Mitophagy, which selectively degrades mitochondria via autophagy, has a significant role in mitochondrial quality control. When autophagy selects mitochondria as a cargo, Atg32 is bound by Atg11. It is shown that the phosphorylation of Atg32, especially phosphorylation of Ser-114 on Atg32, mediates the Atg11–Atg32 interaction and mitophagy.
Abstract
Mitophagy, which selectively degrades mitochondria via autophagy, has a significant role in mitochondrial quality control. When mitophagy is induced in yeast, mitochondrial residential protein Atg32 binds Atg11, an adaptor protein for selective types of autophagy, and it is recruited into the vacuole along with mitochondria. The Atg11–Atg32 interaction is believed to be the initial molecular step in which the autophagic machinery recognizes mitochondria as a cargo, although how this interaction is mediated is poorly understood. Therefore, we studied the Atg11–Atg32 interaction in detail. We found that the C-terminus region of Atg11, which included the fourth coiled-coil domain, interacted with the N-terminus region of Atg32 (residues 100–120). When mitophagy was induced, Ser-114 and Ser-119 on Atg32 were phosphorylated, and then the phosphorylation of Atg32, especially phosphorylation of Ser-114 on Atg32, mediated the Atg11–Atg32 interaction and mitophagy. These findings suggest that cells can regulate the amount of mitochondria, or select specific mitochondria (damaged or aged) that are degraded by mitophagy, by controlling the activity and/or localization of the kinase that phosphorylates Atg32. We also found that Hog1 and Pbs2, which are involved in the osmoregulatory signal transduction cascade, are related to Atg32 phosphorylation and mitophagy.
INTRODUCTION
The mitochondrion is an organelle that produces energy through the use of an electron transport chain and oxidative phosphorylation. On the other hand, mitochondria are the major source of cellular reactive oxygen species (ROS), which cause oxidative damage to mitochondrial DNA, protein, and lipids, and the accumulation of this damage is related to many disorders, such as neurodegenerative diseases, diabetes mellitus, cancer, and aging (Wallace, 2005). Accordingly, appropriate quality control of mitochondria is important to maintain proper cellular homeostasis. Mitochondria have several maintenance systems, including a protein degradation system (Rep and Grivell, 1996; Friguet et al., 2008; Voos, 2009), DNA repair enzymes (Larsson and Clayton, 1995; Bogenhagen, 1999), and phospholipid hydroperoxide glutathione peroxidase (Arai et al., 1999). In addition to these mitochondrial maintenance systems, recent studies from yeast to mammals suggest that mitochondria autophagy (mitophagy) is one of the primary systems that maintains mitochondrial quality by eliminating dysfunctional mitochondria (Elmore et al., 2001; Rodriguez-Enriquez et al., 2004; Priault et al., 2005; Nowikovsky et al., 2007; Narendra et al., 2008; Narendra and Youle, 2011; Twig et al., 2008), although the molecular process of mitophagy is still poorly understood.
Macroautophagy (bulk autophagy) is a nonspecific degradation process of cytoplasmic components that is highly conserved among eukaryotes. After certain cellular stresses, such as nutrient starvation, cytosolic double-membrane vesicles emerge and sequester cytoplasmic proteins and organelles as cargoes, and then the vesicles deliver those cargoes into the lysosome/vacuole (Nakatogawa et al., 2009; Yang and Klionsky, 2010). In contrast to macroautophagy, selective autophagy targets a specific cellular component as a cargo (Johansen and Lamark, 2011). Selective autophagy in yeast includes the cytoplasm-to-vacuole targeting (Cvt) pathway, pexophagy, and mitophagy. The Cvt pathway is an autophagy-like process that delivers the Cvt complex (aminopeptidase I [Ape1] and α-mannosidase complex) and Ape4 from the cytoplasm to vacuoles without delivering any additional known cargo to the vacuoles (Klionsky and Emr, 2000; Yorimitsu and Klionsky, 2005; Yuga et al., 2011), whereas pexophagy and mitophagy are the selective degradation of peroxisomes and mitochondria, respectively, via autophagy (Lemasters, 2005; Oku and Sakai, 2010). Studies on Saccharomyces cerevisiae and other fungi have identified 35 autophagy-related (ATG) genes. Most of the ATG genes are required for both macroautophagy and selective autophagy, whereas some ATG genes, such as ATG11, ATG19, Pichia pastoris ATG30 (PpATG30), and ATG32, play a role only in selective autophagy. Atg19 is a receptor protein for the Cvt pathway, which binds the Cvt complex and then interacts with Atg11, an adaptor protein for selective autophagy. Atg11 recruits the Cvt complex to the preautophagosomal structure/phagophore assembly site (PAS), where sequestering cytosolic vesicles are generated (Shintani et al., 2002). PpAtg30 is also a receptor protein for pexophagy, which localizes to peroxisomes, and is bound by PpAtg11, allowing recruitment of the peroxisomes to the PAS (Farre et al., 2008). Similarly, during mitophagy, Atg11 interacts with the mitochondrial residential receptor Atg32 and recruits mitochondria to the PAS (Kanki et al., 2009c; Okamoto et al., 2009b). Thus, selective autophagy strictly recognizes and degrades the cargo through the cargo-specific receptor and Atg11 interaction. However, it is poorly understood how the receptor–Atg11 interaction, especially the Atg32–Atg11 interaction, is regulated.
Recent studies in mammals revealed that mitophagy is inevitably involved in cellular physiology and disease. For example, during erythroid cell maturation, mitochondria are eliminated by mitochondrial outer membrane protein Nix-related mitophagy (Schweers et al., 2007; Sandoval et al., 2008). Similarly, during adipose tissue differentiation, mitochondria are eliminated by autophagy (Goldman et al., 2010). Loss-of-function mutations of the PARK2 and PARK6 genes, which encode Parkin and PINK1, respectively, cause early-onset Parkinson disease. PINK1 can stably localize on the outer membrane of the impaired mitochondria and recruit Parkin from the cytosol to the mitochondria. Parkin accumulated on mitochondria ubiquitinates mitochondrial proteins and induces mitophagy to degrade impaired mitochondria (Narendra et al., 2008, 2010; Narendra and Youle, 2011). Despite the important role of mitophagy in cellular physiology and disease, the molecular mechanism and regulation of mitophagy are unknown.
To determine the molecular mechanism and the regulation of mitophagy, we studied the Atg11–Atg32 interaction in detail. We found that the C-terminus region of Atg11, which includes the fourth coiled-coil domain of the protein, interacts with the N-terminus region, especially residues of 100–120, of Atg32. We further found that when mitophagy is induced, Ser-114 and Ser-119 on Atg32 are phosphorylated by Ser/Thr protein kinase. The phosphorylation of Atg32, especially phosphorylation of Ser-114 on Atg32, is required for Atg11–Atg32 interaction and mitophagy. These findings suggest that mitophagy induction is strictly regulated by controlling the phosphorylation level of Ser-114 on Atg32.
RESULTS
The C-terminus region of Atg11 interacts with the N-terminus region (51–150) of Atg32
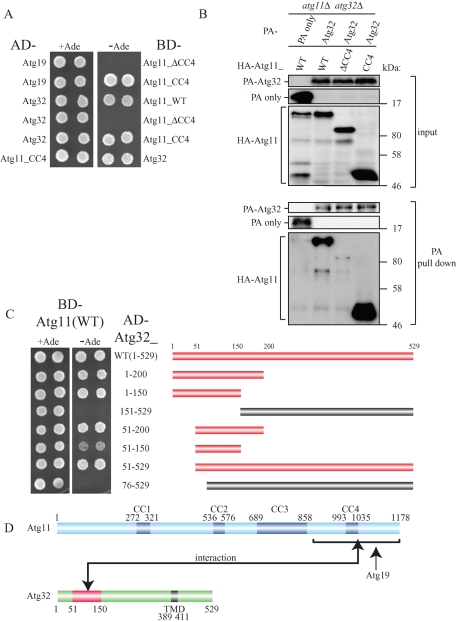
When autophagy selects mitochondria as a cargo, Atg11 interacts with a mitochondrial outer membrane localizing receptor Atg32 and recruits mitochondria to the PAS (Kanki et al., 2009c; Okamoto et al., 2009b). Because it has been reported that the C-terminus region of Atg11, including the fourth coiled-coil domain (CC4), interacts with the Cvt pathway receptor Atg19 (Yorimitsu and Klionsky, 2005), we speculated that the same region of Atg11 interacts with Atg32. To examine this possibility, we prepared a yeast two-hybrid construct to express Atg11 mutants fused with the Gal4 DNA-binding domain (BD-Atg11), activation domain–fused Atg32 (AD-Atg32), and activation domain–fused Atg19 (AD-Atg19). As previously reported, the cells expressing AD-Atg19 and BD-Atg11_CC4 (C-terminus region of Atg11 including CC4) grew on the selective plates, while the cells expressing AD-Atg19 and BD-Atg11_ΔCC4 (N-terminus region of Atg11 lacking CC4) did not grow on selective plates (Figure 1A; Yorimitsu and Klionsky, 2005). Similarly, the cells expressing AD-Atg32 and BD-Atg11_CC4, but not BD-Atg11_ΔCC4, grew on selective plates, suggesting that only the C-terminus region of Atg11 interacts with Atg32 (Figure 1A). To further examine the interaction between truncated Atg11 and Atg32, we expressed the constructs with hemagglutinin (HA)-tagged Atg11_CC4 or HA-tagged Atg11_ΔCC4 with protein A (PA)-tagged Atg32 and performed a protein A affinity pull-down assay with immunoglobulin G (IgG)–Sepharose. As we previously reported, PA-Atg32 coprecipitated a substantial amount of HA-Atg11_wild type (WT; Kanki et al., 2009c; Figure 1B). PA-Atg32 also coprecipitated HA-Atg11_CC4 but not HA-Atg11_ΔCC4 (Figure 1B), suggesting that only the C-terminus region of Atg11 interacts with Atg32.
FIGURE 1:
The N-terminus region (51–150) of Atg32 interacts with the C-terminus region of Atg11. (A) Yeast two-hybrid analysis between Atg19 and Atg11 mutants, and between Atg32 and Atg11 mutants. The PJ69-4A strain was transformed with pGAD and pGBD plasmid, which can express the indicated proteins. Cells were grown on +Ade or –Ade plates at 30°C for 4 d. (B) The C-terminus region of Atg11 associated with Atg32 during nitrogen starvation. The atg11Δ atg32Δ strains expressing PA-tagged Atg32 or PA only and the indicated HA-tagged Atg11 mutants under the control of the CUP1 promoter were grown in SMD medium until the mid–log phase and then starved in SD-N for 1 h. PA-Atg32 was precipitated using IgG–Sepharose from cell lysates. Top two, an immunoblot of total-cell lysates. Bottom two, the IgG precipitates, which were probed with anti-HA and anti-PA antibodies. (C) Yeast two-hybrid analysis between Atg11 and the indicated Atg32 mutants (left). Each bar indicates expressed Atg32 mutants (right). Red bars indicate yeast two-hybrid positive and the gray bars indicate yeast two-hybrid negative. (D) Schematic drawing of Atg11 and Atg32. Atg11 is predicted to have a fourth coiled-coil domain (CC1–CC4). Atg32 has a TMD. The domain of Atg32 required for Atg11 interaction as determined by yeast two-hybrid study is highlighted in red.
To identify the residues of Atg32 that interact with Atg11, we prepared yeast two-hybrid constructs to express the N-terminus– and/or C-terminus–truncated form of Atg32. As shown in Figure 1C, the cells expressing BD-Atg11 and AD-Atg32 mutants including amino acid residues 51–150 (Atg32_WT, 1-200, 1-150, 51-200, 51-150, and 51-529) grew on the selective plates. On the other hand, the cells expressing BD-Atg11 and the AD-Atg32 mutant lacking amino acid residues 51–150 (Atg32_151-529) or the AD-Atg32 mutant lacking 75 amino acids in the N-terminus (Atg32_76-529) did not grow on the selective plates (Figure 1C). These findings suggest that the N-terminus region of Atg32, especially amino acid residues 51–150, is important for the interaction with Atg11 (Figure 1D).
Atg32 is phosphorylated when mitophagy is induced
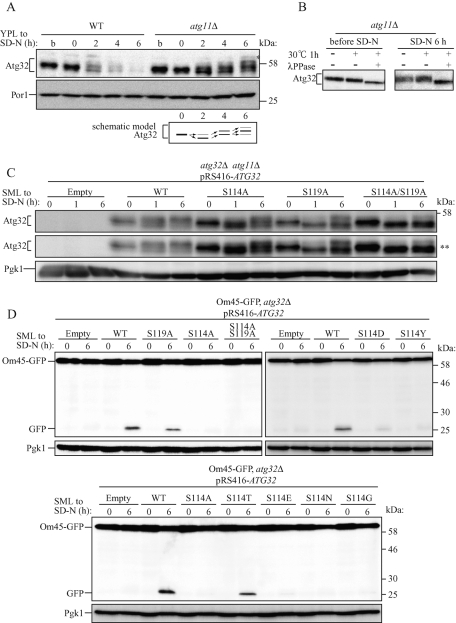
Mitophagy is induced when cells are cultured in lactate medium (YPL) and then shifted to nitrogen starvation medium supplemented with glucose (SD-N), or when cells are cultured in YPL for more than 2 d (allowing growth to the postlogarithmic phase; Kanki and Klionsky, 2008). We found that the molecular mass of Atg32 was changed when mitophagy was induced by nitrogen starvation (Figure 2A) or by allowing cellular growth to the postlogarithmic phase (unpublished data). In WT cells, the molecular mass of Atg32 was decreased by 2 h of nitrogen starvation; Atg32 was degraded following nitrogen starvation (Figure 2A, WT). In atg11Δ or atg1Δ cells, Atg32 was not degraded by nitrogen starvation (Figure 2A, atg11Δ; Supplemental Figure S1A), suggesting that Atg32 is degraded by mitophagy during nitrogen starvation. In atg11Δ cells, the molecular mass of Atg32 was decreased once (at 2 h) and then increased (at 4 and 6 h) during nitrogen starvation (Figure 2A, atg11Δ). To determine whether these molecular mass shifts were due to Atg32 phosphorylation, cellular protein extracts were treated with λ protein phosphatase (λ PPase) and the Atg32 molecular mass was observed. The molecular mass of Atg32 was decreased by λ PPase treatment before and after 6 h of nitrogen starvation (Figure 2B). Therefore we concluded that Atg32 is phosphorylated even in growing conditions in YPL medium, and after nitrogen starvation, Atg32 is dephosphorylated once and then is phosphorylated on two or three residues, depending on the nitrogen starvation period (Figure 2A, schematic model).
FIGURE 2:
Phosphorylation of Atg32, especially phosphorylation of Ser-114 on Atg32, is essential for mitophagy. (A) WT or atg11Δ strains were cultured in YPL medium until the mid–log growth phase (indicated as b) and then shifted to SD-N medium for 0, 2, and 6 h. The amount and the modification of endogenous Atg32 were observed by immunoblotting with anti-Atg32 and anti-Por1 (loading control) antibodies. The variation in molecular weight of Atg32 in the atg11Δ strain is shown as a schematic model. The asterisk indicates a nonspecific band. (B) The atg11Δ strain was cultured in YPL medium until the mid–log growth phase (before SD-N) and then shifted to SD-N for 6 h. Cell lysates were treated with or without λ PPase at 30°C for 1 h. The molecular weight change was observed by immunoblotting with anti-Atg32 antibody. (C) The atg32Δ atg11Δ double-knockout strain transformed with vectors expressing the indicated ATG32 mutants were cultured in YPL medium until the mid–log growth phase and then shifted to SD-N medium for 0, 1, and 6 h. The modification of Atg32 mutants was monitored by immunoblotting with anti-Atg32 and anti-Pgk1 (loading control) antibodies. The asterisks indicate a long exposure. (D) Strains of atg32Δ expressing Om45-GFP were transformed with the indicated Atg32 mutant-expressing vectors. Cells were cultured in SML medium until the mid–log growth phase and then shifted to SD-N for 6 h. GFP processing was monitored by immunoblotting with anti-GFP and anti-Pgk1 (loading control) antibodies.
Phosphorylation of Ser-114 and Ser-119 on Atg32
To determine the phosphorylated residues in Atg32, we created Atg32 single–amino acid mutant expression vectors in which a Ser or Thr residue was altered to Ala (all created mutants are summarized in Supplemental Figure S1B), and those mutants were expressed in ATG32 and ATG11 double-deletion cells (atg32Δ atg11Δ cells), and Atg32 phosphorylation was observed during nitrogen starvation. We found that Atg32 with a mutation of Ser-114 to Ala or Ser-119 to Ala (Atg32S114A and Atg32S119A, respectively) was not efficiently phosphorylated during nitrogen starvation (Figure 2C). Almost complete phosphorylation of Atg32S119A and partial Atg32S114A phosphorylation were inhibited at 1 h of nitrogen starvation compared with WT Atg32 (Atg32WT; Figure 2C, SD-N, 1 h). At 6 h of nitrogen starvation, Atg32S119A phosphorylation was dramatically decreased compared with that of WT, although some phosphorylation remained (Figure 2C and Supplemental Figure S1C). Atg32S114A was phosphorylated to nearly identical levels as those of Atg32WT (Figure 2C and Supplemental Figure S1C). If both Ser-114 and Ser-119 were altered to Ala (Atg32S114A/S119A), phosphorylation was almost completely inhibited, suggesting that phosphorylation observed in Atg32S119A at 6 h of nitrogen starvation is due to the phosphorylation of Ser-114 (Figure 2C and Supplemental Figure S1C). From these findings, we concluded that both Ser-114 and Ser-119 are phosphorylated by induction of mitophagy, although Ser-119 is more efficiently phosphorylated than Ser-114.
Phosphorylation of Atg32, especially phosphorylation of Ser-114 on Atg32, is critically important for mitophagy and for the Atg11–Atg32 interaction
If phosphorylation of Atg32 is necessary for mitophagy, we hypothesized that phosphorylation-deficient Atg32 mutants (Atg32S114A, Atg32S119A, and Atg32S114A/S119A) are not functional for mitophagy. To observe mitophagy, we relied on the Om45–green fluorescent protein (GFP) processing assay, which can monitor mitophagy levels semiquantitatively (Kanki et al., 2009a). Om45 is a mitochondrial outer membrane protein. We found that Om45 tagged with GFP (Om45-GFP) was localized on the mitochondrial outer membrane and accumulated in vacuoles when mitophagy was induced. Om45-GFP that was accumulated in the vacuoles was degraded; however, the GFP was relatively stable within vacuoles and was often released as an intact protein. The level of mitophagy can be semiquantitatively monitored by measuring the amount of GFP processed from Om45-GFP using immunoblotting (Kanki and Klionsky, 2008; Kanki et al., 2009a). We expressed Atg32WT, Atg32S114A, Atg32S119A, and Atg32S114A/S119A in atg32Δ cells expressing Om45-GFP and induced mitophagy by nitrogen starvation or by allowing cellular growth to the postlogarithmic phase. Then we observed mitophagy using the Om45-GFP processing assay. When mitophagy was induced by nitrogen starvation, cells expressing Atg32WT showed substantial levels of GFP processing, whereas cells expressing Atg32S119A showed ∼60% of GFP processing compared with WT. Cells expressing Atg32S114A or Atg32S114A/S119A did not show GFP processing (Figure 2D), suggesting that the S119A mutation partially affects the function of Atg32 for mitophagy and that the S114A mutation completely abolishes the function of Atg32. A similar result was observed if mitophagy was induced by allowing cellular growth up to the postlogarithmic phase (Supplemental Figure S2A). To exclude the possibility that the dysfunction of Atg32S114A is due to mislocalization of this protein, we expressed GFP-Atg32WT and GFP-Atg32S114A in the atg32Δ cells and observed Atg32 localization. As shown in Supplemental Figure S3, both GFP-Atg32WT and GFP-Atg32S114A were colocalized with MitoTracker Red, a probe for mitochondria, suggesting that S114A mutation does not affect Atg32 localization.
To distinguish whether dysfunction of Atg32S114A is due to the critical structural change of Atg32 or inability of Ser-114 phosphorylation, we constructed other Atg32 mutant expression vectors such as mutations of Ser-114 to Asn, Gly, Thr, Tyr, Asp, or Glu (Atg32S114N, Atg32S114G, Atg32S114T, Atg32S114Y, Atg32S114D, or Atg32S114E, respectively) and expressed these mutants in atg32Δ cells expressing Om45-GFP. Then we observed mitophagy. Because alternation of Ser to Asp or Glu can sometimes mimic the phosphorylated form of the protein, we expected that Atg32S114D or Atg32S114E might play a role in mitophagy, but both of these Atg32–mutant-expressing cells, as well as Atg32S114N-, Atg32S114G-, and Atg32S114Y-expressing cells, did not show mitophagy (Figure 2D). Of interest, Atg32S114T, which has the potential to be phosphorylated by Ser/Thr protein kinase, could induce mitophagy (Figure 2D), suggesting that phosphorylation of amino acid residue 114 of Atg32 is essential for mitophagy.
Ser and Thr residues are clustered between Ser-114 and Ser-119 (Supplemental Figure S1B, green square). We further determined whether these Ser or Thr residues have an important role in mitophagy. We expressed Atg32S115A, Atg32S116A, Atg32D117A, and Atg32T118A in atg32Δ cells expressing Om45-GFP and observed mitophagy. All of those Atg32-mutant–expressing cells, however, showed a similar level of mitophagy with that of Atg32WT-expressing cells (Supplemental Figure S2B).
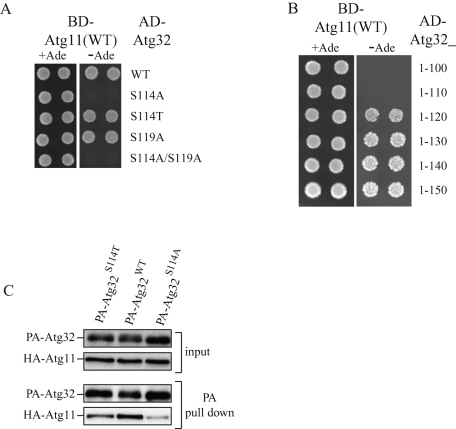
Because phosphorylation of Ser-114 of Atg32 is necessary for mitophagy and because Ser-114 is within the Atg11 interaction region of Atg32 (Figure 1D, residues 51–150), we speculated that phosphorylation of Ser-114 might be necessary for the Atg11–Atg32 interaction. To examine this possibility, we prepared yeast two-hybrid constructs to express Atg32 phosphorylation-site mutants (AD-Atg32S114A, AD-Atg32S119A, AD-Atg32S114T, and AD-Atg32S114A/S119A). As expected, the cells expressing AD-Atg32S114A or AD-Atg32S114A/S119A and BD-Atg11 did not grow, whereas AD-Atg32WT, AD-Atg32S119A, or AD-Atg32S114T and BD-Atg11 did grow on selective plates (Figure 3A), suggesting that phosphorylation of Atg32-Ser-114 or Atg32-Thr114 is important for the Atg11–Atg32 interaction. We further studied the importance of the Ser-114 residue on the Atg11–Atg32 interaction. As shown in Figure 1C, the N-terminus 150 amino acids of Atg32 (Atg32_1-150) interacted with Atg11. We additionally prepared yeast two-hybrid Atg32 constructs to express N-terminus Atg32 including Ser-114 (Atg32_1-120, 1-130, 1-140, and 1-150) or N-terminus Atg32 excluding Ser-114 (Atg32_1-100 and 1-110). As expected, the cells expressing AD-Atg32 including Ser-114 (Atg32_1-120, 1-130, 1-140, and 1-150) and BD-Atg11 grew on the selective plates, whereas AD-Atg32 excluding Ser-114 (Atg32_1-100 and 1-110) and BD-Atg11 did not grow on the selective plates (Figure 3B). This finding further supports the idea that phosphorylation of Ser-114 on Atg32 is important for the Atg11–Atg32 interaction.
FIGURE 3:
Phosphorylation of Ser-114 on Atg32 is critically important for the Atg11–Atg32 interaction. (A, B) Yeast two-hybrid analysis between Atg11 and the indicated Atg32 mutants. (C) The atg11Δ atg32Δ strains expressing PA-tagged Atg32WT, PA-tagged Atg32S114A, or PA-tagged Atg32S114T and HA-tagged Atg11 under the control of the CUP1 promoter were grown in SMD medium until the mid–log phase and then starved in SD-N for 1 h. PA-Atg32 was precipitated using IgG–Sepharose from cell lysates. Top two, an immunoblot of total-cell lysates. Bottom two, the IgG precipitates, which were probed with anti-HA and anti-PA antibodies.
To confirm the importance of Atg32-Ser-114 phosphorylation on the Atg32–Atg11 interaction, we expressed HA-tagged Atg11 and PA-tagged Atg32S114A or PA-tagged Atg32S114T and performed a protein A affinity pull-down assay. PA-Atg32S114A coprecipitated HA-Atg11 to a negligible extent compared with PA-Atg32WT (Figure 3C). As expected, PA-Atg32S114T could coprecipitate HA-Atg11, although the amount of HA-Atg11 coprecipitated with PA-Atg32S114T was smaller than that with PA-Atg32WT (Figure 3C). This finding further supports the idea that phosphorylation of Ser-114 (or Thr-114) in Atg32 is physiologically important for the Atg11–Atg32 interaction.
When mitophagy is induced, Atg11 interacts with Atg32 and recruits mitochondria to the PAS, which is formed on the vacuolar rim. We previously reported that GFP-Atg32WT formed cytosolic GFP puncta at the PAS in an Atg11-dependent manner in the atg1Δ strain during nitrogen starvation (Kanki et al., 2009c). Because phosphorylation of Ser-114 in Atg32 is important for the Atg11–Atg32 interaction, GFP-Atg32S114A or GFP-Atg32S114A/S119A should not form GFP puncta at the PAS. To test this possibility, we expressed GFP-Atg32WT, GFP-Atg32S114A, and GFP-Atg32S114A/S119A in the atg1Δ and atg32Δ double-knockout strain and observed GFP puncta formation during nitrogen starvation. It was difficult to detect cytosolic GFP puncta in GFP-Atg32S114A- or GFP-Atg32S114A/S119A-expressing cells (only 4.5 and 2.0% of cells had puncta on the vacuolar rim during starvation, respectively), whereas cells expressing GFP-Atg32WT showed cytosolic GFP puncta on the vascular rim (36.6% of the cells had puncta on the vacuolar rim; Supplemental Figure S4). This finding also supports the idea that Ser-114 in Atg32 is important for mitophagy.
Phosphorylation of Ser-114 or Ser-119 on Atg32 is not required for Atg32–Atg8 interaction
It was reported that Atg32 interacts with Atg8 through the WQAI motif on Atg32 and that the Atg32–Atg8 interaction also plays role in mitophagy (Okamoto et al., 2009b). Because phosphorylation of Ser-114 on Atg32 is required for mitophagy, we speculated that this phosphorylation might affect Atg32–Atg8 interaction. We examined this possibility by yeast two-hybrid analysis. As shown in Supplemental Figure S5A, cells expressing BD-Atg8 and AD-Atg32 mutants (Atg32S114A, Atg32S119A, and Atg32S114A/S119A), as well as AD-Atg32WT, grew on the selective plates. This finding suggests that phosphorylation of Ser-114 or Ser-119 on Atg32 is not required for Atg32–Atg8 interaction.
Phosphorylation of Atg32 is partially affected by the carbon source in the starvation medium
Mitophagy is induced by nitrogen starvation after preculturing yeast in nonfermentable medium. However, the level of mitophagy induction varies depending on the carbon source supplemented in starvation medium. Mitophagy is efficiently induced by nitrogen starvation in SD-N, whereas mitophagy occurs at a very low level in starvation medium with lactate (SL-N; Kanki and Klionsky, 2008). We speculated that the carbon source that supplements the starvation medium might affect the phosphorylation of Atg32, and as a result, the level of mitophagy induction might vary. To examine this possibility, we observed the phosphorylation of Atg32 in SD-N or SL-N medium after preculturing yeast in SML. As we previously reported, mitophagy occurred at a very low level when mitophagy was induced by SL-N compared with induction by SD-N (Supplemental Figure S5B; Kanki and Klionsky, 2008). Although the phosphorylation of Atg32 was similar after 6 h of nitrogen starvation in both SD-N and SL-N, phosphorylation after 1 h of starvation was very low in SL-N medium compared with that in SD-N (Supplemental Figure S5C). Therefore we concluded that the carbon source supplemented in the starvation medium affects the phosphorylation of Atg32 during starvation in the early period and that the difference in phosphorylation of Atg32, in part, affects the level of mitophagy induced by the starvation medium supplemented with a different carbon source.
Deletion of HOG1 or PBS2 affects both Atg32 phosphorylation and mitophagy
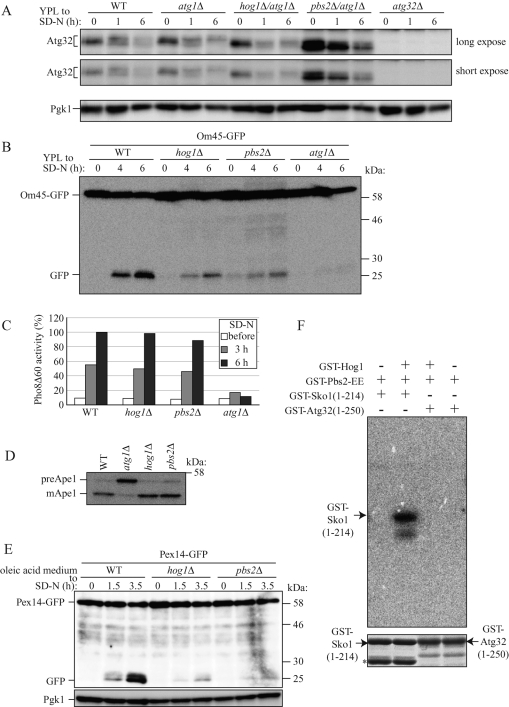
To identify kinases that phosphorylate Atg32 when mitophagy is induced, we investigated kinase or kinase cofactors encoding gene-deleted yeast strains and observed Atg32 phosphorylation when mitophagy was induced by nitrogen starvation. We found two kinases, Hog1 and Pbs2, related to Atg32 phosphorylation. Hog1 is a mitogen-activated protein kinase (MAPK), and Pbs2 is a mitogen-activated protein kinase kinase (MAPKK), and they are involved in the osmoregulatory signal transduction cascade (HOG signaling pathway), which affects gene expression by regulating osmoresponsive transcription factors (Westfall et al., 2004). In both HOG1- and PBS2-deleted cells (hog1Δ and pbs2Δ cells, respectively), phosphorylation of Atg32 was severely but not completely inhibited when mitophagy was induced by nitrogen starvation (Figure 4A and Supplemental Figure S6A). Because phosphorylation of Atg32 is required for mitophagy, we speculated that mitophagy might be defective in hog1Δ and pbs2Δ cells. To test this possibility, we monitored mitophagy using the Om45-GFP processing assay and found that mitophagy was also severely defective in these cells (Figure 4B).
FIGURE 4:
Deletion of HOG1 or PBS2 affects both Atg32 phosphorylation and mitophagy. (A) WT, atg1Δ, and atg32Δ strains and hog1Δ/atg1Δ and pbs2Δ/atg1Δ double-knockout strains were cultured in YPL medium until the mid–log growth phase and then shifted to SD-N medium for 0, 1, and 6 h. Phosphorylation on endogenous Atg32 was monitored by immunoblotting with anti-Atg32 and anti-Pgk1 (loading control) antibodies. (B) Strains deleted for the indicated genes and expressing Om45-GFP were cultured in YPL medium until the mid–log growth phase and then shifted to SD-N for 4 and 6 h. GFP processing was monitored by immunoblotting with anti-GFP antibody. (C) The WT (TKMY236), hog1Δ (TKYM248), pbs2Δ (TKYM249), and atg1Δ (TKYM256) strains were grown in YPD medium and shifted to SD-N for 3 and 6 h. Samples were collected and protein extracts assayed for Pho8Δ60 activity. (D) WT, atg1Δ, hog1Δ, and pbs2Δ strains were cultured in YPD medium and analyzed for prApe1 maturation by immunoblotting with anti-Ape1 antiserum. (E) Strains deleted for the indicated genes and expressing Pex14-GFP were cultured with oleic acid–containing medium for 19 h and then shifted to SD-N for the indicated times and monitored for GFP processing by immunoblotting. (F) In vitro phosphorylation of Sko1(1-214) and Atg32(1-250) by Hog1. Recombinant GST-Sko1(1-214) or GST-Atg32(1-250) was phosphorylated by activated recombinant GST-Hog1 in the presence of [γ-32P]ATP. The labeled proteins were resolved by SDS–PAGE, and an autoradiograph image (top) and Coomassie Brilliant Blue–stained image (bottom) of the gel were taken. The asterisks indicate GST-Sko1 degradation product or nonspecific bands.
We further observed macroautophagy, the Cvt pathway, and pexophagy in hog1Δ and pbs2Δ cells. To observe macroautophagy, we used a Pho8Δ60 activity assay (Noda et al., 1995). Pho8Δ60 is a truncated form of the vacuolar alkaline phosphatase. Deletion of the native signal sequence causes the precursor protein to remain in the cytosol, and it is only delivered to the vacuole by an autophagic mechanism. On delivery, the C-terminal propeptide is removed, resulting in activation of the zymogen, which can be measured enzymatically. We found that both hog1Δ and pbs2Δ cells, as well as WT cells, showed a similar increase in Pho8Δ60-dependent alkaline phosphatase activity following nitrogen starvation (Figure 4C). Consistent with previous reports that nitrogen starvation–induced macroautophagy is normal in the HOG1 deleted strain under normo-osmotic culture conditions (Prick et al., 2006), our finding suggests that macroautophagy is not defective in the hog1Δ and pbs2Δ strains. This finding excludes the possibility that inhibition of mitophagy in hog1Δ and pbs2Δ cells is due to dysfunction of autophagic machinery. To observe the Cvt pathway, we investigated processing of the precursor from Ape1 (preApe1) under steady-state conditions. As shown in Figure 4D, preApe1 in hog1Δ and pbs2Δ cells was processed to comparable levels with those in WT cells, suggesting that the Cvt pathway is normal in hog1Δ and pbs2Δ cells. Finally, we used GFP processing from peroxisomal protein Pex14 tagged with GFP (Pex14-GFP) to observe pexophagy (Reggiori et al., 2005). As shown in Figure 4E, pexophagy was severely defective in both hog1Δ and pbs2Δ cells. These findings suggest that the HOG signaling pathway is not related to the autophagic process but is related to regulation of organelle specific selective autophagy.
Because Hog1 is a kinase, we expected that Hog1 might directly phosphorylate Atg32. To observe this possibility, we challenged with an in vitro Hog1 kinase assay. Purified Hog1 was activated by a purified constitutive active Pbs2-EE mutant, and then purified Sko1 (N-terminus 214 amino acids; positive control) (Proft et al., 2001) or purified Atg32 (N-terminus 250 amino acids) and [γ-32P]ATP were mixed and incubated in vitro. As shown in Figure 4F, Sko1 was phosphorylated by Hog1, whereas Atg32 was not phosphorylated at all. This finding suggests that Atg32 is not directly phosphorylated by Hog1.
We then decided to determine whether components upstream of the HOG signaling pathway are required for mitophagy. Upstream of the HOG signaling pathway, there are two osmosensor complexes, Sho1–Msb2 and Sln1–Ypd1–Ssk1, and a layer of three MAPKKKs (Ssk2, Ssk22, and Ste11), which are responsible for activation of Pbs2 (Clotet and Posas, 2007). We expressed Om45-GFP in sho1Δ, ste11Δ, ssk1Δ, msb2Δ, ssk2Δ, and ssk22Δ cells and observed mitophagy. As shown in Supplemental Figure S6B, none of those mutants affected mitophagy, suggesting that, in the HOG signaling pathway, only Pbs2 and Hog1 are related to mitophagy. This result is consistent with the finding that hyperosmotic stress alone is not sufficient to induce mitophagy (Supplemental Figure S7A).
Finally, we observed whether Hog1 is activated under mitophagy-inducing conditions (Supplemental Figure S7B). As previously reported, Hog1 is phosphorylated (activated) by hyperosmotic stress (YPD + NaCl). Hog1 is also phosphorylated by nitrogen starvation (SD-N) but not by rapamycin (Rap) treatment (YPD + Rap). Of interest, Hog1 was strongly phosphorylated at the early log phase in YPL medium (preculture in YPL). During mitophagy, the phosphorylation of Hog1 was decreased but remained to some extent (SD-N). Surprisingly, when mitophagy was induced by rapamycin, the phosphorylation of Hog1 almost completely disappeared (YPL + Rap). These findings are consistent with the finding that Hog1 does not phosphorylate Atg32 directly and suggest that Hog1 is not the sole factor for Atg32 phosphorylation and mitophagy.
The N-terminus region of Atg32, especially the amino acids 51–100 of Atg32, contributes to stable expression of Atg32 but does not play a role in mitophagy
As shown in Figure 1C, when the first 50 amino acids in the N-terminus of Atg32 were deleted (51–529), this mutant could interact with Atg11 by yeast two-hybrid assay, whereas deletion of 75 amino acids in the N-terminus of Atg32 (76–529) could not. Therefore, we decided to address the function of the N-terminus region of Atg32. We first expressed N-terminus 50, 100, or 150 amino acid–deleted Atg32 mutants (Atg32_51-529, 101-529, or 151-529) in atg32Δ cells expressing Om45-GFP and observed mitophagy during nitrogen starvation. Atg32_51-529–expressing cells showed comparable levels of GFP processing with those of Atg32_WT (Supplemental Figure S8A), suggesting that Atg32_51-529 is functional. Atg32_101-529–expressing cells showed a small amount of GFP processing, whereas Atg32_151-529–expressing cells completely lost GFP processing (Supplemental Figure S8A), suggesting that Atg32_101-529 can still slightly function. We also observed the expression levels of those Atg32 mutants and found that Atg32_101-529 was barely expressed compared with Atg32_WT or Atg32_51-529 (Supplemental Figure S8B). Therefore, we speculate that the N-terminus region of Atg32 might be required for stable expression of Atg32. To test this possibility, we additionally expressed N-terminus 69, 71, 73, or 75 amino acid–deleted Atg32 mutants (Atg32_70-529, 72-529, 74-529, or 76-529) in atg32Δ cells. Depending on the length deleted, the expression level of the Atg32 mutant was decreased, and when 75 amino acids were deleted in the N-terminus (Atg32_76-529), the expression level decreased to the same level of Atg32_101-529 (Supplemental Figure S8B). We then examined the function of these Atg32 mutants for mitophagy using the Om45-GFP processing assay. In response to the reduction of Atg32 mutant expression levels, GFP processing was decreased (compared with results shown in Supplemental Figures S8B and S8C), suggesting that the Atg32 mutant expression level, but not the length of N-terminus residues deleted, affects the level of mitophagy. To determine whether Atg32_76-529 or Atg32_101-529 is functional if the expression level is maintained, we expressed these mutants under the CUP1 promoter, which can express genes more strongly than the Atg32 endogenous promoter. Under the CUP1 promoter, Atg32_76-529 and Atg32_101-529 expressed comparable levels with those of full-length Atg32 expressed under the endogenous promoter (Supplemental Figure S9A). Cells expressing a substantial amount of Atg32_76-529 or Atg32_101-529 showed a similar level of mitophagy to that of full-length-Atg32–expressing cells (Supplemental Figure S9B). In addition, we constructed Atg32 lacking the amino acids 51–75 (Atg32_Δ51-75). Atg32_Δ51-75 was expressed at similar levels to those of full-length Atg32 (Supplemental Figure S9C), and cells expressing Atg32_Δ51-75 showed similar levels of mitophagy to those of full-length-Atg32–expressing cells (Supplemental Figure S9D). From these findings, we concluded that the N-terminus region of Atg32, especially the amino acids 51–100 of Atg32, contributes to stable expression of Atg32 but does not play a role in mitophagy.
The C-terminus region of Atg32, which is located in the inter membrane space, does not play a significant role in mitophagy
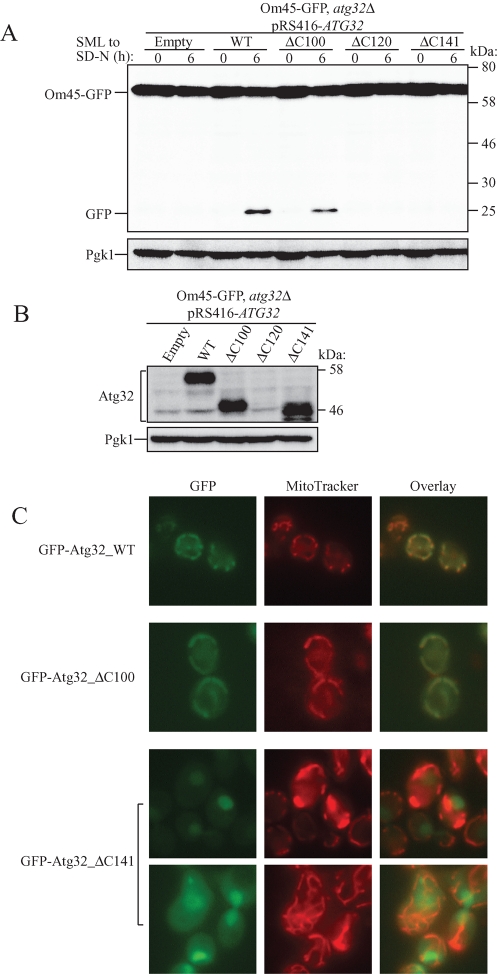
Atg32 is a single-span mitochondrial outer membrane protein with its N- and C-terminal domains oriented toward the cytoplasm and the intermembrane space, respectively. The transmembrane domain (TMD) is predicted to be from Ser-389 to Val-411 (Figure 1D). As shown in the preceding section, the N-terminus part of Atg32 interacts with Atg11, and this is essential for mitophagy. We next decided to address the role of the C-terminus region of Atg32 in mitophagy. We created a 100–amino acid deletion of the C-terminus of Atg32 that maintained the whole TMD (Atg32_ΔC100), a 120–amino acid deletion of the C-terminus of Atg32 that lacked part of the TMD (Atg32_ΔC120), and a 141–amino acid deletion of the C-terminus of Atg32 that completely lacked the TMD (Atg32_ΔC141). To observe mitophagy, we expressed those mutants in atg32Δ cells expressing Om45-GFP and induced mitophagy by nitrogen starvation. Surprisingly, cells expressing Atg32_ΔC100 showed comparable levels of mitophagy to those of full-length Atg32, whereas cells expressing Atg32_ΔC120 or Atg32_ΔC141 showed a complete inhibition of mitophagy (Figure 5A). We then examined the expression level of Atg32 mutants. Both Atg32_ΔC100 and Atg32_ΔC141 were expressed to the same levels as those with full-length Atg32, but Atg32_ΔC120 was barely expressed (Figure 5B). We further observed mitochondrial localization of those Atg32 mutants by tagging GFP on its N-terminus. Similar to full-length Atg32, Atg32_ΔC100 was localized in the mitochondria, but, as expected, Atg32_ΔC141, which completely lacked TMD, was not localized in mitochondria (Figure 5C). From these findings, we concluded that the C-terminus region of Atg32 does not play a significant role in mitophagy, whereas the TMD is essential for Atg32 localization in mitochondria.
FIGURE 5:
Characterization of the C-terminus region of Atg32. (A) Strains of atg32Δ expressing Om45-GFP were transformed with the indicated Atg32 mutant–expressing vectors. Cells were cultured in SML medium until the mid–log growth phase and then shifted to SD-N for 6 h. GFP processing was monitored by immunoblotting with anti-GFP and anti-Pgk1 (loading control) antibodies. (B) The same cells shown in A were cultured in SML medium until the mid–log growth phase. The expression level of Atg32 was observed by immunoblotting with anti-Atg32 and anti-Pgk1 (loading control) antibodies. (C) The wild-type strain transformed with a plasmid expressing the indicated GFP-tagged Atg32 mutants under the control of the CUP1 promoter was cultured in SMD medium until the mid–log growth phase. Cells were labeled with the mitochondrial marker MitoTracker Red and analyzed by fluorescence microscopy.
Discussion
Mitochondria are organelles that supply energy to the cell. However, these organelles are also the major source of cellular ROS. Therefore quality control of mitochondria is important to maintain cellular homeostasis. Selective elimination of mitochondria via autophagy is believed to be a primary mechanism for mitochondrial quality control. Accordingly, it is important to understand the molecular mechanism of mitophagy and its regulation. Recent studies in yeast identified several mitophagy-related genes and showed that when autophagy selects mitochondria as a cargo, mitochondrial receptor protein Atg32 is recognized and bound by adaptor protein Atg11. Atg11 then recruits mitochondria to the PAS, where there is autophagosome uptake in the mitochondria (Kanki and Klionsky, 2009, 2010; Kanki et al., 2009b, 2009c, 2010, 2011; Okamoto et al., 2009a, 2009b). The Atg11–Atg32 interaction is believed to be the initial molecular process of mitophagy. It is unknown how the Atg11–Atg32 interaction is mediated or what signaling regulates it. In this study, we found that the C-terminus region of Atg11 (Atg11_CC4) interacts with the N-terminus region of Atg32 when mitophagy is induced. We also found that Atg32 is phosphorylated mainly on Ser-114 and Ser-119 following mitophagy induction and that the phosphorylation, especially phosphorylation of Ser-114, is necessary for the Atg11–Atg32 interaction and for mitophagy. We attempted to identify the factors that regulate Atg32 phosphorylation, and we found that Hog1 and Pbs2 were related to Atg32 phosphorylation. Hog1 is a MAPK and Pbs2 is a MAPKK, and both are involved in the HOG signaling pathway. Based on the findings that Hog1 does not phosphorylate Atg32 directly (Figure 4F), is not always activated under mitophagy-inducing conditions (Supplemental Figure S7B), and regulates the expression of ∼600 genes (O'Rourke and Herskowitz, 2004), we speculate that Hog1 is one of the factors that affects an unidentified primary mitophagy signaling pathway or affects unidentified kinase(s) that directly phosphorylate Atg32.
This study found that Atg32 was phosphorylated even when cells were cultured in the growing phase (Figure 2, A and B). This unidentified phosphorylation site was dephosphorylated within 30 min by nitrogen starvation (unpublished data). This site phosphorylated in the growing phase is different from Ser-114 and Ser-119 because even Atg32S114A/S119A showed phosphorylation in the growing phase (Figure 2C). Until the current study, the physiological significance of this Atg32 phosphorylation and dephosphorylation on mitophagy was not well understood.
After nitrogen starvation, Atg32 was transiently dephosphorylated as discussed earlier and then phosphorylated on the residues of Ser-114 and Ser-119 (Figure 2, A and C). Although Ser-119 was more efficiently phosphorylated than Ser-114, phosphorylation of Ser-114 was strongly related to mitophagy (Figure 2D). Even if Ser-114 was altered to Thr, the Atg32 mutant (Atg32S114T) was still functional for mitophagy, whereas the change of Ser-114 to Tyr completely disabled Atg32 function (Figure 2D). These findings strongly support our conclusion that a Ser/Thr–specific protein kinase phosphorylates Atg32 and that phosphorylation of the amino acid residue 114 of Atg32 is required for mitophagy.
Contrary to our expectation, neither Atg32S114D nor Atg32S114E was functional for mitophagy (Figure 2D). The substitution Ser to Asp or Glu is commonly used to mimic the phosphorylation status of protein. However, there is no guarantee that an acidic amino acid substitution will replicate the effect of phosphorylation since a phosphate group is considerably larger and carries twice the negative charge of a carboxylate (Cribbs and Strack, 2009). This might be the reason why neither Atg32S114D nor Atg32S114E was functional.
Mitophagy is inhibited even under strong macroautophagy-inducing nitrogen starvation conditions if the nitrogen starvation medium contains nonfermentable carbon as the sole carbon source in which mitochondria are essential for energy production (Kanki and Klionsky, 2008). We in addition found that the phosphorylation of Atg32 was affected by the carbon source supplemented in the starvation medium. These findings suggest that mitophagy is strictly regulated under an unidentified mitophagy-related signaling pathway. Phosphorylation of Ser-114 in Atg32 is likely to be the terminal reaction of this signaling pathway because phosphorylation of Atg32 directly mediates the Atg11–Atg32 interaction, which is the initial physical molecular step delivering mitochondria to vacuoles. The phosphorylation level of Ser-114 in Atg32 may be a rate-limiting factor of mitophagy.
Because Atg32 is evenly spread in the mitochondria (Kanki et al., 2009c; Okamoto et al., 2009b) and because mitochondria carrying phosphorylated Ser-114 in Atg32 are destined to be degraded, it is reasonable that the phosphorylation level of Ser-114 can be restricted to maintain a proper mitochondrial volume. In fact, we found that when mitophagy was induced by nitrogen starvation for 6 h, both degraded mitochondria and Ser-114 phosphorylation were consistently low (Figure 2D, SD-N 0 h compared with the 6 h Om45-GFP signal).
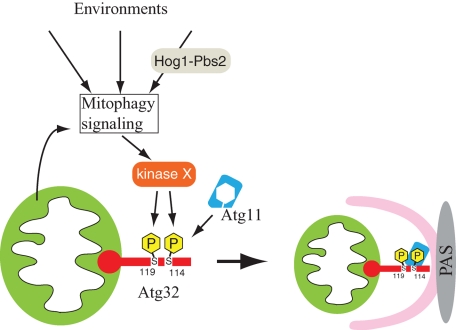
The current model for mitophagy is summarized in Figure 6. When mitophagy is induced, Ser-114 and Ser-119 on Atg32 are phosphorylated. The phosphorylation of Atg32, especially the phosphorylation of Ser-114, mediates the Atg11–Atg32 interaction. Atg11 recruits mitochondria to the PAS, where the autophagosome is generated to enclose the mitochondria. The kinase that directly phosphorylates Atg32 has not been identified yet (Figure 6, kinase X). The mitophagy signaling pathway that connects mitophagy-inducing stimuli derived from mitochondria and/or the cellular environment to kinase X is still unclear. It is possible that several factors related to the mitophagy signaling pathway, such as Hog1 and Pbs2, are involved.
FIGURE 6:
Model of mitophagy in yeast. The initial trigger-inducing mitophagy comes from mitochondria (such as mitochondrial damage) and/or the cellular environment (such as dramatic nutrient change and oxidative stresses). The initial trigger is through an unidentified mitophagy signaling pathway and it reaches kinase X, which is believed to be a Ser/Thr–specific protein kinase. This kinase phosphorylates Ser-114 and Ser-119 on Atg32. Phosphorylation of Atg32, especially phosphorylation of Ser-114, mediates the Atg11–Atg32 interaction. Atg11 recruits mitochondria to the PAS, where the autophagosome is generated to enclose the mitochondria. Hog1 and Pbs2 affect the mitophagy signaling pathway or affect kinase X.
The requirement of Atg32 phosphorylation for mitophagy is similar to pexophagy in Pichia pastoris because phosphorylation of PpAtg30, a receptor protein for pexophagy, is required for PpAtg30–PpAtg11 interaction and pexophagy (Farre et al., 2008). Recently, it was reported that the MAPK Slt2 and upstream MAPK cascade are necessary for pexophagy in Saccharomyces cerevisiae (Manjithaya et al., 2010). In the present study, we found that another MAPK, Hog1, and the upstream Pbs2 were required for mitophagy. Our study also showed that Hog1 was required for pexophagy (Figure 4F), whereas another study reported that it was not required for pexophagy (Manjithaya et al., 2010). Although further studies are required to determine this issue, these findings suggest that there is an interaction between the MAPK cascade and selective autophagy in yeast.
We also studied the role of the first 100 amino acids of the N-terminus and C-terminus of Atg32 on mitophagy. Deletion of 100 amino acids in the C-terminus of Atg32, in which almost all of the intermembrane space domain of Atg32 was deleted, resulted in mitophagy still being functional, suggesting that the C-terminus region of Atg32 does not play a role in mitophagy (at least under our mitophagy-inducing condition). On the other hand, we found that the N-terminus region of Atg32 was required for stable expression of Atg32, but it was not required for mitophagy. When more than 50 amino acids in the N-terminus of Atg32 were deleted, the expression level of Atg32 was decreased, depending on the length of the deleted residues (Supplemental Figure S8B). In response to the reduction of Atg32 mutant expression, induction of mitophagy was also decreased in parallel. This finding is consistent with previous reports that N-acetylcysteine, a scavenger of free radicals, suppresses the expression level of Atg32 (Okamoto et al., 2009b) and mitophagy (Deffieu et al., 2009). Atg32 lacking 100 amino acids in the N-terminus could induce mitophagy comparable with that in WT Atg32 if this mutant was expressed to the same level as that of endogenous WT Atg32 (Supplemental Figure S9, A and B), suggesting that the first 100 amino acids in the N-terminus of Atg32 are not involved in the molecular process of mitophagy. Because the Atg11–Atg32 interaction is essential for mitophagy, these 100 amino acids in the N-terminus of Atg32 can be excluded as the Atg11-interacting domain. Together with the yeast two-hybrid result that 120 amino acids in the N-terminus of Atg32 are sufficient to bind Atg11 (Figure 3B), this suggests that the Atg11 interaction region of Atg32 can be narrowed down as being amino acid residues 100–120.
MATERIALS AND METHODS
Strains and media
The yeast strains used in this study are listed in Supplemental Table S1. Yeast cells were grown in rich medium (YPD: 1% yeast extract, 2% peptone, and 2% glucose), lactate medium (YPL: 1% yeast extract, 2% peptone, and 2% lactate), synthetic minimal medium with glucose (SMD: 0.67% yeast nitrogen base, 2% glucose, and amino acids), or synthetic minimal medium with lactate (SML: 0.67% yeast nitrogen base, 2% lactate, and amino acids). Nitrogen starvation experiments were performed in synthetic minimal medium lacking nitrogen (SD-N: 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose; or SL-N: 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% lactate).
Plasmids
The plasmids for expression of PA-tagged Atg32 (WT), GFP-tagged Atg32 (WT), and HA-tagged Atg11 mutants under the control of the CUP1 promoter, the plasmids for expression of Atg32 (WT) under the endogenous promoter (pRS416-Atg32WT), and yeast two-hybrid plasmid pGBDU-Atg11 mutants, pGAD-Atg19 and pGAD-Atg32 (WT), were described previously (Yorimitsu and Klionsky, 2005; Kanki et al., 2009c). The plasmids expressing Atg32 with Ser or Thr mutation were generated using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) based on WT Atg32 expression plasmids. For the yeast two-hybrid vector pGAD-Atg32 mutants, a DNA fragment encoding N-terminus– or C-terminus–deleted ATG32 was PCR amplified with BglII and SalI restriction sites from yeast genomic DNA and ligated into the BamHI and SalI sites of pGAD-C1 (James et al., 1996). For the N-terminus–deleted Atg32 expression vectors under the control of the endogenous Atg32 promoter, a DNA fragment encoding N-terminus–deleted ATG32 was PCR amplified with the Atg32 promoter sequence from yeast genomic DNA and cotransfected with pRS416-Atg32WT plasmid digested by BamHI and AflII in yeast (SEY6210) to allow recombination between the PCR fragment and digested plasmid. The objective plasmids were purified from the colonies grown on Ura (−) plates. For the C-terminus–deleted Atg32 expression vectors under the endogenous Atg32 promoter, a DNA fragment encoding promoter- and C-terminus–deleted ATG32 was PCR amplified with SpeI and SalI restriction sites from yeast genomic DNA and ligated into the SpeI and SalI sites of the pRS416-CYC1tm vector (pRS416 vector with CYC1 terminator). For the GFP-tagged C-terminus–deleted Atg32 expression vectors under the control of the CUP1 promoter, a DNA fragment encoding C-terminus–deleted ATG32 was PCR amplified with EcoRI and SalI restriction sites from yeast genomic DNA and ligated into the EcoRI and SalI sites of pCuGFP(416) (Kim et al., 2001).
Antibodies
Anti-Atg32 antibody was produced by immunizing rabbits with the recombinant GST-tagged N-terminus (the first 400 amino acids) of Atg32 and affinity purifying the serum with recombinant His-tagged N-terminus (the first 250 amino acids) of Atg32-conjugated Sepharose. The affinity-purified anti-Atg32 antibody does not react with the His-tagged N-terminus (the first 150 amino acids) of Atg32, suggesting that this antibody reacts with residues 151–250. Anti-ApeI antiserum was a gift from Daniel J. Klionsky (University of Michigan, Ann Arbor, MI). Anti-HA antibody (Sigma-Aldrich, St. Louis, MO), anti-Por1 antibody (Invitrogen Molecular Probes, Eugene, OR), anti-Pgk1 antibody (Nordic Immunological Laboratories, Tilburg, Netherlands), anti–protein A antibody (GeneTex, Irvine, CA), anti-Hog1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti–phosphorylated Hog1 antibody (Cell Signaling Technology, Beverly, MA), and anti-GFP antibody (Takara Bio, Otsu, Japan) were used for immunoblotting.
Assays for mitophagy, macroautophagy, the Cvt pathway, and pexophagy
For monitoring mitophagy, the Om45-GFP processing assay was carried out as described previously (Kanki et al., 2009a). For monitoring macroautophagy, the alkaline phosphatase activity of Pho8Δ60 was measured as described previously (Noda et al., 1995). For monitoring the Cvt pathway, cells were cultured in YPD medium up to the mid–log growth phase. The maturation of preApeI was observed by immunoblotting with anti-ApeI antiserum. For monitoring pexophagy, the Pex14-GFP processing assay was carried out as described previously (Reggiori et al., 2005).
Protein A affinity pull-down assay
Cells expressing HA-tagged Atg11 (WT or mutants) and PA-tagged Atg32 (WT or mutants) were cultured in SMD medium until the mid–log growth phase and then shifted to SD-N for 1 h. Cells were collected and lysed with glass beads in IP buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 150 mM KCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], and proteinase inhibitors), and they were centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was mixed with IgG–Sepharose at 4°C for 12 h. The Sepharose was washed by ice-cold wash buffer (50 mM HEPES, pH 7.4, 500 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and 0.1% SDS) five times, and the sample was eluted by SDS–PAGE loading buffer. The elution samples were observed by immunoblotting with anti-HA and anti–protein A antibodies.
λ Protein phosphatase treatment
The atg11Δ strain was cultured in YPL medium until the mid–log phase and then shifted to SD-N for 6 h. Cells were collected and lysed with glass beads in phosphatase buffer (supplied with λ protein phosphatase) with 1 mM PMSF and protease inhibitors. After centrifugation at 10,000 × g for 10 min, the supernatant was incubated with λ protein phosphatase (New England BioLabs, Ipswich, MA) for 1 h at 30°C.
Fluorescence microscopy
Cells expressing GFP-tagged Atg32 mutants were grown in SMD medium until the mid–log growth phase. To label mitochondria, cells were incubated with 1 μM of MitoTracker Red (Invitrogen) at 30ºC for 30 min. After cells were washed with SMD medium, fluorescence signals were visualized on a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan). To observe cytosolic GFP puncta formation, cells were incubated with 20 μg/ml N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4-64; Biotium, Hayward, CA) at 30ºC for 30 min. After being washed with sterilized water, the cells were incubated in SD-N for 2 h. The fluorescence signals were visualized on the A1R confocal laser microscope system (Nikon, Tokyo, Japan).
In vitro kinase assay
For the in vitro kinase assay, we used a method described previously (Bilsland-Marchesan et al., 2000; Proft et al., 2001). N-terminus GST-tagged Hog1, Pbs2-EE (Ser-514 to Glu and Thr-518 to Glu mutant), Sko1 (N-terminus 214 amino acids), and Atg32 (N-terminus 250 amino acids) expression vectors were constructed using pGEX-4T-1 (GE Healthcare, Chalfont St. Giles, United Kingdom) and were expressed in Escherichia coli BL21(DE3). Expressed proteins were purified using Glutathione Sepharose 4B (GE Healthcare). One microgram of recombinant GST–Hog1 was activated by phosphorylation using 0.5 μg of GST–Pbs2-EE in the presence of kinase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 2 mM dithiothreitol) and 20 μM ATP. After 20 min at 30°C, 2.5 μg of GST-Sko1 or GST-Atg32 was added to the previous mixture together with [γ-32P]ATP (0.2 μCi/μl). The mixture was then incubated for 20 min at 30°C. The labeled proteins were resolved by SDS–PAGE. The gel was stained with Coomassie Brilliant Blue, and the gel images were visualized by BAS-2500 autoradiography (Fujifilm, Tokyo, Japan).
Supplementary Material
Acknowledgments
We thank Daniel J. Klionsky (University of Michigan) for providing strains, plasmids, and antiserums and for helpful advice. This work was supported in part by a Grant-in-Aid for Young Scientists (A) (23689032) (T.K.), a Grant-in-Aid for Scientific Research on Priority Areas (22020028) (T.K.) and Scientific Research (A) (22249018) (D.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, as well as support from the Uehara Memorial Foundation (T.K.), the Takeda Science Foundation (T.K.), the Naito Foundation (T.K.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (T.K.), the Kowa Life Science Foundation (T.K.), and Kyushu University Interdisciplinary Programs in Education and Projects in Research Development (T.K.). We appreciate technical support from the Research Support Center, Graduate School of Medical Sciences, Kyushu University.
Abbreviations used:
- Ape1
aminopeptidase I
- Cvt
cytoplasm-to-vacuole targeting
- GFP
green fluorescent protein
- MAPK
mitogen-activated protein kinase
- MAPKK
mitogen-activated protein kinase kinase
- PA
protein A
- PAS
preautophagosomal structure/phagophore assembly site
- ROS
reactive oxygen species.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0145) on July 14, 2011.
REFERENCES
- Arai M, Imai H, Koumura T, Yoshida M, Emoto K, Umeda M, Chiba N, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J Biol Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan E, Arino J, Saito H, Sunnerhagen P, Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF. Repair of mtDNA in vertebrates. Am J Hum Genet. 1999;64:1276–1281. doi: 10.1086/302392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J, Posas F. Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol. 2007;428:63–76. doi: 10.1016/S0076-6879(07)28004-8. [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S. Functional characterization of phosphorylation sites in dynamin-related protein 1. Methods Enzymol. 2009;457:231–253. doi: 10.1016/S0076-6879(09)05013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffieu M, Bhatia-Kissova I, Salin B, Galinier A, Manon S, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem. 2009;284:14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friguet B, Bulteau AL, Petropoulos I. Mitochondrial protein quality control: implications in ageing. Biotechnol J. 2008;3:757–764. doi: 10.1002/biot.200800041. [DOI] [PubMed] [Google Scholar]
- Goldman SJ, Zhang Y, Jin S. Autophagic degradation of mitochondria in white adipose tissue differentiation. Antioxid Redox Signal. 2010;14:1971–1978. doi: 10.1089/ars.2010.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy. 2009a;5:1186–1189. doi: 10.4161/auto.5.8.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky D, Okamoto K. Mitochondria autophagy in yeast. Antioxid Redox Signal. 2011;14:1989–2001. doi: 10.1089/ars.2010.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Atg32 is a tag for mitochondria degradation in yeast. Autophagy. 2009;5:1201–1202. doi: 10.4161/auto.5.8.9747. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol. 2010;75:795–800. doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009b;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009c;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Klionsky DJ. A genomic screen for yeast mutants defective in mitophagy. Autophagy. 2010;6:278–280. doi: 10.4161/auto.6.2.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Manjithaya R, Jain S, Farre JC, Subramani S. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol. 2010;189:303–310. doi: 10.1083/jcb.200909154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. A landmark protein essential for mitophagy: Atg32 recruits the autophagic machinery to mitochondria. Autophagy. 2009a;5:1203–1205. doi: 10.4161/auto.5.8.9830. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009b;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Oku M, Sakai Y. Peroxisomes as dynamic organelles: autophagic degradation. FEBS J. 2010;277:3289–3294. doi: 10.1111/j.1742-4658.2010.07741.x. [DOI] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–1621. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Prick T, Thumm M, Kohrer K, Haussinger D, Vom Dahl S. In yeast, loss of Hog1 leads to osmosensitivity of autophagy. Biochem J. 2006;394:153–161. doi: 10.1042/BJ20051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Grivell LA. The role of protein degradation in mitochondrial function and biogenesis. Curr Genet. 1996;30:367–380. doi: 10.1007/s002940050145. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, He L, Lemasters JJ. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int J Biochem Cell Biol. 2004;36:2463–2472. doi: 10.1016/j.biocel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W. Mitochondrial protein homeostasis: the cooperative roles of chaperones and proteases. Res Microbiol. 2009;160:718–725. doi: 10.1016/j.resmic.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Ballon DR, Thorner J. When the stress of your environment makes you go HOG wild. Science. 2004;306:1511–1512. doi: 10.1126/science.1104879. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011;286:13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.