relatively efficient translocation of both DPAPB and CPY (Cheng et al., 2005). Here we tested translocation of the prl reporters in double-mutant strains that had undergone the adaptation process (Figure 3F). Despite the slow growth rate, the double mutant strains all showed efficient translocation of the prl reporters.
Stabilization of double mutants by overexpression of Sss1p
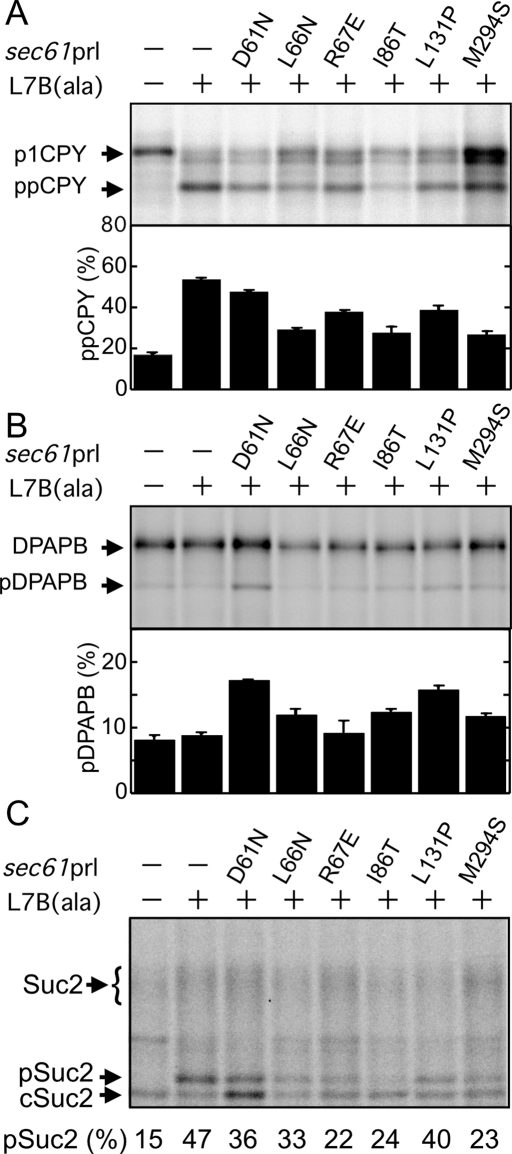
Suppression of the CPY translocation defect of the sec61 L7B(ala) mutant by three of the sec61 prl alleles (R67E, L131P, and M294S) could conceivably be explained by a reduction in precursor flux due to a reduced growth rate at 30°C (for L131P and M294S) or by elevated expression of Sec61p itself (for R67E and L131P). To experimentally address these possibilities, the sec61 L7B(ala) mutant and the sec61 L7B(ala) prl double mutants were transformed with a high-copy plasmid encoding Sss1.
In the presence of excess Sss1p, five of the double mutants (sec61 L7B(ala) combined with L66N, R67E, I86T, L131P, or M294S) had normal growth rates at 30°C (Supplemental Figure S4C). Protein immunoblot analysis indicated that differences in Sec61 content varied less between strains when Sss1p was overexpressed (compare Supplemental Figure S4, B and D). If a reduction in growth rate or an elevation in Sec61p levels is responsible for the ability of the R67E, L131P, and M294S prl alleles to act an intragenic suppressors, we should observe less effective suppression by these alleles in the presence of Sss1p. Instead, all of the tested sec61 prl alleles including D61N were able to partially suppress the CPY translocation defect caused by the sec61 L7B(ala) mutation when excess Sss1p was present (Figure 4A). The three strongest intragenic suppressors correspond to polar amino acid substitutions in the pore ring (I86T and M294S) or the plug domain (L66N). Two of the double mutants (sec61 L7B(ala) combined with D61N or L133P) still showed a minor increase (5–7%) in nonintegrated DPAPB relative to the control strains (Figure 4B). We next asked whether any of the prl mutations could suppress the defect in invertase translocation of the sec61 L7B(ala) mutant (Figure 4C). Several prl alleles (I86T, M294S, and R67E) caused substantial reductions in the accumulation of the invertase precursor. The two double mutants that had a residual defect in DPAPB integration (sec61 L7B(ala) combined with D61N or L131P; Figure 4B) showed the least improvement in invertase translocation relative to sec61 L7B(ala). None of the sec61 double mutants retained an enhanced ability to translocate the prl reporters even when Sss1p was overexpressed (Supplemental Figure S3B).
FIGURE 4:
Overexpression of Sss1p improves the translocation activity of the double-mutant strains. Yeast strains analyzed in Figure 2 were transformed with a high-copy plasmid encoding Sss1p. Translocation of CPY (A), DPAPB (B), and invertase (C) was assayed by pulse labeling. Values shown are the average of two determinations; error bars (A, B) designate individual data points.
Slower translocon gating by sec61 L7 mutants
A translocon-gating assay using the Suc2 series of ubiquitin translocation assay (UTA) reporters (Johnsson and Varshavsky, 1994) was used to compare the in vivo gating kinetics of wild-type and sec61 L7 mutant translocons. The translocon gating assay measures the time required in vivo for the protein translocon to be gated by a ribosome-nascent chain complex (Figure 5B). Rapid folding of the ubiquitin domain in the cytosol allows cleavage of the reporter by a cytosolically localized, ubiquitin-specific protease (UbP) to release the Ura3-HA (U-HA) segment of the reporter (Figure 5B). If translocation of the precursor into the ER lumen (e.g., opening of the lateral and lumenal translocon gates) initiated before the ubiquitin segment emerges from the polypeptide exit tunnel on the large ribosomal subunit, the intact reporter will be translocated into the ER lumen. Increasing the length of the spacer between the signal sequence and the ubiquitin domain (Figure 5A) provides additional time for translocon gating prior to emergence of the Ub domain from the large ribosomal subunit. Quantification of the cleaved and uncleaved forms of the UTA reporters can be used to monitor the in vivo kinetics of translocon gating (Cheng and Gilmore, 2006; Jiang et al., 2008).
FIGURE 5:
L7 mutations cause a delay in translocon gating that is suppressed by the I86T mutation. (A) The Suc2 series of UTA reporters consist of 1) the N-terminal signal sequence of Suc2p (black, Suc2p1–19); 2) 14- to 296-residue spacer segments (cyan); 3) a Ub domain (red); 4) a 42-residue linker (blue) with a processing site (arrowhead) for a UbP; and 5) a Ura3 reporter domain followed by a triple-HA tag (yellow). (B) After RNCs dock onto the Sec61 complex, the UTA reporter is cleaved if the Ub domain folds in the cytosol but remains intact if translocon gating occurs before the Ub domain emerges from the polypeptide exit tunnel on the large ribosomal subunit. (C, E) In vivo cleavage of the Suc2 (C) and Dap2 (E) reporters. Labels designate the intact reporter (e.g., 23) and cleaved (U-HA) reporter domain. (D, F) Spacer-length dependence of Suc2 (D) and Dap2 (F) reporter cleavage (percentage cytosolic Ura3-HA) in wild-type and mutant strains was calculated after quantification of intact and cleaved forms of the reporters. Symbols are the averages of two or three experiments for each strain as noted, with color-coded error bars designating either the SD or the individual data points.
Cleavage of the Suc2 series of UTA reporters decreased rapidly as the spacer length was increased from 14 to 60 residues (Figure 5C). Quantification of the intact (e.g., Suc2-296) and the cleaved (U-HA) forms of the Suc2 UTA reporter yields the spacer length dependence of cleavage (Figure 5D). A biphasic curve is obtained that consists of a gating window and a plateau value. In wild-type cells the majority of Suc2-RNCs gate the Sec61 channel 12–16 s after the signal sequence has emerged from the large ribosomal subunit (gating time = spacer length [23–60 residues] + Ub domain [76 residues)] divided by the protein synthesis elongation rate [∼8 residues/s; Jiang et al., 2008]). Mutations in the SRP or SRP receptor cause a marked elevation in the plateau value, indicating a reduction in precursor flux through the cotranslational pathway. For example (Supplemental Figure S5A), <40% of Suc2-296 RNCs cotranslationally gate the Sec61 complex when the RNC-targeting pathway is impaired by a mutation in the SRP receptor (srp102 K51I; Ogg et al., 1998).
The in vivo kinetics of cotranslational translocation is strikingly different in the sec61 I320-HA mutant, as revealed by significantly reduced levels of intact reporter for all constructs with short and intermediate-length spacers (Figure 5C). Translocon gating in the sec61 I320HA mutant occurs when the spacers are longer, corresponding to a 12-s delay in the average time required for gating of the translocon by a Suc2 RNC (Figure 5D). The sec61 L7B(ala) mutant showed an intermediate length of delay in translocon gating relative to wild-type cells (Figure 5, C and D), which was consistent with the less severe defect in cotranslational translocation of Suc2 (Figure 1G).
Do the sec61 L7 mutations cause a general defect in translocon gating, or is the delay in translocon gating specific for a secretory protein like Suc2? To address this question, we conducted the translocon gating assay using the Dap2 series of UTA reporters (Cheng and Gilmore, 2006) that are derived from DPAPB. As observed previously (Cheng and Gilmore, 2006), most Dap2-RNCs gate the translocon within 22 s after the Dap2 TM span emerges from the large ribosomal subunit (103 residue spacer + 76 residues Ub domain divided by 8 residues/s). Translocon gating by Dap2-RNCs is delayed in the sec61 L7B(ala) mutant, albeit to a lesser extent than observed for Suc2-RNCs (Figure 5, E and F).
We next asked whether the prl mutations suppress the translocation defect of the sec61 L7B(ala) mutant by restoring normal translocon gating kinetics. For this experiment we selected the sec61 I86T L7B(ala) mutation because the I86T allele was the most effective suppressor of the sec61 L7B(ala) mutation as detected by pulse labeling of CPY (Figure 2B). Remarkably, the spacer-length dependence of Suc2 UTA reporter cleavage resembled that of the SEC61 ssh1Δ strain and lacked the gating lag that was displayed by the sec61 L7B(ala) ssh1Δ strain (Figure 5D). Together, the results using the Suc2 UTA and ppCPYΔ4 reporters indicate that the contrasting consequences of the L7B(ala) mutation and the I86T mutation are mutually exclusive and are attenuated when the two mutations are combined.
It has been proposed that SecY prl alleles enhance translocation of prl reporters by changing the equilibrium between the closed and open conformations of the protein translocation channel (Smith et al., 2005). If the transition between the open and the closed conformations of the Sec61 heterotrimer is the primary rate-limiting step in the protein translocation reaction in a wild-type cell, one might expect that Suc2-RNCs would gate the translocon more rapidly in the sec61 I86T mutant. However, if one of the preceding reaction steps in the translocation pathway is rate limiting, one would predict that the translocon gating would not be altered by the prl mutation. Indeed, a translocon gating assay of the sec61 I86T strain did not reveal an additional increase in gating rate but instead showed an elevation in the plateau value that was partially reversed by overexpression of Sss1p (Supplemental Figure S5B). Thus an earlier reaction step in the cotranslational targeting pathway is rate limiting in wild-type cells, whereas the translocon gating event is rate limiting in the sec61 L7B(ala) mutant.
DISCUSSION
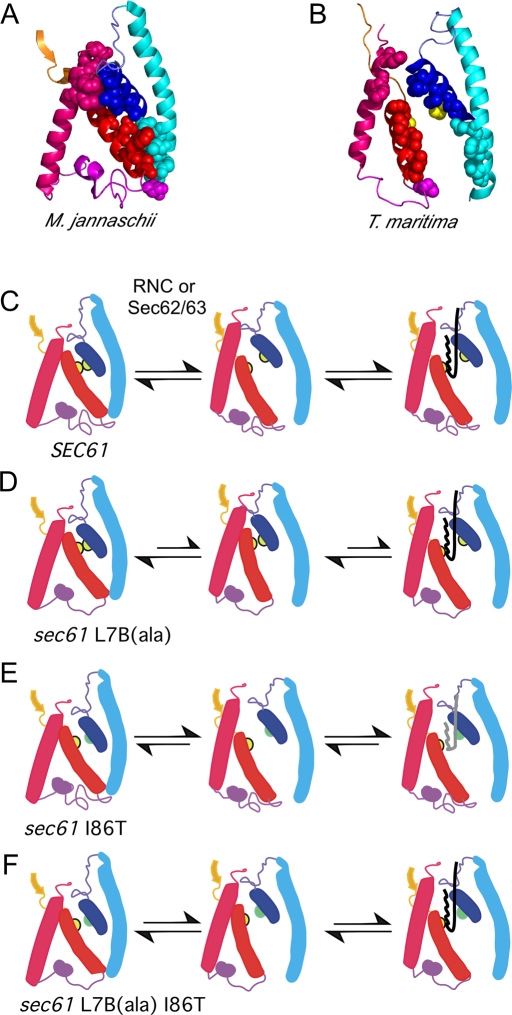
The protein translocation channel is believed to undergo a transition between the resting or closed conformation and an open conformation to allow signal sequence insertion into the SSB site (Figure 6). The conformational change in the vicinity of the SSB site can be best appreciated by examining the structures of translocon lateral gates. A comparison of the closed conformation (Figure 6A) and a partially open conformation (Figure 6B) of prokaryotic translocation channels suggests that the separation of the lateral gate occurs by a rigid-body movement of TMs 6–10 relative to TMs 1–5 (Zimmer et al., 2008). In the case of yeast Sec61, binding of an RNC to the cytosolic face of Sec61 or a presecretory protein to the Sec62/Sec63 complex is proposed to initiate lateral gate opening (Figure 6C). The molecular mechanism of eukaryotic translocation channel gating by cytosolic effectors (RNCs or Sec62/Sec63 complex) has not been elucidated. Here we found that mutations in lumenal loop 7 of Sec61 cause a lag in translocon gating that is diagnostic of delayed opening of the lateral and lumenal translocon gates. The mutations introduced into loop 7 are predicted to break side-chain contacts that link TM7 to TM8 via the minihelix in lumenal loop 7. Weakening of these contacts likely permits uncoordinated movement of TM7 and TM8 during channel gating, thereby delaying signal sequence insertion into the SSB site at the interface between TM2 and TM7 (Figure 6D).
FIGURE 6:
Counteracting effects of the sec61 L7B(ala) and sec61 I86T mutations. (A) Closed conformation of the lateral gate of the M. jannaschii SecY and (B) partially open conformation of the lateral gate of the Thermotoga maritima SecYEG–SecA complex are shown as ribbon diagrams. Lateral gate contact residues are shown as color-coded spheres; pore-ring residues in T. maritima SecY (I86 and I274) are shown as yellow spheres; all other side chains are hidden. The regions shown are TM2 (blue), L2 (slate), TM3 (cyan), a portion of L6 (orange), TM7 (red), L7 (magenta), and TM8 (pink). (C–F) Color-coded models of wild-type and mutant alleles of yeast Sec61p showing closed, partially open, and SSB-occupied conformations of the lateral gate. Pore-ring residues (I86 and M294) are shown as yellow (wild type) or green (I86T) circles. Wild-type (black) and mutant (gray) signal sequences are depicted in the SSB site between TM2 and TM7.
Our results with the sec61 L7 mutants suggest a general mechanism for how many deleterious sec61 alleles reduce protein translocation activity. The sec61 L7 mutations had a more severe defect in translocation of two posttranslational substrates (CPY and Gas1p) than a cotranslational substrate (Suc2). These pathway-dependent differences in defect severity are reminiscent of several previously isolated sec61 alleles, including sec61-2, sec61-3, sec61-41, sec61-87, sec61-32, and sec61-24. The sec61-2 mutation (G214E; Nishikawa et al., 2001) maps to an invariant glycine residue between TM5 and TM6 that is proposed to be within a flexible hinge that permits channel opening (Van den Berg et al., 2004; Gumbart and Schulten, 2007). Consistent with the location of G341 adjacent to the sec61 L7B(ala) patch mutation (Figure 1B), we observed that sec61-3 causes a translocation defect at the permissive temperature that is not explained by reduced Sec61p expression. The sec61-41, sec61-87, sec61-32, and sec61-24 mutations map to residues in TM3, L3, and TM4 (Pilon et al., 1998) adjacent to the N-terminal side of the lateral gate. Many of these classic Sec61 mutations may interfere with the structural transition between the closed and open conformations of Sec61. The less severe impact of L7 mutations on substrates that use the cotranslational pathway is explained by retention of the nascent polypeptide in the immediate vicinity of the translocon pore by contact between the large ribosomal subunit and the cytosolic loops of Sec61. Although translocon gating assays using the Dap2 series of UTA reporters revealed a delay in channel gating, the delay was not as great as observed using the Suc2 series of reporters. This differential delay in gating may explain why the sec61 L7 mutations have little or no impact on translocation of DPAPB.
Smith et al. (2005) proposed that E. coli prl mutations favor the transition to the open conformation of the protein translocation channel by destabilizing the pore ring or the plug domain of SecY. Destabilization of the closed conformation of the channel would allow enhanced insertion of a precursor with a signal sequence mutation (Figure 6E). Molecular dynamics simulations suggest that prl mutations enhance the mobility of the plug domain and increase water penetration into the vicinity of the pore ring (Bondar et al., 2010). It is important to stress that a prl mutation does not “lock” the channel in an open conformation but instead reduces the energetic barrier to channel opening. Our results provide biochemical evidence that support the hypothesis that Sec61 prl mutations act by altering the transition between the closed and open conformations of the protein translocation channel.
Two E. coli SecY prl alleles (prlA4 and prlA6) carry two point mutations in SecY (Smith et al., 2005). The most thoroughly analyzed double mutant is SecY prlA4, which has a polar substitution in a pore-ring residue (I408N) and a second mutation (F286Y) in TM7 that suppresses the lethality caused by the I408N mutation (Duong and Wickner, 1999). The pore-ring mutation is entirely responsible for the prl phenotype, whereas the F286Y mutation causes a severe protein translocation defect if the I408N mutation is not present (Sako and Iino, 1988). Further analysis indicates the F286Y mutation causes a defect in SecA-dependent translocon gating that partially suppresses the strong prl phenotype caused by the I408N mutation (de Keyzer et al., 2002). Thus the prlA4 allele provides precedence for phenotypic suppression by counteracting mutations in a protein translocation channel that impact the transition between the open and closed conformations. Unlike the SecY prlA4, the double mutants we analyzed did not retain the ability to translocate precursors with signal sequence mutations (Figure 6F). In contrast, the sec61 R406E mutation did not suppress, nor was it suppressed by, prl mutations in either the SSH1 or ssh1Δ background, consistent with the evidence that the sec61 R406E mutation interferes with ribosome binding.
The sec61 prl alleles we tested all permitted enhanced translocation of ppCPYΔ4 (Figure 2A). However, the prl alleles showed significant differences in their ability to counteract the L7B(ala) mutation. The combination of the sec61 L7B(ala) mutation and certain prl alleles yielded slow-growing yeast strains that express excess Sec61p. The slow growth rates may be explained by the presence of mutations that destabilize both the open (L7B(ala)) and closed (R67E) conformations of Sec61, thereby reducing assembly of the mutant protein into active translocation channels, particularly at 37°C. This hypothesis is supported by the improvement in growth rate and translocation activity that is afforded upon overexpression of Sss1p. The prl mutations (R67E and D61N) that caused the most severe synthetic growth defects when combined with the L7B(ala) mutation are believed to destabilize the plug domain of Sec61.
Remarkably, the two strongest suppressors (I86T and M294S) of the L7B(ala) mutation are pore-ring residues located in the two lateral gate TM spans (TM2 and TM7) that form the SSB site. We aligned 120 diverse eukaryotic Sec61 and Ssh1 sequences and found that polar amino acid residues are not among observed substitutions at the pore-ring residues in TM2 (I86) and TM7 (M294). Although the I86T and M294S mutations enhance translocation efficiency, these mutations reduce translocation fidelity by reducing the hydrophobicity threshold for sequences that can target a protein to the yeast posttranslational translocation pathway. The balance between translocation efficiency and signal sequence recognition fidelity appears to be maintained by regulating the transition between the open and closed conformations of the protein translocation channel.
The location of the two strongest suppressors relative to the lateral gate suggests that the sec61 L7B(ala) mutation interferes with the separation of TM2 and TM7 to form an open SSB site. The translocon gating assay does not measure a single reaction step, but instead monitors a series of sequential events, including SRP recognition of the signal sequence, RNC binding to the Sec61 complex, lateral and lumenal gate opening, signal sequence insertion into the SSB site, and finally Ub-domain insertion into the transport pore. Mutations in L7 retard the gating kinetics, indicating that one of the later reactions steps has become strongly rate limiting.
The improved translocation activity of the sec61 L7B(ala) I86T mutant is explained by restoration of normal translocon gating kinetics (Figure 5), indicating that the transition between the open and closed conformations of the channel is now more similar to that for wild-type Sec61 (Figure 6F). The results of this study provide insight into how a protein translocation channel makes the transition between the closed, resting conformation and an open conformation that can accommodate a signal sequence in the SSB site.
MATERIALS AND METHODS
Plasmid and strain construction
Standard yeast media (YPAD [yeast extract, bactopeptone, adenine, and dextrose], YPAEG [yeast extract, bactopeptone, adenine, ethanol, and glycerol], and SD [synthetic defined media with dextrose]), supplemented as noted, were used for growth and strain selection (Sherman, 1991). Oligonucleotides encoding HA (YPYDVPDYA)-epitope insertions or amino acid substitutions were used as primers together with the template plasmid pBW11 (Wilkinson et al., 1996) in recombinant PCR reactions to produce the L7 sec61 mutants. The L7 sec61 mutants were characterized in yeast strains that are SSH1 (BWY12) or ssh1Δ (RGY400; Cheng et al., 2005). A plasmid shuffle procedure (Sikorski and Boeke, 1991) was used to replace the plasmid pBW7 (URA3 SEC61) with the LEU2-marked plasmids encoding the L7 sec61 mutants. BWY12 and RGY400 were transformed, and Leu+ prototrophs were selected on SD (synthetic defined media with dextrose) plates supplemented with adenine, tryptophan, and uracil. Several transformants for each sec61 mutant were streaked onto plates containing 5-fluoro-orotic acid and grown for 2 d at 30°C to select for colonies that had lost the pBW7.
The ppCPYΔ2-T7 and ppCPYΔ4-T7 reporters were constructed using recombinant PCR using the yeast PCR1 gene as a template. DNA encoding the T7 epitope tag was inserted in frame before the stop codon. DNA encoding the SSS1 gene was amplified by PCR and cloned into the BamHI/SacI–digested pRS424 or pRS426 to obtain pEM665 and pEM662, respectively.
Immunoprecipitation of radiolabeled proteins
Yeast were grown to midlog phase (0.4–0.6 A600 units) at 30°C in SD media, collected by centrifugation, resuspended in fresh SD media at a concentration of 4 A600 units/ml, and allowed to recover at 30°C for 10 min. For the strain containing the SRP receptor mutation (srp102 K51I), yeast were initially grown to midlog phase (0.4–0.6 A600 units) at 25°C in SD media and then shifted to 37°C for 3 h before being collected, resuspended in fresh SD media and allowed to recover at 37°C prior to pulse labeling. To induce invertase (Suc2p) expression, 4 A600 units of cells were collected by centrifugation and resuspended in 5 ml of SD media containing 0.1% dextrose and incubated for 30 min at 30°C. Cells were pulse labeled for 7 min with 100 µCi of Tran-35S-label/A600. When indicated, cells were pretreated for 30 min with tunicamycin (10 μg/ml) before pulse labeling. All cell labeling, lysis, and subsequent immunoprecipitation of yeast proteins was performed as described previously (Jiang et al., 2008). The prl reporters (ppCPYΔ2-T7 and ppCPYΔ4-T7) were immunoprecipitated using antisera specific for the T7 epitope tag (Covance, Berkeley, CA). Immunoprecipitated proteins were resolved by PAGE in SDS and quantified using Molecular Imager FX (Bio-Rad Laboratories, Hercules, CA) or Fluorescent Image Analyzer FLA-5000 (FujiFilm, Tokyo, Japan). Ubiquitin translocation assays using the Suc2 or Dap2 series of UTA reporters were quantified as described previously (Cheng and Gilmore, 2006).
Supplementary Material
Acknowledgments
We thank Randy Schekman (University of California, Berkeley) for providing antisera to Sec61p, Sec63p, and Suc2. This work was supported by National Institutes of Health Grant GM35687.
Abbreviations used:
- CPY
carboxypeptidase Y
- DPAPB
dipeptidylaminopeptidase B
- ER
endoplasmic reticulum
- HA tag
hemagglutinin epitope tag
- p1CPY
endoplasmic reticulum form of carboxypeptidase
- ppCPY
prepro-carboxypeptidase
- RNC
ribosome-nascent chain
- SRP
signal recognition particle
- SSB
signal sequence binding
- TM
transmembrane span
- Ub
ubiquitin
- UbP
ubiquitin-specific protease
- U-HA
Ura3-hemagglutinin segment
- UTA
ubiquitin translocation assay
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0070) on July 7, 2011.
REFERENCES
- Bankaitis VA, Bassford PJ., Jr Proper interaction between at least two components is required for efficient export of proteins to the Escherichia coli cell envelope. J Bacteriol. 1985;161:169–178. doi: 10.1128/jb.161.1.169-178.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Bondar AN, del Val C, Freites JA, Tobias DJ, White SH. Dynamics of SecY translocons with translocation-defective mutations. Structure. 2010;18:847–857. doi: 10.1016/j.str.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Gilmore R. Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat Struct Mol Biol. 2006;13:930–936. doi: 10.1038/nsmb1146. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon EC, Gilmore R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keyzer J, van der Does C, Swaving J, Driessen AJ. The F286Y mutation of PrlA4 tempers the signal sequence suppressor phenotype by reducing the SecA binding affinity. FEBS Lett. 2002;510:17–21. doi: 10.1016/s0014-5793(01)03213-6. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 1999;18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault Y, Blondel M-O, Deshaies R, Schekman R, Képes F. The yeast SSS1 gene is essential for secretory protein translocation and encodes a conserved protein of the endoplasmic reticulum. EMBO J. 1993;12:4083–4093. doi: 10.1002/j.1460-2075.1993.tb06092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault Y, Feldheim D, Blondel M-O, Schekman R, Képes F. SSS1 encodes a stabilizing component of the Sec61 subcomplex of the yeast protein translocation apparatus. J Biol Chem. 1994;269:27478–27485. [PubMed] [Google Scholar]
- Finke K, Plath K, Panzer S, Prehn S, Rapoport TA, Hartmann E, Sommer T. A second trimeric complex containing homologues of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Gumbart J, Schulten K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Cheng Z, Mandon EC, Gilmore R. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J Cell Biol. 2008;180:1149–1161. doi: 10.1083/jcb.200707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1994;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- Mandon EC, Trueman SF, Gilmore R. Translocation of proteins through the Sec61 and SecYEG channels. Curr Opin Cell Biol. 2009;21:501–507. doi: 10.1016/j.ceb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetret JF, Hegde RS, Aguiar M, Gygi SP, Park E, Rapoport TA, Akey CW. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetret JF, et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Mizuta K, Warner JR. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol Biol Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DTW, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S-I, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Barz WP, Walter P. A functional GTPase domain, but not its transmembrane domain, is required for function of the SRP receptor b-subunit. J Cell Biol. 1998;142:341–354. doi: 10.1083/jcb.142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Poritz MA, Walter P. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Pilon M, Romisch K, Quach D, Schekman R. Sec61p serves multiple roles in secretory precursor binding and translocation into the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:3455–3473. doi: 10.1091/mbc.9.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako T, Iino T. Distinct mutation sites in prlA suppressor mutant strains of Escherichia coli respond either to suppression of signal peptide mutations or to blockage of staphylokinase processing. J Bacteriol. 1988;170:5389–5391. doi: 10.1128/jb.170.11.5389-5391.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:1–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned genes to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- van der Wolk JP, Fekkes P, Boorsma A, Huie JL, Silhavy TJ, Driessen AJ. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 1998;17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson BM, Critchley AJ, Stirling CJ. Determination of the transmembrane topology of yeast Sec61p; an essential component of the ER translocation complex. J Biol Chem. 1996;271:25590–25597. doi: 10.1074/jbc.271.41.25590. [DOI] [PubMed] [Google Scholar]
- Wilkinson BM, Esnault Y, Craven RA, Skiba F, Fieschi J, Képès F, Stirling CJ. Molecular architecture of the ER translocase probed by chemical crosslinking of Sss1p to complementary fragments of Sec61p. EMBO J. 1997;16:4549–4559. doi: 10.1093/emboj/16.15.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke S, Dunnwald M, Albertsen M, Johnsson N. Recognition of a subset of signal sequences by Ssh1p, a Sec61p-related protein in the membrane of endoplasmic reticulum of yeast Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2223–2232. doi: 10.1091/mbc.01-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.