The vacuolar ATPase complex in yeast contains two isoforms of subunit a that dictate the subcellular localization of the V-ATPase enzyme. The most recent common ancestor of subunit a (Anc.a) is reconstructed, and its function and localization in modern Saccharomyces cerevisiae are characterized. Anc.a is able to replace both subunit a isoforms.
Abstract
The vacuolar-type, proton-translocating ATPase (V-ATPase) is a multisubunit enzyme responsible for organelle acidification in eukaryotic cells. Many organisms have evolved V-ATPase subunit isoforms that allow for increased specialization of this critical enzyme. Differential targeting of the V-ATPase to specific subcellular organelles occurs in eukaryotes from humans to budding yeast. In Saccharomyces cerevisiae, the two subunit a isoforms are the only difference between the two V-ATPase populations. Incorporation of Vph1p or Stv1p into the V-ATPase dictates the localization of the V-ATPase to the vacuole or late Golgi/endosome, respectively. A duplication event within fungi gave rise to two subunit a genes. We used ancestral gene reconstruction to generate the most recent common ancestor of Vph1p and Stv1p (Anc.a) and tested its function in yeast. Anc.a localized to both the Golgi/endosomal network and vacuolar membrane and acidified these compartments as part of a hybrid V-ATPase complex. Trafficking of Anc.a did not require retrograde transport from the late endosome to the Golgi that has evolved for retrieval of the Stv1p isoform. Rather, Anc.a localized to both structures through slowed anterograde transport en route to the vacuole. Our results suggest an evolutionary model that describes the differential localization of the two yeast V-ATPase isoforms.
INTRODUCTION
The vacuolar-type, proton-translocating ATPase (V-ATPase) is a multisubunit molecular machine responsible for organelle acidification in eukaryotes (Forgac, 2007). The V-ATPase enzyme couples hydrolysis of ATP with proton translocation across membranes using a conserved rotary mechanism (Marshansky and Futai, 2008). The electrochemical gradient generated by the V-ATPase is required for a diverse set of biological processes, including vesicular trafficking, exocytosis, membrane fusion, ion homeostasis, development, and pH regulation (Kane 2006; Forgac 2007). The V-ATPase has also been implicated in a number of disease models, including cancer cell migration (Martínez-Zaguilán et al., 1999), osteopetrosis (Frattini et al., 2000), viral entry into cells (Gruenberg and van der Goot, 2006), and renal tubular acidosis (Karet et al., 1999).
The budding yeast Saccharomyces cerevisiae serves as an excellent model system to study the V-ATPase because the enzyme complex is not required for viability. Yeast uses the proton gradient generated by the V-ATPase to maintain intracellular pH levels and regulate ion homeostasis (Kane, 2006). Toxic levels of metals are sequestered within the yeast vacuole by proton-exchange antiporter pumps (Klionsky et al., 1990). Disruption of V-ATPase function results in a loss of vacuolar acidification and renders cells sensitive to excess levels of divalent cations such as calcium or zinc. The yeast V-ATPase complex contains 14 different subunits, some of which are present in multiple copies (Forgac, 2007). The enzyme contains two subdomains: the cytosolic V1 portion is responsible for hydrolysis of ATP, whereas the membrane-bound V0 portion transports protons across the lipid bilayer (Graham et al., 2003). The V1 domain contains subunits A, B, C, D, E, F, G, and H, whereas the V0 domain contains subunits a, d, e, c, c′, and c′′. All of these subunits are required for V-ATPase function. Assembly of the V0 subdomain occurs within the endoplasmic reticulum (ER) and requires the presence of five dedicated assembly factors: Vma21p, Vma22p, Vma12p, Pkr1p, and Voa1p (Hirata et al., 1993; Hill and Stevens, 1995; Malkus et al., 2004; Davis-Kaplan et al., 2006; Ryan et al., 2008). These ER-resident proteins aid in chaperoning components of the V0 and are required for full enzyme function. Regulation of V-ATPase function can occur through rapid, reversible dissociation of the V1 and V0 subcomplexes, differential assembly, or differences in the localization of the enzyme complex (Kane, 2006; Forgac, 2007).
Yeast contain two different V-ATPase complexes, depending on the incorporation of one of two isoforms of the V0 subunit a, Vph1p or Stv1p (Figure 1A; Manolson et al., 1992, 1994). The subunit composition of the V-ATPase is identical in these two populations, yet there are dramatic functional and phenotypic differences for each protein complex (Manolson et al., 1994; Qi and Forgac, 2007). In yeast, subunit a is responsible for targeting of the V-ATPase to distinct subcellular compartments; complexes containing Vph1p (Vph1p-complex) reside on the vacuolar membrane, whereas Stv1p-containing complexes (Stv1p-complex) are found within the Golgi/endosomal network (Manolson et al., 1994). The cytosolic, N-terminal portion of Stv1p and Vph1p contains the targeting information necessary for trafficking of the V-ATPase to these organelles (Kawasaki-Nishi et al., 2001a). Complexes containing the Stv1p isoform are transported to the late endosome and are actively retrieved back to the Golgi (Kawasaki-Nishi et al., 2001a). Aside from the difference in localization, the Stv1p and Vph1p complexes have been shown to differ in transcript expression, protein abundance, assembly, coupling efficiency, and reversible dissociation. The Stv1p-complex is found at much lower protein levels, has a lower ATP coupling efficiency, and does not undergo reversible dissociation of V1 and V0 on glucose starvation compared with the Vph1p-complex (Manolson et al., 1992, 1994; Kawasaki-Nishi et al., 2001b; Perzov et al., 2002).
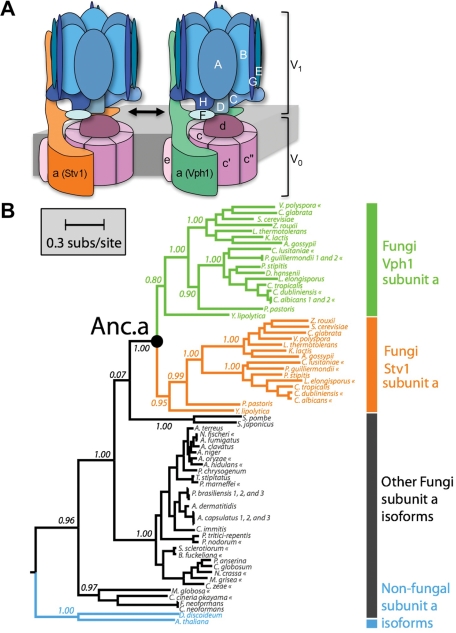
FIGURE 1:
Model and phylogeny of the two isoforms of the yeast V-ATPase complex. (A) The 14-subunit complex differs by the incorporation of the two isoforms of subunit a (Stv1p or Vph1p). This subunit a isoform determines the subcellular localization of the V-ATPase in S. cerevisiae. (B) The maximum likelihood phylogeny of fungal V-ATPase subunit a. The tree was rooted using subunit a sequences from D. discoideum (Amoebozoa) and A. thaliana (Plantae) as outgroups. The position of the reconstructed fungal ancestral subunit a protein is labeled as “Anc.a.” Extant taxa marked with « indicate predicted subunit a sequences according to GenBank annotations, whereas all other sequences have been functionally assayed. Branch support values are approximate-likelihood ratio test values based on a Shimodaira-Hasegawa–like procedure. Statistical support for the Anc.a sequence can be found in Supplemental Figure S1.
There is a relative paucity of information describing the evolutionary mechanisms responsible for the presence of multiple isoforms of various V-ATPase subunits. Phylogenetic analysis suggests that gene duplication events played a significant role in the evolution of subunit a and the entire V-ATPase enzyme in eukaryotes (Müller and Grüber, 2003; Cross and Müller, 2004). Characterization of the two subunit a isoforms within budding yeast revealed a number of key biochemical differences in regulation, assembly, and trafficking between the V-ATPase complexes. However, it is unclear how subunit a in S. cerevisiae evolved from a preduplicated, single isoform into the two contemporary Vph1p and Stv1p subunits that have distinct localization patterns.
Eukaryotes outside of fungi also have multiple genes and many isoforms of various V-ATPase subunits, including subunit a (Forgac, 2007). For instance, Caenorhabditis elegans has four isoforms of subunit a (Oka et al., 2001b), mice have four isoforms, including one present on the plasma membrane (Oka et al., 2001a; Toyomura et al., 2003), and Paramecium tetraurelia is reported to have 17 distinct subunit a isoforms (Wassmer et al., 2006). The increasing number of separate isoforms allows for a high degree of specialization in the subcellular localization of the V-ATPase, as well as cell-specific expression and regulation (Toei et al., 2010).
Although most eukaryotes use several distinct V-ATPase isoforms of subunit a, there are groups of species that have only a single isoform. How are eukaryotes with only one V-ATPase complex able to properly acidify various cellular compartments? Fungal species basal to the clade of budding yeast belong to a group that contains only a single subunit a isoform (Chavez et al., 2006). However, attempts to characterize homologues of subunit a from Arabidopsis thaliana and Schizosaccharomyces pombe within budding yeast for V-ATPase function have been unsuccessful (Aviezer-Hagai et al., 2000; unpublished results) because difficulties in this horizontal evolutionary approach often result from differences in genetic background (Harms and Thornton, 2010). This approach does not address 1) sequence changes that have accumulated over evolutionary time that did not contribute to protein function and 2) the effects of epistatic interactions between mutational changes that have occurred along lineage-specific evolutionary trajectories (Harms and Thornton, 2010).
To address these questions, we reconstructed the most recent common ancestor of Vph1p and Stv1p and tested its function as the only isoform of subunit a in S. cerevisiae. The ancestral subunit a (Anc.a) functionally replaces both Vph1p and Stv1p through acidification of both the Golgi/endosomal network and vacuole. Anc.a displays a dual localization pattern to both of these cellular compartments. In addition, the dual localization of Anc.a does not require the retromer complex that is necessary for retrieval of the Stv1p V-ATPase from the endosome to the late Golgi. Rather, it is likely that Anc.a localizes to both structures through slowed anterograde transport en route to the yeast vacuole.
RESULTS
Anc.a functions in extant S. cerevisiae as part of a hybrid V-ATPase complex
It is likely the two subunit a isoforms in yeast (Vph1p and Stv1p; Figure 1A) evolved from a gene duplication event within the fungal clade (Figure 1B). We tested the function of the most recent common ancestor of Vph1p and Stv1p (referred to as Anc.a) in extant S. cerevisiae. The sequence of this ancient protein was determined by calculating the maximum likelihood sequence from the phylogeny of a large set of subunit a isoforms from modern species (Supplemental Table S1 and Figure S1). Anc.a shares a high degree of sequence identity to the C-termini of both Stv1p and Vph1p (Figure 2A). However, Anc.a is only 38 and 53% identical to the N-termini of Stv1p and Vph1p, respectively (Figure 2A). Secondary structure predictions of the N-terminal, cytosolic domains of Vph1p, Stv1p, and Anc.a revealed a very similar overall structure of the three proteins (Figure 2B).
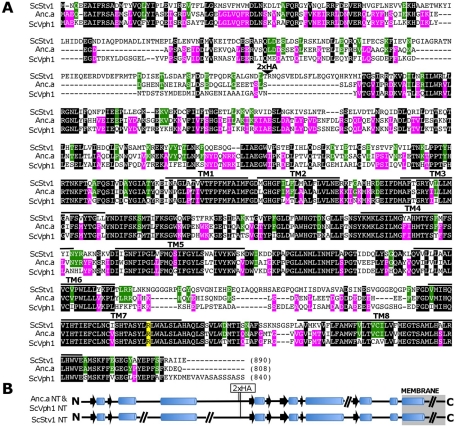
FIGURE 2:
Alignment of S. cerevisiae Stv1p, Vph1p, and Anc.a amino acid sequences. (A) Identical residues among all three proteins are shown against a black background. Residues identical between only Vph1p and Anc.a are shown against a magenta background. Residues identical between Stv1p and Anc.a are shown against a green background. The alignment was performed using the CLUSTALW program (Thompson et al., 1994). Potential transmembrane domains are marked with black bars above the alignment [modeled from Wang et al. (2008)]. The critical arginine residues (Stv1p R795, Vph1p R735, and Anc.a R737) are marked against a yellow background. The position of the double-HA epitope tag is indicated by a triangle (beginning after residue 171 for Vph1p, 227 for Stv1p, and 185 for Anc.a). (B) Secondary structure prediction of the N-termini of Stv1p, Vph1p, and Anc.a by the PSIPRED program (University College, London). α-Helices are shown as cylinders, β-sheets are shown as black arrows, and coiled regions are shown as black lines. The breaks in the lines represent short insertions that have no predicted structure.
Because the expression level of the ancestral subunit a is unknown, we chose to test the expression of an integrated copy of Anc.a under control of either the STV1 or VPH1 promoter; the coding region of Anc.a replaced the entire coding region of either yeast gene. We assayed the ancestral isoform in yeast lacking both contemporary Vph1p and Stv1p on media buffered with either excess calcium or zinc (Figure 3A). Yeasts deleted for both isoforms do not acidify their vacuoles and are unable to survive on media containing excess metals (Manolson et al., 1994). Cells deleted for STV1 (stv1Δ) are not sensitive to either calcium or zinc, as they still contain a functioning vacuolar isoform. However, yeast lacking VPH1 (vph1Δ) grow in the presence of 100 mM Ca2+ but cannot survive on media containing 3.5 mM Zn2+. Resistance to calcium depends on proton antiporters present on both the vacuole and Golgi apparatus; cells containing an acidified Golgi/endosome are therefore able to survive on excess calcium (Rudolph et al., 1989; Miseta et al., 1999). Resistance to zinc depends on the presence of proton/zinc antiporters present on the vacuolar membrane (MacDiarmid et al., 2000). Expression of Anc.a (which replaced the endogenous, integrated copy of either STV1 or VPH1) under control of either promoter was sufficient to promote growth on toxic levels of calcium or zinc (Figure 3A). Western blots to the HA epitope tag revealed the difference in steady-state levels of Anc.a under control of either the STV1 or VPH1 promoter; strains differed in levels of Anc.a protein by ∼20-fold (Figure 3B). The levels of the V1 subunit Vma2p did not change in either of these strains. These results indicate that Anc.a allows for V-ATPase function on the yeast vacuole.
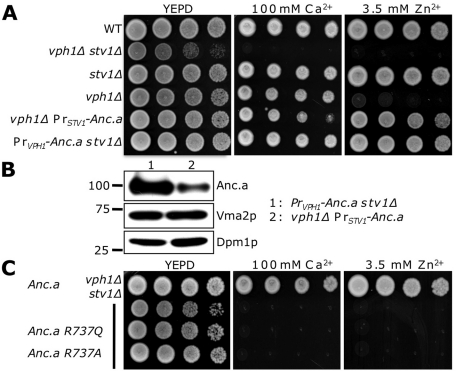
FIGURE 3:
Ancestral subunit a complements a loss of both VPH1 and STV1. (A) Exponentially growing cultures of wild-type (WT; SF838-1Dα), vph1Δ stv1Δ (GFY271), stv1Δ (KEBY4), vph1Δ (LGY120), vph1Δ PrSTV1-Anc.a (GFY251), and PrVPH1-Anc.a stv1Δ (GFY250) strains were spotted onto rich media and media containing either 100 mM calcium or 3.5 mM zinc. (B) Western blot analysis of strains (vph1Δ stv1Δ) expressing epitope-tagged Anc.a protein (GFY250 and GFY251). Samples were separated by SDS–PAGE, transferred to nitrocellulose, and probed with antibodies to the HA epitope tag and V1 subunit, Vma2p. Antibodies to Dpm1p served as a loading control. The position of the closest molecular marker is indicated. (C) Yeast deleted for VPH1 and STV1 (GFY271) were transformed with vectors expressing Anc.a (pGF245), Anc.a R737Q (pGF342), or Anc.a R737A (pGF341) and tested on media containing calcium and zinc as described in A. Vectors used were under control of the VPH1 promoter.
Our ancestral reconstruction included 134 residues that were “poorly” supported by our maximum likelihood phylogenetic algorithm (posterior probability, <0.8) and had plausible secondary alternate states (posterior probability, >0.2). Posterior probability (PP) is a measure of the confidence of each ancestral state expressed as a probability (Hanson-Smith et al., 2010). Given the statistical support for alternative amino acids within the Anc.a sequence, we sampled 50 independent, single–amino acid substitutions to Anc.a; sites were chosen randomly across the entire protein sequence. Point mutations were introduced into the protein sequence of Anc.a, and each alternative state was tested in vph1Δ stv1Δ yeast by growth assays on calcium and zinc. We determined that our reconstruction was robust to uncertainty, as all 50 changes to Anc.a still allowed for full complementation (Supplemental Table S2).
Previous work highlighted the importance of the arginine residue at site 735 in Vph1p and the corresponding arginine at site 795 in Stv1p (Kawasaki-Nishi et al., 2001c). This amino acid is essential for proton translocation and is highly conserved in eukaryotic sequences. Complementation by Anc.a in vph1Δ stv1Δ yeast was fully dependent on the presence of this critical arginine residue within the C-terminus. It is also strongly supported in our reconstruction with a PP of 1.0. Substitution of Vph1p arginine 735 for glutamine (which allows for V0 assembly) or alanine (which disrupts V0 assembly) was shown to completely abolish V-ATPase activity (Kawasaki-Nishi et al., 2001c). When these mutational changes were introduced into Anc.a in cells lacking VPH1 and STV1 we observed no complementation on media containing either calcium or zinc (Figure 3C), yet Western blots revealed that the Anc.a R737Q mutant protein was stable (unpublished data). This demonstrates that Anc.a functions within a hybrid V-ATPase complex and is required for proton translocation.
The yeast isoforms of subunit a are subject to degradation on loss of Vma21p or any of the other essential assembly factors (Graham et al., 1998; Hill and Cooper, 2000). Stability of the Anc.a protein was also fully dependent on the presence of the conserved, dedicated V-ATPase assembly factor Vma21p (Supplemental Figure S2). These data demonstrate that assembly of the hybrid ancestral V-ATPase complex requires the modern-day yeast assembly machinery.
Anc.a functionally replaces yeast Stv1p and Vph1p
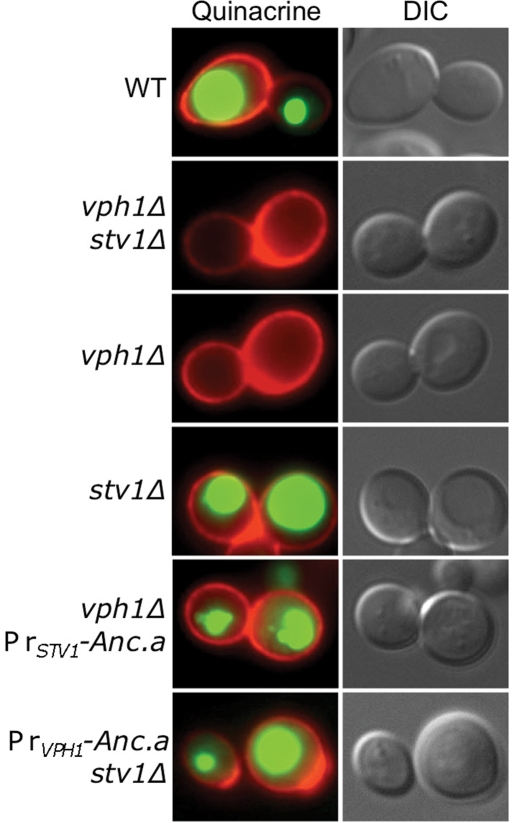
We assayed whether expression of Anc.a properly acidified the yeast vacuole by staining with the dye quinacrine. Quinacrine is a weakly basic, fluorescent dye that accumulates within acidified compartments. Staining of vph1Δ or vph1Δ stv1Δ mutants showed that these cells do not acidify their vacuoles (Figure 4). Yeast lacking the Golgi-isoform, Stv1p, properly acidify their vacuoles, as they contain the vacuolar isoform, Vph1p. Expression of Anc.a was sufficient to confer wild-type levels of acidification in cells lacking both STV1 and VPH1 (Figure 4). These data suggest that the ancestral protein can function in place of the vacuolar isoform.
FIGURE 4:
Expression of Anc.a results in acidification of the yeast vacuole. Cultures of wild-type (WT; SF838-1Dα), vph1Δ stv1Δ (GFY271), vph1Δ (LGY120), stv1Δ (KEBY4), vph1Δ PrSTV1-Anc.a (GFY251), and PrVPH1-Anc.a stv1Δ (GFY250) strains were stained with quinacrine (green; acidified compartments) and concanavalin A–tetramethylrhodamine (red; stains the yeast cell wall) and viewed by fluorescence or DIC microscopy.
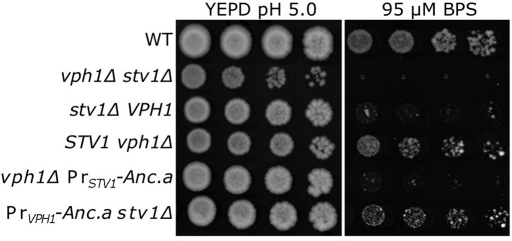
Quinacrine staining is not sensitive enough to visualize Golgi/endosomal acidification in yeast. In addition, conventional growth assays (on elevated calcium or zinc) do not differentiate between wild-type yeast and yeast lacking STV1, as the vacuolar isoform compensates for a loss of the Stv1p-containing complex (Figure 3A). However, a recent report described a phenotypic difference between wild-type yeast and stv1Δ mutant yeast on media containing the iron chelator bathophenanthrolinedisulfonic acid (BPS; Jo et al., 2009). Acidification of the Golgi/endosomal network is critical for iron metabolism/homeostasis (Philpott and Protchenko, 2008). We used low-iron conditions to determine whether Anc.a could functionally substitute for Stv1p in vivo. Cells lacking the V-ATPase (vph1Δ stv1Δ) were fully sensitive to media containing 95 μM BPS (Figure 5). Although yeast lacking Vph1p survived under these conditions, cells lacking the Golgi isoform, Stv1p, were fully sensitive. Expression of Anc.a under control of the STV1 promoter was not sufficient to promote growth on media buffered with BPS. However, expression of Anc.a under control of the VPH1 promoter conferred growth under these low-iron conditions (Figure 5). These results demonstrate that Anc.a is also sufficient to substitute for the Golgi isoform, Stv1p.
FIGURE 5:
Expression of Anc.a complements the low-iron growth defect of stv1Δ yeast. Cultures of wild-type (WT; SF838-1Dα), vph1Δ stv1Δ (GFY271), vph1Δ (LGY120), stv1Δ (KEBY4), vph1Δ PrSTV1-Anc.a (GFY251), and PrVPH1-Anc.a stv1Δ (GFY250) strains were grown to saturation in YEPD, pH 5.0, plus 25 μM BPS, backdiluted into YEPD, pH 5.0, plus 100 μM BPS, and grown for 6 h before being spotted onto rich media or media containing 95 μM BPS.
Anc.a localizes to both the Golgi/endosome and vacuole in yeast
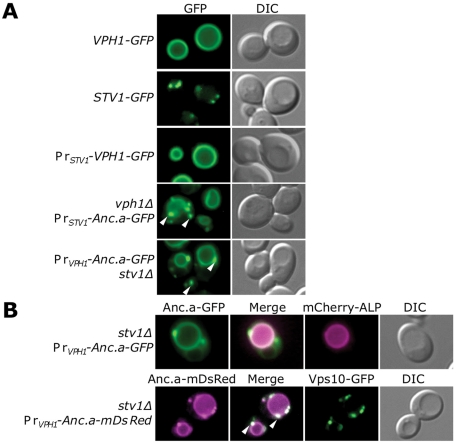
Because Anc.a could complement the loss of either Stv1p or Vph1p, we assayed the cellular localization of the ancestral V-ATPase complex in yeast. Tagging of subunit a isoforms with GFP at the C-terminus did not compromise V-ATPase function (unpublished data). Vph1p-GFP localized to the limiting membrane of the vacuole, whereas Stv1p-GFP localized to punctate structures (Figure 6A). Stv1p colocalizes with the late-Golgi marker A-ALP and, to a lesser extent, the endosomal marker Pep12p (Kawasaki-Nishi et al., 2001a). A-ALP is a reporter protein containing a fusion between the cytosolic portion of Ste13p and yeast alkaline phosphatase encoded by the PHO8 gene (Nothwehr et al., 1997). The majority of Stv1p resides within the late Golgi, although it was shown to cycle through the endosome (Kawasaki-Nishi et al., 2001a). Overexpression of Stv1p results in its missorting to the vacuolar membrane (Manolson et al., 1994). Because the ancient expression level of the ancestral isoform is unknown, we tested whether altering the expression level of the contemporary yeast subunit Vph1p would result in a different localization pattern. We tested the localization of Vph1p–green fluorescent protein (GFP) under control of the weaker STV1 promoter. Whereas the intensity of the fluorescence signal was greatly decreased, we found no difference in the localization of Vph1p-containing V-ATPase complex, as it was still present on the vacuolar membrane. Of interest, Anc.a localized to both punctate structures and the vacuolar membrane (Figure 6A). The localization pattern did not change when Anc.a was expressed under either the STV1 or VPH1 promoter, suggesting that localization is not dependent on expression levels for this isoform.
FIGURE 6:
Anc.a localizes to the vacuole membrane and Golgi/endosomal network. (A) Yeast cultures expressing VPH1-GFP (GFY308), STV1-GFP (GFY310), VPH1-GFP at the STV1 locus (GFY307), vph1Δ PrSTV1-Anc.a-GFP (GFY253), and PrVPH1-Anc.a-GFP stv1Δ (GFY255) were visualized by fluorescence or DIC microscopy. White arrows label several of the punctate structures seen in strains expressing Anc.a protein. (B) Cultures of PrVPH1-Anc.a-GFP stv1Δ (GFY255) were transformed with a vector expressing mCherry-ALP and visualized by fluorescence microscopy. The merged image shows the overlay of the GFP (green) and mCherry (magenta) channels as a white signal. Cultures of stv1Δ PrVPH1-Anc.a-mDsRed (GFY314) were transformed with a CEN-based plasmid expressing Vps10p-GFP (under control of the VPS10 promoter) and visualized by fluorescence microscopy. The merged image between the mDsRed (magenta) and GFP (green) channels presents overlap as a white color. White arrows indicate the presence of several distinct, punctate structures in the merged image.
We next determined the precise cellular compartments that contained the ancestral V-ATPase. ALP served as a marker for the vacuolar membrane. ALP traffics to the vacuole using a different route (ALP pathway) than the V-ATPase (vacuolar carboxypeptidase Y encoded by the PRC1 gene [CPY] pathway; Raymond et al., 1992; Cowles et al., 1997; Piper et al., 1997; Stepp et al., 1997). In Figure 6B we show colocalization of mCherry (a mutated form of monomeric red fluorescent protein)-tagged ALP with Anc.a-GFP on the limiting membrane of the vacuole. Vps10p, the receptor for CPY, cycles between the late Golgi and endosome and serves as a marker for the late Golgi (Marcusson et al., 1994; Cooper and Stevens, 1996). We assayed the localization of Anc.a-mDsRed (amonomeric form of a red fluorescent protein from the genus Discosoma) in yeast also expressing Vps10p-GFP (Figure 6B). Vps10p-GFP localized to punctate structures, as expected, and the punctate structures containing Anc.a colocalized with Vps10p. These data demonstrate that the Anc.a-containing V-ATPase complex is localized to both the trans-Golgi network and the vacuolar membrane.
The Anc.a V-ATPase complex uses slowed anterograde trafficking to the vacuole membrane
We investigated the different trafficking mechanisms that might describe the dual localization pattern of Anc.a. It is possible that a portion of Anc.a protein could be retrieved from the endosome back to the Golgi (similar to yeast Stv1p). Alternatively, rather than retrograde transport, Anc.a could use slowed anterograde transport en route to the vacuole. Either of these models would account for the steady-state localization of Anc.a to both vacuolar and Golgi/endosomal compartments.
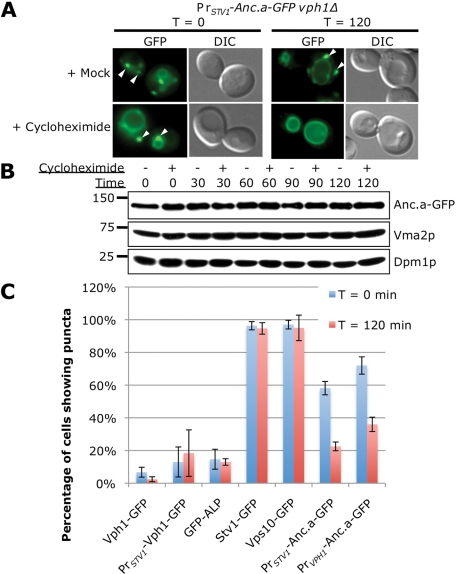
To differentiate between these models, we treated cells expressing Anc.a-GFP with cycloheximide to inhibit protein synthesis yet still allow for continued vesicular transport of cargo proteins. Cargo that is actively retrieved from the endosome to the Golgi should continue to localize to the Golgi/endosome upon treatment with cycloheximide. However, proteins lacking a retrieval signal should be transported to the vacuole after protein synthesis is inhibited. Yeast treated with cycloheximide showed a dramatic shift in the localization pattern of Anc.a-GFP (Figure 7A). Whereas the mock treatment displayed dual localization to both the vacuolar membrane and punctate structures, cells treated with cycloheximide only showed strong GFP signal on the vacuolar membrane. Control treatments did not show any detectible difference in localization pattern. We also tested to ensure that the shift in localization was not due to a decrease in steady-state protein levels of Anc.a-GFP. Western blots demonstrated that there was no significant change in Anc.a protein levels during the cycloheximide treatment compared with control samples (Figure 7B). Finally, we quantified the localization of Anc.a-GFP compared with a number of other resident Golgi proteins (Stv1p and Vps10p) or resident vacuolar proteins (Vph1p and ALP) upon treatment with cycloheximide (Figure 7C). Cells were scored for the presence of the fluorescent proteins in punctate structures. On drug treatment, ALP, Vph1p, or Vph1p expressed from the STV1 locus did not show any shift in the percentage of cells containing puncta (Figure 7C). Similarly, we did not see any strong shift in the localization pattern for the late-Golgi membrane proteins Stv1p and Vps10p. However, there was a dramatic difference in the localization pattern of Anc.a-GFP under either the VPH1 (50% reduction) or STV1 (61% reduction) promoter. In addition, the fluorescence intensity of the remaining puncta was also decreased (unpublished data). These experiments indicate that the trafficking of Anc.a likely involves slow, anterograde transport en route to the vacuole and is likely not to involve active retrieval/retention within the Golgi/endosome.
FIGURE 7:
Anc.a localization shifts to the vacuole membrane after treatment with cycloheximide. (A) An exponentially growing culture of PrSTV1-Anc.a-GFP vph1Δ (GFY253) was treated with either 0.02% DMSO (mock) or 20 μg/ml cycloheximide. Cells were visualized both immediately after and 2 h after treatment by fluorescence and DIC microscopy. White arrows indicate the presence of multiple puncta. (B) Cultures of PrSTV1-Anc.a-GFP vph1Δ (GFY253) were treated with DMSO or cycloheximide as described in A. Whole-cell extracts were prepared at 30-min intervals, separated by SDS–PAGE, and probed with anti-HA, Vma2p, and Dpm1p antibodies. (C) Strains containing the following GFP-tagged constructs were treated with DMSO or cycloheximide: VPH1-GFP (GFY308), STV1-GFP (GFY310), VPH1-GFP at the STV1 locus (GFY307), wild-type yeast expressing GFP-PHO8 (ALP) or VPS10-GFP (pGF35), vph1Δ PrSTV1-Anc.a-GFP (GFY253), and PrVPH1-Anc.a-GFP stv1Δ (GFY255). The percentage of cells showing punctate fluorescent staining was quantified. Between 150 and 400 cells were counted for each control strain, and 400–600 cells were counted for stains containing Anc.a. Experiments were performed in triplicate, and the error is expressed as the SD.
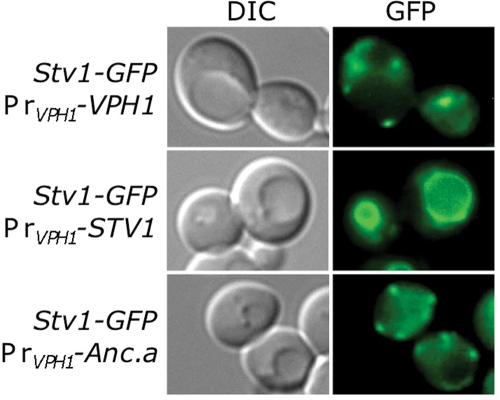
Finally, since Anc.a also localizes to the Golgi/endosomal network, we tested whether Anc.a would use the same sorting machinery as the Stv1p isoform. The retrieval machinery that sorts the Stv1p-containing V-ATPase is saturable; overexpression of Stv1p results in its mislocalization to the yeast vacuole (Manolson et al., 1994). Therefore we tested whether expression of Anc.a would saturate the Stv1p-sorting machinery and cause endogenous levels of Stv1p to be missorted to the vacuolar membrane. Stv1p-GFP localized to punctate structures when VPH1 was also present (Figure 8, row 1). When VPH1 was replaced with an untagged copy of STV1 (resulting in Stv1p overexpression), the endogenous copy of Stv1p-GFP was mislocalized to the vacuole (Figure 8, row 2). Expression of Anc.a from the VPH1 locus did not cause a shift in the localization of endogenous Stv1p-GFP (Figure 8, row 3). Finally, overexpression of Anc.a did not cause miscolocalization of Stv1-GFP (unpublished data). These data imply that trafficking of Anc.a does not use the same sorting machinery as the Stv1p isoform.
FIGURE 8:
Anc.a does not perturb Stv1p sorting in yeast. Strains expressing STV1-GFP that contained either VPH1 (GFY315), STV1, (GFY316), or Anc.a (GFY317) integrated at the VPH1 locus were visualized by fluorescence and DIC microscopy.
Disruption of endosome-to-Golgi retrograde transport causes mislocalization of the Stv1 isoform but does not perturb Anc.a localization
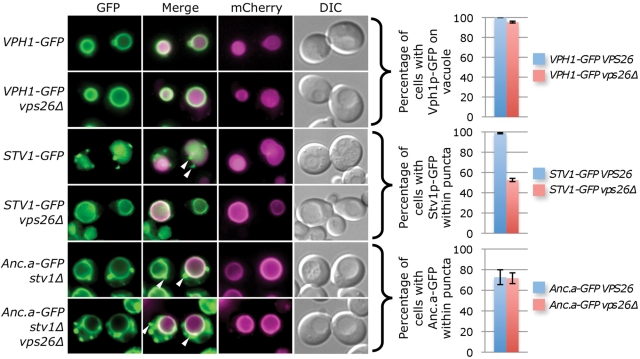
We tested whether the sorting of Anc.a required active retrieval from the late endosome to the Golgi by analyzing mutants of the yeast “retromer” complex. This conserved molecular machine has been shown to be required for maintaining the steady-state localization of a number of resident Golgi proteins, including Vps10p, Ste13p, and Kex2p, by actively retrieving cargo from the late endosome to the Golgi (Nothwehr and Hindes, 1997; Seaman et al., 1998; Seaman, 2005). We disrupted retrograde transport by deleting VPS26 and assayed the localization of Vph1p, Stv1p, and Anc.a in these cells (Figure 9). As expected, Vph1p-GFP localized to the vacuole membrane even when the retromer was deleted. However, the distribution of Stv1p-GFP was dramatically altered in vps26Δ cells; fluorescence signal shifted from discrete cellular puncta (Golgi/endosome) to the limiting membrane of the vacuole. In comparison, there was no change in the localization pattern for cells expressing Anc.a-GFP when the retromer was disrupted. Similar results were obtained when VPS29, a second retromer subunit, was deleted (unpublished data). Finally, there was no change in the steady-state levels of subunit a protein when the retrograde transport was disrupted, suggesting that mislocalization of Stv1p to the vacuole was not due to overexpression (unpublished data). These data demonstrate that 1) the modern Stv1p isoform requires retrieval from the late endosome to the Golgi and 2) the transport/sorting of Anc.a is independent of retromer function.
FIGURE 9:
Disruption of endosome-to-Golgi retrograde transport causes mislocalization of the Stv1 isoform but does not perturb the dual localization of Anc.a. Yeast strains VPH1-GFP (GFY308), STV1-GFP (GFY310), and stv1Δ PrVPH1-Anc.a-GFP (shown as Anc.a-GFP stv1Δ; GFY255) were deleted for VPS26, one of the essential components of the yeast retromer complex, transformed with a vector expressing mCherry-ALP (pGF242), and visualized by fluorescence and DIC microscopy. Cells were scored for the presence of GFP fluorescence on either 1) the vacuole membrane (Vph1p-GFP) or 2) within distinct punctate structures (Stv1p-GFP and Anc.a-GFP). Cells that did not contain Stv1p-GFP signal within puncta displayed vacuole membrane localization. Experiments were performed in triplicate, and the error is expressed as the SD. White arrows indicate several puncta that do not colocalize with mCherry-ALP signal. Overlay of the GFP and mCherry channels is presented as white signal within the merge image.
We also assayed the localization of Anc.a in strains disrupted for other components of vesicular trafficking. To test whether Anc.a traffics from the Golgi directly to the vacuole using the ALP pathway, we deleted APS3, a component of the AP-3 machinery (Cowles et al., 1997), and assayed the localization of Anc.a (Supplemental Figure S3). There was no change in the distribution of Anc.a-GFP in these mutant cells. These results are consistent with previous observations that the V-ATPase complex is transported to the vacuole using the late endosome (Raymond et al., 1992). To test the requirement of endosomal transport for Anc.a sorting, we deleted VPS27, a gene required for trafficking through the late endosome. Deletion of this type of vacuolar protein sorting gene (“class E”) causes the formation of an aberrant multivesicular body (MVB) and traps protein cargo en route to the vacuole and cargo cycling between the Golgi and endosome (Bowers and Stevens, 2005). Stv1p, Vph1p, and Anc.a all localized to the aberrant MVB in vps27Δ yeast (Supplemental Figure S3; unpublished data). The requirement for two other classes of genes required for post-Golgi sorting of Anc.a were also assayed. The dynamin-like GTPase VPS1 is required for post-Golgi trafficking, yet protein cargo has been found to reach the vacuole in vps1Δ yeast via the plasma membrane (Wilsbach and Payne, 1993; Nothwehr et al., 1995). In addition, VPS51, a component of the Golgi-associated retrograde protein (GARP) complex, is required for retrograde transport from the early endosome (Reggiori et al., 2003). Deletion of either VPS1 or VPS51 did not alter the localization of Anc.a, Stv1p, or Vph1p (Supplemental Figure S3; unpublished data). Endogenous levels of Stv1p-GFP were not mislocalized to the vacuole in yeast lacking GARP components Vps51p, Vps52p, or Vps53p (unpublished data). These data indicate that loss of these classes of trafficking components is not sufficient to disrupt the dual localization pattern of the hybrid Anc.a V-ATPase complex in yeast.
The information for the dual localization for Anc.a is present within both the N- and C-terminal domains
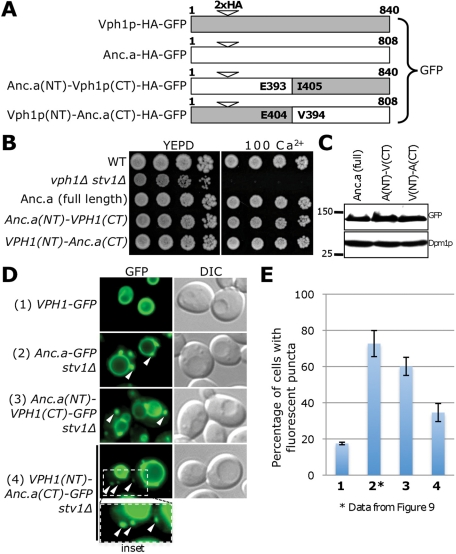
Previous work provided insight into the sorting of the modern V-ATPase isoforms; the N-terminal subdomain of Vph1p or Stv1p is sufficient to facilitate the subcellular localization of the V-ATPase (Kawasaki-Nishi et al. 2001a). We chose to investigate which domain of the ancestral isoform was sufficient to cause a dual localization pattern to both the vacuole and Golgi/endosome, using a similar experimental strategy. We tested reciprocal protein fusions between the amino and carboxyl subdomains of Anc.a and Vph1p (Figure 10A). The two protein chimeras were stably expressed in yeast cells and displayed strong complementation phenotypes in stv1Δ vph1Δ mutants (Figure 10, B and C). Surprisingly, both chimeras displayed a dual localization pattern to the vacuole membrane and cellular puncta. The Anc.a(NT)–Vph1p(CT) fusion appeared very similar to the localization pattern of full-length Anc.a yet also had fluorescence signal present within the endoplasmic reticulum, indicating a partial V0 assembly and/or ER exit defect for this chimera. The Vph1p(NT)–Anc.a(CT) chimera showed a marked decrease in both the number of cells showing puncta and the fluorescence intensity of these structures (Figure 10, D and E). These data indicate that either subdomain of Anc.a is sufficient to cause a dual localization pattern (as part of protein chimeras), yet there is also a stronger localization phenotype associated with the N-terminus of Anc.a.
FIGURE 10:
The localization information for Anc.a is found within both the N- and C-terminal domains. (A) Reciprocal chimeras were constructed using the N- and C-terminal domains of Anc.a and Vph1p. The position of the double-HA tag is at residue 171 for Vph1p and residue 185 for Anc.a. All constructs contain a C-terminal GFP tag and are under control of the VPH1 promoter. The position chosen for the junction between the cytosolic and lumenal domains was a conserved glutamic acid residue (E404 in Vph1p and E393 in Anc.a). (B) Growth assays were performed on strains expressing full-length Anc.a (GFY255), the Anc.a(NT)–Vph1p(CT) chimera (GFY361), or the Vph1p(NT)–Anc.a(CT) chimera (GFY362) compared with control strains on 100 mM calcium plates. Strains expressing Anc.a or Anc.a chimeras (under PrVPH1 control) were also deleted for STV1. (C) Yeast strains from B were assayed by Western blot using anti-GFP antibodies; Dpm1p served as a loading control. (D) Localization was visualized by fluorescence and DIC microscopy and compared with control strains. Vph1p, Anc.a, and the two protein chimeras are under control of the VPH1 promoter. White arrows indicate the presence of distinct cellular puncta. The insert at the bottom (GFY362) slightly enlarges and overexposes the fluorescence signal. (E) Yeast strains from D (using the same numbering scheme) were scored for the percentage of cells displaying fluorescent puncta. Images scored for VPH1-GFP and PrVPH1-VPH1(NT)-Anc.a(CT) stv1Δ were taken at slightly higher (but identical) exposure levels. The quantification for the control strain PrVPH1-Anc.a-GFP stv1Δ (GFY255) is from Figure 9. Experiments were performed in triplicate, and error is expressed as the SD.
DISCUSSION
Like other large, multiprotein complexes, the V-ATPase enzyme has undergone radical architectural changes through evolutionary time while maintaining a conserved enzymatic function. The high degree of homology between specific V-ATPase subunit pairs such as V1 subunits A and B, V0 subunit a isoforms, and V0 proteolipid ring subunits c, c′, and c′′ across all eukaryotes suggests that gene duplication events have played a significant role in the increasing complexity of this multiprotein enzyme (Müller and Grüber, 2003; Cross and Müller, 2004). Within the V0 subdomain, subunit a is unique within budding yeast because it is the only complex-specific subunit isoform of the V-ATPase. In S. cerevisiae, Stv1p and Vph1p are solely responsible for the phenotypic differences seen between the two isoforms of the V-ATPase (Toei et al., 2010). One of the most dramatic differences can be seen in the distinct subcellular localizations of Stv1p-complex (Golgi/endosome) and Vph1p-complex (vacuole).
We sought to describe the evolutionary origins of the Stv1p and Vph1p isoforms in budding yeast using ancestral gene reconstruction. Previous work was unsuccessful at characterizing other V-ATPase subunit a isoforms from various eukaryotes within budding yeast (Aviezer-Hagai et al., 2000; unpublished results). This type of horizontal approach fails to provide the specific genetic background for mutational changes that have accumulated within extant nodes along different evolutionary trajectories (Harms and Thornton, 2010). Ancestral reconstruction incorporates mutational information from modern sequences to infer the most probable ancestral state of a gene (Thornton, 2004; Dean and Thornton, 2007).
Several evolutionary models of gene duplication events followed by subfunctionalization might describe the phenotypes of contemporary subunits Stv1p and Vph1p (Innan and Kondrashov, 2010). For instance, it is possible that the ancestral subunit a was exclusively localized to either the vacuole or the Golgi apparatus, and the derived function of the duplicated subunit was to occupy a second compartment. Our in vivo analyses of Anc.a suggests that the preduplicated subunit a functioned on both the Golgi/endosomal and vacuolar compartments. Surprisingly, modern yeast are able to properly assemble the ancestral subunit as part of a hybrid V-ATPase complex. Furthermore, yeast allow for efficient transport of the V-ATPase and in vivo acidification despite differences in the primary amino acid sequence among Anc.a, Stv1p, and Vph1p. These results highlight the conservation of overall structural identity of subunit a, which makes contacts with V0 and V1 subunits and interacts with various assembly factors (Landolt-Marticorena et al., 2000; Wilkens et al., 2004; Zhang et al., 2008; Qi and Forgac, 2008; Ediger et al., 2009). This is also evident when examining the support values for the Anc.a reconstruction; despite 23.9% of all sites having <80% posterior probability (Supplemental Figure S1), our reconstruction resulted in a high degree of complementation and function in vivo.
Of interest, trafficking of the Anc.a-containing V-ATPase does not use the extant, Stv1p-sorting machinery. Similar to other resident Golgi cargo proteins (such as Vps10p, Kex2p, and Ste13p), the Stv1p-containing V-ATPase complex also requires retrieval from a late endosomal component mediated by the retromer complex. Stv1p sorting does not appear to require retrieval from the early endosome mediated by the GARP complex. The dual localization of Anc.a within modern yeast could result from retrieval from a post-Golgi compartment or some other trafficking mechanism. However, our results suggest that Anc.a transport functions independent of the retromer and likely involves slowed anterograde transport through the biosynthetic pathway. Our data suggest that Anc.a transport is being slowed at the level of the Golgi; Anc.a-containing V-ATPase complexes do not accumulate within the ER, indicating that ER-to-Golgi transport is not affected. Furthermore, Anc.a colocalizes with the late Golgi marker Vps10p, and localization of Anc.a does not require the function of the GARP complex (retrieval from the early endosome) or the retromer complex (retrieval from the late endosome). Therefore it seems likely that diminished interactions of Anc.a with modern yeast vesicular transport machinery cause slowed movement out of the Golgi complex.
Great progress has been made in describing the mechanisms of membrane traffic (vesicle budding, transport, tethering, and fusion) within eukaryotic cells (Lee et al., 2004; Cai et al., 2007; Bröcker et al., 2010; Dancourt and Barlowe, 2010). However, it is still unclear what transport machinery interacts with the V-ATPase to assist in cargo recognition, packaging, and movement to various organelles. Cargo might be directly recognized (by coat proteins), as in the case of the COPII cargo adaptor complex (Cai et al., 2007) or Vps35p within the retromer complex (Nothwehr et al., 2000). Alternatively, the V-ATPase may require the presence of additional sorting receptors, such as the Erv proteins within budding yeast that improve the efficiency of cargo packaging (Dancourt and Barlowe, 2010). Although Anc.a is able to properly assembly and function within the context of the yeast V-ATPase, it is unclear whether modern transport machinery in-trans will maintain proper contacts with the hybrid enzyme complex. Many of the components of vesicular trafficking pathways that function at the level of the Golgi/endosome (including the transport protein particle, conserved oligomeric Golgi, endosomal sorting complex required for transport, and GARP complexes) are highly conserved among eukaryotes, including humans (Field and Dacks, 2009; Barrowman et al., 2010; Bonifacino and Hierro, 2011; Reynders et al., 2011). However, a large amount of evolutionary change (measured by phylogenetic branch lengths) has occurred along the trajectories leading from Anc.a to either Stv1p or Vph1p (Figure 1B). It is likely that the Vph1p isoform evolved to improve its transport through the biosynthetic pathway to exclusively localize to the vacuole membrane, whereas the Stv1p isoform evolved a dependence on retromer-mediated endosomal retrieval for its steady-state Golgi localization. Further investigation will require identification of the transport machinery that directly associates with the V-ATPase. It will be of interest to determine whether inefficient anterograde trafficking out of the Golgi may have provided a selective advantage for the ancestral V-ATPase complex.
The sorting information inherent to the Stv1p and Vph1p subunits was shown to be present within the N-terminal subdomain (Kawasaki-Nishi et al., 2001a). Protein chimeras between the ancestral isoform and modern isoforms have also provided insight into the mechanism of transport for Anc.a. Fusions between Anc.a and Stv1p were not informative due to problems with V-ATPase assembly with the Anc.a-Stv1p hybrid proteins and/or higher levels of expression of the hybrid proteins (unpublished data). However, fusions between Anc.a and Vph1p have shown that either of the ancestral domains is sufficient to produce a dual localization pattern. Consistent with previous observations (Kawasaki-Nishi et al., 2001a), the N-terminal domain of Vph1p causes an increase in the efficiency of transport to the vacuole. In addition, either domain of the ancestral isoform can impair efficient transport from the Golgi to the vacuole. The stronger localization phenotype associated with the cytosolic portion of Anc.a highlights the importance of this region within subunit a, which appears solely responsible for dictating trafficking of the V-ATPase in modern yeast. Our results support a unique mechanism for the trafficking of the ancestral subunit a–containing V-ATPase that is independent of retromer-mediated retrieval and likely involves slowed forward transport out of the Golgi.
Given the localization of Anc.a in yeast, as well as previous insights into the biochemical differences between the contemporary yeast subunits, we propose a model of subfunctionalization for subunit a following the gene duplication event that took place within the fungal lineage. A number of other factors contributed to the evolution of Stv1p and Vph1p that allowed for the shift in V-ATPase localization. A common evolutionary trend following subfunctionalization is the difference in gene expression of the two duplicated genes (Force et al., 1999). Differences in the relative levels of transcript and/or differential regulation can lead to alterations in spatial and temporal control of the two genes following the duplication event. In the case of Stv1p and Vph1p, previous work determined the difference in mRNA transcripts (fivefold) between the two genes (Manolson et al., 1994). However, this is not sufficient to completely explain the difference in steady-state levels between yeast Stv1p and Vph1p (Vph1p is ∼20-fold higher). One possibility includes differences in V0 assembly between the two isoforms. On a loss of Vph1p, there is a reproducible increase in the levels of Stv1p protein (Perzov et al., 2002; unpublished data). In addition, preliminary results have shown that there is a difference in the steady-state levels between Anc.a and Stv1p protein when under the control of the STV1 promoter; Anc.a shows a higher protein level (unpublished data). This suggests a possible reduction in assembly for the Stv1p isoform following the duplication event from Anc.a. Because our results demonstrate that Anc.a functions on the vacuole, it is possible that the expression of Anc.a was similar to that of modern VPH1. Therefore a reduction in the levels of Stv1p had to accompany a shift in the localization of the evolving Stv1p isoform to the Golgi/endosome because the modern Stv1p-sorting machinery is saturable (Manolson et al., 1994).
It is unknown why the two postduplication gene products were maintained in budding yeast, whereas other fungi contain only a single subunit a isoform. However, work on other eukaryotes has provided insight into the importance of V-ATPase specialization using subunit a. This involves both the localization of this critical enzyme to specific subcellular compartments (including the plasma membrane) and a variety of additional functional roles for subunit a, including membrane fusion, pH sensing, development, and V-ATPase regulation (Adams et al., 2006; Marshansky, 2007; Baars et al., 2007; Forgac, 2007). There may have been a selective fitness advantage to evolving a dedicated Golgi/endosomal isoform of the V-ATPase in budding yeast. The importance of iron uptake and metabolism under varying environmental conditions is evident in almost every organism (Philpott and Protchenko, 2008). Yeast may have evolved the Stv1p isoform to specifically acidify the Golgi and endosomal compartments to allow for essential processes, including iron homeostasis, that use an acidified endosomal network.
This work also provides unique insight into how organisms with only a single isoform of subunit a, and therefore a single population of V-ATPase enzyme, are able to successfully acidify the diverse set of intracellular compartments seen in all eukaryotes. Little is known about the mechanisms governing the assembly, transport, and function of the V-ATPase enzyme in organisms that only contain one subunit a gene. These species present an intriguing problem associated with specialization of a single V-ATPase enzyme and subsequent transport to multiple cellular compartments. For example, the slime mold Dictyostelium discoideum only has a single V-ATPase complex, which is present in both the endolysosomal and contractile vacuole systems, and yet the precise mechanism for trafficking is unclear (Liu and Clarke, 1996; Clarke et al., 2002, 2010). Within the fungal clade, our phylogenetic analysis revealed that both model organisms S. pombe and Neurospora crassa only have a single subunit a gene. Biochemical analysis suggests that the V-ATPase functions within the various organelles of the secretory pathway in S. pombe, including the vacuole (Okorokov et al., 2001; Iwaki et al., 2004). In addition, live imaging of N. crassa showed that the V-ATPase localizes to both vacuolar structures and novel organelles within the hyphal tip (Bowman et al., 2009). Our characterization of Anc.a in budding yeast describes one possible evolutionary model that might be applicable to other species. Additional ancestral reconstructions, coupled with further study in fungal species such as S. pombe and N. crassa, will provide insight into the evolutionary origins of Anc.a transport.
The process of ancestral gene reconstruction has been a powerful tool providing insight into the evolution of enzyme–substrate specificity and the directionality of evolutionary change (Dean and Thornton, 2007; Ortlund et al., 2007; Bridgham et al., 2009). Anc.a serves as a useful platform to perform future structural and mutational analyses to address questions regarding other biochemical differences between Stv1p and Vph1p, including assembly, coupling efficiency, and in vivo dissociation. Further experiments will also characterize the evolutionary trajectory of the Stv1p-dependent sorting signal(s). Preliminary analyses have found that residues necessary for Stv1p trafficking are not found within the Anc.a N-terminus (G. Cronan and T.H. Stevens, unpublished results). Anc.a also provides a scenario in which it is the sole V-ATPase isoform, similar to other fungal species, such as N. crassa and S. pombe. Additional experimentation will provide insight into how organisms with a single isoform are able to efficiently traffic the V-ATPase to multiple cellular compartments. Anc.a has provided a model for the evolutionary origins of the two yeast isoforms. Future ancestral reconstructions within yeast and other eukaryotic systems will allow for a more complete understanding of the molecular mechanisms responsible for the diverse set of specialized V-ATPase isoforms seen across the tree of life.
MATERIALS AND METHODS
In silico reconstruction of ancestral protein sequences
Our method for reconstructing ancestral protein sequences generally follows the strategy described by Thornton (2004), with specific details as follows. The methodology involves 1) collecting amino acid sequences from modern species of the query protein, 2) inferring a phylogenetic history describing the relationship between species, and 3) using a maximum likelihood (ML) method to predict the most probable ancestral state of the protein sequence of interest (the experimental strategy is reviewed in Thornton [2004], Dean and Thornton [2007], and Harms and Thornton [2010]). Briefly, an amino acid residue is predicted (with a certain statistical confidence) to be present within an ancestral sequence based on the proportion of occurrence within the sequences of modern species and their specific phylogenetic relationship.
GenBank was queried for all fungal V-ATPase subunit a protein sequences; 68 homologous isoforms were returned (Supplemental Table S1). We also retrieved the nonfungal protein sequences for subunit a in D. discoideum and A. thaliana to use as a phylogenetic outgroup. Sequences were aligned using PRANK, version 0.081202 (Loytynoja and Goldman, 2005, 2008). Phylogenetic inference was performed using the ML criterion to optimize a probabilistic model of amino acid substitution (Felsenstein, 1981). The best-fitting model for our sequence alignment is the Whelan–Goldman matrix with gamma-distributed rate variation (+G) and proportion of invariant sites (+I), according to the Akaike information criterion as implemented in PROTTEST (Whelan and Goldman, 2001; Abascal et al., 2005). We used PhyML, version 3.0, to infer the ML topology, branch lengths, and model parameters (Guindon and Gascuel, 2003). The tree topology was optimized using the best result from nearest-neighbor interchange and subtree pruning and regrafting (using PhyML's implementation). All other model parameters were optimized using the limited-memory Broyden-Fletcher-Goldfarb-Shanno algorithm (Liu and Nocedal, 1989), which we implemented as an in-house C extension to PhyML. Phylogenetic branch support was calculated as the approximate likelihood ratio based on a Shimodaira-Hasegawa–like procedure (Anismova and Gascuel, 2006).
ML ancestral states were reconstructed at each site for all ancestral nodes in the ML phylogeny using a set of Python scripts called Lazarus (Hanson-Smith et al., 2010), which wraps PAML, version 4.1 (Yang, 2007). Lazarus parsimoniously placed ancestral gap characters according to Fitch's algorithm (Fitch, 1971). We characterized the overall support for the reconstructed Anc.a sequence by binning the posterior probability of the ML state at each ancestral site into 5%-sized bins and then counted the proportion of total sites within each bin (Supplemental Figure S2).
Plasmids and yeast strains
Standard molecular biology procedures were used in this study (Sambrook and Russel, 2001). Plasmids that were used can be found in Table 1. To construct pGF35, plasmid pAAC200 (Cooper and Stevens, 1996) was digested with StuI. Next a PCR product of the C-terminal portion of the VPS10 open reading frame (ORF) fused to GFP, the S. pombe HIS5 cassette (Yeast GFP Collection; Invitrogen, Carlsbad, CA), and VPS10 3′ untranslated region was obtained. Using homologous recombination and in vivo ligation, the PCR fragment and gapped vector were cotransformed into wild-type (BY4741) yeast and selected for growth on media lacking histidine to create pGF35. The Anc.a sequence was synthesized by GenScript (Piscataway, NJ) with a yeast codon bias. A double hemagglutinin (HA) epitope tag was included after site 171 in Anc.a. This corresponds to the same position as the double-HA tags found in both Stv1p (site 219) and Vph1p (site 185; Figure 2A; Kawasaki-Nishi et al., 2001a). The Anc.a gene was subcloned to a single-copy, CEN-based yeast vector and tagged with either the ADH terminator sequence (247 base pairs) and NatR drug resistance cassette (Goldstein and McCusker, 1999) or the GFP::ADH::HygR cassette (amplified from pGF123) using in vivo ligation. A second round of in vivo ligation was used to place the Anc.a gene (with or without the GFP tag) under control of either the STV1 promoter (500 base pairs) or VPH1 promoter (380 base pairs) to create pGF243-pGF246. To create pGF341-pGF342, the VPH1 promoter and Anc.a gene (containing codons 1–736) were PCR amplified (from pGF245), propagated in pCR4Blunt-TOPO (Invitrogen), and subcloned to pRS316. Similarly, the C-terminal portion of Anc.a (codons 737–stop), including the ADH::NatR cassette, was cloned into a TOPO vector. The R737Q or R737A mutation was introduced into primers that contained 40 base pairs of sequence homology to the Anc.a-coding region and downstream vector sequence. In vivo ligation was used to recreate a full-length Anc.a gene containing the two mutational changes. For construction of pGF242, the mCherry tag (Shaner et al., 2004) was amplified, cloned into TOPO, subcloned to a CEN vector, and tagged with ADH::NatR using in vivo ligation. The mCherry tag was placed behind the promoter of VPH1 (380 base pairs) and included a start codon. Subsequent subcloning removed the ADH::NatR marker. The PHO8 gene (ALP) was amplified from genomic DNA (SF838-1Dα), propagated in TOPO, subcloned to pRS316, and tagged with ADH::NatR as previously described. Finally, the PHO8::ADH::NatR sequence was amplified and in vivo ligated behind the prVPH1::mCherry sequence to create pGF242. Constructs were verified by diagnostic PCR and DNA sequencing.
TABLE 1:
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pRS415 | CEN, LEU2 | Simons et al. (1987) |
| pRS316 | CEN, URA3 | Sikorski and Hieter (1989) |
| GFP-ALP | pRS426 PrCPY::GFP::ALP | Cowles et al. (1997) |
| pGF35 | pRS315 VPS10::GFP::Sp-HIS5 | This study |
| pGF244 | pRS415 PrSTV1::Anc.a::2xHA::ADH::NatR | This study |
| pGF245 | pRS415 PrVPH1::Anc.a::2xHA::ADH::NatR | This study |
| pGF243 | pRS415 PrSTV1::Anc.a::2xHA::GFP::ADH::HYGR | This study |
| pGF246 | pRS415 PrVPH1::Anc.a::2xHA::GFP::ADH::HYGR | This study |
| pGF342 | pRS316 PrVPH1::Anc.a::2xHA::ADH::NatR R737Q | This study |
| pGF341 | pRS316 PrVPH1::Anc.a::2xHA::ADH::NatR R737A | This study |
| pGF242 | pRS316 PrVPH1::mCherry::ALP::ADH::NatR | This study |
Yeast strains can be found in Table 2. Yeast strains GFY270 and GFY271 were created by first switching the drug resistance cassette of vph1Δ::KanR (LGY120) to either vph1Δ::HygR or vph1Δ::NatR. The HygR or NatR cassettes were amplified by PCR, transformed into LGY120, and selected for hygromycin (Hyg) or nourseothricin (Nat) resistance and loss of kanamycin (Kan) resistance. Next the STV1 locus from KEBY4 (stv1Δ::KanR) was amplified with 500 base pairs of flanking, untranslated sequence and transformed into vph1Δ::HygR or vph1Δ::NatR to create GFY270 or GFY271, respectively. To construct GFY251, the Anc.a gene was integrated into the genome at the STV1 locus; the parent yeast strain was GFY270. The Anc.a gene (including the double-HA tag) was amplified from pGF244, including the full promoter of STV1 and the ADH::NatR cassette. Following treatment with the restriction enzyme DpnI, the PCR product was transformed into vph1Δ::HygR stv1Δ::KanR yeast and selected for Nat resistance and the loss of Kan resistance. The drug resistance cassettes all contain 239 base pairs of the same downstream terminator, allowing for homologous recombination. Yeast strains GFY250, GFY253, GFY255, and GFY256 were all created using a similar method. The vma21Δ::KanR deletion cassette was PCR amplified from pLG139 and transformed into GFY256 to generate GFY300. GFY308 was constructed by first tagging VPH1::2xHA (pSKN12; Kawasaki-Nishi et al., 2001a) with a GFP::ADH::HygR cassette using in vivo ligation to create pGF336. The VPH1 promoter, coding sequence, and GFP::HygR cassette were amplified and transformed into vph1Δ::KanR yeast (LGY120) as previously described to create GFY308.
TABLE 2:
Yeast strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| SF838-1Dα | MATα ura3-52 leu2-3112 his4-519 ade6 pep4-3 gal2 | Rothman and Stevens (1986) |
| KEBY4 | SF838-1Dα stv1Δ::KanR | Kawasaki-Nishi et al. (2001a) |
| LGY120 | SF838-1Dα vph1Δ::KanR | Davis-Kaplan et al. (2006) |
| GFY270 | SF838-1Dα vph1Δ::HygR stv1Δ::KanR | This study |
| GFY271 | SF838-1Dα vph1Δ::NatR stv1Δ::KanR | This study |
| GFY251 | SF838-1Dα vph1Δ::HygR PrSTV1::Anc.a::2xHA::ADH::NatR | This study |
| GFY250 | SF838-1Dα PrVPH1::Anc.a::2xHA::ADH::NatR stv1Δ::KanR | This study |
| GFY253 | SF838-1Dα vph1Δ::NatR PrSTV1::Anc.a::2xHA::GFP::ADH::HygR | This study |
| GFY255 | SF838-1Dα PrVPH1::Anc.a::2xHA::GFP::ADH::HygR stv1Δ::KanR | This study |
| GFY256 | SF838-1Dα PrVPH1::Anc.a::2xHA::GFP::ADH::HygR | This study |
| GFY308 | SF838-1Dα VPH1::2xHA::GFP::ADH::HygR | This study |
| GFY310 | SF838-1Dα STV1::GFP::ADH::NatR | This study |
| GFY325 | SF838-1Dα STV1::2xHA::GFP::ADH::HygR | This study |
| GFY307 | SF838-1Dα PrSTV1::VPH1::GFP::ADH::HygR | This study |
| GFY314 | SF838-1Dα stv1Δ::KanR PrVPH1::Anc.a::2xHA::mDsRed::ADH::NatR | This study |
| GFY315 | SF838-1Dα STV1::GFP::KanR VPH1::2xHA::ADH::NatR | This study |
| GFY316 | SF838-1Dα STV1::GFP::KanR PrVPH1::STV1::2xHA::ADH::NatR | This study |
| GFY317 | SF838-1Dα STV1::GFP::KanR PrVPH1::Anc.a::2xHA::ADH::NatR | This study |
| GFY256 | SF838-1Dα PrVPH1::Anc.a::2xHA::GFP::ADH::HygR | This study |
| GFY300 | SF838-1Dα PrVPH1::Anc.a::2xHA::GFP::ADH::HygR vma21Δ::KanR | This study |
| GFY396 | SF838-1Dα VPH1::2xHA::GFP::ADH::HygR vps26Δ::KanR | This study |
| GFY387 | SF838-1Dα STV1::GFP::ADH::NatR vps26Δ::KanR | This study |
| GFY384 | SF838-1Dα PrVPH1::Anc.a::2xHA::GFP::ADH::HygR stv1Δ::KanR vps26Δ::KanR | This study |
| GFY361 | SF838-1Dα PrVPH1::Anc.a::2xHA(1-393)::VPH1(405-840)::GFP::ADH::HygR stv1Δ::KanR | This study |
| GFY362 | SF838-1Dα PrVPH1::VPH1::2xHA(1-404)::Anc.a(394-808)::GFP::ADH::HygR stv1Δ::KanR | This study |
| vps27Δ | BY4741 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps27Δ::KanR | Genome Deletion Collection |
| vps1Δ | BY4741 vps1Δ::KanR | Genome Deletion Collection |
| aps3Δ | BY4741 aps3Δ::KanR | Genome Deletion Collection |
| vps51Δ | BY4741 vps51Δ::KanR | Genome Deletion Collection |
For GFY310, STV1 (from pJG2) was tagged with GFP::ADH::NatR (amplified from pGF02) using in vivo ligation. STV1::GFP::NatR was then integrated into stv1Δ::KanR yeast (KEBY4). For GFY325, STV1::2xHA from pSKN11 (Kawasaki-Nishi et al., 2001a) was tagged with GFP::ADH::HygR (amplified from pGF67) using in vivo ligation. GFP-tagged Stv1 was then integrated into stv1Δ::KanR yeast (KEBY4). For GFY307, VPH1::GFP::HygR was amplified without any promoter sequence from pGF336 and in vivo ligated downstream of the STV1 promoter to create pGF332. Finally, the VPH1 locus, including the GFP tag and drug cassette, was amplified and integrated at the STV1 locus in LGY120 yeast. For construction of GFY314, the sequence of mDsRed was amplified from pERGmDsRed (Binns et al., 2006), propagated into TOPO, subcloned to pRS316, and tagged with ADH::NatR. A PCR fragment of mDsRed::ADH::NatR was then generated with a forward primer containing homology to the coding region of Anc.a, and the product was transformed into GFY255 to exchange the GFP tag for mDsRed. For GFY315-317, the drug marker in GFY310 was first switched to KanR. Next VPH1 was deleted with the HygR cassette (using genomic DNA from GFY270). Finally, VPH1::2xHA::ADH::NatR (from pGF339), STV1::2xH::ADH::NatR (from pGF337), and Anc.a::2xHA::ADH::NatR (from pGF245) were PCR amplified and integrated at the VPH1 locus to create GFY315, GFY316, and GFY317, respectively. Strains GFY396 and GFY387 were constructed by amplifying the vps26Δ::KanR deletion cassette from the haploid genome deletion collection (Invitrogen) and transforming the PCR product into GFY308 and GFY325, respectively. GFY255 was transformed with a PCR fragment of the NatR cassette to replace the KanR cassette at the STV1 locus. A second transformation was used to delete VPS26 as previously described to create GFY384.
To create strains GFY361 and GFY362 containing chimeras between Anc.a and VPH1, the following method was used. A triple in vivo ligation was used to fuse 1) the VPH1 promoter, 2) the double HA-tagged, N-terminus of either Anc.a (codons 1–393) or VPH1 (codons 1–404), and 3) the C-terminus of VPH1 (codons 405–840) or Anc.a (codons 394–808) tagged with GFP::ADH::HygR on a CEN-based vector. The chimeras were PCR amplified (including promoter and drug resistance cassette) and integrated at the VPH1 locus in GFY271 as previously described. Strains from the Genome Deletion Collection (Invitrogen) were confirmed by performing genomic DNA preparations and a PCR reaction with primers 800 base pairs upstream of the ORF and internal to the KanR cassette. All yeast strains were confirmed by diagnostic PCR, Western blots (when applicable), and growth tests.
Culture conditions
Yeast was cultured in 1% yeast extract, 2% peptone, and 2% dextrose (YEPD), pH 5.0, buffered using 50 mM succinate/phosphate plus 0.01% adenine, or synthetic drop-out media containing dextrose and supplemented with amino acids. Growth assays were performed by spotting 3–5 μl of exponentially growing cells onto agar media. Fivefold serial dilutions were used; the first spot corresponds to an OD600 of ∼0.8–1.0. For growth tests on media containing bathophenanthrolinedisulfonic acid (BPS; Sigma-Aldrich, St. Louis, MO), cells were grown overnight to saturation in YEPD, pH 5.0, plus 25 μM BPS. Cultures were backdiluted to an OD600 of 0.25–0.30 in YEPD, pH 5.0, plus 100 μM BPS for 6 h before being spotted (2–3 μl) onto agar media. Conditions used include YEPD, pH 5.0, YEPD + 100 mM Ca2+, YEPD + 3.5 mM Zn2+, and YEPD + 95 μM BPS. Plates were incubated at 30C for 2–5 d.
Whole-cell extract preparation and immunoblotting
Extracts from yeast cells were harvested as previously described (Ryan et al., 2008). Briefly, cultures were growth to saturation overnight in rich media, diluted to an OD600 of 0.25, and grown for an additional 4–6 h to an optical density of 1.0. Cells from 10 ml of this culture were harvested, resuspended into Thorner buffer (8 M urea, 5% SDS, and 50 mM Tris, pH 6.8), and vortexed with glass beads for 8–10 min. A modified Lowry assay was used to determine the amount of protein within each extract (Markwell et al., 1978). Equal loads of protein were probed by SDS–PAGE, moved to nitrocellulose, and incubated with antibodies. Antibodies included monoclonal anti-HA (Sigma-Aldrich), anti-Vma2p (13D11B2; Molecular Probes, Invitrogen), anti-Dpm1p (5C5; Invitrogen), polyclonal, full-length anti-GFP (Clontech Laboratories, Mountain View, CA), and secondary horseradish peroxidase–conjugated anti–mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Western blots were visualized using enhanced chemiluminescence detection.
Fluorescence microscopy
Cells were stained with quinacrine as previously described (Flannery et al., 2004). Briefly, cells were grown overnight in YEPD, pH 5.0, plus adenine and then backdiluted to an OD600 of 0.25 in YEPD (unbuffered) for 4–6 h. A 1-ml amount of exponentially growing yeast was harvested, incubated on ice for 5 min, and resuspended in 200 μM quinacrine, 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.6, and 50 μg/ml concanavalin A–tetramethylrhodamine (Invitrogen) in YEPD for 10 min at 30°C. Cells were washed three or four times in 100 mM HEPES plus 2% glucose (on ice) and then visualized by fluorescence microscopy within 30 min of quinacrine treatment.
For visualization of fluorescently tagged proteins (GFP, mCherry, mDsRed), strains were grown overnight in the appropriate media (minimal media if containing a vector) and then backdiluted to an OD600 of 0.25 in rich media at pH 5.0. Exponentially growing cells were harvested by centrifugation, washed once with water, and visualized by fluorescence and differential interference contrast (DIC) microscopy.
For yeast treated with cycloheximide, cultures were grown overnight in rich media, backdiluted to an OD600 of 0.25, and grown at 30°C for 4 h to a density of ∼0.7–0.8. Ten-milliliter samples were treated with 20 μg/ml cycloheximide (Sigma-Aldrich) or 0.02% dimethyl sulfoxide (DMSO) (mock treatments) and incubated at 30°C for 0–2 h. At specific time points, 1 ml of cells was centrifuged, washed with water, and visualized on the microscope. Cycloheximide-treated cells were quantified by scoring the subcellular localization for 150–400 cells for control strains (Vph1p-GFP, Vph1p-GFP under control of the STV1 promoter, Stv1p-GFP, GFP-ALP, or Vps10p-GFP) and 400–600 cells for experimental strains (Anc.a-GFP under control of either the VPH1 or STV1 promoter). A similar method was used for quantification (150–500 cells) of strains expressing GFP-tagged Vph1p, Stv1p, Anc.a (Figure 9), or Vph1p-Anc.a chimeras (Figure 10). All experiments were performed in triplicate, and error is expressed as the SD.
All images were taken using a 100× objective on an Axioplan 2 fluorescence microscope (Carl Zeiss, Thornwood, NY) and analyzed using Axiovision software (Carl Zeiss) and Photoshop CS, version 8.0 (Adobe, San Jose, CA).
Supplementary Material
Acknowledgments
We thank Laurie Graham and Emily Coonrod for reading the manuscript and Joe Thornton (University of Oregon; Howard Hughes Medical Institute) for discussions. This work was supported by National Institutes of Health Grant GM38006 (to T.H.S.) and Training Grant S T32 GM007257 (to G.C.F.).
Abbreviations used:
- ALP
yeast alkaline phosphatase encoded by the PHO8, gene
- Anc.a
reconstructed, ancestral subunit a of vacuolar H+-ATPase within the fungal clade
- BPS
bathophenanthrolinedisulfonic acid
- CPY
vacuolar carboxypeptidase Y encoded by the PRC1 gene
- GFP
green fluorescent protein
- HA
hemagglutinin
- mCherry
a mutated form of monomeric red fluorescent protein
- mDsRed
monomeric form of a red fluorescent protein from the genus Discosoma
- ML
maximum likelihood
- PP
posterior probability
- V-ATPase
vacuolar-type, proton-translocating ATPase
- YEPD
yeast extract peptone dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0244) on July 7, 2011.
REFERENCES
- Abascal F, Zardoya R, Posada D. Prottest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adams DS, Robinson KR, Fukumoto T, Yuan S, Albertson RC, Yelick P, Kuo L, McSweeney M, Levin M. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development. 2006;133:1657–1671. doi: 10.1242/dev.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anismova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Aviezer-Hagai K, Nelson H, Nelson N. Cloning and expression of cDNAs encoding plant V-ATPase subunits in the corresponding yeast null mutants. Biochim Biophys Acta. 2000;1459:489–498. doi: 10.1016/s0005-2728(00)00188-2. [DOI] [PubMed] [Google Scholar]
- Baars TL, Petri S, Peters C, Mayer A. Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol Biol Cell. 2007;18:3873–3882. doi: 10.1091/mbc.E07-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol. 2010;11:759–763. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 2011;21:159–167. doi: 10.1016/j.tcb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bowman BJ, Draskovic M, Freitag M, Bowman EJ. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell. 2009;8:1845–1855. doi: 10.1128/EC.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C, Engelbrecht-Vandré S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol. 2010;20:R943–R952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Chavez C, Bowman EJ, Reidling JC, Haw KH, Bowman BJ. Analysis of strains with mutations in six genes encoding subunits of the V-ATPase: eukaryotes differ in the composition of the V0 sector of the enzyme. J Biol Chem. 2006;281:27052–27062. doi: 10.1074/jbc.M603883200. [DOI] [PubMed] [Google Scholar]
- Clarke M, Köhler J, Arana Q, Liu T, Heuser J, Gerisch G. Dynamics of the vacuolar H+-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J Cell Sci. 2002;115:2893–2905. doi: 10.1242/jcs.115.14.2893. [DOI] [PubMed] [Google Scholar]
- Clarke M, Maddera L, Engel U, Gerisch G. Retrieval of the vacuolar H+-ATPase from phagosomes revealed by live cell imaging. PLoS One. 2010;5:e8585. doi: 10.1371/journal.pone.0008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cross RL, Müller V. The evolution of the A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio. FEBS Lett. 2004;576:1–4. doi: 10.1016/j.febslet.2004.08.065. [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- Davis-Kaplan SR, Compton MA, Flannery AR, Ward DM, Kaplan J,, Stevens TH, Graham LA. PKR1 encodes an assembly factor for the yeast V-type ATPase. J Biol Chem. 2006;281:32025–32035. doi: 10.1074/jbc.M606451200. [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediger B, Melman SD, Pappas DL, Finch M, Applen J, Parra KJ. The tether connecting cytosolic (N terminus) and membrane (C terminus) domains of yeast V-ATPase subunit a (Vph1) is required for assembly of V0 subunit d. J Biol Chem. 2009;284:19522–19532. doi: 10.1074/jbc.M109.013375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Field MC, Dacks JB. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicular coat proteins, and nuclear pore complexes. Curr Opin Cell Biol. 2009;21:4–13. doi: 10.1016/j.ceb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. [Google Scholar]
- Flannery AR, Graham LA, Stevens TH. Topological characterization of the c, c′, and c′′ subunits of the vacuolar ATPase from the yeast Saccharomyces cerevisiae. J Biol Chem. 2004;279:39856–39862. doi: 10.1074/jbc.M406767200. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Frattini A, et al. Defects inTCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Graham LA, Flannery AR, Stevens TH. Structure and assembly of the yeast V-ATPase. J Bioenerg Biomembr. 2003;35:301–312. doi: 10.1023/a:1025772730586. [DOI] [PubMed] [Google Scholar]
- Graham LA, Hill KJ, Stevens TH. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, van der Goot FG. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hanson-Smith V, Kolaczkowski B, Thornton JW. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol Biol Evol. 2010;27:1988–1999. doi: 10.1093/molbev/msq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr Opin Struct Biol. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Cooper AA. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KJ, Stevens TH. Vma22p is a novel endoplasmic reticulum-associated protein required for assembly of the yeast vacuolar H(+)-ATPase complex. J Biol Chem. 1995;270:22329–22336. doi: 10.1074/jbc.270.38.22329. [DOI] [PubMed] [Google Scholar]
- Hirata R, Umemoto N, Ho MN, Ohya Y, Stevens TH, Anraku Y. VMA12 is essential for assembly of the vacuolar H+-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J Biol Chem. 1993;268:961–967. [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Iwaki T, Goa T, Tanaka N, Takegawa K. Characterization of Schizosaccharomyces pombe mutants defective in vacuolar acidification and protein sorting. Mol Genet Genomics. 2004;271:197–207. doi: 10.1007/s00438-003-0971-7. [DOI] [PubMed] [Google Scholar]
- Jo WJ, Kim JH, Oh E, Jaramillo D, Holman P, Loguinov AV, Arkin AP, Nislow C, Giaever G, Vulpe CD. Novel insights into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae. BMC Genomics. 2009;10:130. doi: 10.1186/1471-2164-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karet FE, et al. Mutations in the gene encoding B1 subunit of the H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet. 1999;21:84–90. doi: 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of protein transport and ATP hydrolysis. J Biol Chem. 2001a;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S, Nishi T, Forgac M. Yeast V-ATPase complexes containing different isoforms of the 100 kDa a-subunit differ in coupling efficiency and in vivo dissociation. J Biol Chem. 2001b;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S, Nishi T, Forgac M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc Natl Acad Sci USA. 2001c;98:12397–12402. doi: 10.1073/pnas.221291798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt-Marticorena C, Williams KM, Correa J, Chen W, Manolson MF. Evidence that the NH2 terminus of Vph1p, and integral subunit of the V0 sector of the yeast V-ATPase, interacts directly with the Vma1p and Vma13p subunits of the V1 sector. J Biol Chem. 2000;275:15449–15457. doi: 10.1074/jbc.M000207200. [DOI] [PubMed] [Google Scholar]
- Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Math Program. 1989;45:503–528. [Google Scholar]
- Liu T, Clarke M. The vacuolar proton pump of Dictyostelium discoideum: molecular cloning and analysis of the 100 kDa subunit. J Cell Sci. 1996;109:1041–1051. doi: 10.1242/jcs.109.5.1041. [DOI] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;5885:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkus P, Graham LA, Stevens TH, Schekman R. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol Biol Cell. 2004;15:5075–5091. doi: 10.1091/mbc.E04-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. STV1 gene encodes functional homologue of the 95-kDa yeast vacuolar H+-ATPase subunit Vph1p. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:578–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Markwell MA, Haas SM, Beiber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Marshansky V. The V-ATPase a2-subunit as a putative endosomal pH-sensor. Biochem Soc Trans. 2007;35:1092–1099. doi: 10.1042/BST0351092. [DOI] [PubMed] [Google Scholar]
- Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zaguilán R, Raghumand N, Lynch RM, Bellamy W, Martinez GM, Rojas B, Smith D, Dalton WS, Gillies RJ. pH and drug resistance. I. Functional expression of plasmalemmal V-type H+-ATPase in drug-resistant human breast carcinoma cell lines. Biochem Pharmacol. 1999;57:1037–1046. doi: 10.1016/s0006-2952(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Miseta A, Kellermayer R, Aiello DP, Fu L, Bedwell DM. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 1999;451:132–136. doi: 10.1016/s0014-5793(99)00519-0. [DOI] [PubMed] [Google Scholar]
- Müller V, Grüber G. ATP synthases: structure, function and evolution of unique energy converters. Cell Mol Life Sci. 2003;60:474–494. doi: 10.1007/s000180300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuole membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Ha SA, Bruinsma P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J Cell Biol. 2000;151:297–310. doi: 10.1083/jcb.151.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AE. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Oka T, Murata Y, Namba M, Yoshimizu T, Toyomura T, Yamamoto A, Sun-Wada GH, Hamasaki N, Wada Y, Futai M. a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J Biol Chem. 2001a;276:40050–40054. doi: 10.1074/jbc.M106488200. [DOI] [PubMed] [Google Scholar]
- Oka T, Toyomura T, Honjo K, Wada Y, Futai M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem. 2001b;276:33079–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- Okorokov LA, Silva FE, Okorokova Façanha AL. Ca2+ and H+ homeostasis in fission yeast: a role of Ca2+/H+ exchange and distinct V-H+-ATPases of the secretory pathway organelles. FEBS Lett. 2001;505:321–324. doi: 10.1016/s0014-5793(01)02852-6. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perzov N, Padler-Karavani V, Nelson H, Nelson N. Characterization of yeast V-ATPase mutants lacking Vph1p or Stv1p and the effect on endocytosis. J Exp Biol. 2002;205:1209–1219. doi: 10.1242/jeb.205.9.1209. [DOI] [PubMed] [Google Scholar]
- Philpott CC, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Forgac M. Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J Biol Chem. 2007;282:24742–24751. doi: 10.1074/jbc.M700663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Forgac M. Function and subunit interactions of the N-terminal domain of subunit a (Vph1p) of the yeast V-ATPase. J Biol Chem. 2008;283:19274–19282. doi: 10.1074/jbc.M802442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde transport from the early endosome and Cvt vesicle completion. J Biol Chem. 2003;278:5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders E, Foulquier F, Annaert W, Matthijs G. How Golgi glycosylation meets and needs trafficking: the case of the COG complex. Glycobiology. 2011;21:853–863. doi: 10.1093/glycob/cwq179. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of the Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Ryan M, Graham LA, Stevens TH. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol Biol Cell. 2008;19:5131–5142. doi: 10.1091/mbc.E08-06-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Seaman MN. Recycle your receptors with retromer. Trends Cell Biol. 2005;15:68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer A. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nature. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- Toei M, Saum R, Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomura T, Murata Y, Yamamoto A, Oka T, Sun-Wada GH, Wada Y, Futai M. From lysosomes to the plasma membrane: localization of vacuolar type H+-ATPase with the a3 isoform during osteoclast differentiation. J Biol Chem. 2003;278:22023–22030. doi: 10.1074/jbc.M302436200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Toei M, Forgac M. Analysis of the membrane topology of transmembrane segments in the C-terminal hydrophobic domain of the yeast vacuolar ATPase subunit a (Vph1p) by chemical modification. J Biol Chem. 2008;283:20696–20702. doi: 10.1074/jbc.M803258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T, Kissmehl R, Cohen J, Plattner H. Seventeen a-subunit isoforms of paramecium V-ATPase provide high specialization in localization and function. Mol Biol Cell. 2006;17:917–930. doi: 10.1091/mbc.E05-06-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wilkens S, Inoue T, Forgac M. Three-dimensional structure of the vacuolar ATPaseLocalization of subunit H by difference imaging and chemical cross-linking. J Biol Chem. 2004;279:41942–41949. doi: 10.1074/jbc.M407821200. [DOI] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Vps1p, a membrane of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 1993;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish-Trier E, Heck AJ, Kane PM, Wilkens S. Structure of the yeast vacuolar ATPase. J Biol Chem. 2008;283:35983–35995. doi: 10.1074/jbc.M805345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.