The findings presented indicate that polyamine-regulated CUG-binding protein 1 and microRNA-222 modulate cyclin-dependent kinase 4 (CDK4) translation at least in part by altering the recruitment of CDK4 mRNA to processing bodies.
Abstract
The amino acid–derived polyamines are organic cations that are essential for growth in all mammalian cells, but their exact roles at the molecular level remain largely unknown. Here we provide evidence that polyamines promote the translation of cyclin-dependent kinase 4 (CDK4) by the action of CUG-binding protein 1 (CUGBP1) and microRNA-222 (miR-222) in intestinal epithelial cells. Both CUGBP1 and miR-222 were found to bind the CDK4 mRNA coding region and 3′-untranslated region and repressed CDK4 translation synergistically. Depletion of cellular polyamines increased cytoplasmic CUGBP1 abundance and miR-222 levels, induced their associations with the CDK4 mRNA, and inhibited CDK4 translation, whereas increasing the levels of cellular polyamines decreased CDK4 mRNA interaction with CUGBP1 and miR-222, in turn inducing CDK4 expression. Polyamine-deficient cells exhibited an increased colocalization of tagged CDK4 mRNA with processing bodies; this colocalization was abolished by silencing CUGBP1 and miR-222. Together, our findings indicate that polyamine-regulated CUGBP1 and miR-222 modulate CDK4 translation at least in part by altering the recruitment of CDK4 mRNA to processing bodies.
INTRODUCTION
The natural polyamines (spermidine, spermine, and their precursor putrescine) are organic cations found in all eukaryotic cells and are implicated in the control of multiple signaling pathways and distinct cellular functions (Casero and Pegg, 2009; Eisenberg et al., 2009). The intracellular polyamine concentrations are maintained at a cell type–specific level that depends on the dynamic balance among polyamine biosynthesis, degradation, and transport (Gerner and Meyskens, 2004). Cellular polyamine content increases rapidly in cells stimulated to grow and divide, whereas decreasing cellular polyamines stops cell-cycle progression and causes growth arrest in the G1 phase (Gerner and Meyskens, 2004; Casero and Marton, 2007; Persson, 2009). Since the recognition that polyamines are absolutely required for mammalian cell growth, the targeting of their function and metabolism has been an attractive strategy for antiproliferative therapy (Casero and Marton, 2007; Palmer and Wallace, 2010). Previous studies from our laboratory (Wang and Johnson, 1991; Wang et al., 1991; Li et al., 2002; Xiao et al., 2007, 2010; Liu et al., 2009; Zou et al., 2010) and others (Ray et al., 2003; Seiler and Raul, 2007) show that, in normal intestinal mucosa, growth and repair after injury require the supply of polyamines to the dividing cells in the crypts, while reducing cellular polyamines by inhibiting ornithine decarboxylase (ODC), the rate-limiting enzyme in polyamine biosynthesis (Gerner and Meyskens, 2004), represses intestinal epithelial cell (IEC) proliferation and delays wound healing in vivo and in vitro.
Posttranscriptional processes, particularly altered mRNA turnover and translation, are major mechanisms by which mammalian cells control gene expression in response to various stressful stimuli (Lackner and Bahler, 2008; Shen and Pili, 2008). Changes in mRNA stability and translation are governed by two types of trans-acting factors that directly interact with the mRNA: RNA-binding proteins (RBPs) and noncoding RNAs such as microRNAs (miRNAs) (Bolognani and Perrone-Bizzozero, 2008; Bartel, 2009). RBPs and miRNAs bind to cis-elements on the mRNA, frequently at the 3′-untranslated regions (3′-UTRs), and regulate stability and translation rates of target transcripts (Baltimore et al., 2008; Shen and Pili, 2008; Jackson et al., 2010). The CUG-binding protein 1 (CUGBP1) is a novel RBP that binds to given mRNAs through GU-rich elements (GREs) commonly found in the 3′-UTRs (Vlasova et al., 2008; Sen et al., 2009). The interaction of CUGBP1 with its target mRNAs enhances mRNA decay and represses translation of some target transcripts (Vlasova and Bohjanen, 2008; Zhang et al., 2008), although in some instances CUGBP1 promotes mRNA translation (Iakova et al., 2004). Recently, several studies showed that RBPs and miRNAs can function jointly in the regulation of a shared target mRNA (Abdelmohsen et al., 2008; Kim et al., 2009). In this regard, the RBP HuR is shown to recruit let-7/RISC to repress c-Myc mRNA expression (Kim et al., 2009), whereas the RBP Dnd-1 (dead end 1) inhibits miRNA access to target mRNA (Kedde et al., 2007).
Cyclin-dependent kinase 4 (CDK4) controls how cells enter into and are committed to progress through the cell cycle by phosphorylating, and thereby inhibiting the activity of, retinoblastoma protein (Rb) (Reed, 1997; Tsutsui et al., 1999). In quiescent cells, Rb associates with and inactivates E2F activity, but this Rb-dependent repression of E2F is relieved upon phosphorylation of Rb by CDKs (Reed, 1997). CDK1 was thought to be the only essential cell-cycle CDK (Santamaria et al., 2007), but cells derived from CDK4–/– mice exhibit a prolonged transition from G1 to S phase after serum stimulation (Tsutsui et al., 1999). Specific inhibition of CDK4 activity delays cell-cycle progression and results in growth arrest in the G1 phase, similar to the phenotype of polyamine-deficient IECs (Xiao et al., 2007). Here we provide evidence that CUGBP1 and miR-222 jointly bind the CDK4 mRNA and repress CDK4 translation synergistically. CUGBP1 was found to interact with both the 3′-UTR and coding region (CR) of the CDK4 mRNA, but miR-222 only binds to the CDK4 CR. Moreover, polyamine depletion inhibited CDK4 translation by inducing cytoplasmic CUGBP1 and miR-222 levels, whereas increased levels of cellular polyamines stimulated CDK4 expression by decreasing CDK4 mRNA associations with CUGBP1 and miR-222.
RESULTS
Polyamines inhibit CUGBP1 and miR-222 expression
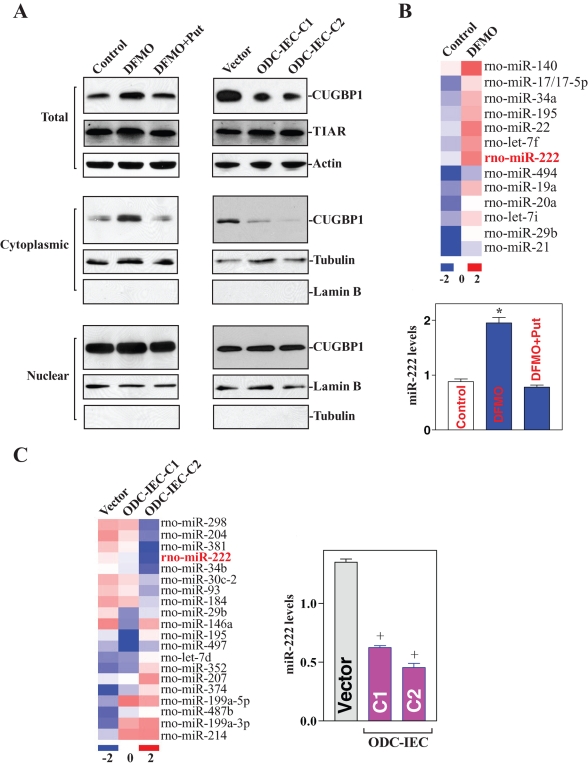
We sought to identify translational repressors implicated in the cellular response to polyamines. First, a survey of RBPs implicated in the response of IECs (IEC-6) revealed that CUGBP1 levels declined as cellular polyamine levels increased. The cellular polyamine content was lowered by addition of DFMO (d,l-α-difluoromethylornithine) to the culture medium to inhibit the enzyme ODC. IEC cultures were divided, and one-half was supplemented with the polyamine putrescine to monitor the specific effect of DFMO. Consistent with our previous studies (Li et al., 2002; Xiao et al., 2007), DFMO treatment for 4 d almost completely depleted cellular polyamines; the levels of putrescine and spermidine were undetectable, and spermine was decreased by ∼60% (unpublished data). To increase the levels of cellular polyamines, two clonal populations of IECs stably expressing ODC (ODC-IEC) were used. These stable ODC-IECs exhibited high levels of ODC protein, greater than 50-fold in ODC enzyme activity, and elevated cellular polyamines putrescine, spermidine, and spermine (by ∼12-fold, ∼2-fold, and ∼25% when compared with control populations, respectively; unpublished data). As shown in Figure 1A, left, depletion of cellular polyamines by DFMO increased total CUGBP1 protein level without affecting the abundance of T-cell intracellular antigen-1 related (TIAR), another RBP; CUGPB1 levels increased in the cytoplasm but did not change substantially in the nucleus. Supplementation with exogenous putrescine reversed the DFMO-triggered changes in total CUGBP1 expression and its subcellular redistribution, as did spermidine supplementation (unpublished data). In contrast, increased levels of cellular polyamines by ODC overexpression decreased total and cytoplasmic CUGBP1 abundances but failed to alter the levels of nuclear CUGBP1 protein in stable ODC-IECs (Figure 1A, right). This effect was not simply due to clonal variation, as two stable clones, ODC-IEC-C1 and ODC-IEC-C2, showed similar results. Consistently, increased levels of cellular polyamines did not affect TIAR protein levels. To monitor the quality and abundance of the nuclear and cytoplasmic fractions, we examined the levels of lamin B (a nuclear protein) and β-tubulin (a cytoplasmic protein), respectively, and determined that there was no contamination between cytoplasmic and nuclear fractions. These results indicate that lowering cellular polyamines increased the cytoplasmic levels of CUGBP1, whereas increasing polyamines reduced its cytoplasmic abundance.
FIGURE 1:
Changes in CUGBP1 and miR-222 expression after altering the levels of cellular polyamines. (A) Changes in the levels and cellular distribution of CUGBP1 and TIAR proteins after decreasing cellular polyamines by inhibiting ODC with DFMO (left) or after increasing polyamine levels by ectopic ODC overexpression (right) in stable ODC-IEC cells. Whole-cell, cytoplasmic, and nuclear lysates were prepared for Western blotting. Equal loading was monitored by β-actin in total lysates, β-tubulin in cytoplasmic fractions, and lamin B in nuclear fractions. (B) Changes of global miRNA profile (top) as measured by miRNA array and the levels of miR-222 (bottom) as examined by Q-PCR analysis after polyamine depletion by treatment with DFMO. Values are means ± SE of data from three separate experiments. *p < 0.05 compared with control cells and cells exposed to DFMO plus putrecine (Put). (C) Changes of global miRNA profile (left) and miR-222 levels (right) in stable ODC-IEC cells. +p < 0.05 compared with cells transfected with a control vector.
Second, we determined the effect of cellular polyamines on global miRNA expression by miRNA array analysis. Total RNAs were labeled using miRCURY Hy3/Hy5 power labeling kit and hybridized on the miRCURY LNA Array (Exiqon, Denmark). A comparison of the miRNA expression profiles in untreated relative to polyamine-deficient cells (DFMO) revealed several miRNAs that increased after polyamine depletion, including miR-222, miR-195, miR-140, and miR-29b (Figure 1B, top), whereas other miRNAs, such as miR-503 and miR-322, decreased (unpublished data). In contrast, increasing cellular polyamines decreased the levels of some miRNAs, such as miR-222 and miR-29b (Figure 1C, left), but it increased expression of some miRNAs, such as miR-207 and miR-503 (unpublished data). Although a sizable subset of miRNAs showed altered abundance in IECs after modulating polyamines, we focused on miR-222, based on its strong dependence on polyamine abundance and its predicted interaction with the CDK4 mRNA. Real-time quantitative PCR (Q-PCR) analysis to confirm changes in the levels of miR-222 after altering the levels of cellular polyamines revealed that miR-222 levels increased by polyamine depletion (Figure 1B, bottom) but decreased in ODC-IECs, which contained high polyamine levels (Figure 1C, right). Taken together, these findings show that polyamines negatively regulate expression of CUGBP1 and miR-222 in normal IECs.
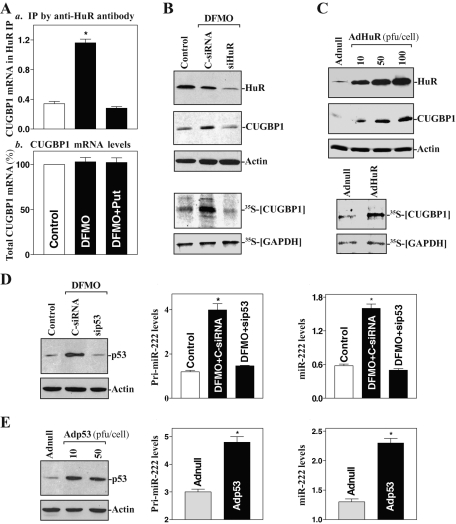
Third, we investigated the mechanisms underlying the regulation of CUGBP1 and miR-222 expression by polyamines. Because polyamine depletion is shown to increase the abundance of cytoplasmic HuR (a well-characterized effector of polyamines) through AMPK-regulated events (Zou et al., 2006, 2008), we determined whether polyamines regulate CUGBP1 expression via HuR. As shown in Figure 2A, polyamine depletion by DFMO increased the concentration of the [HuR/CUGBP1 mRNA] complex, which was prevented by exogenous putrescine given together with DFMO; neither intervention changed total CUGBP1 mRNA levels. HuR silencing prevented an increased level of newly synthesized CUGBP1 protein in polyamine-deficient cells (Figure 2B), although it failed to alter total CUGBP1 mRNA content (unpublished data). Ectopic HuR overexpression also stimulated CUGBP1 expression by increasing the rate of newly synthesized CUGBP1 protein (Figure 2C), but it did not affect total CUGBP1 mRNA levels (unpublished data). These results indicate that CUGBP1 levels are controlled at the translation level and that polyamine depletion increases the CUGBP1 expression by enhancing HuR-mediated CUGBP1 translation. To define the regulation of miR-222 expression by polyamines, we determined whether polyamines regulate miR-222 biogenesis via p53, a transcription factor implicated in the transcriptional up-regulation of numerous miRNAs (Hermeking, 2007). Previous studies from several laboratories, including ours, have shown that p53 levels are modulated by polyamines (Li et al., 2001b; Gerner and Meyskens, 2004) and that decreasing the levels of cellular polyamines increases nuclear p53 levels through HuR-mediated stabilization of p53 mRNA (Li et al., 2001b; Zou et al., 2006). The current results further demonstrate that increased endogenous p53 by polyamine depletion was associated with an induction in the levels of primary (pri-) miR-222 (Figure 2D, center) and mature miR-222 (Figure 2D, right), whereas p53 silencing (Figure 2D, left) prevented the increased levels of both pri-miR-222 and mature miR-222. Furthermore, ectopic p53 overexpression also increased pri-miR-222 and mature miR-222 levels (Figure 2E). Because processing of pri- and precursor miRNAs is rapid, and because assessment of pri-miRNA levels is thought to provide an approximate measure of miRNA gene transcription (Krol et al., 2010), our results suggest that polyamines repress miR-222 expression by inhibiting p53-mediated transcription of the miR-222 gene.
FIGURE 2:
Polyamine depletion induces CUGBP1 translation via HuR and enhances miR-222 expression by increasing p53-mediated gene transcription. (A) Association of endogenous CUGBP1 mRNA with endogenous HuR as measured by RNP IP/Q-PCR analysis (a) and total CUGBP1 mRNA levels as measured by Q-PCR assays (b) in cells exposed to DFMO or DFMO plus Put for 4 d. Values are means ± SE of data from three separate experiments. *p < 0.05 compared with control cells and cells exposed to DFMO plus Put. (B) Changes in CUGBP1 expression after HuR silencing in polyamine-deficient cells. Cells were transfected with a specific HuR-directed siRNA (siHuR) or a control siRNA (C-siRNA); 48 h later, newly synthesized CUGBP1 protein (bottom) was measured by IP with anti-CUGBP1 antibody. (C) Changes in CUGBP1 (top) and newly synthesized CUGBP1 protein (bottom) after HuR overexpression. Cells were infected with the recombinant adenoviral vector encoding HuR cDNA (AdHuR) and control vector (Adnull) for 48 h, and then cell lysates were collected for various assays. (D) Effect of p53 silencing by transfection with a p53 mRNA-directed siRNA (sip53) on pri-miR-222 (center) and mature miR-222 (right) levels as measured by Q-PCR analysis in polyamine-deficient cells. (E) Changes in pri-miR-222 and mature miR-222 levels after p53 overexpression by infection with the recombinant adenoviral vector encoding p53 cDNA (Adp53). *p < 0.05 compared with cells infected with Adnul.
Polyamines enhance CDK4 mRNA translation
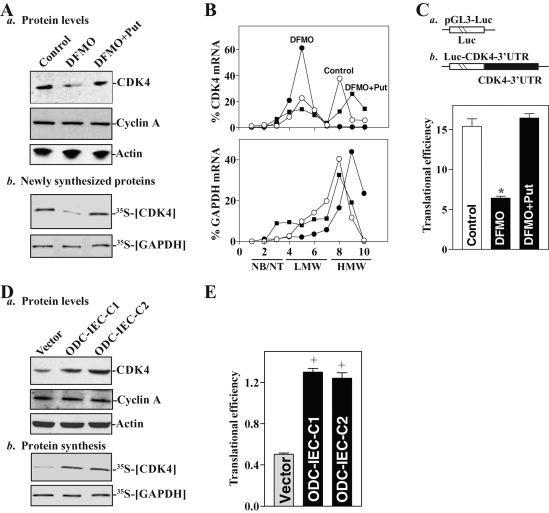
With the induction in cytoplasmic CUGBP1 and miR-222 levels following polyamine depletion, CDK4 expression levels decreased significantly, an effect that was completely rescued by addition of exogenous putrescine (Figure 3Aa). In contrast, polyamine depletion did not alter the levels of cyclin A. To test whether this reduction in the CDK4 protein expression was due, at least in part, to a decrease in CDK4 translation, we examined changes in the rate of new CDK4 protein synthesis after polyamine depletion. Cells were incubated with l-[35S]-methionine and of l-[35S]cysteine for 30 min, whereupon newly translated CDK4 was visualized by immunoprecipitation (IP). The brief incubation period was chosen to minimize the contribution of CDK4 degradation in our analysis. As shown in Figure 3Ab, newly synthesized CDK4 decreased markedly in DFMO-treated cells, whereas exogenous putrescine restored the rate of new CDK4 synthesis to normal levels in the presence of DFMO. Inhibition of CDK4 protein synthesis by polyamine depletion was specific, as neither global protein translation (Liu et al., 2009) nor nascent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) synthesis was altered in DFMO-treated cells. To further define the role of polyamines in the regulation of CDK4 translation, we examined the relative distributions of CDK4 mRNA in individual fractions from polyribosome gradients after polyamine depletion. Although polyamine depletion did not affect global polysomal profiles (unpublished data) as reported previously (Chen et al., 2008; Zou et al., 2010), the abundance of CDK4 mRNA associated with actively translating fractions 8–10 decreased dramatically in DFMO-treated cells with a shift of CDK4 transcripts to low-translating fractions 4–5 (Figure 3B, top). This redistribution of CDK4 mRNA in fractions across the gradient in DFMO-treated cells was prevented by exogenous putrescine. In contrast, housekeeping GAPDH mRNA distributed similarly in all three groups (Figure 3B, bottom). To examine whether the translational effect of polyamines on CDK4 mRNA was exerted through the GU-rich elements (GREs), we used a firefly luciferase reporter gene construct containing the CDK4 GRE within the 3′-UTR (Luc-CDK4-3′UTR) and negative control vector pGL3-Luc (Figure 3C, schematic). A plasmid expressing Renilla luciferase was also cotransfected as an internal control for normalization of firefly luciferase. To distinguish translational output from changes in mRNA turnover, the luciferase activity assays were normalized to luciferase-reporter mRNA levels to assess the translational efficiency. Polyamine depletion by DFMO treatment inhibited CDK4 translation as indicated by a decrease in the Luc-CDK4 3′-UTR reporter activity (Figure 3C, bottom). Combined DFMO and putrescine treatment prevented the decrease in reporter construct translation, rendering it similar to what was observed in control cells. These results indicate that decreasing the levels of cellular polyamines inhibits CDK4 expression at least partially by repressing its translation through the CDK4 3′-UTR.
FIGURE 3:
Changes in CDK4 mRNA translation after increasing or decreasing the levels of cellular polyamines. (A) Changes in the levels of total CDK4 and cyclin A proteins (a) and newly synthesized CDK4 protein (b) after polyamine depletion by DFMO. Newly synthesized CDK4 protein was measured by IP with anti-CDK4 antibody; signals were visualized using a PhosphorImager. (B) Distributions of CDK4 mRNA (top) and housekeeping GAPDH mRNA (bottom) in each gradient fraction of polysomal profiles after polyamine depletion. After fractionation through sucrose gradients, total RNA was isolated from different fractions; the levels of CDK4 and GAPDH mRNAs were measured by Q-PCR analysis and plotted as a percentage of the total CDK4 or GAPDH mRNA levels in each sample. NB, not bound to polysomes; NT, not translated; LMW, low-molecular-weight polysomes; HMW, high-molecular-weight polysomes. (C) Changes in CDK4 translation efficiency as measured by analysis of CDK4 3′-UTR luciferase reporter (schematic), after cotransfection with a Renilla luciferase reporter. Value are means ± SE of data from three separate experiments. *p < 0.05 compared with controls and cells exposed to DFMO plus Put. (D) Changes in the levels of CDK4 and cyclin A proteins (a) and newly synthesized CDK4 protein (b) in stable ODC-IEC cells. (E) Changes in CDK4 translation efficiency as measured by using CDK4-3′-UTR luciferase reporter assays. +p < 0.05 compared with cells infected with a control vector.
We also examined the effect of increasing the levels of cellular polyamines on CDK4 translation in ODC-IEC cells. As shown in Figure 3Da, increased polyamines by ODC overexpression enhanced total CDK4 protein expression in stable ODC-IEC cells when compared with that observed in cells transfected with the control vector lacking ODC cDNA. This induction in CDK4 expression by increasing cellular polyamines was also partially due to an increase in CDK4 mRNA translation as indicated by an increase in the levels of newly synthesized CDK4 protein (Figure 3Db) and Luc-CDK4-3′-UTR reporter gene activity (Figure 3E). Increasing cellular polyamines failed to induce GAPDH translation because there were no differences in the levels of nascent GAPDH synthesis between stable ODC-IEC cells and cells transfected with control vector. Data obtained from two clones of stable cell lines, ODC-IEC-C1 and ODC-IEC-C2, were consistent. These results indicate that the increased levels of cellular polyamines stimulate CDK4 translation.
CUGBP1 binds to the CDK4 mRNA and represses its translation
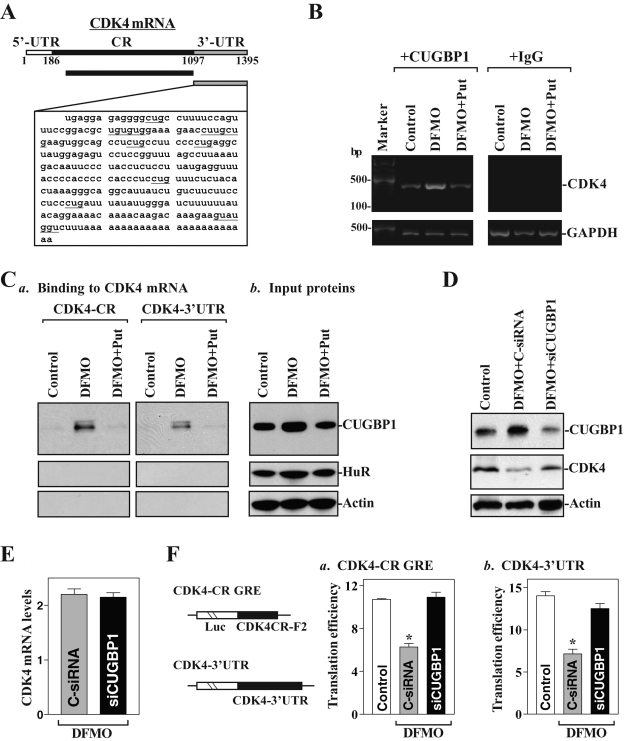
To define the role of CUGBP1 in the regulation of CDK4 mRNA translation, the following three experiments were performed. First, given the predicted affinity of CUGBP1 for the CUG-rich 3′-UTR of the CDK4 mRNA (Figure 4A), we first examined whether CUGBP1 bound the CDK4 mRNA and whether this association could be regulated by cellular polyamines by performing ribonucleoprotein (RNP) IP assays using anti-CUGBP1 antibody under conditions that preserved RNP integrity. The results presented in Figure 4B show that CDK4 PCR products were enriched in CUGBP1 samples compared with control immunoglobulin G (IgG) samples and that polyamine depletion increased CDK4 mRNA abundance in the RNP complex. Exogenous putrescine given together with DFMO prevented the increased association of CUGBP1 with the CDK4 mRNA. In this study, GAPDH mRNA was also examined as a negative control, because this highly abundant transcript is present as a low-level contaminant in the IP materials, thus serving to monitor the evenness of sample input, as reported previously (Wang et al., 2010; Zou et al., 2010).
FIGURE 4:
Binding of cytoplasmic CUGBP1 to CDK4 transcripts and its regulatory effect on CDK4 translation in polyamine-deficient cells. (A) Schematic representation of CDK4 mRNA and the GU-rich sequences in its 3′-UTR. (B) Association of endogenous CUGBP1 with endogenous CDK4 mRNA was tested by RNP IP analysis; RNA was isolated, and RT-PCR products of CDK4 were examined by RT-PCR analysis. (C) Representative CUGBP1 immunoblots after pull-down using biotinylated CDK4 CR (a, left) or 3′-UTR transcripts (a, right) and cytoplasmic lysates and input proteins (b). β-actin, negative control in pull-down materials. (D) Effect of CUGBP1 silencing on CDK4 protein expression levels. Whole-cell lysates were harvested 48 h after cells were transfected with either control siRNA (C-siRNA) or siCUGBP1. (E) Levels of total CDK4 mRNA in polyamine-deficient cells after CUGBP1 silencing as measured by Q-PCR analysis. Values are means ± SE of data from three samples. (F) Changes in CDK4 translation efficiency as measured by analysis of CDK4-CR GRE or CDK4 3′-UTR luciferase reporter (schematic), after cotransfection with a Renilla luciferase reporter. Values are means ± SE of data from three separate experiments. *p < 0.05 compared with controls and cells exposed to DFMO plus Put.
Second, [CUGBP1/CDK4 mRNA] associations were further characterized by using biotinylated transcripts spanning the CDK4 3′-UTR and CR in RNA pull-down assays using cell lysates prepared from control cells and from cells treated with DFMO alone or DFMO plus putrescine. The CDK4 3′-UTR directly interacted with CUGBP1; the binding intensity increased significantly when using cell lysates prepared from polyamine-deficient cells, but it returned to normal levels when cells were treated with DFMO plus putrescine (Figure 4Ca, right). Unexpectedly, CUGBP1 also bound to the CDK4 CR, and this interaction was also increased by polyamine depletion and was prevented by exogenous putrescine (Figure 4Ca, left). Neither the 3′-UTR nor the CR of CDK4 interacted with the RBP HuR in any of the three groups, although the associations of HuR with the p53 (Zou et al., 2006) and XIAP (Zhang et al., 2009) mRNAs increased in polyamine-deficient cells. The CDK4 mRNA did not associate with β-actin, which served as negative control in this experiment. None of the proteins interacted with a control biotinylated RNA spanning the 3′-UTR of the housekeeping gene GAPDH (unpublished data). To examine whether binding of CUGBP1 to the CDK4 3′-UTR or CR is mediated through the specific binding site containing GREs, biotinylated transcripts partially spanning the CDK4 3′-UTR or CR were prepared, and their associations with CUGBP1 were measured in pull-down assays. CUGBP1 was found to bind to the F2 (containing a putative GRE) but not to the F1, F3, and F4 in the CDK4 CR, although the F1 and F3 also contained possible GREs (Supplemental Figure 1). In addition, CUGBP1 bound the F5 and F6 in the CDK4 3′-UTR, and both contained GREs. These findings strongly suggest that CUGBP1 binds CDK4 mRNA through both the 3′-UTR and CR and that the levels of the complex [CUGBP1/CDK4 mRNA] are increased by polyamine depletion. Nonetheless, it remains to be investigated whether the increased CUGBP1 association with the CDK4 mRNA in polyamine-deficient cells results primarily from altered cytoplasmic CUGBP1 abundance or from increased CUGBP1 binding affinity.
Third, we determined whether increased [CUGBP1/CDK4 mRNA] associations repressed CDK4 mRNA translation following polyamine depletion. As shown in Figure 4D, silencing CUGBP1 by transfection with siRNA targeting the CUGBP1 mRNA (siCUGBP1) significantly restored CDK4 expression in polyamine-deficient cells. This restoration in CDK4 expression by CUGBP1 silencing resulted primarily from the induction of CDK4 mRNA translation, as CUGBP1 silencing did not alter CDK4 mRNA stability (Supplemental Figure 2) and its total levels in DFMO-treated cells (Figure 4E). To determine whether repression of CDK4 mRNA translation by CUGBP1 is mediated through interaction with the CDK4 CR, 3′-UTR, or both, a firefly luciferase reporter construct containing the CDK4 GRE within the CR (F2) (CDK4-CR GRE) was generated (Figure 4F, schematic). CUGBP1 silencing not only increased the levels of CDK4-3′-UTR luciferase reporter gene activity (Figure 4F, right) but it also restored the reporter gene activity of the CDK4-CR GRE to normal levels (Figure 4F, left) in polyamine-deficient cells. These results indicate that CUGBP1 associates with both the 3′-UTR and CR of CDK4 and that increased cytoplasmic CUGBP1 following polyamine depletion represses CDK4 translation by interacting with the GREs within both the CR and 3′-UTR of CDK4.
miR-222 represses CDK4 mRNA translation by interacting with the CDK4 CR
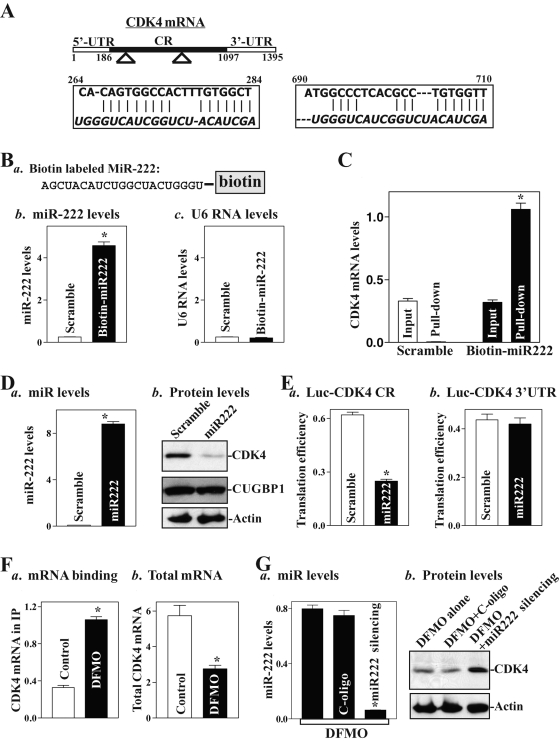
Using the program RNA22, we found that there are two predicted binding sites of miR-222 within the CR of the CDK4 mRNA (Figure 5A), suggesting the potential role of miR-222 in the regulation of CDK4 expression. To test this possibility, we first examined the association of miR-222 with the CDK4 mRNA by RNA pull-down assays using biotin-labeled miR-222 (custom made by Dharmacon, shown in Figure 5Ba). To assess the transfection efficiency, the levels of miR-222 and small nuclear RNA U6 (which served as control) were examined by Q-PCR analysis 48 h after the transfection. As shown in Figure 5Bb, cells transfected with the biotin-labeled miR-222 for 48 h exhibited elevated miR-222 levels but displayed no changes in RNA U6 levels (Figure 5Bc). When the presence of CDK4 mRNA in the materials pulled down by biotin–miR-222 was examined, the levels of CDK4 mRNA were highly enriched in the materials from cells transfected with the biotin-labeled miR-222 but not from cells transfected with scrambled control miRNA (Figure 5C). In contrast, there were no changes in the levels of CUGBP1 or GAPDH mRNAs in the pull-down materials between biotin-labeled miR-222–transfected cells and cells transfected with scrambled miRNA (unpublished data). These results strongly suggest that miR-222 interacts with the CDK4 mRNA and forms the [miR-222/CDK4 mRNA] complex. To define the functional consequence of [miR-222/CDK4 mRNA] association, miR-222 levels were increased by transfection with the miR-222 precursors (premiR-222). As shown in Figure 5Da, transfection with premiR-222 increased miR-222 levels remarkably; this induction was specific, as it failed to induce the levels of RNA U6 (unpublished data). Importantly, increased levels of miR-222 by the premiR-222 transfection decreased CDK4 protein levels (Figure 5Db), although they did not alter the levels of CDK4 mRNA (unpublished data). Ectopic miR-222 overexpression did not alter CUGBP1 expression levels. To examine whether miR-222 inhibited CDK4 expression by repressing its translation, and if this inhibitory effect is mediated through the CDK4 CR, 3′-UTR, or both, fractions of the CDK4 CR and its 3′-UTR were subcloned into the pmirGLO dual-luciferase miRNA target expression vector to generate pmirGLO-CDK4-CR and pmirGLO-CDK4-3′-UTR reporter constructs. miR-222 overexpression decreased the levels of CDK4-CR luciferase reporter activity (Figure 5Ea), but it did not inhibit the activity of CDK4-3′-UTR reporter activity (Figure 5Eb), indicating that increasing the levels of miR-222 represses CDK4 mRNA translation through interaction with the CDK4 CR rather than with its 3′-UTR.
FIGURE 5:
miR-222 directly interacts with CDK4 mRNA through its CR. (A) Schematic representation of CDK4 mRNA depicting two predicted target sites for miR-222 in the CDK4 CR. Alignment of the CDK4 mRNA sequences with miR-222: top strand, CDK4 mRNA; bottom strand, miR-222. (B) Levels of biotinylated miR-222 after transfection for 48 h: (a) schematic representation of biotinylated miR-222; (b) miR-222 levels as measured by Q-PCR analysis; (c) U6 RNA levels. Values are means ± SE from three separate experiments. *p < 0.05 compared with cells transfected with control scrambled RNA. (C) Binding of biotinylated miR-222 to CDK4 mRNA in pull-down materials. *p < 0.05 compared with input control. (D) Changes in the levels of CDK4 and CUGBP1 proteins after ectopic overexpression of miR-222: (a) levels of miR-222 as measured by Q-PCR analysis 48 h after transfection; (b) representative immunoblots of CDK4. (E) Changes in CDK4 translation efficiency as measured by analysis of CDK4-CR (a) and CDK4 3′-UTR (b) luciferase reporter. (F) Polyamine depletion enhances CDK4 mRNA association with miR-222: (a) binding of CDK4 mRNA to miR-222; (b) total CDK4 mRNA levels. (G) Effect of miR-222 silencing on CDK4 expression in polyamine-deficient cells: (a) binding of miR-222 to CDK4 mRNA; (b) CDK4 protein levels.
The results presented in Figure 5Fa further show that polyamine depletion increased miR-222 association with the CDK4 mRNA, although the levels of total CDK4 mRNA were decreased in DFMO-treated cells (Figure 5Fb). To determine the role of increased levels of [miR-222/CDK4 RNA] complex in the repression of CDK4 translation after polyamine depletion, we lowered miR-222 activity by transfecting the corresponding oligomer (antagomir) targeting miR-222 (anti–miR-222). Transfection of DFMO-treated cells with anti–miR-222 oligo reduced the levels of miR-222 dramatically (Figure 5Ga) and markedly increased CDK4 expression (Figure 5Gb). Neither miR-222 nor CDK4 protein levels in DFMO-treated cells were affected by transfection with a control oligo (C-oligo). These results indicate that increased levels of miR-222 following polyamine depletion also contribute to inhibition of CDK4 translation.
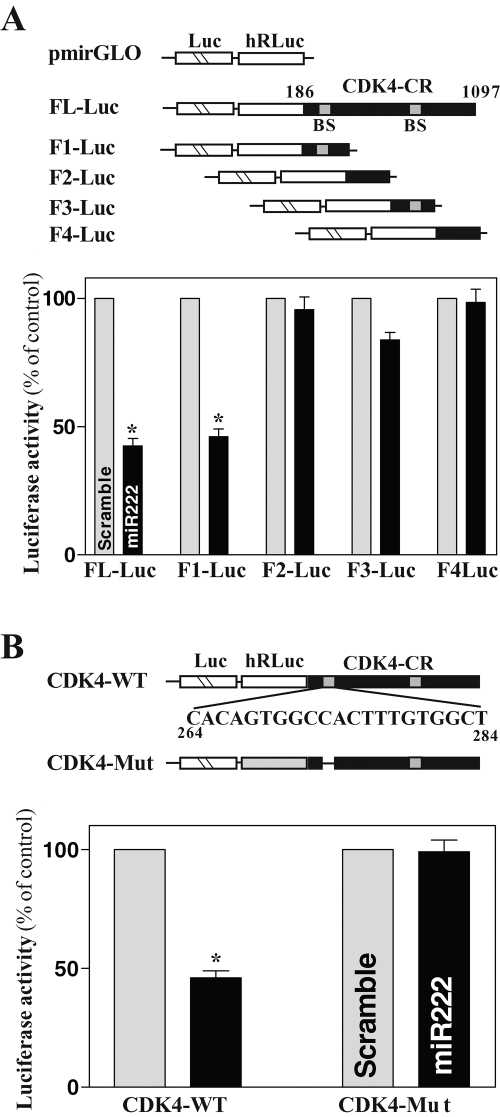
To define the specific binding site of miR-222 in the CDK4 CR, we prepared various reporter constructs that expressed chimeric RNA containing the luciferase and partial transcripts spanning the CDK4 CR with or without the potential binding site (Figure 6A, schematic). Ectopic miR-222 overexpression was found to decrease the levels of luciferase reporter gene activity when cells were transfected with the FL-Luc (containing full-length CDK4 CR) or F1-Luc (containing a predicted interaction region) but not with the F2-Luc or F4-Luc (in which the potential binding sites were deleted). When cells were transfected with the F3-Luc (that also contained a potential binding site), increasing the levels of miR-222 just slightly reduced the reporter activity. The internal deletion-mutation of the site located at the F1 of the CDK4 CR was also performed, in which the nucleotides spanning positions 266–286 of the CDK4 CR were eliminated (Figure 6B, schematic). As shown in Figure 6B (left), CDK4 repression by miR-222 overexpression was completely prevented when this specific binding site was deleted from the CDK4 CR. These results indicate that miR-222 interacts with CDK4 mRNA via the specific binding site at 266–286, thus repressing CDK4 translation.
FIGURE 6:
Changes in activities of CDK4 CR luciferase reporters after deletion of miR-222-binding site. (A) Effect of 5′-deletion of CDK4 CR on its luciferase reporter activity. Left, schematic of plasmids of different chimeric firefly luciferase CDK4 CR reporters. BS, predicted miR-222-binding site. Right, levels of CDK4 CR luciferase reporter activity. Twenty-four hours after transfection with premiR-222, cells were cotransfected with CDK4 CR F-Luc constructs and a Renilla luciferase reporter. Levels of firefly and Renilla luciferase activities were assayed 24 h later. Results were normalized to the Renilla luciferase activity and expressed as the means ± SE of data from three separate experiments. *p < 0.05 compared with cells transfected with control scrambled oligomer. (B) Effect of deletion of specific miR-222–binding site (schematic) in CDK4 CR on luciferase reporter activity after miR-222 overexpression.
miR-222 and CUGBP1 recruit the CDK4 mRNA to processing bodies
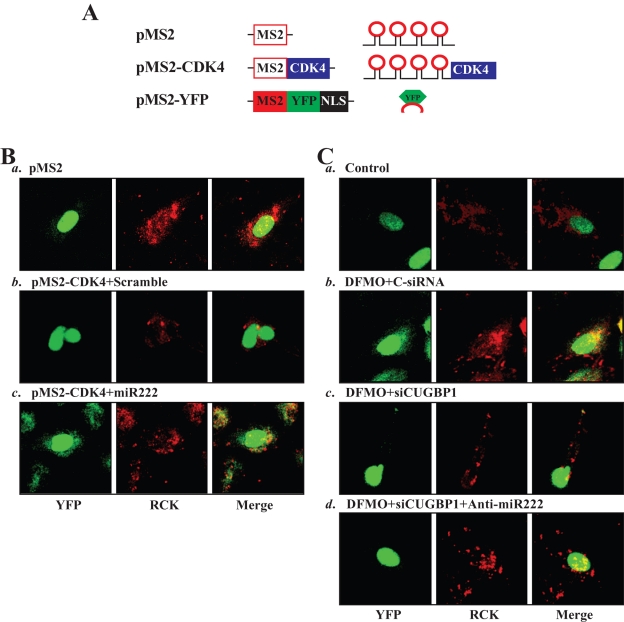
Increasing evidence shows that nontranslating mRNAs accumulate in processing bodies (PBs), where these transcripts are sorted for degradation and/or translational repression (Buchan and Parker, 2009; Lee et al., 2010a). To determine whether increased levels of CUGBP1 and miR-222 repress CDK4 translation by enhancing the recruitment of the CDK4 mRNA to PBs following polyamine depletion, we studied the subcytoplasmic localization of CDK4 mRNA. The reporter construct pMS2-CDK4, which expressed a chimeric RNA (MS2-CDK4) comprising the CDK4 CR (which contained binding sites for CUGBP1 and miR-222) and 12 tandem MS2 RNA hairpins (Figure 7A), was prepared. Cotransfection of pMS2-CDK4 together with plasmid pMS2-YFP, which expressed the chimeric fluorescent protein MS2-YFP with a nuclear localization signal (NLS), allowed us to track the subcellular localization of the chimeric MS2-CDK4 RNA (as the complex MS2-YFP/MS2-CDK4) as well as the control MS2 RNA (as the complex MS2-YFP/MS2) by confocal microscopy. Signals of the PB marker RCK were detected in the same cells. As shown, the control MS2 RNA appeared to be exclusively nuclear in all of the transfected cells (Figure 7Ba) due to the presence of the NLS in the chimeric protein MS2-YFP. In contrast, some MS2-CDK4 RNA was retained in the cytoplasm, colocalizing with some RCK signals in control cells transfected with scrambled miRNA (colocalization results in yellow signals in the merged image in Figure 7Bb, right). Interestingly, miR-222 overexpression by transfection with premiR-222 increased the colocalization of MS2-CDK4 RNA and RCK signals (Figure 7Bc), suggesting that miR-222 enhanced the association of CDK4 mRNA with PBs. Furthermore, the colocalization of MS2-CDK4 RNA and RCK signals increased in polyamine-deficient cells when compared with that observed in control cells (Figure 7C, a vs. b). This induced colocalization of MS2-CDK4 RNA and RCK signals in DFMO-treated cells declined when CUGBP1 was silenced (Figure 7C, b vs. c) and was almost completely lost when both CUGBP1 and miR-222 were lowered (Figure 7C, b vs. d). Together, these results support the notion that miR-222 and CUGBP1 repress CDK4 mRNA translation at least partially by recruiting the CDK4 transcripts to PBs following polyamine depletion.
FIGURE 7:
Colocalization of CDK4 mRNA with PBs after miR-222 overexpression and following polyamine depletion. (A) Schematic of the plasmids used for the visualization of CDK4 mRNA. PMS2 and pMS2-CDK4 expressed MS2 and MS2-CDK4 RNAs, each containing 12 tandem MS2 hairpins; pMS2-YFP expressed a fusion fluorescent protein (MS2-YFP) capable of detecting MS2-containing RNA. (B) Images of CDK4 mRNA colocalization with PBs in cells overexpressing miR-222. Using confocal microscopy, MS2 and MS2-CDK4 mRNA were visualized using MS2-YFP (green fluorescence); red, RCK (PB marker) signals; yellow, colocalized red and green signals. (C) Images of CDK4 mRNA colocalization with PBs in polyamine-deficient cells after CUGBP1 silencing or silencing both CUGBP1 and miR-222. CDK4 mRNA was detected using pMS2-CDK4 as explained in (B); three experiments were performed that showed similar results.
CUGBP1 and miR-222 repress CDK4 translation synergistically and modulate IEC proliferation
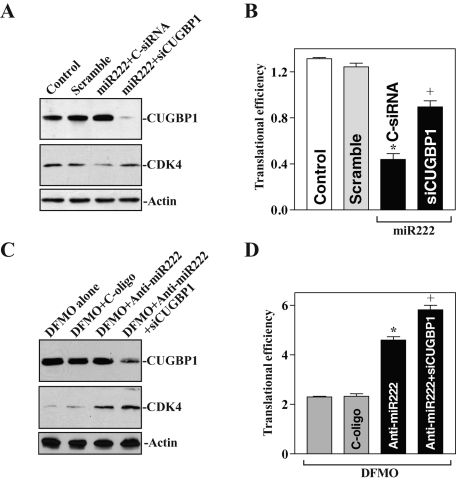
To determine whether CUGBP1 and miR-222 regulate CDK4 mRNA translation synergistically, we first examined the effect of CUGBP1 silencing on CDK4 translation in cells overexpressing miR-222. As shown in Figure 8A, transfection with premiR-222 decreased CDK4 protein levels, although it failed to alter the levels of endogenous CUGBP1. This inhibition, however, was partially abolished when CUGBP1 was silenced by siCUGBP1 transfection. Consistently, CUGBP1 silencing also prevented miR-222–induced repression of CDK4 translation as indicated by a decrease in the levels of CDK4 CR luciferase reporter gene activity (Figure 8B), even though cotransfection with premiR-222 and siCUGBP1 did not affect CDK4 mRNA levels (unpublished data). In addition, CUGBP silencing did not affect the levels of miR-222 (Supplemental Figure 3). Second, we examined changes in CDK4 translation in polyamine-deficient cells after reducing miR-222 or after reducing both miR-222 and CUGBP1 together. Although CDK4 expression in DFMO-treated cells was marginally rescued by miR-222 silencing alone, this restoration of CDK4 translation was further increased when the levels of both miR-222 and CUGBP1 were reduced (Figure 8, C and D). In contrast, transfection with anti–miR-222 or siCUGBP1 alone or by cotransfection of anti–miR-222 with siCUGBP1 did not alter CDK4 mRNA levels in polyamine-deficient cells (unpublished data). These results indicate that CUGBP1 and miR-222 repress CDK4 translation synergistically following polyamine depletion.
FIGURE 8:
CUGBP1 and miR-222 synergistically repress CDK4 translation. (A) Changes in the levels of CDK4 protein expression after miR-222 overexpression with or without CUGBP1 silencing. After cells were cotransfected premiR-222 and control-siRNA (C-siRNA) or siRNA targeting CUGBP1 (siCUGBP1) for 48 h, CUGBP1 and CDK4 protein levels were examined. (B) CDK4 translation efficiency was measured by analysis of CDK4 CR luciferase reporter in cells described in (A). Values are the means ± SE of data from three separate experiments. *p < 0.05 compared with controls and cells transfected with control scrambled oligomer. +p < 0.05 compared with cells cotransfected with premiR-222 and C-siRNA. (C) Effects of reducing miR-222 alone or reducing both miR-222 and CUGBP1 on CDK4 protein expression in polyamine-deleted cells. After cells were treated with DFMO for 4 d, they were transfected with control oligomers, anti-miR-222 oligos, or anti-miR-222 oligos plus siCUGBP1; protein levels were measured 48 h after transfection. (D) CDK4 translation efficiency in cells described in (B).
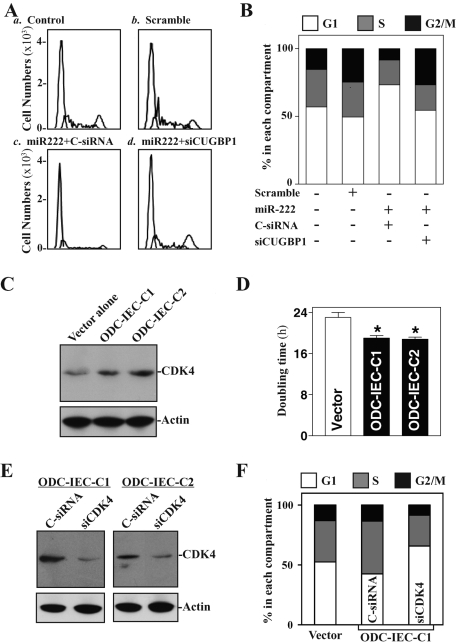
Finally, we studied the consequences of polyamine-regulated CDK4 expression via CUGBP1 and miR-222 upon IEC proliferation. As shown in Figure 9, A and B, increasing the levels of miR-222 resulted in growth arrest in G1 phase in IEC-6 cells, whereas this inhibition of cell proliferation was significantly diminished by CUGBP1 silencing. Furthermore, decreasing the levels of CUGBP1 and miR-222 by increasing cellular polyamines in ODC-IECs (Figure 1) was associated with increases in the levels of CDK4 expression (Figure 9C) and cell proliferation (Figure 9D). The doubling time of cell growth in stable ODC-IECs decreased, which was accompanied by an induction in the S phase population (Figure 9F). CDK4 silencing by transfecting ODC-IEC-C1 cells with siRNA targeting CDK4 mRNA (siCDK4) prevented the polyamine-induced cell proliferation as indicated by a decrease in populations in the S and G2/M phases, along with increases in G1 cells. A similar inhibitory effect of CDK4 silencing on cell proliferation in ODC-IEC-C2 cells was also observed (unpublished data). These findings indicate that polyamines stimulate IEC proliferation at least partially by enhancing CDK4 expression via down-regulation of CUGBP1 and miR-222.
FIGURE 9:
Effect of CUGBP1/miR-222–regulated CDK4 expression on cell proliferation. (A) Flow cytometric analysis of cell-cycle distribution after miR-222 overexpression with or without CUGBP1 silencing. IEC-6 cells were transfected with control scrambled oligomer, premiR-222 alone, or premiR-222 plus siCUGBP1; 48 h later, cells were subjected to fluorescence-activated cell sorting analysis. (B) The relative G1, S, and G2/M compartments were also calculated from data described in (A). Data are representative of three separate experiments. (C and D) Changes in the levels of CDK4 protein expression (C) and doubling time of cell growth (D) in ODC-IEC populations. (E and F) Effect of CDK4 silencing on cell growth in stable ODC-IEC cells. After cells were transfected with either control siRNA (C-siRNA) or siRNA targeting CDK4 mRNA (siCDK4) for 48 h, the levels of CDK4 protein (E) and cycle distribution (F) were examined. Three separate experiments were performed that showed similar results.
DISCUSSION
All mammalian cells depend on polyamines for normal growth and proliferation (Gerner and Meyskens, 2004; Casero and Marton, 2007; Casero and Pegg, 2009; Eisenberg et al., 2009), but the exact roles of polyamines at the molecular level are not well understood. In this study, we highlight the novel functions of the RBP CUGBP1 and miR-222 in the regulation of CDK4 translation and provide evidence that polyamines modulate CDK4 protein synthesis by altering the association of the CDK4 mRNA with CUGBP1 and miR-222. CUGBP1 interacted with both the CR and 3′-UTR of the CDK4 mRNA, but miR-222 interacted only with the CDK4 CR. Polyamine depletion increased cytoplasmic levels of CUGBP1 and miR-222, induced their associations with the CDK4 mRNA, and repressed CDK4 translation, whereas increasing the levels of cellular polyamines decreased the interactions of CDK4 mRNA with CUGBP1 and miR-222, thus inducing CDK4 protein synthesis.
Our results indicate that DFMO-mediated depletion of cellular polyamines increased levels of CUGBP1 protein, which was predominantly located in the cytoplasm. The specificity of these effects was demonstrated by the addition of exogenous putrescine, because it completely prevented the increase in cytoplasmic CUGBP1 abundance. Polyamines were previously shown to influence multiple distinct signaling pathways leading to alterations in gene transcription (Celano et al., 1989; Patel and Wang, 1997; Xiao et al., 2007), posttranscriptional events (Butcher et al., 2007; Zhang et al., 2009), protein phosphorylation (Liu et al., 2003, 2009; Zhang et al., 2004), and protein trafficking, including the nuclear import of transcription factors such as nuclear factor κB (Li et al., 2001a), Smads (Liu et al., 2003), and JunD (Li et al., 2002; Xiao et al., 2007). Recently, we found that polyamines also modulate subcellular trafficking of RBP HuR and that decreasing the levels of cellular polyamines increases cytoplasmic levels of HuR without effect on whole-cell HuR content (Zou et al., 2006, 2008). To define the mechanisms whereby polyamines influence CUGBP1 and miR-222 expression, our results provide evidence that regulation of CUGBP1 by polyamines occurs at the translational level via HuR (Figure 2). Polyamine depletion increased the [HuR/CUGBP1 mRNA] complex and stimulated CUGBP1 translation, whereas HuR silencing repressed CUGBP1 expression in polyamine-deficient cells. Our findings also show that polyamines down-regulate miR-222 expression at least in part by repressing the p53-mediated increases in pri-miR-222 levels. We further propose that p53 may promote transcription of pri-miR-222, but this hypothesis awaits experimental testing.
The results reported here also show that CUGBP1 and miR-222 jointly bind to the CDK4 mRNA and that these associations are inhibited by polyamines. On the basis of the present findings, we propose that CUGBP1 and miR-222 associate with the CDK4 mRNA through a distinct nonoverlapping binding site and that there are no common sites for both CUGBP1 and miR-222 in the CDK4 mRNA. CUGBP1 was found to bind both the CDK4 CR and its 3′-UTR, but miR-222 interacted only with the CDK4 CR. Experiments mapping the CDK4 CR and deletion-mutation revealed that the binding site of CUGBP1 was located in the F2 fragment of the CDK4 CR (Supplemental Figure 1), whereas the functional binding site of miR-222 was identified within the F1 of CDK4 CR (Figure 6). Although the CDK4 mRNA does not contain any high-affinity CUGBP1-binding site such as the canonical GRE (defined as UGUUUGUUUGU), there are several GU repeats and CUG repeats in the 3′-UTR and CR of the CDK4 transcript, which were also recently recognized as the GRE and interacted with CUGBP1 (Tsuda et al., 2009; Lee et al., 2010b; Rattenbacher et al., 2010). In contrast, there are two predicted miR-222 binding sites within the CDK4 CR, but only the sequence (spanning positions 264–284) was functional, because repression of CDK4 by miR-222 was completely prevented when this binding site was mutated (Figure 6).
One interesting facet of this study is that CUGBP1 and miR-222 repress CDK4 translation synergistically. Induced [CUGBP1/CDK4 mRNA] complex inhibits CDK4 translation in polyamine-deficient cells, as CUGBP1 silencing increased CDK4 protein expression levels but it failed to alter CDK4 mRNA content. Our observations are consistent with studies from others who showed that CUGBP1 binds to specific GRE-bearing target mRNAs and represses their translation and/or stimulates degradation (Wang et al., 2007; Zhang et al., 2008). CDK4 mRNA association with miR-222 also represses CDK4 translation, because the CDK4 expression and CDK4-CR–driven luciferase reporter activity decreased in cells overexpressing miR-222 but increased in miR-222–silenced cells. Importantly, CDK4 inhibition by miR-222 was potentiated by CUGBP1, because CUGBP1 silencing partially prevented miR-222–mediated CDK4 repression, whereas silencing both CUGBP1 and miR-222 had a stronger effect on restoring CDK4 translation in polyamine-deficient cells compared with miR-222 silencing alone.
The exact mechanisms underlying the inhibition of CDK4 translation by induced CUGBP1 and miR-222 following polyamine depletion are unclear at present, but our results support the notion that CUGBP1 and miR-222 repress CDK4 translation at least in part by recruiting CDK4 transcripts to PBs, where nontranslated mRNAs accumulate. Polyamine depletion increased the colocalization of CDK4 mRNA with PBs, which was prevented by silencing CUGBP1 and miR-222 (Figure 7). How PBs repress the translation of resident CDK4 mRNA in polyamine-deficient cells also remains unknown. PBs are shown, however, to contain many translational repressors, such as decapping enzymes (DCP1 and DCP2) and RBPs (TIAR and TIA), mRNA deadenylation factors (e.g., the CCR4-CAF-1-Not complex), activators of decapping (Dhh1/RCK/p54, Pat1, Scd6/RAP55, Edc3 and the Lasm-7 complex), and exonucleases (Franks and Lykke-Andersen, 2008; Balagopal and Parker, 2009; Buchan and Parker, 2009). The mRNAs within PBs are believed to be subject to RNP remodeling to modulate their subsequent turnover and translational rates (Franks and Lykke-Andersen, 2008; Buchan and Parker, 2009).
Finally, our findings may have important implications in the control of normal intestinal epithelial renewal under biological conditions. The mammalian intestinal epithelium is a rapidly self-renewing tissue in the body, and its homeostasis is preserved through strict regulation of IEC proliferation, growth arrest, and apoptosis (Seiler and Raul, 2007). The epithelium of the human small intestine undergoes ∼1011 mitoses per day, and this rapid cell proliferation is shown to absolutely require polyamines (Wang and Johnson, 1991; Wang et al., 1991). Because intracellular polyamines are tightly regulated by stress stimulation and the status of cell growth, the current findings suggest that the polyamine-mediated activation of CDK4 expression by targeting CUGBP1 and miR-222 in IECs can directly regulate the growth of the intestinal mucosa in vivo and thereby contributes to maintaining the integrity of the intestinal epithelium.
MATERIALS AND METHODS
Chemicals and cell culture
Tissue culture medium and dialyzed fetal bovine serum (FBS) were obtained from Invitrogen (Carlsbad, CA), and biochemicals were obtained from Sigma (St. Louis, MO). The antibodies recognizing CUGBP-1, CDK4, and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and BD Biosciences (Sparks, MD), and the secondary antibody conjugated to horseradish peroxidase was obtained from Sigma. DFMO (α-difluoromethylornithine) was purchased from GENZYME (Cambridge, MA). Pre-miR miRNA precursor and anti-miR miRNA inhibitor of miR-222 were purchased from Ambion (Austin, TX). Biotin-labeled miRNA-222 was custom made by Dharmacon (Lafayette, CO), and siCUGBP1 was purchased from Santa Cruz Biotechnology.
The IEC-6 cell line, derived from normal rat intestinal crypt cells (Quaroni et al., 1979), was used at passages 15–20; cells were maintained in DMEM supplemented with 5% heat-inactivated FBS. ODC-IEC cells were developed as described in our previous studies (Zou et al., 2006; Wang et al., 2010) and expressed a more stable ODC variant with full enzyme activity. They were maintained in Eagle's minimum essential medium with 10% heat-inactivated FBS and 50 μg/ml gentamicin as described previously (Liu et al., 2003).
Plasmid construction
CUGBP1 expression vector was purchased from Origene (Rockville, MD). The chimeric firefly luciferase reporter construct of the CDK4 CR or 3′-UTR was generated as described previously (Zou et al., 2010). The full-length CDK4 CR or its 3′-UTR and the different GRE fragments were amplified and subcloned into the pGL3-Luc plasmid (Promega, Madison, WI) at the Xba1 site to generate various chimeric pGL3-Luc-CDK4-CR or CDK4-3′-UTR reporter constructs. The sequence and orientation of the fragment in the luciferase reporter were confirmed by DNA sequencing and enzyme digestion. Transient transfections were performed using Lipofectamine reagent and performed as recommended by the manufacturer (Invitrogen, Carlsbad, CA). The luciferase reporter constructs were transfected into cells along with phRL-null, a Renilla luciferase control reporter vector from Promega, to monitor transfection efficiencies as described previously (Liu et al., 2009). Luciferase activity was measured using the Dual Luciferase Assay System, and the levels of the pGL3-Luc-CDK4-CR or CDK4-3′-UTR luciferase activity were normalized to Renilla luciferase activity and were further compared with the levels of luciferase mRNA in every experiment. The CDK4-CR and CDK4-3′-UTR were also subcloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) to generate the pmirGLO-CDK4-CR and pmirGLO-CDK4-3′-UTR. Both pcDNA-MS2 and pcDNA-MS2-YFP plasmids were described previously (Lee et al., 2010a), and the fragment of CDK4 CR-F2 (280–690) was inserted into pcDNA-MS2 at the XhoI site. All primer sequences for generating these constructs are provided in Supplemental Table 1.
RT followed by real-time PCR analysis and miRNA array analysis
Total RNA was isolated by using the RNeasy mini kit (Qiagen, Valencia, CA) and used in RT and PCR amplification reactions as described (Zou et al., 2010). The levels of GAPDH PCR product were assessed to monitor the evenness in RNA input in RT-PCR samples. Real-time Q-PCR analysis was performed using 7500 Fast Real-Time PCR Systems with specific primers, probes, and software (Applied Biosystems, Foster City, CA).
For miRNA array studies, total RNA was purified with a miRCURY RNA isolation kit (Exiqon, Vedbaek, Denmark), and a miRCURY LNA array of miRNA profiling services was performed by Exiqon. The levels of miRNA-222 were also quantified by Q-PCR by using Taqman MicroRNA assay; small nuclear RNA (snRNA) U6 was used as endogenous control.
Western blotting analysis
Whole-cell lysates were prepared using 2% SDS, sonicated, and centrifuged (12,000 rpm) at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS–PAGE (7.5% acrylamide). After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing CUGBP1 or CDK4; following incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
Analysis of newly translated protein and polysome analysis
New synthesis of CDK4 protein was measured by l-[35S]methionine and l-[35S]cysteine incorporation assays as described (Liu et al., 2009). Cells were incubated with 1 mCi (1 Ci = 37 GBq) l-[35S]methionine and l-[35S]cysteine per 60-mm plate for 20 min, whereupon cells were lysed using RIPA buffer. IPs were carried out for 1 h at 4ºC using either a polyclonal antibody recognizing c-Myc or IgG1 (BD PharMingen, San Diego, CA). Following extensive washes in TNN buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% NP-40), the immunoprecipitated material was resolved by 10% SDS–PAGE, transferred onto polyvinylidene fluoride filters, and visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Polysome analysis was performed as described (Chen et al., 2008). Briefly, cells at ∼70% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide, then lifted by scraping in 1 ml of PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCl, pH 7.6, 1% Triton X-100, 1 mg/ml heparin, and 0.1 mg/ml cycloheximide) and lysed on ice for 10 min. Nuclei were pelleted (10,000 × g, 10 min) and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weights. The eluted fractions were prepared with a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm using a UV-6 detector (ISCO, Louisville, KY). After RNA in each fraction was extracted with 8 M guanidine–HCl, the levels of each individual mRNA were quantified by RT-Q-PCR in each of the fractions, and their abundance was represented as a percentage of the total mRNA in the gradient.
Biotin pull-down assays and RNP IP analysis
The synthesis of biotinylated transcripts and analysis of RBPs bound to biotinylated RNA were done as previously described (Zhang et al., 2009). cDNA from IEC-6 cells was used as a template for PCR amplification of the CR and 3′-UTR of CDK4. The 5′ primers contained the T7 RNA polymerase promoter sequence [(T7), CCAAGCTTCTAATACGACTC ACTATAGGGAGA]. All sequences of oligonucleotides for preparation of full-length CDK4 CR, 3′-UTR, and various short RNA probes for mapping the CDK4-CR or 3′-UTR are described in Supplemental Table 1. PCR-amplified products were used as templates to transcribe biotinylated RNAs by using T7 RNA polymerase in the presence of biotin-cytidine 5′-triphosphate as described (Wang et al., 2010). Biotinylated transcripts (6 μg) were incubated with 120 μg of cytoplasmic lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) and analyzed by Western blot analysis.
To assess the association of endogenous CUGBP1 with endogenous CDK4 mRNA, IP of RNP complexes was performed as described (Wang et al., 2010). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 μg) IP antibody (IgG, anti-CUGBP1). RNA in IP materials was used in RT followed by PCR and Q-PCR analysis to detect the presence of CDK4 and GAPDH mRNAs.
Biotin-labeled miR-222 pull-down assays
Biotin-labeled miR-222 was transfected into cells for 48 h, and then whole-cell lysate was collected. Cell lysates were mixed with Streptavidin-Dynal beads (Invitrogen) and incubated at 4°C on a rotator overnight. After the beads were washed thoroughly, the bead-bound RNA was isolated and subjected to RT followed by Q-PCR analysis. Input RNA was extracted and served as control.
Immunofluorescence staining
Immunofluorescence was performed as described (Lee and Wilusz, 2010). Cells were fixed using 3.7% formaldehyde, and the rehydrated samples were incubated overnight at 4ºC with primary antibody anti-RCK diluted 1:300 (vol/vol) in blocking buffer and then incubated with secondary antibody conjugated with Alexa Fluor-594 (Molecular Probes, Eugene, OR) for 2 h at room temperature. Images were processed using an Axio Observer microscope with LSM 510 Meta image-processing software (Zeiss, Thornwood, NY).
Cell-cycle analysis
Isolation and staining of nuclei from cells after different treatments were performed with a CycleTest Plus DNA Reagent Kit (BD Biosciences, San Jose, CA). After propidium iodide was stoichiometrically bound to the nuclei, the samples were run on a flow cytometer.
Statistics
Values are means ± SE from three to six samples. Autoradiographic results were repeated three times. The significance of the difference between means was determined by analysis of variance. The level of significance was determined by using Duncan's multiple-range test (Harter, 1960).
Supplementary Material
Acknowledgments
This work was supported by Merit Review Grants (to J.N.R and J.Y.W.) from the U.S. Department of Veterans Affairs and by National Institutes of Health Grants DK57819, DK61972, and DK68491 (to J.Y.W.). M. Gorospe is supported by the National Institute on Aging–Intramural Research Program, National Institutes of Health. J.Y.W. is a Senior Research Career Scientist, Medical Research Service, U.S. Department of Veterans Affairs.
Abbreviations used:
- CDK4
cyclin-dependent kinase 4
- CR
coding region
- CUGBP1
CUG-binding protein 1
- DFMO
D,L-α-difluoromethylornithine
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IEC
intestinal epithelial cell
- IP
immunoprecipitation
- miRNA
microRNA
- ODC
ornithine decarboxylase
- PB
processing body
- Q-PCR
quantitative PCR
- Rb
retinoblastoma protein
- RBPs
RNA-binding proteins
- RNP
ribonucleoprotein
- TIAR
T-cell intracellular antigen-1 related protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0069) on July 7, 2011.
REFERENCES
- Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher NJ, Broadhurst GM, Minchin RF. Polyamine-dependent regulation of spermidine-spermine N1-acetyltransferase mRNA translation. J Biol Chem. 2007;282:28530–28539. doi: 10.1074/jbc.M701265200. [DOI] [PubMed] [Google Scholar]
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J. 2009;421:323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celano P, Baylin SB, Casero RA., Jr Polyamines differentially modulate the transcription of growth-associated genes in human colon carcinoma cells. J Biol Chem. 1989;264:8922–8927. [PubMed] [Google Scholar]
- Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol Biol Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Harter JL. Critical values for Duncan's new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]
- Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Lackner DH, Bahler J. Translational control of gene expression from transcripts to transcriptomes. Int Rev Cell Mol Biol. 2008;271:199–251. doi: 10.1016/S1937-6448(08)01205-7. [DOI] [PubMed] [Google Scholar]
- Lee EK, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010a;17:732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010b;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wilusz J. Translational symphony in (hnRNP) C major for APP. Nat Struct Mol Biol. 2010;17:675–676. doi: 10.1038/nsmb0610-675. [DOI] [PubMed] [Google Scholar]
- Li L, Liu L, Rao JN, Esmaili A, Strauch ED, Bass BL, Wang JY. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through P21 after polyamine depletion. Gastroenterology. 2002;123:764–779. doi: 10.1053/gast.2002.35386. [DOI] [PubMed] [Google Scholar]
- Li L, Rao JN, Bass BL, Wang JY. NF-kappaB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2001a;280:G992–G1004. doi: 10.1152/ajpgi.2001.280.5.G992. [DOI] [PubMed] [Google Scholar]
- Li L, Rao JN, Guo X, Liu L, Santora R, Bass BL, Wang JY. Polyamine depletion stabilizes p53 resulting in inhibition of normal intestinal epithelial cell proliferation. Am J Physiol Cell Physiol. 2001b;281:C941–C953. doi: 10.1152/ajpcell.2001.281.3.C941. [DOI] [PubMed] [Google Scholar]
- Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell. 2009;20:4885–4898. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Santora R, Rao JN, Guo X, Zou T, Zhang HM, Turner DJ, Wang JY. Activation of TGF-beta-Smad signaling pathway following polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1056–G1067. doi: 10.1152/ajpgi.00151.2003. [DOI] [PubMed] [Google Scholar]
- Palmer AJ, Wallace HM. The polyamine transport system as a target for anticancer drug development. Amino Acids. 2010;38:415–422. doi: 10.1007/s00726-009-0400-2. [DOI] [PubMed] [Google Scholar]
- Patel AR, Wang JY. Polyamines modulate transcription but not posttranscription of c-myc and c-jun in IEC-6 cells. Am J Physiol. 1997;273:C1020–C1029. doi: 10.1152/ajpcell.1997.273.3.C1020. [DOI] [PubMed] [Google Scholar]
- Persson L. Polyamine homoeostasis. Essays Biochem. 2009;46:11–24. doi: 10.1042/bse0460002. [DOI] [PubMed] [Google Scholar]
- Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell cultures from rat small intestineCharacterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RM, McCormack SA, Covington C, Viar MJ, Zheng Y, Johnson LR. The requirement for polyamines for intestinal epithelial cell migration is mediated through Rac1. J Biol Chem. 2003;278:13039–13046. doi: 10.1074/jbc.M208741200. [DOI] [PubMed] [Google Scholar]
- Reed SI. Control of the G1/S transition. Cancer Surv. 1997;29:7–23. [PubMed] [Google Scholar]
- Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Caceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- Seiler N, Raul F. Polyamines and the intestinal tract. Crit Rev Clin Lab Sci. 2007;44:365–411. doi: 10.1080/10408360701250016. [DOI] [PubMed] [Google Scholar]
- Sen S, Talukdar I, Webster NJ. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol. 2009;29:871–880. doi: 10.1128/MCB.01709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Pili R. Posttranscription regulation of prostate cancer growth. Cancer J. 2008;14:46–53. doi: 10.1097/PPO.0b013e318162108a. [DOI] [PubMed] [Google Scholar]
- Tsuda K, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Hesabi B, Moons DS, Pandolfi PP, Hansel KS, Koff A, Kiyokawa H. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol Cell Biol. 1999;19:7011–7019. doi: 10.1128/mcb.19.10.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Johnson LR. Polyamines and ornithine decarboxylase during repair of duodenal mucosa after stress in rats. Gastroenterology. 1991;100:333–343. doi: 10.1016/0016-5085(91)90200-5. [DOI] [PubMed] [Google Scholar]
- Wang JY, Johnson LR, Tsai YH, Castro GA. Mucosal ornithine decarboxylase, polyamines, and hyperplasia in infected intestine. Am J Physiol. 1991;260:G45–G51. doi: 10.1152/ajpgi.1991.260.1.G45. [DOI] [PubMed] [Google Scholar]
- Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J. 2010;426:293–306. doi: 10.1042/BJ20091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Passaniti A, Wang JY. Induced JunD in intestinal epithelial cells represses CDK4 transcription through its proximal promoter region following polyamine depletion. Biochem J. 2007;403:573–581. doi: 10.1042/BJ20061436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Zou T, Liu L, Yu TX, Zhu XY, Donahue JM, Wang JY. Induced ATF-2 represses CDK4 transcription through dimerization with JunD inhibiting intestinal epithelial cell growth after polyamine depletion. Am J Physiol Cell Physiol. 2010;298:C1226–C1234. doi: 10.1152/ajpcell.00021.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Rao JN, Guo X, Liu L, Zou T, Turner DJ, Wang JY. Akt kinase activation blocks apoptosis in intestinal epithelial cells by inhibiting caspase-3 after polyamine depletion. J Biol Chem. 2004;279:22539–22547. doi: 10.1074/jbc.M314337200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem J. 2008;409:389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.