Msn2 and Msn4 transcription factors play a major role in yeast response to a variety of stress conditions. A systematic approach to the identification of Msn2/4 activators or suppressors shows that the majority of the Msn2 protein regulatory network acts to fine-tune its activity following yeast exposure to diverse stress conditions.
Abstract
The Msn2 and Msn4 transcription factors play major roles in the yeast general stress response by mediating the transcription of hundreds of genes. Despite extensive information on Msn2/4–mediated gene expression profiles, much less is known regarding the network of proteins that regulate its activity. Here we describe a systematic approach designed to examine the roles of 35 Msn2/4 partners in regulating Msn2/4 transcriptional activity in the face of four different environmental conditions. Our analysis indicates that single deletions of 26 Msn2/4 partners significantly affect Msn2/4 transcription activity under four different conditions. The low functional redundancy of the Msn2 regulatory network indicates that Msn2/4 activity is finely tuned by many of Msn2/4 partners to provide an optimized stress response through differential activation, nuclear localization, degradation, and chromatin remodeling. Our specific analysis of Msn2 activity showed that a relatively large number of partners act to suppress Msn2 activity under nonstress conditions through independent mechanisms, including cytoplasmic retention, proteosome-mediated Msn2 degradation, and chromatin remodeling. Such negative regulation is crucial to minimize the cost of uncontrolled stress response gene expression and ensures a high growth rate in the absence of stress.
INTRODUCTION
All cells possess the ability to respond to extreme changes in environmental conditions capable of threatening viability. Cell response mechanisms include environmental sensing and signal transduction pathways that lead to significant alterations in gene expression programs. Induction or repression of gene expression under stress conditions enables fast adaptation to diverse conditions, resulting in increased cell fitness and survival (Gasch et al., 2000). In yeast, several transcription factors (TFs) that promote the transcription of hundreds of different genes in response to diverse stress conditions have been identified (Estruch, 2000). Whereas some TFs promote the transcription of specific sets of genes, thus enabling adaptation to specific stresses, others promote the transcription of many genes in response to a wide variety of environmental stresses (Estruch, 2000).
Extensive studies on stress response gene expression have been performed, largely focusing on genome-wide transcription profiles (Gasch et al., 2000; Causton et al., 2001). By contrast, much less is known regarding regulation mechanisms that control stress response TF activity. Such mechanisms can involve posttranslational modification of TFs that can affect TF localization or activation (Kobor and Greenblatt, 2002). It is also clear that complex TF regulation mechanisms are mediated by a dense network of TF-interacting partners that activate or suppress transcription under diverse environmental conditions (Tarassov et al., 2008). Hence the development of a comprehensive approach for the systematic characterization of partners that either activate or suppress TF activity could prove to be highly useful for revealing novel transcriptional control mechanisms and for enhancing our understanding of how cells integrate and translate diverse environmental signals. The joining of such an approach with genome-wide expression analysis could, furthermore, provide a more comprehensive view of global transcription regulation mechanisms.
Msn2 and Msn4 are two homologous master regulators that play a major role in the yeast general stress response program by transcribing hundreds of genes following exposure to diverse stress conditions (Boy-Marcotte et al., 1998; Gorner et al., 2002; Hasan et al., 2002; Kandror et al., 2004). It was shown that Msn2 and Msn4 clearly affect gene induction in a differential manner and that this regulation is both gene and condition specific (Hohman and Mager, 2003; Berry and Gasch, 2008). Msn2/4 are located at the heart of a protein network that involves numerous physical and genetic interactions with various kinases, phosphatases, transporters, and chromatin remodelers. Previous research analyzed the regulatory roles of specific Msn2/4 partners, focusing mainly on kinases and phosphatases (Gorner et al., 1998; Kaida et al., 2002; De Wever et al., 2005). These, however, were sporadic studies that do not provide a comprehensive view of the function of the Msn2/4 protein regulatory network. Specifically, little is known regarding the roles assumed by many Msn2/4 partners in regulating Msn2/4 activity under multiple stress conditions or whether Msn2/4 partners are functionally redundant or instead serve distinct roles in activating or suppressing Msn2/4 transcription activity under diverse conditions.
With these points in mind, we undertook a systematic approach to examine how the Msn2/4 regulatory network controls gene expression activity under multiple stress conditions (Figure 1). To do so, we examined the effects of single deletions of 35 Msn2/4 partners on Msn2/4–mediated gene expression following exposure to three different stress conditions and in the absence of stress. In addition, we analyzed the phenotypic effects of some of these deletions on yeast stress survival, growth rates, and Msn2 nuclear localization. This comprehensive analysis allowed us to identify 26 Msn2/4 partners that serve as either activators or suppressors of Msn2/4–mediated gene expression and allowed us to assemble a uniform framework for the comparison of different Msn2 regulators. We found that the Msn2/4 network exhibits similar response behavior following exposure to different stress conditions, indicating that these stress signals are sensed upstream of the Msn2/4 regulatory network. Our data suggest that suppression of Msn2/4 activity in the absence of stress is achieved by several independent control mechanisms, including Msn2/4 phosphorylation and proteosome degradation. Our focused analysis of Msn2 activity allowed for the identification of redundancy between Msn2 regulators and isoform specificity in the well-studied Msn2 regulation by cAMP–protein kinase A (PKA) (Gorner et al., 1998; Smith et al., 1998). Further analysis revealed the epistatic relationships between Msn2 regulators, highlighting the importance of Msn2 dephosphorylation for its activation under diverse stress conditions.
FIGURE 1:
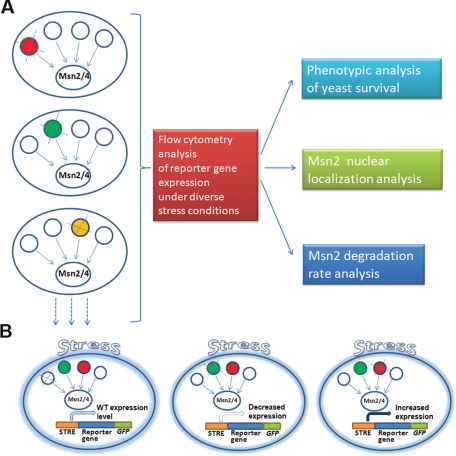
A systematic approach for the identification of Msn2 partners that regulate Msn2 gene expression activity under different stress conditions. (A) Yeast strains deleted of Msn2 partners were generated on the background of a strain expressing the HSP12-GFP, the TPS2-GFP, or the PNC1-GFP reporter gene. Following exposure to diverse environmental conditions, reporter gene expression levels were analyzed by flow cytometry. Selected deletion strains with increased or decreased Hsp12-GFP expression relative to the nondeleted strain were further analyzed for survival, Msn2 nuclear localization, and Msn2 protein degradation. (B) Deletion of Msn2 partners can result in no change (left), in decrease (middle), or in increase (right) in reporter gene expression level. Deletion of Msn2 partners that resulted in decrease or increase in the reporter gene expression level allowed for identification of the absent protein as an Msn2 activator (green) or suppressor (red), respectively.
RESULTS
The experimental approach used to examine the Msn2/4 regulatory network
To systematically examine the Msn2/4 regulatory network, Msn2/4 partners were identified by extensive literature and database searches (e.g., the Saccharomyces Genome Database and bioGRID; Stark et al., 2006). We focused on the analysis of proteins that are in physical or genetic interaction with Msn2 and represent major biological pathways that can potentially regulate Msn2/4 activity. Overall, we focused on 35 Msn2 partners meeting these criteria. A subset of these proteins are also Msn4 interactors, representing four essential and 31 nonessential proteins participating in diverse biological pathways. The 35 Msn2/4 partners include 14 kinases, three phosphatases, seven chromatin remodelers, and two chaperones (see Supplemental Table S1). Of the 35 interactors, 10 are in physical interaction with Msn2 and 25 are in genetic interaction. We analyzed these partners using targeted gene deletion of the nonessential partners and the Decreased Abundance by mRNA Perturbation (DAmP) fusion strategy (Breslow et al., 2008) to reduce the expression level of the essential partners. We initially analyzed the effects of partner deletion in S288c-derived strains containing both MSN2 and MSN4. As such, any effects of partner deletion in these strains can be collectively attributed to increases/decreases in Msn2/4 activity. To facilitate fast and quantitative analysis of the effect of each deletion on Msn2/4 activity, these deletions were also generated on the background of strains containing the HSP12 gene fused to green fluorescent protein (GFP) (HSP12-GFP) (Figures 1 and 2 and Supplemental Table S2). HSP12 is a general stress response gene encoding a plasma membrane protein, and its expression depends on Msn2/4 activity (Supplemental Figure S1A; Pacheco et al., 2009). A subset of these deletions was also generated on the background of two other Msn2/4-regulated reporter genes, TPS2-GFP and PNC1-GFP, and on the background of a control PFK2-GFP housekeeping gene (Supplemental Figure S1). We used flow cytometry to analyze the different reporter gene expression levels in the deletion strains, following exposure to four environmental conditions, including heat, oxidative, and osmotic stresses (for examples see Supplemental Figure S2). Decrease or increase in the reporter gene expression levels in the different deletion strains relative to the parent strain allowed for identification of partners that either activate or suppress Msn2/4 activity following exposure to the different stress conditions (Figure 1B).
FIGURE 2:
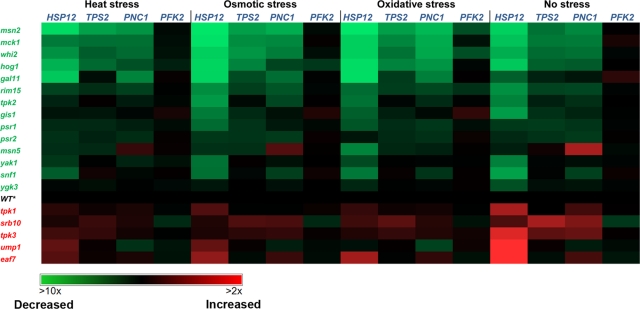
Msn2 activity in 35 Msn2 partner–deleted strains generated on the S288c background analyzed by flow cytometry measurements of Hsp12-GFP reporter gene expression. The Hsp12-GFP expression level in the different strains was measured following yeast exposure to four different stress conditions and normalized to the wild-type strain (see Supplemental Figure S2 for examples and Supplemental Table S2 for raw data). The yeast cells were exposed to 40 min of stress, including oxidative stress (0.6 mM H2O2), osmotic stress (0.5 M NaCl), and heat stress (temperature shift from 30 to 37°C). Msn2 partner deletions that result in at least a 30% decrease or increase in Hsp12-GFP expression levels relative to the nondeleted strain led to the identification of the absent protein as a Msn2 activator or suppressor, highlighted in green and red, respectively. The glc7, cyr1, tor2, and cdc25 mutant strains contained a DaMP fusion (see Materials and Methods and Supplemental Table S5).
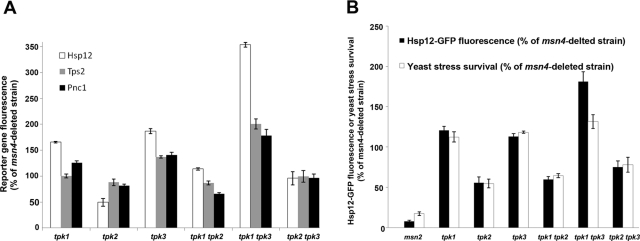
To verify that Msn2/4 partner deletion affected Hsp12-GFP expression through a missing direct or indirect interaction with Msn2, 18 of the partner-deleted strains were examined using the TPS2-GFP and PNC1-GFP reporter genes (Figure 3 and Supplemental Table S3). These strains were generated on the background of msn4 deletion in a W303-derived strain to examine the effects of the deletions on Msn2 without any background Msn4 activity. We found very good correlation between the Hsp12-GFP expression levels of the different deletions generated on the background of the S288c- and W303-derived strains (Supplemental Figure S3). These reporter genes were chosen due to their induced expression following exposure to three different stress conditions and the presence of at least four stress response elements (STREs) in their promoters (Supplemental Figure S4; Martinez-Pastor et al., 1996). In addition, these genes were shown to be directly regulated by Msn2/4 both at the mRNA levels (Gasch et al., 2000) and by direct Msn2/4 interactions using chromatin immunoprecipitation combined with microarray (ChIP-on-chip) experiments (Supplemental Table S4). The generation of the partner deletions on the background of PFK2-GFP made it possible to obtain negative controls for the stress-induced reporter gene experiments. PFK2 is a metabolic housekeeping enzyme containing no STRE elements in its promoter and is not induced following exposure to the three stress conditions examined (Figure 3). Previous work has shown the PFK2 is induced under different stress conditions, including nitrogen starvation, diauxic shift, and stationary phase (Gasch et al., 2000). We found that the deletion of many of the partners affected the expression of all three Msn2/4 reporter genes (i.e., HSP12, TPS2, and PCN1) but did not significantly affect PFK2 reporter gene expression (Figure 3). Such a correlation between the expression levels of the three reporter genes (Supplemental Figure S5) strongly suggests that partner deletions affect reporter gene expression through missing interactions with Msn2 rather than through other, nonrelated factors. The lack of effects on the PFK2 expression level indicates that the mild stress conditions used in the flow cytometry experiment do not significantly affect cell physiology and demonstrate the specificity of Msn2 in mediating stress-induced gene expression. These mild conditions (see Figure 2 and Materials and Methods for details) were previously used to induce Msn2/4 activity without significantly affecting cell viability (Gasch et al., 2000).
FIGURE 3:
Msn2 activity in 18 Msn2 partner–deleted strains analyzed by flow cytometry measurements of HSP12-GFP, TPS2-GFP, or PNC1-GFP reporter gene expression. The expression levels of the PFK2-GFP housekeeping gene is used as a negative control for the experiments. The 18 partner-deleted strains were generated on the background of W303 msn4-deleted strain to specifically analyze the effects of partner deletion on Msn2 activity. Gene expression levels in the different strains were measured following yeast exposure to four different stress conditions, as described in the legend to Figure 2, and normalized to the msn4-deleted strain (WT*) strain (see Supplemental Table S3 for raw data). Msn2 partner deletions that result in at least a 30% decrease or increase in Hsp12-GFP expression levels relative to the reference strain led to the identification of the absent protein as an Msn2 activator or suppressor, highlighted in green and red, respectively.
The effect of the different deletions is not condition specific
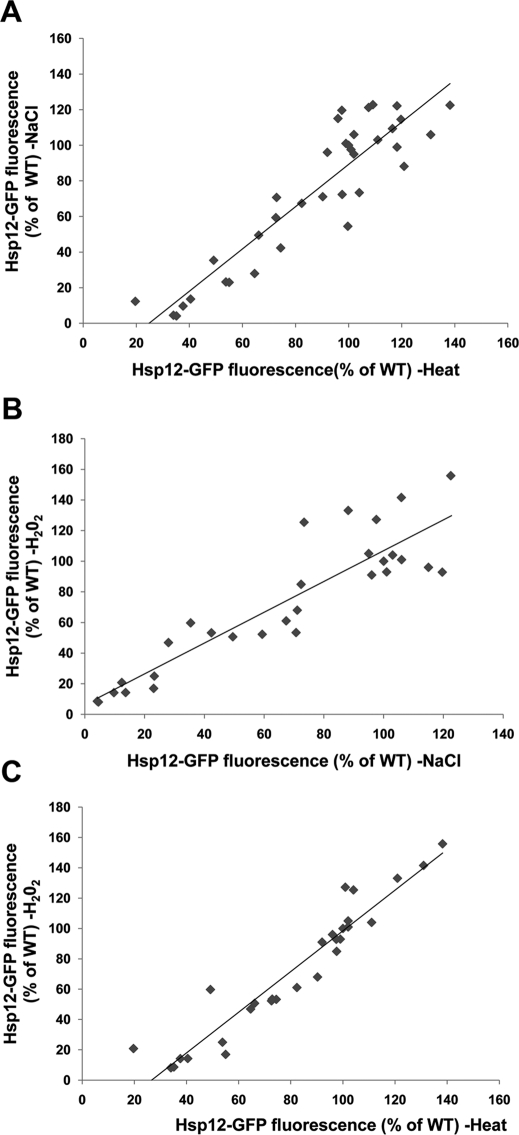
Msn2/4 are activated following yeast exposure to a wide variety of environmental conditions, such as nitrogen or carbon starvation, osmotic stress, DNA-damaging agents, ethanol, and heat stress (Gasch et al., 2000; Causton et al., 2001). It was previously shown that different cellular sensors monitor the extracellular, as well as the intracellular, yeast environment (e.g., GPR1 monitors glucose sensing and WPR1 monitors osmotic stress; Zaman et al., 2008), leading to stress-dependent activation of Msn2/4 transcriptional activity. However, it is not clear how diverse environmental signals are integrated to activate Msn2/4 activity. To explore whether Msn2/4 regulation depends on the type of stress condition or whether diverse stresses are integrated uniformly by Msn2/4 regulators, we examined Msn2/4 activity in the different deletion strains following exposure to three stress conditions as well as in the absence of stress (Figure 2). We found tight correlation between the changes in reporter gene expression levels following exposure to oxidative, heat, or osmotic stress (Figure 4). The condition-independent effects of the different partner deletions, including effects on gene expression noted in the absence of stress (Figure 2), indicate that their missing activity is a prerequisite for the activation of Msn2/4 under all examined conditions. Overall, these results suggest that different stress signals are translated by Msn2/4 partners in a uniform manner to regulate the transcriptional activity of Msn2/4 and that there is little partner specificity.
FIGURE 4:
Correlations between Hsp12-GFP expression following exposure of the different deletion strains to osmotic stress (NaCl) and heat (37°C) (A), oxidative stress (H2O2) and osmotic stress (NaCl) (B), and oxidative (H2O2) stress and heat (37°C) (C). The r values of the linear fit for the data in A–C are 0.83, 0.81, and 0.9, respectively. Each point represents the Hsp12-GFP expression level of a specific strain in the face of two different environmental conditions. The high correlation between the Hsp12-GFP expression levels of the different strains indicates that different partners regulate Msn2 similarly following exposure to different conditions. Data were derived from analysis of deletion strains generated on the S288c background (Supplemental Table S2).
Effects of Msn2 partner deletions on yeast stress survival
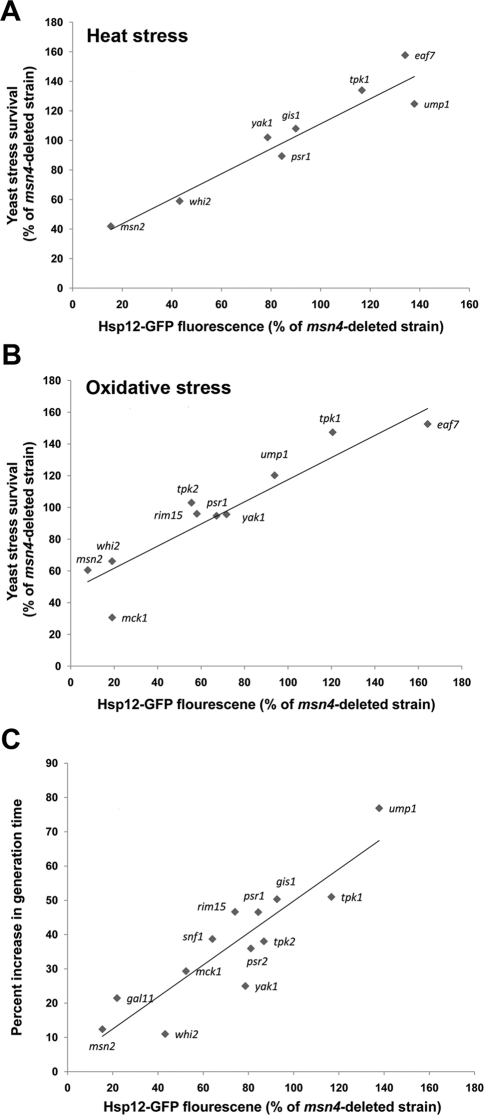
Msn2/4 activity was shown to protect yeast cells from the detrimental effects of extreme stress (Martinez-Pastor et al., 1996). Indeed, it was previously shown that upon double deletion of MSN2 and MSN4, yeast survival is significantly reduced following exposure to diverse stress conditions (Martinez-Pastor et al., 1996; Hasan et al., 2002; Kandror et al., 2004). To examine the survival phenotypes of the deletion W303-derived strains in response to different stress conditions, selected msn4-deleted strains were exposed to extreme heat or oxidative stress and their survival was measured. We found that strains deleted of Msn2 partners that serve as Msn2 activators or suppressors showed significantly decreased or increased stress survival, respectively (Figure 5, A and B). We found that the level of stress survival of the different deletion strains following exposure to extreme stress conditions is tightly correlated to the level of Hsp12-GFP reporter gene expression (r = 0.89 and 0.84 for Figure 5, A and B, respectively). These results show that Hsp12-GFP reporter gene expression levels can serve as an excellent predictor of the overall yeast stress survival phenotype. These results are, moreover, in agreement with microarray data indicating that HSP12, TPS2, and PNC1 are part of a large cluster of general stress response genes that are expressed following yeast exposure to diverse stress conditions (Gasch et al., 2000).
FIGURE 5:
Yeast stress survival and generation time are significantly affected by deletion of several Msn2 partners. Linear correlations between yeast stress survival and Hsp12-GFP expression levels in the different deletion strains relative to the msn4-deleted strain following exposure to heat stress (A) or oxidative stress (B). The r values for the linear fit in A and B are 0.89 and 0.84, respectively. (C) Correlation between the increase in yeast generation time and Hsp12-GFP fluorescence of the different deletion strains (r = 0.8). The increase in generation time was calculated by comparing the strain growth rate at 30°C to that observed at 37°C (see Materials and Methods for a detailed description). Hsp12-GFP fluorescence values were obtained by flow cytometry analysis of the different strains in the absence of stress. All partner deletions were generated on the background of the W303 msn4-deleted strain.
The trade-off between Msn2 activity and yeast growth rate
To test whether the extent of Msn2 activity is correlated to the rate of yeast growth, selected W303 deletion strains, generated on the background of msn4 deletion, were grown in the presence or absence of mild heat stress. Mild heat stress results in an activation of Msn2 transcriptional activity, thus allowing for examination of the effects of Msn2 activity on growth rate. Accordingly, for each strain, the increase in generation time was calculated by comparing the growth rate of the different strains in the presence or absence of mild heat stress to correct for any affects of the deletions on basal growth rates. We observed close linear correlation (r = 0.8) between Msn2 activity (indicated by the Hsp12 expression level) and the increase in yeast generation time following exposure to mild heat stress (Figure 5C). This correlation highlights the trade-off between Msn2 activity and yeast growth rate and the high importance of mechanisms suppressing Msn2 activity in the absence of stress. We found that strains deleted of Msn2 suppressors display a slow growth rate, even in the absence of stress (see later discussion of Figure 8; unpublished data).
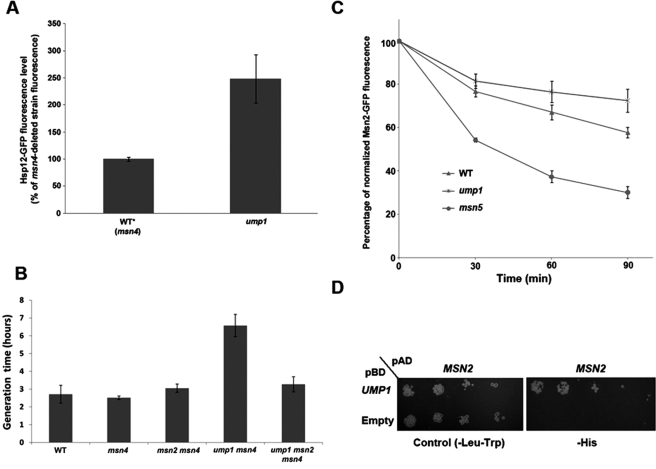
FIGURE 8:
Deletion of UMP1 increases Hsp12-GFP expression levels in the absence of stress and reduces Msn2 degradation and yeast growth rates. (A) A significant increase in Hsp12-GFP expression is seen in the ump1-deleted strain in the absence of stress relative to the WT* strain containing only the msn4 deletion. (B) The slow growth rate of the ump1-deleted strain is suppressed by deletion of the MSN2 gene under nonstress growth conditions. Comparison of the generation times of the different strains indicates significant difference between the ump1 msn4 double-deleted strain and the ump1 msn2 msn4 triple-deleted strain. (C) Reduced degradation rate of Msn2-GFP in the ump1-deleted strain relative to the wild-type or msn5-deleted strains. The decrease in the Msn2-GFP fluorescence level was monitored following cycloheximide addition using flow cytometry. (D) Msn2 is in physical interaction with Ump1, as indicated by yeast two-hybrid analysis. MSN2 was fused to the DNA-activating domain (AD), whereas UMP1 was fused to the DNA-binding domain (BD). All strains, except the Y2H strains, are W303 derivatives. All data presented in A and C represent the average of four independent repeats of each experiment. The data presented in B represent an average of eight independent repeats.
Msn2/4 activators and suppressors participate in diverse cellular processes
Our deletion strain analysis revealed that of 35 partners examined, deletion of 26 partners significantly decreased or increased Hsp12-GFP expression relative to the level noted in the parent strain (Figure 2). We defined an Msn2/4 partner as an activator or suppressor if its deletion led to more than 30% decrease or increase in the reporter gene expression level, respectively, following exposure to at least one condition (Figures 2 and 3). The majority of Msn2/4 activators include genes encoding for protein kinases (MCK1, RIM15, YAK1, SNF1, and HOG1) and phosphatases (PSR1, PSR2, and GLC7). We also found that the deletion of WHI2, encoding for a scaffold protein known to bind Msn2 and Psr1 (Kaida et al., 2002), results in a dramatic decrease in Msn2/4 activity (see later discussion). We found that mck1 deletion almost completely abolished Msn2/4 activity, indicating that this kinase is the most prominent Msn2/4 kinase activator. Analysis of Msn2 nuclear localization on the background of the mck1 deletion indicated a significance decrease in such localization (Supplemental Figure S6), which can result in low Msn2 transcriptional activity. In addition, we observed that the RPD3 and HDA3 genes, encoding for chromatin remodelers, are strictly required for Msn2/4 activity. Deletion of either of these genes results in decreased Hsp12-GFP expression to the same level as seen in an msn2 msn4 double-deleted strain.
We found that deletion of several partners results in increased Msn2/4 activity in the absence or presence of stress conditions relative to the parent strain (Figure 2). As previously shown, we observed that Msn2/4 suppressors belong to three different pathways, including the cAMP-PKA pathway, the TOR pathway, and the proteasome degradation pathway (Gorner et al., 1998; Smith et al., 1998; Beck and Hall, 1999; Durchschlag et al., 2004; Lallet et al., 2004). We found that deletion of either the TOR1 or TOR2 gene, belonging to the TOR pathway, and RAS2, CDC25, CYR1, TPK1, or TPK3 gene, belonging to the cAMP-PKA pathway, significantly elevated Msn2/4 activity following exposure to the different conditions. In addition, the deletion of the UMP1 gene, encoding for a proteasome maturation factor, significantly increased Msn2/4 activity (see later discussion for details). The suppression of Msn2/4 activity was maximal in the absence of stress (Figure 2), indicating the effectiveness of such mechanisms in preventing Msn2/4 activity under normal growth conditions. We found that in these deletion strains, Msn2/4 can be still activated following exposure to the three stress conditions. These results suggest that several parallel suppression mechanisms act in concert to produce maximal suppression of Msn2/4 activity in the absence of stress.
Next we examined the effects of overexpression of Msn2/4 interactors on the expression levels of Hsp12-GFP following yeast exposure to osmotic or heat stress and under nonstress conditions (Supplemental Figure S7). We specifically analyzed the effects of overexpression of Whi2 and Psr1, proteins that function as Msn2 activators, and of Srb10, Tpk3 and Ras2, proteins that function as Msn2 suppressors in strains containing the msn4 deletion (Figure 2). We also examined the effects of overexpression of these partners on the background of the msn2 msn4 double-deleted strain to detect any nonspecific effects on Hsp12-GFP reporter gene expression. We found that the overexpression of Whi2 or Psr1 leads to a significant increase in Hsp12-GFP expression levels, especially under nonstress conditions (Supplemental Figure S7). In contrast, we found that the overexpression of Ras2 leads to a significant decrease in Hsp12-GFP expression levels. These results are in excellent agreement with the actions of these partners as Msn2 activators (Whi2 and Psr1) or suppressors (Ras2).
PKA catalytic subunits display isoform specificity in regulating MSN2 activity
The cAMP-PKA regulatory effect on Msn2/4 was extensively studied , showing that PKA activity suppresses Msn2/4 activity (Boy-Marcotte et al., 1998; Gorner et al., 1998; Smith et al., 1998; Lee et al., 2008). However, the roles of specific TPK isoforms in regulating Msn2/4 activity remain unknown due to the use of the triple tpk1 tpk2 tpk3 mutant strain to abolish all PKA activity (Boy-Marcotte et al., 1998; Gorner et al., 1998; Smith et al., 1998). Our systematic approach allowed for analysis of the role of each TPK isoform in the regulation of Msn2 activity (Figure 3). We examined the effects of single deletions of the TPK1, TPK2, or TPK3 genes on Hsp12-GFP, Tps2-GFP, and Pnc1-GFP expression levels and yeast stress survival in W303 strains containing the msn4 deletion. We observed a significant increase in the different reporter gene expression levels and yeast stress survival in the tpk1- or tpk3-deleted strain, indicating that these isoforms are the most important PKA subunits for the suppression of Msn2/4 activity. In contrast, we observed that upon deletion of the TPK2 gene, a decrease in reporter gene expression level and stress survival was observed (Figure 6). Next we examined the relationships between the different TPK isoforms by analyzing the effects of double TPK deletions on Hsp12-GFP expression and yeast survival following exposure to oxidative stress (Figure 6). We observed that double deletion of the different TPK isoforms yielded additive effects on Hsp12-GFP expression and yeast stress survival, indicating a lack of genetic interactions between these isoforms.
FIGURE 6:
Different effects of deletion of TPK isoforms on Msn2 activity and yeast stress survival. (A) Hsp12-GFP, Tps2-GFP, and Pnc1-GFP expression levels in the different single and double TPK deletion strains following exposure to multiple stress conditions. Reporter gene expression levels were normalized to that of the msn4-deleted strain. (B) Survival and Hsp12-GFP expression levels of the different TPK isoform-deleted strains following exposure to extreme and mild oxidative stress, respectively. Survival and Hsp12-GFP expression levels were normalized to the levels of the msn4-deleted strain. All gene deletions were generated on the background of the W303 msn4-deleted strain.
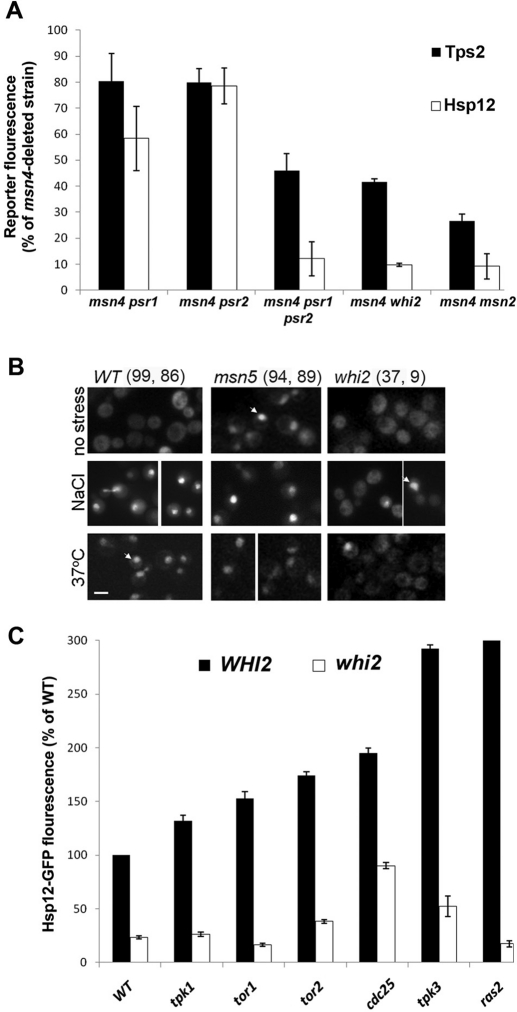
Deletion of WHI2 suppresses the effect of mutations in the cAMP-PKA and TOR pathways
Whi2 is a scaffold protein that was shown to physically bind both Msn2 and Psr1 phosphatase in the plasma membrane (Kaida et al., 2002). Msn2 was shown to be hyperphosphorylated in the whi2-deleted strain under nonstress conditions, indicating the lack of phosphatase activity (Kaida et al., 2002). In accordance with these findings, we observed that deletion of WHI2 results in significantly decreased Hsp12, Tps2, and Pnc1 expression under all environmental conditions tested (Figure 3). In addition, deletion of either the PSR1 or PSR2 genes, generated on the background of the msn4-deleted W303 strain, results in mild effects on Msn2-mediated gene expression. The double deletion of PSR1 and PSR2, however, results in a dramatic decrease in Msn2 activity (Figure 7A). These results indicate that Psr1 and Psr2 are partially redundant in regulating Msn2 activity. To further characterize the involvement of Whi2 in Msn2 regulation, we monitored the levels of Msn2-GFP nuclear localization in response to heat and osmotic stress on the background of the whi2-deleted strain. Such a deletion resulted in significantly impaired Msn2 nuclear localization under these conditions, leading to up to 10-fold reduction in the number of cells containing nuclear-localized Msn2 (Figure 7). In addition, we examined whether overexpression of Msn2 in a whi2-deleted strain background can restore Hsp12-GFP expression levels. We found that such overexpression leads to a much higher Hsp12-GFP expression level than seen in the wild-type strain (Supplemental Figure S8), indicating that Msn2 overexpression can efficiently suppress the effect of whi2 deletion.
FIGURE 7:
Whi2 is a central Msn2/4 regulator controlling Msn2 nuclear localization and transcription activity. (A) Changes in Hsp12-GFP and Tps2-GFP in the whi2, psr1, and psr2 deletion strains relative to the msn4-deleted strain examined under nonstress conditions. (B) Nuclear localization of Msn2-GFP is significantly reduced in a whi2-deleted strains following exposure to heat or osmotic stress. The msn5-deleted strain served as a positive control. The percentage of cells containing nuclear-localized Msn2-GFP are highlighted above each column following exposure to osmotic stress (left) or heat stress (right). For each strain, 200 cells were analyzed, and the percentage of cells containing at least a twofold increase in nuclear fluorescence was counted as positive. Arrows indicate representative nuclear localization of Msn2-GFP. All strains analyzed in A and B were generated on the W303 background. (C) Relationships between Whi2 and several Msn2 suppressors indicating that Whi2 activity is independent of the PKA or the TOR pathway. The analyzed strains were generated on the S288c background.
We next tested for the existence of epistatic relationships between WHI2 and genes from the cAMP-PKA and TOR pathways that suppress Msn2 activity. To examine such relationships, strains containing the whi2 tor1, whi2 tor2, whi2 tpk1, whi2 tpk3, whi2 ras2, and whi2 cdc25 double mutants were generated on the background of an S288c-derived strain expressing the HSP12-GFP reporter. We found that these strains exhibit dramatically reduced Hsp12-GFP fluorescence relative to the single deletions of the different suppressors (Figure 7C). These results show that Whi2 is essential for Msn2 activity and that such activity is dominant and independent of either the PKA or the TOR pathway.
Ump1 suppresses Msn2 activity by promoting its degradation
Ump1 is a proteasome maturation factor that functions as a chaperone to promote the assembly of the 20S proteasome and is conserved from yeast to human (Le Tallec et al., 2007). We found that deletion of the UMP1 gene significantly increased Hsp12-GFP levels (Figures 2, 3, and 8A) and reduced yeast growth rates in the absence of stress (Figure 8B). These results indicate that Ump1 functions as an Msn2 suppressor by decreasing Msn2 cellular levels to minimize its activity under nonstress conditions. To examine whether the decrease in growth rate of the ump1-deleted strain is related to Msn2, we also assessed the growth rate of a strain deleted of both UMP1 and MSN2. Of interest, the growth rate of this strain was much higher than the growth rate of the ump1-deleted strain, indicating suppression of the slow-growth phenotype (Figure 8B). As Ump1 is a proteasome maturation factor, our results suggest that the slow growth rate of the ump1-deleted strain is associated with the inability of the cell to degrade Msn2 or proteins expressed by Msn2. To test this hypothesis, we monitored the degradation rate of Msn2-GFP in the ump1-deleted strain relative to that rate observed in the parent and msn5-deleted strains following cycloheximide addition, a reagent that blocks protein biosynthesis (Ennis and Lubin, 1964). We observed that in the ump1-deleted strain, the Msn2-GFP degradation rate was much lower than the rate observed in the parent and msn5-deleted strains, consistent with previous observations (Lallet et al., 2004; Figure 8C) and in agreement with the function of Ump1 as a proteasome maturation factor (Le Tallec et al., 2007). A high degradation rate of Msn2 was previously observed in msn5-deleted cells due to reduced Msn2 nuclear export (Durchschlag et al., 2004). Finally, to address the physical interaction between Ump1 and Msn2, we used yeast two hybrid (Y2H) analysis (Fields and Sternglanz, 1994) and found that Ump1 physically interacts with Msn2 (Figure 8D). This result provides direct support for the previous finding that Ump1 colocalizes with Msn2 in the nucleus (Erkina et al., 2008).
DISCUSSION
In this study, we developed a systematic approach to examine the effects of deletion of each of 35 Msn2 partners on Msn2/4–mediated gene expression activity in cells encountering different environmental challenges. This approach allowed for the identification of partners that function as either activators or suppressors of Msn2 activity and provides a uniform framework for comparison between different regulators (Figure 2). Overall, the output of our systematic approach is a functional mapping of the Msn2/4 partner interaction network to reveal regulators that control Msn2/4 activity through different mechanisms, such as phosphorylation or dephosphorylation, nuclear localization, and degradation. The approach is based on quantitative measurement of changes in Msn2/4 target gene expression levels following systematic deletion of the different regulators (Figure 1). We chose to analyze Hsp12 expression level as a reporter of Msn2/4 activity due to dependence of the encoding gene on Msn2/4 and the significant increase in Hsp12 expression on yeast exposure to the three examined stress conditions. The high dynamic range of Hsp12 expression is critical to permit sensitive measurement of minor decreases or increases in expression on deletion of the different partners (Supplemental Figure S2 and Figure 2). In addition, 18 partner-deleted strains were analyzed, along with Hsp12-GFP and two other Msn2-dependent reporter genes, TPS2 and PNC1, and examined on the background of the msn4-deleted strain. Such analysis allowed us to assess deletion effects on Msn2 activity without any background Msn4 activity. The high correlation noted between the responses of the different reporter genes strongly suggests that partner deletions affect reporter gene expression through missing interactions with Msn2. The small effects of deletions and environmental conditions on the expression of the PFK2 control gene indicates that these stress conditions do not result in global, nonspecific effects on cell physiology. The systematic approach described in this study can be extended to a genome-wide approach to facilitate identification of all of the Msn2/4 regulators in the yeast proteome. Indeed, previous approaches employing yeast, Caenorhabditis elegans, and Drosophila demonstrated the usefulness of high-throughput screens in providing important information regarding protein networks regulating diverse biological processes (Friedman and Perrimon, 2006; Lehner et al., 2006; Boone et al., 2007; Perrimon and Mathey-Prevot, 2007).
In our study, we found that of 35 partners examined, deletion of 26 partners significantly affected Msn2/4 transcriptional activity (Figure 2). Analysis of the different interactors revealed that 25 of these interactors are in genetic interaction with Msn2 and the rest are in physical interaction (Table S1). The deletion of the physical interactors of Msn2 can affect Msn2 activity through the same pathway, whereas the deletion of the genetic interactors of Msn2 can affect its activity through parallel pathways. Thus, this analysis indicates that the majority of Msn2 partners affect its activity indirectly through parallel pathways. The large fraction of partners that interact with Msn2/4 to either activate or suppress Msn2/4 activity indicates low functional redundancy between the different partners in regulating Msn2/4 transcriptional activity. Previous studies indicated the robustness of protein–protein interaction networks to genetic perturbations (e.g., on partner inactivation) due to functional redundancy of different network partners (Wagner, 2005; Barkai and Shilo, 2007). The low functional redundancy in the Msn2/4 regulatory network suggests that this network is finely tuned to achieve an optimized response to different stress conditions and to provide several regulatory mechanisms that ensure Msn2/4 activation only in the presence of stress. We found that partner deletion similarly affected Msn2/4 transcriptional activity, regardless of the type of stress condition applied (Figure 4). This result indicates that the Msn2/4 regulatory network functions as a general stress response network that translates the three different stress signals to uniformly activate Msn2/4. The specificity of the yeast stress response may thus stem from the combinatorial activities of Msn2/4, together with stress-specific TFs that can together reshape global gene expression programs following exposure to different stress conditions (Estruch, 2000; Gasch et al., 2000; Causton et al., 2001). One such example could be Yak1, a protein kinase that was recently found to serve as a common regulator of the Hsf1 transcription factor, and Msn2/4, acting through PKA activity (Lee et al., 2008).
The roles of kinases and phosphatases in regulating Msn2/4 activity was described in this and previous studies (Smith et al., 1998; Kaida et al., 2002; De Wever et al., 2005). Other recent studies indicated the link between PKA and Msn2 activity by demonstrating that abolition of PKA activity results in accumulation of Msn2 in the nucleus and increased Msn2 activity (Gorner et al., 1998; Smith et al., 1998). However, little is known of the roles of specific TPK isoforms in regulating Msn2 activity. It was shown that strains deficient in all three TPK isoforms are nonviable, whereas strains containing any one of the three isoforms are viable (Broach and Deschenes, 1990). Despite this apparent redundancy, previous studies showed that the three TPK isoforms play different roles in pseudohyphal growth and in the regulation of iron uptake (Robertson and Fink, 1998; Robertson et al., 2000). Our results indicate that the Tpk1 and Tpk3 isoforms are the most dominant subunits that lead to the suppression of Msn2 activity (Figure 6). In contrast, Tpk2 serves as a partial Msn2 activator, with its deletion resulting in a mild decrease in Msn2 transcriptional activity (Figure 6). We also analyzed the roles of Whi2 and Psr1/2 in activating Msn2 and showed that their activity is independent of the PKA or the TOR pathway (Figure 7). These results indicate that parallel pathways exist for controlling Msn2 activity and that a fine balance in the different phosphorylation patterns of Msn2/4 affects its transcriptional activity. In agreement with this hypothesis, it was previously shown that Msn2 is hyperphosphorylated following exposure to different stress conditions and that such hyperphosphorylation is stress dependent (Garreau et al., 2000).
A central question regarding the Msn2-mediated stress response asks what control mechanisms suppress Msn2 activity in the absence of stress. Cytoplasmic localization of Msn2 in the absence of stress ensures that the majority of Msn2 is unavailable for transcription. However, the presence of a small amount of nuclear Msn2 in the absence of stress can still result in basal expression of stress response genes that can reduce cell growth rates (Figure 8). Our analysis of the 35 regulators allowed for the identification of suppressors that further minimize Msn2/4 transcriptional activity in the absence of stress (Figure 2). Specifically, we found that suppression of Msn2/4 activity in the absence of stress can involve Msn2/4 degradation by the proteasome and Msn2/4 phosphorylation by the PKA or the TOR pathway (Figure 2). Previous work demonstrated that nuclear degradation of Msn2 is an important mechanism for decreasing Msn2 nuclear levels following the termination of stress (Durchschlag et al., 2004; Lallet et al., 2004) and that impaired proteasome function results in prolonged Msn2 abundance on the HSP12 promoter (Erkina et al., 2008). Our data suggest that Msn2 degradation takes place constantly, even in the absence of stress, and that this is a highly important mechanism for suppressing Msn2 activity under normal growth conditions (Figure 8). The importance of Msn2 suppression under normal growth conditions is highlighted by the trade-off between Msn2 activity and yeast growth rate (Figure 5C). This result is in agreement with previous work showing that overexpression of Msn2 or the expression of a hyperactive Msn2 mutant resulted in a reduced yeast growth rate (Gorner et al., 1998; Smith et al., 1998). On exposure to different stresses, the level of nuclear Msn2 dramatically increases (Gorner et al., 1998), rendering Msn2 suppression mechanisms ineffective.
In summary, we have described a simple and efficient approach for the characterization of Msn2 regulators that control Msn2 transcriptional activity by phosphorylation, dephosphorylation, nuclear localization, and degradation. This approach can be used for the study of other TFs that regulate diverse processes in yeast in response to different environmental signals. Whereas much previous work focused on downstream transcription events to reveal detailed transcription regulation programs mediated by TF-promoter binding events (Lee et al., 2002; Kim et al., 2009), the regulatory mechanisms described in this study reveal regulation mechanisms that control the availability of an active TF to promote promoter binding and gene expression. The future combination of these approaches can significantly broaden our view of global gene expression programs and shed new light on key regulators that control such programs.
MATERIALS AND METHODS
Strains and plasmids
The Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table S5. To construct the MSN2Δ and HIS3Δ deletion strains, we replaced the complete MSN2 and HIS3 genes with antibiotic resistance cassettes in W303-1A. The resistance cassettes for G418 and hygromycin B were amplified from plasmids pFA6a-kanMX6 and pAG32, respectively, with primers containing the flanking regions of MSN2 and HIS3 coding sequence, as required. The deletion cassettes were integrated by homologous recombination to generate the his3::hphMX4 and msn2::kanMX6 strains. The HSP12-GFP, TPS2-GFP, and PNC1-GFP fusions were generated by genomic integration of PCR fragments amplified from strains containing the target genes fused to GFP (Invitrogen, Carlsbad, CA), respectively, into strains W303-1A his3::hphMX4 and msn2::kanMX6, giving rise to the strains GS2–4 (Supplemental Table S5). Strain GS1 was generated by genomic integration of a PCR deletion fragment amplified from plasmid pAG32 into the W303-1B strain to generate msn4::hphMX4. Strains AYC1-12 were obtained by mating between strains GS1 and GS2, GS3 or GS4, followed by sporulation, tetrad dissection, and selection on appropriate media and confirmed by PCR using genomic DNA as template (Supplemental Table S5). Strains AIC1-20, APM1-20, and ATM1-20 containing deletions of Msn2 partner-encoding genes were generated by homologous recombination of a plasmid pAG25-amplified PCR deletion fragment to AYC7, AYC3, and AYC11, respectively (Supplemental Table S5). Strains ASD1–3 were generated by homologous recombination of a plasmid pAG25-amplified PCR deletion fragment into strain W303-1A, followed by transformation with the centromeric plasmid containing the MSN2-GFP fusion under the control of a constitutive ADH1 promoter and a LEU2 marker (a gift from C. Schuller). All genomic integrations were confirmed by PCR using appropriate oligonucleotides. Transformations, sporulation, and tetrad dissections were performed using standard yeast methodologies. Strains APK1-20 and APK1-40 (Supplemental Table S5) were generated using the synthetic genetic array approach as previously described (Tong et al., 2001). Briefly, the query strain AQS1 and strains of the yeast deletion or DAmP Yeast Library (Open Biosystems, Thermo Fisher Scientific, Huntsville, AL; Breslow et al., 2008) were mated on yeast-extract peptone dextrose (YPD) containing 10 g/l yeast extract, 20 g/l peptone, and 20 g/l dextrose, and diploids were selected on synthetic defined (SD) medium containing 6.7 g/l yeast nitrogen base, 20 g/l dextrose, 5.4 g/l Na2HPO4, 8.6 g/l NaH2PO4*H2O, and dropout solution of amino acids −His +G418. Diploids were then sporulated, and haploids were selected first on SD −His −Arg −Lys −Ura +canavanine +thialysine, and then a second round of haploid selection was performed on the same selection media. Haploids were further selected by two rounds of growth on SD −His −Arg −Lys −Leu +canavanine +thialysine +G418 media to identify those containing both HSP12::HSP12-GFP-HIS3MX6 and the desired gene deletion. For strains containing double deletions the same procedure was used, except that the query strain was AQS2, diploid selection was done on YPED media +G418 +clonNAT, and final haploid selection was done on SD −His +G418 +clonNAT.
To generate plasmids for overexpression of different partners and Msn2, the plasmid containing MSN2-GFP under the control of the ADH1 promoter was digested with the Sall and NotI restriction enzymes to release the Msn2-GFP gene fusion. The different partner-encoding genes were amplified from strain W303 genomic DNA and inserted into the digested vector by homologous recombination to yield plasmids overexpressing the different genes under the control of a constitutive ADH1 promoter. To generate a plasmid for overexpression of Msn2, the pMsn2-GFP plasmid was digested with NotI to release the GFP gene and recombined to generate the circular plasmid. All clones was verified by PCR and sequenced.
Hsp12-GFP, Tps2-GFP, PNC1-GFP, and PFK2-GFP expression analysis
Strains were grown overnight in 5 ml of synthetic complete (SC) medium, diluted 1:50 to OD600 0.1 in the same medium, and grown for an additional 6 h. Each culture was then divided and challenged with different conditions. The conditions used were: 0.6 mM H2O2 (oxidative stress), 0.5 M NaCl (osmotic stress), and temperature shift from 30 to 37°C (heat stress). Reference samples to which no stress was applied served as control. Following 40 min of stress application, the samples were analyzed by flow cytometry (FACSCalibur, BD Biosciences, Franklin Lakes, NJ), and the Hsp12-GFP, Tps2-GFP, Pnc1-GFP, and Pfk2-GFP fluorescence intensity of 10,000 yeast cells was measured. We found that 40 min after stress application, the fluorescent levels of the reporter genes were close to maximal (see Supplemental Figure S9 for examples). The median fluorescence intensity of each strain sample was calculated from the flow cytometry histograms, with the fluorescence intensity of each strain following exposure to the different stress conditions being normalized to that of the msn4-deleted strain measured under the same condition.
Viability assay
The different strains were grown overnight in 5 ml of SC medium and then diluted 1:50 to OD600 0.1 and grown for an additional 6 h at 30°C to OD600 0.4. Oxidative stress was induced by adding H2O2 (2 mM, final concentration) to the cultures. For heat stress, the temperature was shifted from 30 to 50°C. An aliquot of 70 μl of each strain was taken at 0, 20, 40, and 60 min after stress application and diluted 1:5000, 1:1000, and 1:500, and then 70 μl of each dilution was plated onto YPD plates. The plates were incubated for 2 d at 30°C, and colonies were counted and averaged. The viability of each strain was calculated as the ratio of the number of colonies at the different time points to the number of colonies on the reference plates (i.e., time 0). The survival of each strain was normalized to that of the msn4-deleted strain at the same time point for assessment of the effect of deletion on yeast stress survival.
Growth rate experiments
The different strains were grown overnight in YPD medium and diluted 1:100 to OD600 0.05 in the same medium. Cultures were grown in 96-well plates (96w; Greiner Bio-One, Frickenhausen, Germany) at 30 or 37°C for 15 h with constant shaking. Absorbance at 600 nm was recorded using an Infinite M200 plate reader (Tecan, Männedorf, Switzerland). To eliminate problems arising from evaporation during the growth rate experiments, we added 30 μl of mineral oil to 160 μl of growth media. To obtain growth rate estimations, we considered that part of the curve reflecting the logarithmic (exponential) growth phase (usually after 5–12 h of growth). We used linear regression following logarithmic transformation of the ODt = OD00.2t/τ equation to obtain Log2 ODt = Log2OD0 + t/τ; the slope value of the linear fit is 1/τ. The percentage increase in generation time was calculated by comparing the growth rate at 30°C to that at 37°C, according to the following equation:
MSN2 degradation experiments
Deletion strains carrying the pMSN2-GFP plasmid (Supplemental Table S5) were grown overnight in SC medium lacking leucine, diluted 1:50 to OD600 0.1 in the same media, and grown for an additional 6 h. Cycloheximide was added to the cultures to a final concentration of 400 μg/ml, and the fluorescence of each strain was recorded 30, 60, and 90 min following cycloheximide addition by flow cytometry (FACSCalibur). The percentage of degradation of Msn2-GFP was calculated as the ratio of Msn2-GFP fluorescence at the different time points to the Msn2-GFP fluorescence level determined prior to cycloheximide addition.
MSN2 localization analysis
For Msn2 localization experiments, cells expressing Msn2-GFP were grown overnight, diluted to OD600 0.1, and grown for an additional 5–6 h prior to stress application. Msn2-GFP localization was monitored on a Zeiss Axiovert 200M–based Nipkow spinning disk confocal microscope (UltraView ESR; PerkinElmer, Cambridge, United Kingdom), supplemented with an electron-multiplying charge-coupled device camera. For osmotic stress and starvation experiments, cells were applied to a flow chamber (∼10 μl) formed between a microscope slide and a coverslip, mounted with double stick tape, and sealed with wax to avoid evaporation. For heat stress experiments, 1 ml of cell culture was applied to a 35-mm Petri dish and incubated at 37°C in a temperature-controlled microscope chamber. Nuclear localization of Msn2 was monitored as a function of time at 1-min intervals. At each time point, 40–200 cells were analyzed, and the percentage of cells presenting nuclear localization of Msn2 was determined.
Yeast two-hybrid analysis
Yeast two-hybrid analysis was performed using the Yeast Two-Hybrid Phagemid Vector Kit (Stratagene, Santa Clara, CA), following the manufacturer's instructions. The pAD-MSN2 plasmid was used as bait, and plasmid pBD-UMP1 was used as prey. The pBD empty plasmid that contains no insert was used as a negative control to ensure that yeast growth was due to the Msn2–Ump1 interaction (for additional examples, see Fridman et al. [2010]). The YRG2 host strain (Stratagene) was cotransformed with plasmids pAD-MSN2 and pBD-UMP1 using the LiAC method. Single transformants were grown in liquid SC –Leu –Trp to OD600 10, washed twice with distilled, deionized water, and diluted to an initial OD600 of 0.3. A series of 10-fold serial dilutions was then spotted onto selective SC –Leu –Trp –His plates and incubated at 30C for 3 d.
Supplementary Material
Acknowledgments
We are grateful to the T. Pilpel, N. Barkai, and M. Schuldiner laboratories, Weizmann Institute of Science, Rehovot, Israel, and to the M. Kupiec laboratory, Tel Aviv University, Tel Aviv, Israel, for providing yeast strains. We thank Christoph Schuller for providing the Msn2-GFP–containing plasmid; Orna Dahan for helping with yeast dissection, tetrad analysis, and critical reading of the manuscript; and Anat Ben-Zvi, Dan Mishmar, and other colleagues at Ben-Gurion University for discussion and advice. We thank the European Research Council Ideas Program (A.A.) and Israeli Science Foundation Grants 1496/06 (A.A.) and 1043/09 (L.G.) for support.
Abbreviations used:
- GFP
green fluorescent protein
- PKA
protein kinase A
- SD
synthetic defined
- STRE
stress response element
- TF
transcription factor
- YPD
yeast-extract peptone dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-1007) on July 14, 2011.
REFERENCES
- Barkai N, Shilo BZ. Variability and robustness in biomolecular systems. Mol Cell. 2007;28:755–760. doi: 10.1016/j.molcel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol. 1998;180:1044–1052. doi: 10.1128/jb.180.5.1044-1052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR, Deschenes RJ. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 2005;24:4115–4123. doi: 10.1038/sj.emboj.7600871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durchschlag E, Reiter W, Ammerer G, Schuller C. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J Biol Chem. 2004;279:55425–55432. doi: 10.1074/jbc.M407264200. [DOI] [PubMed] [Google Scholar]
- Ennis HL, Lubin M. Cycloheximide: aspects of inhibition of protein synthesis in mammalian cells. Science. 1964;146:1474–1476. doi: 10.1126/science.146.3650.1474. [DOI] [PubMed] [Google Scholar]
- Erkina TY, Tschetter PA, Erkine AM. Different requirements of the SWI/SNF complex for robust nucleosome displacement at promoters of heat shock factor and Msn2- and Msn4-regulated heat shock genes. Mol Cell Biol. 2008;28:1207–1217. doi: 10.1128/MCB.01069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000;24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Fridman Y, Palgi N, Dovrat D, Ben-Aroya S, Hieter P, Aharoni A. Subtle alterations in PCNA-partner interactions severely impair DNA replication and repair. PLoS Biol. 2010;8:e1000507. doi: 10.1371/journal.pbio.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- Garreau H, Hasan RN, Renault G, Estruch F, Boy-Marcotte E, Jacquet M. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology. 2000;146:2113–2120. doi: 10.1099/00221287-146-9-2113. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schuller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan R, Leroy C, Isnard AD, Labarre J, Boy-Marcotte E, Toledano MB. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol Microbiol. 2002;45:233–241. doi: 10.1046/j.1365-2958.2002.03011.x. [DOI] [PubMed] [Google Scholar]
- Hohman S, Mager WH, editors. Yeast Stress Responses. Berlin: Springer-Verlag; 2003. [Google Scholar]
- Kaida D, Yashiroda H, Toh-e A, Kikuchi Y. Yeast Whi2 and Psr1-phosphatase form a complex and regulate STRE-mediated gene expression. Genes Cells. 2002;7:543–552. doi: 10.1046/j.1365-2443.2002.00538.x. [DOI] [PubMed] [Google Scholar]
- Kandror O, Bretschneider N, Kreydin E, Cavalieri D, Goldberg AL. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol Cell. 2004;13:771–781. doi: 10.1016/s1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- Kim HD, Shay T, O'Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim Biophys Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- Lallet S, Garreau H, Poisier C, Boy-Marcotte E, Jacquet M. Heat shock-induced degradation of Msn2p, a Saccharomyces cerevisiae transcription factor, occurs in the nucleus. Mol Genet Genomics. 2004;272:353–362. doi: 10.1007/s00438-004-1063-z. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Lee P, Cho BR, Joo HS, Hahn JS. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol Microbiol. 2008;70:882–895. doi: 10.1111/j.1365-2958.2008.06450.x. [DOI] [PubMed] [Google Scholar]
- Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Pacheco A, Pereira C, Almeida MJ, Sousa MJ. Small heat-shock protein Hsp12 contributes to yeast tolerance to freezing stress. Microbiology. 2009;155:2021–2028. doi: 10.1099/mic.0.025981-0. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Mathey-Prevot B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics. 2007;175:7–16. doi: 10.1534/genetics.106.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LS, Causton HC, Young RA, Fink GR. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc Natl Acad Sci USA. 2000;97:5984–5988. doi: 10.1073/pnas.100113397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Wagner A. Distributed robustness versus redundancy as causes of mutational robustness. Bioessays. 2005;27:176–188. doi: 10.1002/bies.20170. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.