Abstract
CAC1/RLF2 encodes the largest subunit of chromatin assembly factor I (CAF-I), a complex that assembles newly synthesized histones onto recently replicated DNA in vitro. In vivo, cac1/rlf2 mutants are defective in telomeric silencing and mislocalize Rap1p, a telomere-binding protein. Here, we report that in cells lacking CAF-I the silent mating loci are derepressed partially. MATa cac1 cells exhibit an unusual response to α-factor: They arrest and form mating projections (shmoos) initially, but are unable to sustain the arrest state, giving rise to clusters of shmooing cells. cac1 MATa HMLa HMRa strains do not form these shmoo clusters, indicating that derepression of HMLα causes the shmoo cluster phenotype in cac1 cells. When SIR3 is reintroduced into sir1 sir3 cells, HML remains derepressed indicating that SIR1 is required for the re-establishment of silencing at HML. In contrast, when SIR3 is reintroduced into cac1 sir3 cells, silencing is restored to HML, indicating that CAF-I is not required for the re-establishment of silencing. Loss of the other CAF-I subunits (Cac2p and Cac3p/Msi1p) also results in the shmoo cluster phenotype, implying that loss of CAF-I activity gives rise to this unstable repression of HML. Strains carrying certain mutations in the amino terminus of histone H4 and strains with limiting amounts of Sir2p or Sir3p also form shmoo clusters, implying that the shmoo cluster phenotype is indicative of defects in maintenance of the structural integrity of silent chromatin. MATa cac− sir1 double mutants have a synergistic mating defect, suggesting that the two silencing mechanisms, establishment and maintenance, function cooperatively. We propose a model to explain the distinctions between the establishment and the maintenance of silent chromatin.
Keywords: Silencing, chromatin, Sir proteins, histone acetylation, Rap1 localization factors
In differentiated cells, two identical genomic sequences can sometimes be found in two distinct states of expression. For example, in female mammals, one of the two X chromosomes is inactivated, whereas the other remains fully active (Latham 1996). Similarly, chromosomal imprinting ensures that a specific locus, when inherited from one parent, is completely inactive, whereas the same locus inherited from the other parent is completely active (Ferguson-Smith 1996). The imprinted state of the locus is inherited through many mitotic divisions and is generally reset only during meiosis. Inappropriate genomic imprinting can cause serious developmental defects, and several human genetic disorders are caused by mutations affecting imprinted genes (Hall 1990; Lalande 1996). Although the molecular mechanisms by which X inactivation and genomic imprinting are initiated and maintained are not well understood, the inactive X chromosome is in a highly condensed heterochromatic state and a similar chromatin state may occur at silenced, imprinted loci (John and Surani 1996).
One of the best studied examples of silencing occurs at the HM loci in the budding yeast Saccharomyces cerevisiae. S. cerevisiae has three mating type loci. Mating type genes expressed from the MAT locus normally determine the yeast mating type, either a or α, in haploid cells. Haploid cells normally respond to the mating pheromone of the opposite mating type by arresting in late G1 and forming mating projections (shmoos). In addition, wild-type strains have functional but transcriptionally repressed mating information at the HM loci, HML and HMR. If the HM loci become derepressed in haploid cells, both a and α mating information is expressed and the cells do not arrest growth or form mating projections in response to mating pheromones. Thus, by monitoring the pheromone response of haploid cells one can infer the expression state of the HM loci.
The silent state of the HM loci is attributable to a specialized form of chromatin that is the yeast version of metazoan heterochromatin (Grunstein 1995; Braunstein et al. 1996). Genes within the HM loci are inaccessible to DNA modification enzymes, RNA polymerases II and III, and excision repair enzymes (for review, see Fox and Rine 1996). The acetylation state of histones H3 and H4 in the nucleosomes of silent chromatin is different from that of bulk chromatin or of the active MAT locus; at the HM loci, histone H4 is hypoacetylated except on lysine-12 (Braunstein et al. 1996). This is similar to the acetylation pattern conferred on newly synthesized histone H4 by the cytoplasmic histone acetyltransferase Hat1p (Kleff et al. 1995; Parthun et al. 1996). A number of mutations in acetylated lysines in the amino termini of histones H3 and H4 weaken silencing at the HM loci or at telomeres (for review, see Grunstein 1995). Thus, histone acetylation may play an important role in the inheritance of chromatin expression states.
The Sir complex proteins (composed of Sir2p, Sir3p, and Sir4p and not including Sir1p) are structural components of yeast heterochromatin that associate with histones (Hecht et al. 1996; Strahl-Bolsinger et al. 1997). Loss of any one of these Sir complex proteins abrogates silencing completely (Rine and Herskowitz 1987). Sir3p and Sir4p interact with one another genetically (Ivy et al. 1986; Marshall et al. 1987) and in two-hybrid screens (Moretti et al. 1994), and all three Sir complex proteins can be isolated in complexes with each other (Moazed et al. 1997; Strahl-Bolsinger et al. 1997). Histones H3 and H4 coprecipitate with Sir3p (Hecht et al. 1996), and mutations in the amino termini of either H3 or H4 that affect silencing in vivo also affect the interaction of H3 and H4 with the Sir complex in vitro (Hecht et al. 1995).
The concentration of Sir complex proteins is critical for silencing. Changes in the stoichiometry of Sir complex proteins alters silencing (Ivy et al. 1986; Marshall et al. 1987; Sussel et al. 1993). The Sir complex proteins localize to a number of perinuclear foci that are often associated with silent telomeric DNA (Gotta et al. 1996). These foci are thought to reflect subnuclear domains of high Sir complex concentration in which silent chromatin is localized (Gotta et al. 1996, 1997). The HM loci and telomeres compete for Sir proteins, and the proximity of the HM loci to telomeres contributes to HM silencing (Buck and Shore 1995; Maillet et al. 1996).
The DNA sequences at the HM loci differ from the sequences at the MAT locus in that each HM locus is flanked by two silencers, E and I. Each E or I silencer contains an autonomously replicating (ARS) consensus sequence that is bound by the origin recognition complex (ORC) (Bell et al. 1993). In addition, each silencer contains a binding site for the ARS-binding factor 1 (Abf1p) or a binding site for the repressor/activator protein 1 (Rap1p). The E and I silencers, as well as the individual binding sites and the factors that bind them directly, have redundant functions. In most situations, one silencer is sufficient to silence an HM locus and any two of the three individual sites within a silencer are sufficient for HM silencing (Brand et al. 1987; Mahoney and Broach 1989; McNally and Rine 1991). Specific mutations in the sites (or in the factors that bind them) reduce the redundancy of HMR silencing and can reveal the roles of silencing factors such as Rap1p (Sussel and Shore 1991), ORC (Bell et al. 1993; Micklem et al. 1993; Loo et al. 1995a), and Abf1p (Loo et al. 1995b; Fox et al. 1997).
The study of situations in which silencing is weakened, but not abrogated, has provided important insights into the mechanisms by which silencing occurs. sir1 mutants exhibit epigenetic silencing of HML. In a subset of the sir1 cells, HML is fully repressed and the repressed state is inherited in most of their progeny; in the remaining sir1 cells, HML is fully derepressed and the derepressed state is inherited (Pillus and Rine 1989). Sir1p interacts physically with both Orc1p and with Sir4p (Triolo and Sternglanz 1996). Sir1p, when tethered to the HML locus in the absence of a silencer, is sufficient to direct silencing (Chien et al. 1993). Deletion of the ORC-binding site also causes defects in the establishment of silencing, which lead to derepression of the HM loci in a subset of the mutant cells (Mahoney et al. 1991; Sussel et al. 1993). Thus, Sir1p contributes to the establishment of silencing in wild-type cells by interacting with ORC and recruiting structural components of silent chromatin, such as Sir4p, to the silent loci.
Pillus and Rine (1989) proposed that there are two steps in HM silencing: (1) maintenance of the current state of the silent chromatin, and (2) re-establishment of the repressed state when HML becomes derepressed. Although deletion of SIR1 and mutation of single sites within the HM loci cause defects in the re-establishment of silencing, they do not affect the ability to inherit the repressed chromatin state (Pillus and Rine 1989; Mahoney et al. 1991). Derepression of HMR (by inactivation of a temperature-sensitive Sir3 protein) can be restored only after passage through S phase (Miller and Nasmyth 1984), indicating that the re-establishment of silencing requires passage through S phase. Conversely, Holmes and Broach (1996) demonstrated that if the cis-silencer is excised from the chromosome, the repressed state of the chromatin can be maintained during α-factor arrest, but cannot be inherited efficiently. Taken together, these studies indicate that the establishment, maintenance, and inheritance of silencing all contribute to the formation of fully silenced HM loci.
Mammalian chromatin assembly factor I (CAF-I) was identified by its ability to assemble histones into nucleosomes in a DNA replication-dependent manner in vitro (Stillman 1986). CAF-I assembles preferentially histones H3 and H4 with the acetylation pattern of newly synthesized cytoplasmic histones (Smith and Stillman 1991; Kaufman et al. 1995; Verreault et al. 1996). S. cerevisiae CAF-I is encoded by CAC1, CAC2, and CAC3 (Kaufman et al. 1997). CAC1, the largest subunit of CAF-I, is identical to RLF2, a gene that we identified in a screen for mutants defective in telomere-related functions (Enomoto et al. 1994, 1997), and CAC3 is identical to MSI1, a gene identified in high-copy suppressor screens (Ruggieri et al. 1989; Hubbard et al. 1992). All three cac mutant strains display similar phenotypes; cells grow well but are defective in telomeric silencing, the segregation of TEL + CEN plasmids, and Rap1p localization (Enomoto et al. 1997; Kaufman et al. 1997). Similar phenotypes have been observed in strains carrying mutations in either SIR2, SIR3, or SIR4, in strains carrying mutant alleles of histones H3 and H4, and in strains carrying rap1s mutations (Enomoto et al. 1994). Because many of these genes are involved in HM silencing, as well as telomeric silencing, we examined the role of CAF-I in HM silencing.
In this paper we show that CAF-I contributes to the maintenance, but not the re-establishment, of silencing at the HM loci. In cac− mutants, we observed a transient loss of α-factor response, at the individual cell level. Cells form mating projections and divide slowly on α-factor, forming clusters of shmooing cells. The formation of shmoo clusters requires α-mating information at HML, indicating that this α-factor response reflects a defect in the maintenance of HML silencing. We have investigated the relationship between the maintenance and the re-establishment of silencing at HML by analyzing the roles of CAF-I, histones, Sir complex proteins, and Sir1p using α-factor confrontation assays.
Results
cac1 mutations affect HML silencing
Many of the factors that contribute to telomere position effect also contribute to silencing at the two HM loci HML and HMR. Although cac1/rlf2 mutants are defective in the repression of telomere adjacent genes, quantitative mating assays did not detect a mating defect in cac− strains (Enomoto et al. 1997; Kaufman et al. 1997). We tested HML repression in cac1 cells using an α-factor response assay that is more sensitive to derepression of HMLα than are quantitative mating assays. Exponentially growing MATa cells were resuspended in liquid medium containing α-factor and the proportion of dividing cells (cells with one or more growing buds) and the proportion of arrested cells (unbudded cells with or without mating projections) was determined. Three hours after the addition of α-factor, 1% of the wild-type cells were dividing, whereas 10–20% of the cells in cac1 strains (cac1-Δ1 and cac1-1) were dividing. This difference between wild-type and cac1 cells suggested that either HML is slightly derepressed in all cac1 cells or that HML is derepressed in a population of the cac1 cells. For comparison, in an isogenic MATa sir1 strain, 32% of the cells were dividing in α-factor.
CAC1, together with the ORC-binding site, contributes to silencing at HMR
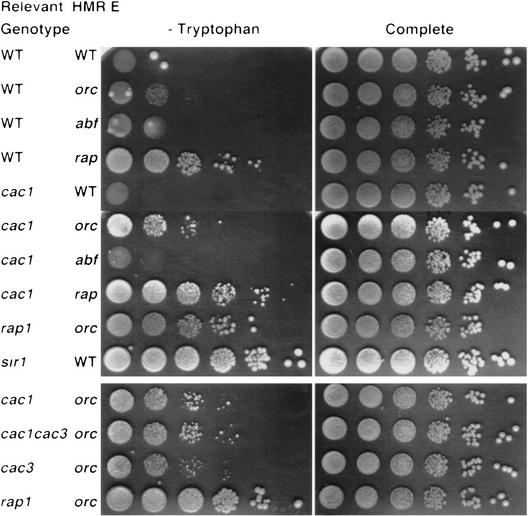
To measure the role of CAC1 at HMR, we used an HMR::TRP1 construct in which the a1 and a2 genes are replaced by the TRP1 gene (Hardy et al. 1992). Assays that measure expression of HMR::TRP1 are more sensitive to low levels of HMR derepression than are mating assays. We constructed a series of isogenic strains carrying the cac1-Δ1 allele and either HMR::TRP1 (including the intact silencer) or derivatives missing binding sites for either ORC, Abf1p, or Rap1p. We compared the ability of these strains to grow on medium lacking tryptophan with the growth of isogenic CAC1 strains (Fig. 1). Consistent with published results (e.g., Sussel and Shore 1991), only the hmr::TRP1 strain missing the Rap1p site grew on medium lacking tryptophan (Fig. 1). In the cac1 series of strains, Trp+ colonies also appeared in the strain lacking the ORC site (Fig. 1), suggesting that CAC1 contributes to silencing at HMR. In addition, this result suggests that CAC1 and the ORC-binding site in HMR E are necessary together for HMR silencing.
Figure 1.
CAF-I contributes to the repression of HMR. Cells with the indicated genotype at the CAC1, CAC3, RAP1, or SIR1 locus and deleted for the listed sites within HMR E::TRP1 were plated in 10-fold serial dilutions onto medium lacking tryptophan (left) or complete medium (right). Colonies were photographed after 2 days at 30°C. (Top) Strains used: WT WT, YJB959; WT orc YJB955; WT abf1, YJB1143; WT rap1, YJB1104; cac1 WT, YJB1960; cac1 orc YJB958; cac1 abf1, YJB1139; cac1 rap1, YJB1101; rap1-12 orc, YJB1638; and sir1 WT, YJB2006. (Bottom) Strains used in a separate experiment photographed after 3 days at 30°C were: cac1 orc, YJB958; cac3 orc, YJB2011; cac1 cac3 orc, YJB2009; and rap1-12 orc, YJB1638.
Interestingly, on medium lacking tryptophan, colonies of cac1 hmr::TRP1 strains lacking the ORC site are smaller than colonies formed by either sir4 HMR::TRP1 (data not shown) or sir1 HMR::TRP1 mutants (Fig. 1). sir4 strains are derepressed completely at the HM loci, whereas sir1 strains include two populations of cells, those that are repressed and those that are derepressed (Pillus and Rine 1989). Like sir1 mutants, strains carrying the rap1-12 mutation (which also causes derepression of hmr::TRP1 in strains lacking the ORC site) give rise to a population of cells that grows without tryptophan and each of these forms a colony that is larger than the cac1 colonies (Fig. 1). Because on complete medium colonies of cac1 hmr:TRP1 strains lacking the ORC site are similar in size to colonies of isogenic wild-type, sir1, or rap1-12 strains (Fig. 1), the “minicolony” phenotype observed for cac1 cells on medium lacking tryptophan is related specifically to expression of the TRP1 allele in the hmr locus. Because we do not observe a population of large Trp+ colonies in these strains, this observation also implies that, in contrast to sir1 mutants, either none of the cac1 cells are derepressed completely when hmr::TRP1 is missing the ORC site, or derepressed cells and their descendants do not remain derepressed as long as sir1 mutant cells.
cac1 cells exhibit an unusual budding shmoo response to α-factor
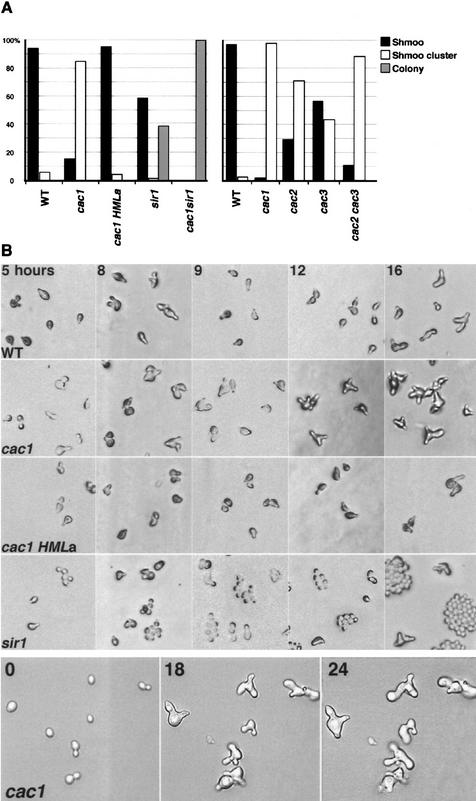
MATa cells expressing only a mating information form mating projections, termed shmoos, and arrest in the G1 stage of the cell cycle in response to α-factor. However, α-factor does not affect the growth and division of cells expressing both a and α mating type genes. To understand the α-factor response of cac1 strains, we assayed the response of individual MATa cac1 cells to prolonged α-factor treatment on solid medium. After 18 hr at 23°C, 94% of the wild-type cells arrested as shmoos in response to the α-factor treatment (Fig. 2A). In sir1 strains, two populations of cells were observed; ∼60% of the cells arrested as shmoos, and ∼40% of the cells divided actively and formed colonies of round yeast cells (Fig. 2A). The cac1 strains exhibited an entirely different response to α-factor (Fig. 2). After 18 hr, the vast majority of the cac1 cells (85%) had formed clusters of cells with multiple shmoo-like projections extending in different directions (Fig. 2A). We have observed similar clusters of shmooing cells in cac1 strains in a number of genetic backgrounds (data not shown). In all cases, the vast majority of cac1 cells formed these unusual shmoo clusters on α-factor at a time that wild-type cells were arrested as individual shmoo cells. Individual mating projections contained a nucleus (as determined by DAPI staining) and eventually could be separated by micromanipulation (data not shown), indicating that the mating projections are buds that give rise to individual cells. Eventually (∼12 hr later than cac1 cells) wild-type cells formed similar clusters of shmoo cells. After longer periods of time, small colonies of shmooing cac1 cells are evident. The presence of shmoos at the colony edges indicated that the dividing cells in the colony were not resistant to α-factor. Furthermore, it demonstrated that the α-factor in the medium was still active.
Figure 2.
α-Factor response of cac strains on solid media. Yeast cells were spread onto α-factor–YPD plates and maintained at 23°C. Cells were analyzed at indicated times after exposure to α-factor. (A) Analysis of yeast cell populations after 18 hr on α-factor. More than 100 cells per strain were analyzed. (Shmoo) Individual cells that formed mating projections and remained arrested; (shmoo cluster) individual cells that formed multiple mating projections and eventually divided at least once; (colony) cells that formed colonies of round cells and did not appear to respond to α-factor. (Left) Strains used: WT, YJB276; cac1, YJB469; cac1 HMLa, YJB2057; sir1, YJB335; and sir1 cac1, YJB744. (Right) Strains used: WT, YJB195; cac1, YJB1838; cac2, YJB1803; cac3, YJB1581; and cac2 cac3, YJB1865. (Left) χ2 tests indicated that the difference between WT and cac1 HMLa strains was not significant, whereas differences between all other pairwise combinations were significant. (Right) All pairwise combinations were significantly different except that the cac1 and the cac2 cac3 strains were not significantly different. (B) Analysis of cells over time. (Top four rows) Populations of cells; (bottom row) the same individual cells photographed at indicated times after exposure to α-factor.
To analyze the dynamics of shmoo cluster formation in cac1 cells, we observed cells after different times on α-factor (Fig. 2B) and followed individual cells by time-lapse microscopy (Fig. 2B, bottom row). Virtually all MATa cac1 cells responded with an initial period of cell cycle arrest. Within the first 3–5 hr on α-factor at 23°C they formed mating projections at a time when α-factor-resistant sir1 cells were dividing. However, after 8–9 hr of arrest, virtually all of the cac1 cells formed a second shmoo-like projection. This second projection continued to grow and a third projection, often projecting perpendicular to the surface of the medium, appeared on most cells by 12 hr. At 16 hr, a fourth projection appeared on many of the cac1 cells, whereas a few of the arrested wild-type shmoos began to form a second projection. α-Factor-resistant sir1 cells divided approximately once every 2 hr. In contrast, in cac1 cells, new mating projections appeared approximately once every 4 hr, and eventually, each mating projection gave rise to an individual cell. This suggests that cac1 cells arrested in response to α-factor, but eventually resume and complete a cell cycle. We term the groups of cells with multiple mating projections shmoo clusters and the individual cells that arise from each mating projection budding shmoos.
The budding shmoo phenotype occurs in virtually all cac1 cells, indicating that, unlike sir1 cells, cac1 cells do not exist in two distinct epigenetic states. Rather, our observations suggest that cac1 cells are all in a similar state that responds to α-factor initially, but cannot sustain the α-factor response over time.
cac1 α-factor-resistant colonies are attributable to expression of α genes from HML
When MATa cells encounter α-factor, they must commit to a new developmental program by repressing pathways that lead to continued cell division and by activating pathways required for cell cycle arrest and polarized growth toward the α-factor source. Later on, the process of recovery or adaptation to α-factor stimulation is induced. The most parsimonious explanation for the appearance of shmoo clusters in MATa cac1 strains exposed to α-factor and for the formation of small colonies of cac1 hmr::TRP1 strains lacking the ORC site on medium lacking tryptophan, is that silencing of the HM loci is weakened in cac1 strains. If this is the case, cac1 cells lacking α1 or α2 genes should remain arrested on α-factor. However, an alternative possibility is that the loss of CAF-I activity alters directly the transcriptional efficiency of genes that either control cell division or that affect the adaptation of cells to α-factor. If this is the case, cac1 cells lacking α1 and α2 genes should form shmoo clusters in response to α-factor. To distinguish between these two alternatives, we analyzed the α-factor response of strain YJB2057 (MATa cac1 HMRa HMLa), in which a mating information was substituted for α mating information at HML. YJB2057 did not form a second mating projection; only individual shmoo cells were observed (Fig. 2A). Like wild-type strains, these cells remained arrested for >18 hr. This result indicates that HMLα information is required for the budding shmoo phenotype in MATa cac1 strains and implies that the unusual α-factor response of MATa cac1 strains is attributable to weakened repression of HML in cells lacking Cac1p. Because the cac1 cells continue to shmoo and arrest, our results suggest that in all cac1 cells, HML oscillates between the repressed state and the derepressed state.
cac1 mutations enhance the mating defect of sir1 strains
Pillus and Rine (1989) demonstrated that in sir1 strains, silencing of a derepressed HML locus can be reestablished at a rate of 4 × 10−3 changes in state per cell generation. In contrast, if the HML locus is repressed, its repressed state can be maintained in >90% of the cells in the absence of Sir1p. We analyzed the response of MATa cac1 sir1 cells to α-factor to determine whether the residual HML repression in cac1 single mutants is dependent on Sir1p functions. In other words, when cac1 mutants become derepressed after incubation on α-factor, is the re-establishment of α-factor responsiveness in the next cell cycle dependent on Sir1p function? We reasoned that if residual HML silencing in cac1 mutants as independent of Sir1p re-establishment, then the proportion of α-factor-resistant cells in the double mutant population should remain similar to the number of α-factor-resistant cells in the sir1 single mutant population. On the other hand, if HML repression is relieved transiently in a cac1 mutant, and if Sir1p was required for re-establishment of the repressed state of HML, then we would expect an increase in the proportion of α-factor-resistant colonies in the cac1 sir1 double mutants. Consistent with the latter expectation, virtually all of the MATa cac1 sir1 cells were resistant to α-factor (Fig. 2A). The proportion of α-factor-resistant cells in the double mutant strains was significantly greater than the proportion of α-factor-resistant cells in the sir1 single mutant strain in two different strain backgrounds (data not shown). In addition, quantitative mating assays confirmed that MATa cac1 sir1 strains have reduced mating efficiency (Fig. 4, below). It is paradoxical that almost 100% of cac1 sir1 cells form “colonies” rather than shmoos or shmoo clusters on α-factor, yet the mating efficiency of cac1 sir1 strains is reduced only 10- to 100-fold (depending on the strain background). We think this difference is due to some of the cac1 sir1 colonies having elongated cells, which we presume to be mating competent. Thus, we conclude that derepression caused by the cac1 mutation requires Sir1p to reestablish repression in cells where HML becomes derepressed.
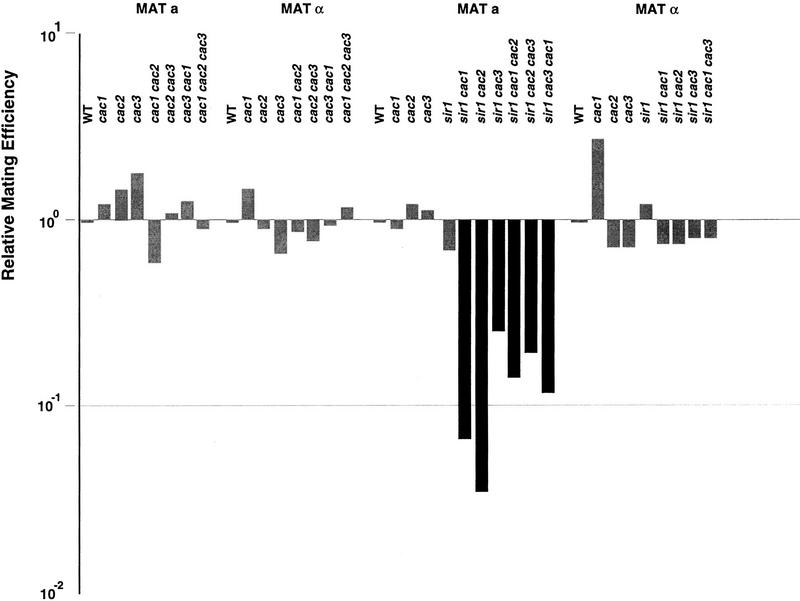
Figure 4.
Mutation of CAF-I subunits causes subtle MATa mating defects. At least four individual quantitative mating assays were performed for each strain. The median value of the assays is shown. All values are normalized to the isogenic wild type. Solid bars indicate that results were statistically different from wild type at the P < 0.05 level. Strains (MATa, MATα): WT, YJB195, YJB209; cac1, YJB1838, YJB1578; cac2, YJB1803, YJB1599; cac3, YJB1581, YJB1836; cac1 cac2, YJB1804, YJB1802; cac2 cac3, YJB1865, YJB1864; cac1 cac3, YJB1862, YJB1863; sir1, YJB1940, YJB1941; sir1 cac1, YJB1962, YJB1961; sir1 cac2, YJB2000, YJB2034; sir1 cac3, YJB1945, YJB1946; sir1 cac1 cac2, YJB2044; sir1 cac2 cac3, YJB2048; sir1 cac1 cac3, YJB2007, YJB1993.
CAF-I is not required for the re-establishment of silencing
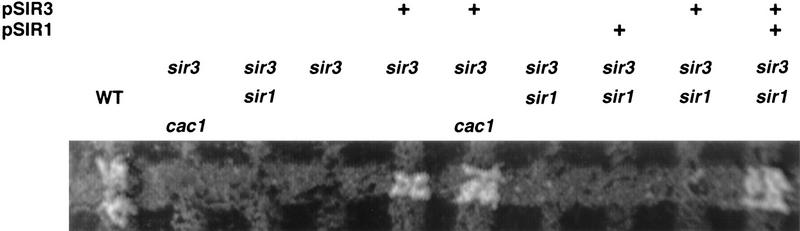
The re-establishment of silencing at a derepressed HM locus occurs readily in wild-type cells but is a very rare event in sir1 cells (Pillus and Rine 1989). We compared the role of cac1 and sir1 in the re-establishment of silencing by monitoring the state of HML in sir3 strains in which Sir3p expression was restored by transformation with a centromere plasmid-carrying SIR3 (pSIR3). In all cases, sir3 cells not carrying pSIR3 did not mate (Fig. 3) and did not respond to α-factor (data not shown). These strains were then transformed with pSIR3 to provide a single copy of the SIR3 gene expressed from its own promoter. In the otherwise wild-type sir3 pSIR3 strain, mating competence was readily restored and, when exposed to α-factor, arrested shmoo cells appeared. As expected, as SIR1 is an important contributor to the re-establishment of HML silencing, the opposite result was seen in the isogenic sir1 strain (Fig. 3); sir1 sir3 pSIR3 cells did not mate (Fig. 3) and, when exposed to α-factor, did not give rise to arrested shmoo cells. Cotransformation with both pSIR3 and a centromere plasmid-carrying SIR1 (pSIR1) in this strain restored mating competence and α-factor responsiveness, indicating that it was the lack of Sir1p that limited the ability of this strain to restore HML to the repressed state. In contrast, transformation of the cac1 sir3 cells with pSIR3 led to the appearance of mating competent cells and these cells arrested as shmoos when exposed to α-factor, indicating that HML was restored to the silent state. These results clearly demonstrate that cac1 is not required for the re-establishment of silencing when HML has been derepressed.
Figure 3.
SIR1 is required, and CAC1 is not required, for the re-establishment of HML silencing. Plasmids pSIR3 (pSE334 or pJR273) and pSIR1 (pJR910) were introduced (indicated by +) into strains carrying sir3 and the other indicated mutations. Two days after transformation, transformants were allowed to mate for 18 hr with a Matα tester strain (TD1). Diploids were then selected by replica plating onto SDC medium lacking adenine and histidine. Strains used were sir3 sir1, YJB2471; sir3 cac1, YJB2109; sir3, YJB2544; WT, YJB195.
The HM silencing defect in cac1 strains is attributable to the loss of CAF-I function
CAC1 encodes the large subunit of CAF-I, a trimeric complex that includes Cac1p/Rlf2p, Cac2p, and Cac3p/Msi1p (Kaufman et al. 1997). To determine whether the HM silencing defect in cac1 mutants was attributable to the absence of CAF-I function, we constructed strains carrying mutations in one, two, or all three genes encoding CAF-I subunits and performed quantitative mating assays on both MATa and MATα mutant strains. Like cac1 mutants, the single, double, and triple cac mutants of both mating types were able to mate with wild-type efficiency in quantitative mating assays (Fig. 4). However, all of the single and double cac mutants have measurable MATa mating defects in combination with sir1 (Fig. 4). Single and double cac mutants in combination with sir1 behaved like the sir1 cac1 strain; the MATa strains mated with reduced efficiency, whereas the MATα strain mating was not significantly different from the wild-type, sir1, or cac single mutant strains (Fig. 4).
Like MATa cac1 strains, the MATa cac2 and MATa cac3 strains produced shmoo clusters by 18 hr at 23°C (see Fig. 2A). Interestingly, cac2 and cac3 mutations caused less severe silencing defects (61% and 42% shmoo clusters, respectively) than did cac1 mutants (97% shmoo clusters), which encodes the largest CAF-I subunit (see Fig. 2A). This may occur because CAC2 and CAC3 both encode small proteins that include WD40 repeats (Verreault et al. 1996; Kaufman et al. 1997), which may be partially redundant with one another. Consistent with this idea, the silencing defect of cac2 cac3 double mutants (89% shmoo clusters) is not significantly different from that of cac1 mutants (see Fig. 2A).
To determine whether loss of any one of the CAF-I components also causes derepression of hmr::TRP1 strains lacking the ORC site, we analyzed the Trp phenotype of strains carrying either cac3 alone or as a double mutant with cac1-Δ1 (see Fig. 1). The cac3 ORC site mutant grew slower and formed smaller colonies than the cac1 ORC site mutant, although the number of Trp+ colonies was similar in both strains (see Fig. 1). This effect of cac3 on hmrΔA::TRP1 silencing is reminiscent of the mini-colony phenotype of cac3 strains relative to cac1 and cac2 strains in telomeric silencing assays (Kaufman et al. 1997). The cac1 cac3 mutant was derepressed to the same degree as the cac1 single mutant, suggesting that cac3 mutants may retain some CAF-I function that is lost in cac1 mutants. Taken together, our results indicate that loss of CAF-I function, rather than the loss of Cac1p alone, causes derepression of both HML (in sir1 strains) and HMR (when the ORC site is missing).
Can defects in histones give rise to shmoo clusters?
CAF-I is unlikely to be a structural component of silent chromatin, because CAF-I localizes to replication foci in mammalian cells (Krude 1995) and overexpressed epitope-tagged Cac1p localizes to nuclear foci that do not colocalize with Rap1p (Enomoto et al. 1997), a structural component of the silencers. In vitro studies identified CAF-I as an activity that preferentially assembles specifically acetylated histones H3 and H4 into nucleosomes on recently replicated DNA. Yet genes encoding CAF-I subunits are not essential for yeast cell viability, indicating that alternative chromatin assembly mechanisms must function in cac mutants (Kaufman et al. 1997). If weakened silencing at the HM loci in cac1 mutants is attributable to subtle alterations in the nucleosomes assembled at these loci, we hypothesized that mutations in CAF-I and certain mutations in histones H3 and H4 should give rise to similar phenotypes. To test this hypothesis, we analyzed the α-factor response of strains carrying mutations in either histone H3 or histone H4.
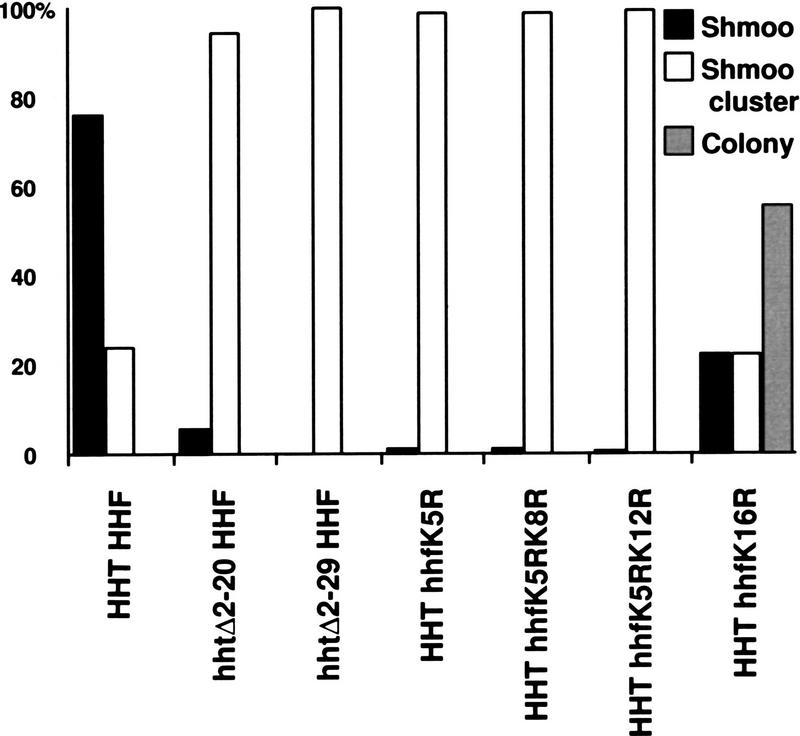
A number of mutations in lysine 16 of histone H4 virtually eliminate HM silencing (e.g., histones H4 K16A and H4 K16Q), whereas H4 K16R causes a small, but measurable reduction in mating of MATa strains (Johnson et al. 1990; Megee et al. 1990; Park and Szostak 1990). Other mutations in the amino termini of histones H3 and H4, such as deletion of the entire H3 amino terminus or mutation of H4 lysines 5, 8, or 12 to arginine, have little, if any, effect on silencing, when monitored by quantitative mating assays (Johnson et al. 1990; Megee et al. 1990; Park and Szostak 1990; Morgan et al. 1991; Thompson et al. 1994). As expected, we found that strains carrying the histone H4 K16A and H4 K16Q mutations did not arrest at all in response to α-factor (data not shown), and therefore, were not informative with regard to the mechanism by which HM silencing was defective. Also consistent with published data, the H4 K16R strain included a population of cells that did not respond to α-factor. However, like cac1 strains, strains carrying mutations in the amino terminus of histone H4 (K5R, K5R K8R, K5R K12R, and K16R; Megee et al. 1990), or strains carrying deletions in the histone H3 amino terminus (Δ2-20 and Δ2-29; Morgan et al. 1991) that are competent to mate gave rise to shmoo clusters in response to α-factor (Fig. 5). The fact that changes within the amino terminus of histones H3 or H4 are sufficient to give rise to the shmoo cluster phenotype implies that this phenotype reflects subtle defects in the structural integrity of silenced chromatin. These results also suggest that silencing defects in the histone mutant strains (which carry a wild-type allele of CAC1), are similar to the silencing defects in strains lacking CAF-I (which carry only wild-type histone alleles).
Figure 5.
α-Factor response of strains with mutations in the amino termini of histones H3 or H4. Yeast cells were treated as described in Fig. 2A. χ2 tests indicated that HHT HHF and HHT hhfK16R were significantly different from each other and from the other histone mutants. Strains used: HHT HHF, YJB2166; hhtΔ2-20 HHF, YJB2167; hhtΔ2-29 HHF, YJB2168; HHT hhfK5R, YJB2169; HHT hhfK5R K8R, YJB2170; HHT hhfK5R K12R, YJB2171; and HHT hhfK16R, YJB2172.
Is the budding shmoo phenotype sensitive to Sir complex protein concentration?
The stoichiometry of Sir complex proteins is critical for silencing (Ivy et al. 1986; Marshall et al. 1987), and the concentration of Sir complex proteins is likely limiting, as under a number of conditions the telomeres and HM loci compete for Sir complex proteins (Buck and Shore 1995). Our working hypothesis is that silencing is maintained by the efficient assembly (by CAF-I) of nucleosomes and the strong association of these CAF-I-assembled nucleosomes with Sir complex proteins, which render the underlying DNA inaccessible to enzymes. This hypothesis predicts that the efficiency of silencing should be dependent on the concentration of Sir complex proteins. We tested whether providing an additional copy of each SIR gene could improve the repression of the HML locus in cac1 mutants. MATa cac1 strain YJB469 was transformed with a plasmid carrying either SIR1, SIR2, SIR3, or SIR4 or with the vector (YCplac33) alone, and the response to α-factor was monitored by time-lapse microscopy. All MATa cac1 cells carrying only the vector plasmid formed shmoo clusters within 18 hr of exposure to α-factor; no individual shmoo cells were observed. Similarly, all MATa cac1 cells expressing an extra copy of SIR1 formed shmoo clusters on α-factor, indicating that Sir1p was not limiting in MATa cac1 cells. In contrast, the presence of an extra copy of either SIR2, SIR3, or SIR4 improved the silencing and α-factor response of MATa cac1 cells; 20–30% of the cells arrested to form individual shmoos.
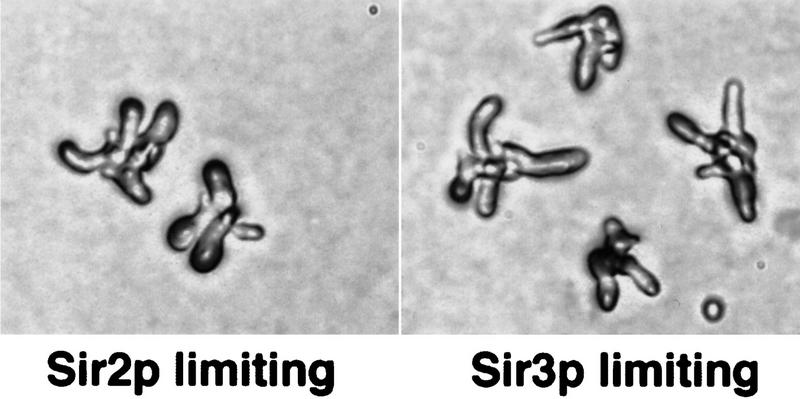
The ability of additional Sir2p, Sir3p, or Sir4p to restore α-factor arrest to cac1 cells is consistent with the idea that cac1 mutants have subtle defects in the maintenance of the heterochromatin at HML, perhaps attributable to a limiting amount of Sir complex proteins in the complex. If this is the case, reduced concentrations of Sir complex proteins in otherwise wild-type cells should give rise to phenotypes similar to those seen in strains lacking CAF-I activity. To determine whether limiting Sir complex protein concentration can give rise to shmoo clusters, we used sir mutant strains that expressed the wild-type SIR gene from the GAL10 promoter. Cells were pregrown on raffinose, which permitted sufficient expression of the SIR genes to repress HML. The strains were then released into glucose medium (to repress the SIR gene expression) and plated either immediately or after 2, 4, 6, or 8 hr onto α-factor plates containing glucose. The response to α-factor was then monitored by time-lapse microscopy. In cultures in which either SIR2 expression or SIR3 expression was repressed (by growth on glucose for 8 hr before α-factor exposure), shmoo clusters appeared (Fig. 6). Furthermore, the proportion of cells giving rise to shmoo clusters (and α-factor-resistant colonies) increased with increasing time of glucose repression of either SIR2 or SIR3 before α-factor exposure (data not shown). Thus, limiting the amount of Sir2p or Sir3p in otherwise wild-type cells is sufficient to weaken silencing and generate the budding shmoo phenotype.
Figure 6.
Limiting amounts of Sir2p or Sir3p weaken the maintenance of silencing at HML. Strains limiting for Sir2p (YJB285 [pAR14]) or Sir3p (YJB397 [pAR16]) were generated by pregrowth on raffinose, transfer to glucose for 8 hr, and then plating on α-factor lacking leucine and containing glucose. Elongated shmoo clusters that arose in these cultures are shown.
Discussion
Complete HM silencing requires CAF-I
Mutations in CAF-I subunits cause derepression of the HM loci. Sensitive assays that detect derepression of the HM loci, such as α-factor arrest or HMR::TRP1 expression, reveal silencing defects in cac mutant strains at both HML and HMR. The unusual budding shmoo phenotype observed in MATa cac1 cells grown on α-factor plates provides a new tool for analyzing the mechanisms of silencing. Derepression of the HMLα locus causes the budding shmoo phenotype seen in cac1 cells, because we do not see shmoo clusters in a MATa cac1 HMLa strain.
Sir1p and CAF-I contribute to different aspects of mating silencing
Mutation of either SIR1 or CAC1 causes subtle mating defects. However, there are important differences between sir1 and cac1 mutant strains. cac1 and sir1 mutants have different relationships with the sites in the HMRE silencer. The silencing defect of sir1 cells is enhanced when the Abf1 site is deleted from HMRE (Chien et al. 1993). In contrast, the silencing defect of cac1 cells is enhanced only in strains lacking the ORC site at HMRE. Most important, sir1 cells exist in one of two distinct epigenetic states that can be detected by their responses to α-factor: (1) the derepressed state that forms colonies, and (2) the repressed state that arrests as shmoos. In contrast, cac1 cells are not found in two distinct states; they all appear to form shmoo clusters with similar kinetics. Moreover, sir1 and cac1 cells respond very differently when SIR3 is restored to sir3 cells (Fig. 3). Clearly, SIR1 is required for the re-establishment of the silent state at HML, whereas CAC1 is not required for the re-establishment process.
We propose that the budding shmoo phenotype reflects a transient defect in the maintenance, rather than the re-establishment of HML silencing. The following observations lead us to this proposal. First, the budding shmoo phenotype does not occur in cac1 mutants that lack α information at HML, indicating that it is derepression of HMLα that gives rise to the phenotype. Second, in hmr::TRP1 strains lacking the ORC site, cac1 colonies are smaller than sir1 or sir4 colonies on medium lacking tryptophan. Similarly, clusters of MATa cac1 cells dividing on α-factor are much smaller than MATa sir4 or MATa sir1 colonies. The smaller size of cac1 colonies is consistent with the idea that, in cac1 cells, both HML and HMR are partially, rather than completely, derepressed. Third, the shmoo–bud–shmoo cycle observed in cac1 cells suggests that HML derepression occurs because the HML locus in each cac1 cell oscillates between the repressed state (because cells initially respond to α-factor) and the derepressed state (because cells form new mating projections/buds at a time when wild-type cells have not recovered from α-factor arrest). Fourth, this oscillation between the two states of HML, and the loss of this oscillation in most MATa cac1 sir1 cells, implies that Sir1p facilitates the re-establishment of repression at the HML locus in cac1 cells. Finally, alterations in either the quantity or quality of the major components of yeast heterochromatin (Sir complex proteins and histones, respectively) give rise to the budding shmoo phenotypes, suggesting that the shmoo cluster response to α-factor reflects subtle defects in the structural integrity of the heterochromatin itself. Thus, we propose that Cac1p (and CAF-I) are required for the structural integrity, or maintenance, of yeast silent chromatin.
A model for silencing at HM loci
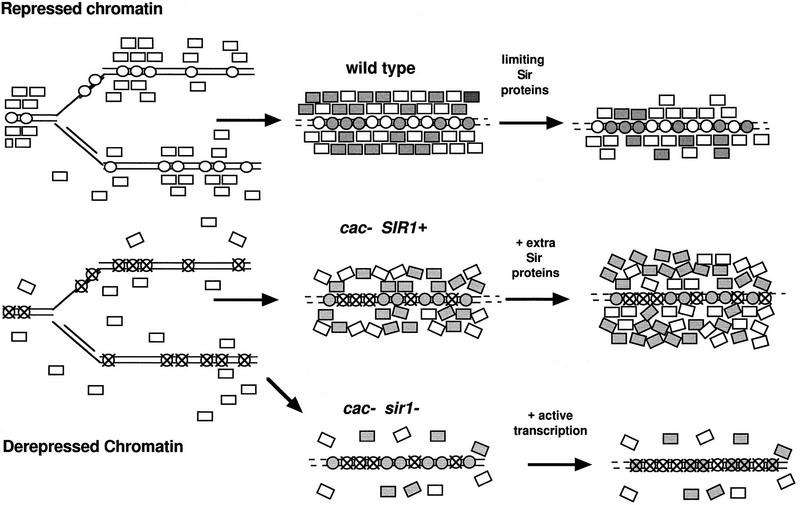
Previously, we proposed that CAF-I is required for telomeric silencing because it promotes efficient nucleosome assembly and permits rapid association of Sir complex proteins with telomeres and telomere-adjacent regions (Enomoto et al. 1997). The study of HM silencing in cac1 mutants allows us to extend this model to the maintenance of silencing at the HM loci as well. We propose that nucleosomes assembled from appropriately acetylated histones form a solid “foundation” on which a strong “wall” of silent chromatin is built (Fig. 7). The Sir complex proteins are essential “bricks” in this silencing wall. We propose that DNA and nucleosomes protected by a stable Sir protein wall remain completely inaccessible to enzymes including histone acetylases and histone deacetylases. In addition, the wall of Sir complex proteins may become unstable if the foundation of nucleosomes is weakened by the presence of histones with inappropriately acetylated amino termini (Fig. 7).
Figure 7.
Model for the mechanism by which CAF-I contributes to formation of stable heterochromatin. Heterochromatin is represented as a “wall” of Sir complex proteins (Sir2p, Sir3p, and Sir4p) built on a foundation of nucleosomes (circles) composed of appropriately acetylated histones. After replication, existing nucleosomes (white circles) are randomly distributed between daughter strands of DNA. (Top) CAF-I assembles newly synthesized nucleosomes (gray circles) into chromatin. Existing Sir complex proteins (white rectangles), as well as newly synthesized Sir complex proteins (gray rectangles), associate with the nucleosomes to form a wall of proteins that restrict accessibility to the DNA. If Sir complex proteins are limiting, the wall is thinner or weaker. (Middle) If derepression of the locus occurs, nucleosomes with the “active” acetylation patterns generated during the previous cell cycle (white circles with X) form an unstable foundation that does not associate as tightly with the Sir complex proteins. In the absence of CAF-I, these nucleosomes are recycled onto daughter strands and a fragile wall of Sir complex proteins (recruited by Sir1p) is subject to “leaking” or eventual derepression. If extra Sir complex proteins are provided, the wall can become thicker and thus, more stable. (Bottom) In cac sir1 double mutants, an unstable foundation (attributable to the lack of CAF-I) and limited recruitment of Sir complex proteins (attributable to the lack of Sir1p) leads to more derepression of the locus.
Role of CAF-I in the assembly of silent chromatin
We propose that CAF-I contributes to the association of Sir complex proteins with the heterochromatin domain in two ways. First, CAF-I ensures that replication-coupled nucleosome assembly occurs soon after the replication fork has passed through the heterochromatin. Second, CAF-I ensures that the nucleosomes are assembled from the appropriately acetylated histones, forming a solid foundation for Sir complex propagation.
Timing of nucleosome assembly
We posit that CAF-I-mediated nucleosome assembly facilitates the rapid association of Sir complex proteins whose local concentration is likely to be high immediately after replication. Because Rap1p localization and presumably Sir complex localization as well are perturbed in cac mutants (Enomoto et al. 1997), it appears that the concentration of heterochromatin proteins at telomeres (and presumably at the HM loci) is reduced in cells lacking CAF-I, and that these proteins are distributed more randomly throughout the nucleus. If chromatin assembly is delayed, the local Sir complex concentration near silencers may decrease as a result of diffusion of the proteins over the time required for assembly of nucleosomes. This would lead to the formation of a silencer wall with fewer bricks (Fig. 7). Because the efficiency of silencing is a function of competition between transcription activators and the silent chromatin components such as Sir3p (Renauld et al. 1993), a wall composed of fewer Sir proteins at HML would be less effective at restricting the accessibility of the transcriptional machinery to the α-genes at HML.
Assembly of nucleosomes from appropriately acetylated histones
How is the repressed chromatin state inherited and how does CAF-I contribute to that inheritance? Sogo et al. (1986) demonstrated that preexisting nucleosomes segregate randomly after DNA replication. Thus, the replicated chromatin is composed of histones present in the previous cell cycle as well as newly synthesized histones. Because the acetylation pattern of histone H4 in silent chromatin (Braunstein et al. 1996) resembles the acetylation pattern conferred on histone H4 by the cytoplasmic histone acetyltransferase Hat1p (Kleff et al. 1995; Parthun et al. 1996), we propose that the acetylation pattern of histones within silent chromatin remains unaltered and old nucleosomes from the silent domain can be recycled within the silent domain after replication. The incorporation of these recycled nucleosomes (that resemble nucleosomes assembled from newly synthesized histones) into silent chromatin would facilitate the inheritance of the silent chromatin state (Fig. 7).
Active chromatin is characterized by a histone acetylation pattern that is different from newly synthesized or silent chromatin. Mammalian CAF-I preferentially assembles newly synthesized, cytoplasmic histones (Verreault et al. 1996). Assuming that yeast CAF-I has a similar activity, we propose that the role of CAF-I is to ensure that only appropriately acetylated histones are assembled into silent chromatin (Fig. 7). CAF-I may exclude histones with the “active” acetylation pattern from being recycled into silent chromatin, which is especially relevant when an HM locus has become derepressed in the previous cell cycle.
Shmoo clusters appear when structural components of heterochromatin are altered
Our experiments with histone H3 and histone H4 mutants support the idea that defects in the nucleosome foundation lead to defects in the maintenance of silencing. Mutations that alter the histone amino termini give rise to budding shmoo cells, suggesting that a defect in the ability of histones H3 or H4 to be acetylated is sufficient to cause a problem with the maintenance of silencing.
Our model suggests that CAF-I ensures that local Sir2p, Sir3p, and Sir4p concentrations are elevated enough to permit assembly of a strong silencer and that in the absence of CAF-I, the local concentration of Sir complex proteins decreases, because of simple diffusion of the proteins over time (Fig. 7). Our observation that an additional copy of either Sir2p, Sir3p, or Sir4p improves silencing is consistent with the idea that in strains lacking CAF-I activity, the concentration of Sir complex proteins at the silencers is suboptimal for silencing. Also consistent with this idea, decreasing the concentration of Sir2p or Sir3p in CAC1 cells results in the appearance of budding shmoos, indicating that, in otherwise wild-type cells, suboptimal Sir complex protein concentrations are sufficient to give rise to silencing defects like those seen in cac1 strains.
Role of CAF-I in the Sir1 independent maintenance of silencing
Our studies indicate that CAF-I and Sir1p act synergistically to silence HML in MATa cells. In SIR1 cells, if HML becomes derepressed re-establishment of silencing occurs, presumably because Sir1p can attract more Sir complex proteins and can nucleate the formation of a silencer when the local concentration of Sir complex proteins decreases below the threshold for Sir1p independent maintenance. In most sir1Δ cells, repressed HM loci remain silent (Pillus and Rine 1989). Because this is not true in sir1Δ cac1Δ cells, we presume that CAF-I-assembled nucleosomes contribute to the ability of recycled and new Sir complex proteins to associate with the heterochromatin and to form a silencer wall of sufficient “thickness” (Fig. 7). In sir1Δ cells, derepressed HM loci remain active, presumably because once expression of an HM locus occurs, histone acetylation patterns are altered by the transcriptional machinery, and the local concentration of Sir complex proteins decreases (Maillet et al. 1996). In this case, recycled histones would not have the appropriate acetylation pattern (Fig. 7) and the local concentration of Sir complex proteins would not be sufficient to attract newly synthesized Sir complex proteins to the region. We assume that sir1Δ cac1Δ cells become derepressed more frequently (because of the lack of CAF-I) and, once derepressed, silencing cannot be reestablished (because of the lack of Sir1p).
Materials and methods
Plasmids and strains
Plasmids pJR910, pJR69, pJR273, and pJR368, carrying SIR1, SIR2, SIR3, and SIR4, respectively, were provided by Jasper Rine, University of California, Berkeley. Yeast strains used in this study are listed in Table 1. Strains were constructed by standard crosses within isogenic genetic backgrounds. SIR1 was disrupted in W303 using pJR533 (Kimmerly and Rine 1987). To construct strain YJB2057, pJH132 (carrying GAL-HO) was induced to switch HMLα to HMLa in a sir4 strain (Klar et al. 1981), allowing it to mate as a MATα cell. A Leu+ segregant from this cross, which was unable to switch mating type in the presence of GAL-HO expression, was selected. The strain was then cured of pJH132. pSIR3 (pSE334) includes the complete SIR3 gene in YCPlac111 (Gietz and Sugino 1988).
Table 1.
Yeast strains used in this study
| Strain names
|
Genotype
|
Source
|
|---|---|---|
| S150B-2 | ||
| YJB276 | MATa; leu2-3,112; ura3-52; trpl-289; his3Δ; ade2Δ, [cir+] | Berman lab |
| YJB277 | MATα; leu2-3,112; ura3-52; trp1-289; his3Δ; ade2Δ [cir+] | Berman lab |
| YJB485 | 276 cac1-1 (rlf2-1,Enomoto et al. 1997) | Berman lab |
| YJB335 | 276 sir1::LEU2 | Berman lab |
| YJB744 | 276 HIS+ cac1-1 sir1::LEU2 | this study |
| YJB1289 | 277 lys2; VR–ADE2–TEL | this study |
| YJB469 | 276 cac1Δ1 (=rlf2−Δ1::LEU2;Enomoto et al. 1997) | Berman lab |
| YJB2057 | 276 HMLa cac1-Δ1 | this study |
| YJB285 | 276 sir2::HIS3 | Berman lab |
| YJB397 | 276 sir3::TRP1 | Berman lab |
| W303 | ||
| YJB195 | MATa ura3-1, ade2-1, his3-11, leu 2-3,112, can1-100, trp1-1 | Berman lab |
| YJB209 | MATα ura3-1, ade2-1, his3-11, leu 2-3,112, can1-100, trp1-1 | Berman lab |
| YJB959 | 195 HMR::TRP1 | D. Shore |
| YJB955 | 195 hmrΔA::TRP1 (missing ORC site) | D. Shore |
| YJB1143 | 195 hmrΔB::TRP1 (missing Abf1 (site) | D. Shore |
| YJB1104 | 195 hmrΔE::TRP1 (missing Rap1 site) | D. Shore |
| YJB1960 | 195 cac1-Δ1 HMR::TRP1 | this study |
| YJB958 | 195 cac1-Δ1 hmrΔA::TRP1 | this study |
| YJB1139 | 195 cac1-Δ1 hmrΔB::TRP1 | this study |
| YJB1101 | 195 cac1-Δ1 hmrΔE::TRP1 | this study |
| YJB1638 | 195 rap1-12 hmrΔA::TRP1 | D. Shore |
| YJB2006 | 195 sir1::HIS3 HMR::TRP1 | this study |
| YJB2011 | 195 msil::hisG–URA3–hisG hmrΔA::TRP1 | this study |
| YJB2009 | 195 cac1-Δ1 msil::hisG–URA3–hisG hmrΔA::TRP1 | this study |
| YJB1838 | 195 cac1-Δ1 | this study |
| YJB1578 | 209 cac1-Δ1 | this study |
| YJB1803 | 195 cac2::TRP1 | this study |
| YJB1599 (=pky086) | 209 cac2::TRP1 | P. Kaufman |
| YJB1581 | 195 msil::hisG | P. Kaufman |
| YJB1836 | 209 msil::his G–URA3–hisG | this study |
| YJB1804 | 195 cac1-Δ1 cac2::TRP1 | this study |
| YJB1802 | 209 cac1-Δ1 cac2::TRP1 | this study |
| YJB1865 | 195 cac2::TRP1 msil::hisG–URA3–hisG | this study |
| YJB1864 | 209 cac2::TRP1 msil::hisG–URA3–hisG | this study |
| YJB1862 | 195 msil::hisG–URA3–hisG cac1-Δ1 | this study |
| YJB1863 | 209 msil::hisG–URA3–hisG cac1-Δ1 | this study |
| YJB1919 | 195 cac1-Δ1 cac2::TRP1 msil::his G–URA3–hisG | this study |
| YJB1918 | 209 cac1-Δ1 cac2::TRP1 msil::hisG–URA3–hisG | this study |
| YJB1940 | 195 sir1::HIS3 | this study |
| YJB1941 | 209 sir1::HIS3 | this study |
| YJB1962 | 195 sir1::HIS3 cac1-Δ1 | this study |
| YJB1961 | 209 sir1::HIS3 cac1-Δ1 | this study |
| YJB2000 | 195 sir1::HIS3 Δcac2::TRP1 | this study |
| YJB2034 | 209 sir1::HIS3 Δcac2::TRP1 | this study |
| YJB1945 | 195 sir1::HIS3 Δmsil::hisG–URA3–hisG | this study |
| YJB1946 | 209 sir1::HIS3 Δmsil::hisG–URA3–hisG | this study |
| YJB2044 | 195 sir1::HIS3 Δcac1-Δ1 cac2::TRP1 | this study |
| YJB2048 | 195 sir1::HIS3 Δcac2::TRP1 msil::hisG–URA3–hisG | this study |
| YJB2007 | 195 sir1::HIS3 Δmsil::hisG–URA3–hisG cac1-Δ1 | this study |
| YJB1993 | 209 sir1::HIS3 Δmsil::hisG–URA3–hisG cac1-Δ1 | this study |
| YJB2109 | 195 cac1-Δ1 sir3::TRP1 | this study |
| YJB2471 | 195 sir1::HIS3 sir3::TRP1 | this study |
| YJB2544 | 195 sir3::TRP1 | this study |
| MX4-22A (S288C) | ||
| YJB2166(=MSY552) | MATa ura3-52 lys2Δ201 leu2-3,112 Δ(HHT1 HHF1) Δ(HHT2 HHF2) pMS337[CEN ARS LEU2 HHT1 HHF1] | M.M. Smith |
| YJB2167 (MSY343) | 2166 hht1-1 (Δ2–20) | M.M. Smith |
| YJB2168 (MSY344) | 2166 hht1-2 (Δ2–29) | M.M. Smith |
| YJB2169 (MSY541) | 2166 hhf1-21 (KR5) | M.M. Smith |
| YJB2170 (MSY613) | 2166 hhf1-14 (KR5KR8) | M.M. Smith |
| YJB2171 (MSY641) | 2166 hhf1-15 (KR5KR12) | M.M. Smith |
| YJB2172 (MSY742) | 2166 hhf1-13 (KR16) | M.M. Smith |
| Miscellaneous | ||
| A364A | MATa ade1 ura1 gal1 ade2 tyr1 his7 lys2 | L. Hartwell |
| B364B | MATα ade1 ura1 gal1 ade2 tyr1 his7 lys2 | L. Hartwell |
| TD1 | MATα his4-38 ura3-52 trp1-289 | Berman lab |
Quantitative mating assay
To assay mating of specific strains, 105 cells were mixed with 106 tester cells for 4.5 hr at 30°C on solid complete synthetic medium (Rose et al. 1990). Mating mixtures were excised from the solid medium, resuspended in sterile water, and serial dilutions of the mixtures were plated on appropriate solid media to select for diploids or for haploid parents. Four assays were performed for each strain. The mating competence of the mutants (proportion of diploids to total cells) was expressed as a proportion of the mating competence of wild-type cells, which were always included in the same sets of experiments. A rank sum test (Snedecor and Cochran 1980) was performed on the ratio of diploid/total for each experiment in pairwise combination with all other strains tested in the same experiment.
α-Factor response
Liquid assays were performed by incubating the relevant strains in YPAD (Rose et al. 1990) containing 500 ng/ml of α-factor (Sigma, St. Louis, MO) for 3 hr. Time lapse assays were performed on the appropriate solid YPAD medium. Five microliters of α-factor (200 μg/ml) was placed on a 5 mm diameter region of the plate. The appropriate MATa strains were streaked across this region. Just after streaking (time = 0), areas containing well-separated cells were identified and marked by puncturing the agar surface nearby with a dissecting needle. The location and cell shape were detected and recorded with an Olympus BX-40 Photomicroscope III, equipped with a CoolCAM liquid-cooled three-chip color CCD camera (Cool Camera Co.) and captured using Image Pro Plus version 1.3 software (Media Cybernetics). Cells were incubated at 23°C and then scored at different times after streaking. The χ2 test of goodness to fit (Snedecor and Cochran 1980) was performed by taking the distribution of wild type into three classes as the null model and testing each strain against this model. Similarly, pairs of mutant strains were tested against one another. Samples were considered significantly different at the p < 0.01 level.
Limiting Sir protein experiments
sir2 (YJB285) and sir3 (YJB397) strains carrying either GAL-SIR2 (pAR14) or GAL-SIR3 (pAR16) (Holmes et al. 1997), respectively, were pregrown on raffinose overnight, which allowed sufficient expression of the GAL-SIR constructs to maintain HM silencing. Cultures were diluted into SDC–Leu medium containing 2% glucose for 2, 4, 6, or 8 hr, to repress Sir complex protein expression, before assaying α-factor response on solid medium containing glucose as described above.
Acknowledgments
We thank J. Broach, P. Kaufman, J. Rine, D. Shore, and M.M. Smith for providing yeast strains and plasmids. We also thank Mark Sanders, of the University of Minnesota Imaging Center, for assistance with time-lapse video microscopy, Sue Wick for use of her microscope, camera, and image collection system, and Cathy Asleson, Janna Beckerman, Kathleen Conklin, Jodi Lew, Steve Johnston, and P.T. Magee for critical reading of the manuscript. This work was supported by the National Institutes of Health (GM38626).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL berma003@tc.umn.edu; FAX (612) 625-1738
References
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Brand AH, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes & Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- Chien C-t, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–555. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Enomoto S, Longtine MS, Berman J. TEL + CEN antagonism on plasmids involves telomere repeat sequence tracts and gene products that interact with chromosomal telomeres. Chromosoma. 1994;103:237–250. doi: 10.1007/BF00352248. [DOI] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes & Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC. Imprinting moves to the center. Nature Genet. 1996;14:119–121. doi: 10.1038/ng1096-119. [DOI] [PubMed] [Google Scholar]
- Fox CA, Rine J. Influences of the cell cycle on silencing. Curr Opin Cell Biol. 1996;8:354–357. doi: 10.1016/s0955-0674(96)80009-3. [DOI] [PubMed] [Google Scholar]
- Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3 and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3343–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M, Hecht A, Fisher-Adams G, Wan J, Mann RK, Strahl-Bolsinger S, Laroche T, Gasser S. The regulation of euchromatin and heterochromatin by histones in yeast. J Cell Sci. 1995;19:29–36. doi: 10.1242/jcs.1995.supplement_19.4. [DOI] [PubMed] [Google Scholar]
- Hall JG. Genomic imprinting: Review and relevance to human diseases. Am J Hum Genet. 1990;46:857–873. [PMC free article] [PubMed] [Google Scholar]
- Hardy CFJ, Balderes D, Shore D. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol Cell Biol. 1992;12:1209–1217. doi: 10.1128/mcb.12.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Holmes SC, Broach JR. Silencers are required for inheritance of the repressed state in yeast. Genes & Dev. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- Holmes SG, Rose AB, Steuerle K, Saez E, Sayegh S, Lee YM, Broach JR. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 1997;145:605–614. doi: 10.1093/genetics/145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Yang XL, Carlson M. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics. 1992;130:71–80. doi: 10.1093/genetics/130.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John RM, Surani MA. Imprinted genes and regulation of gene expression by epigenetic inheritance. Curr Opin Cell Biol. 1996;8:348–353. doi: 10.1016/s0955-0674(96)80008-1. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and Histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: A molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes & Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Kimmerly WJ, Rine J. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol Cell Biol. 1987;7:4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJS, Strathern JN, Hicks JB. A position–effect control for gene transposition: State of expression of yeast mating-type genes affects their ability to switch. Cell. 1981;25:517–524. doi: 10.1016/0092-8674(81)90070-2. [DOI] [PubMed] [Google Scholar]
- Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Lalande M. Parental imprinting and human disease. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- Latham KE. X chromosome imprinting and inactivation in the early mammalian embryo. Trends Genet. 1996;12:134–138. doi: 10.1016/0168-9525(96)10017-2. [DOI] [PubMed] [Google Scholar]
- Loo S, Fox CA, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell-cycle progression, and DNA-replication. Mol Biol Cell. 1995a;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Laurenson P, Foss M, Dillin A, Rine J. Roles of ABF1, NPL3 and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995b;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Broach JR. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989;9:4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Marquardt R, Shei G-J, Rose AB, Broach JR. Mutations in the HMLE silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes & Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Marshall M, Mahoney D, Rose A, Hicks JB, Broach JR. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: Essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley JFX. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- Miller AM, Nasmyth KA. Role of DNA replication in the repression of silent mating-type loci in yeast. Nature. 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: A SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of Sir proteins interacts with the silencer and telomere-binding protein Rap1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Szostak JW. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and SIR3 dosage. Genes & Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Ruggieri R, Tanaka K, Nakafuku M, Kaziro Y, Toh-e A, Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor GW, Cochran WG. Statistical methods. 7th ed. Ames, IA: The Iowa State University Press; 1980. [Google Scholar]
- Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: Isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Vannier D, Shore D. Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3919–3928. doi: 10.1128/mcb.13.7.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]