In the dimorphic fungus Ustilago maydis, Rac1 and its activator Cdc24 are essential for hyphal tip growth. Rac1 is shown to stimulate Cla4 kinase, which in turn triggers destruction of Cdc24. Expression of stabilized Cdc24 interferes with cell polarization, indicating that negative feedback regulation of Cdc24 is critical for tip growth.
Abstract
Dimorphic switching from budding to filamentous growth is a characteristic feature of many pathogenic fungi. In the fungal model organism Ustilago maydis polarized growth is induced by the multiallelic b mating type locus and requires the Rho family GTPase Rac1. Here we show that mating type–induced polarized growth involves negative feedback regulation of the Rac1-specific guanine nucleotide exchange factor (GEF) Cdc24. Although Cdc24 is essential for polarized growth, its concentration is drastically diminished during filament formation. Cdc24 is part of a protein complex that also contains the scaffold protein Bem1 and the PAK kinase Cla4. Activation of Rac1 results in Cla4-dependent degradation of the Rac1-GEF Cdc24, thus creating a regulatory negative feedback loop. We generated mutants of Cdc24 that are resistant to Cla4-dependent destruction. Expression of stable Cdc24 variants interfered with filament formation, indicating that negative feedback regulation of Cdc24 is critical for the establishment of polarized growth.
INTRODUCTION
Highly polarized growth occurs in many eukaryotic cells and requires precise temporal and spatial control of the cytoskeleton, vesicular trafficking, and membrane fusion. Apical growth of fungal cells serves as an excellent system in which to study the molecular mechanisms of cell polarization and the functional contributions of Rho-family GTPases (Etienne-Manneville and Hall, 2002). These proteins act as molecular switches and exist in two conformations—the active, GTP-bound form and the inactive, GDP-bound form. Rho GTPases are activated by guanine nucleotide exchange factors (GEFs) that catalyze the exchange of GDP for GTP (Rossman et al., 2005). Inactivation of GTPases is mediated by GTPase-activating proteins (GAPs) that enhance the low intrinsic GTPase activity (Bernards and Settleman, 2004). In the yeast Saccharomyces cerevisiae, the essential Rho protein Cdc42p acts as central regulator of polarity (Johnson and Pringle, 1990; Etienne-Manneville, 2004). Cdc42p and its GEF Cdc24p interact with the scaffold Bem1p and form a ternary complex that also contains the downstream effector of Cdc42p, the PAK kinase Cla4p (Peterson et al., 1994; Gulli et al., 2000; Bose et al., 2001). Many studies have addressed the question of how this complex regulates yeast morphogenesis (Gulli et al., 2000; Bose et al., 2001; Kozubowski et al., 2008). Localization and activity of Cdc24p are regulated at different levels (Gulli and Peter, 2001). Cdc24p is subject to cell cycle–dependent nuclear sequestration (Toenjes et al., 1999; Nern and Arkowitz, 2000; Shimada et al., 2000), oligomerization (Mionnet et al., 2008), and autoinhibition (Shimada et al., 2004). In addition, Cdk- and Cla4-dependent phosphorylation of Cdc24 has been observed, but the biological function of these modifications is still elusive (Gulli et al., 2000; Bose et al., 2001; Moffat and Andrews, 2004; Cole et al., 2009; Wai et al., 2009).
The dimorphic basidiomycetous fungus Ustilago maydis is an excellent model system in which to study the molecular mechanisms of fungal pathogenesis and hyphal growth (Bölker, 2001; Steinberg and Perez-Martin, 2008; Brefort et al., 2009). Haploid cells grow by budding, whereas dikaryotic cells, which result from fusion of compatible cells, display highly polarized growth. This dimorphic switch is controlled by the b mating type locus and is critical for virulence (Banuett, 1995). Here we address the regulation of the Rac1-specific GEF Cdc24 during filamentous growth. We observe that Cla4-dependent phosphorylation targets Cdc24 for destruction. We provide evidence that this negative feedback loop plays an important role during polarized growth.
RESULTS
b mating type–induced polarized growth involves Cdc24-dependent activation of Rac1
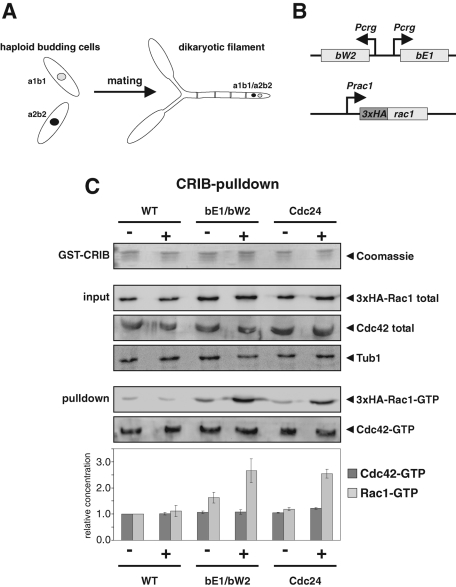
Sexual and pathogenic development in the plant pathogenic fungus U. maydis requires the morphogenetic transition from budding to filamentous growth (Figure 1A). This dimorphic switch is controlled by the multiallelic b mating type locus. The b locus encodes a pair of homeodomain transcription factors, which form heterodimers if derived from different alleles (Kämper et al., 1995). Expression of an active bE/bW heterodimer is sufficient to trigger filamentous growth in haploid cells (Brachmann et al., 2001). Many genes have been identified that are regulated by the bE/bW heterodimeric transcription factor (Heimel et al., 2010; Wahl et al., 2010). However, the molecular mechanism of filament formation is largely unknown. In U. maydis, polarized growth depends on the Rho GTPase Rac1, whereas the closely related GTPase Cdc42, which acts as central regulator of polarity in yeast, triggers cell separation (Mahlert et al., 2006). Expression of Rac1 is not upregulated during b mating type–dependent dimorphic switching (Mahlert et al., 2006). To test whether bE/bW expression results in stimulation of Rac1 activity, we determined the level of active Rac1-GTP in a pull-down assay using the Cdc42/Rac1 interaction and binding (CRIB) domain of the U. maydis Smu1 kinase (Leveleki et al., 2004; Smith et al., 2004). The genomic copy of Rac1 was tagged at its N-terminus with a 3xHA epitope and expressed in strain AB31. AB31 carries the bE1 and bW2 mating type proteins under control of a regulatable promoter (Figure 1B; Brachmann et al., 2001). If grown under inducible conditions, these cells switch from budding to filamentous growth (Figure 2A). We observed that in filaments the level of active Rac1-GTP was significantly enhanced, whereas the level of active Cdc42-GTP was unaffected (Figure 1C).
FIGURE 1:
Mating type–dependent dimorphic switching involves activation of Rac1. (A) Schematic representation of U. maydis dimorphic switching. Haploid cells of different mating type fuse at their tips to form a dikaryotic filament. (B) Strain AB31 expresses the active combination of bW2 and bE1 under control of the arabinose inducible crg promoter. The endogenous copy of Rac1 was tagged by an N-terminal 3xHA tag. (C) The levels of active Rac1-GTP and Cdc42-GTP were determined by CRIB pull-down assays before (−) and after (+) induction (4 h) of either bE1/bW2 or Cdc24 overexpression. GSH agarose beads were loaded with GST-CRIB and incubated with protein extracts from cultures grown under noninducing (−) and inducing (+) conditions. The amount of GST-CRIB is indicated by Coomassie staining. Rac1, Cdc42, and tubulin were detected with antibodies. The diagram shows the mean values of three independent determinations. Standard deviations are indicated.
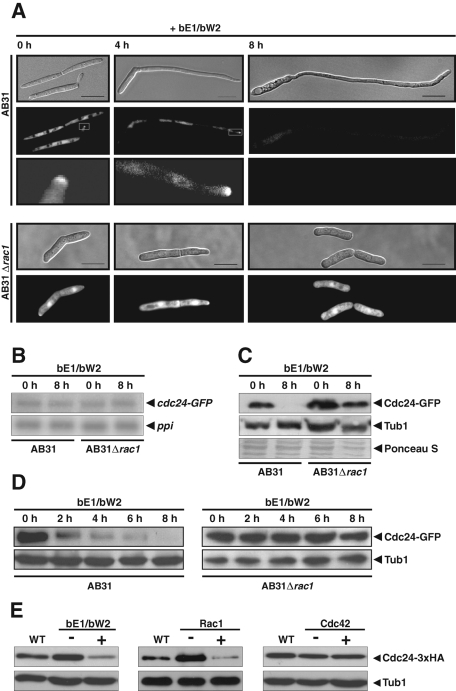
FIGURE 2:
Cdc24 is degraded upon mating type–dependent dimorphic switching. (A) Localization of Cdc24 was followed by constitutive expression of Cdc24-GFP fusion protein in strains AB31 and AB31Δrac1. These strains express the active bE1/bW2 heterodimer under control of the arabinose-inducible crg promoter. DIC and fluorescence micrographs were taken before induction (0 h) and 4 and 8 h after induction of bE1/bW2 expression. Bars, 10 μm. (B) Northern analysis of cdc24-GFP mRNA levels in strains AB31 and AB31Δrac1 before (0 h) and after induction (8 h) of the bE1/bW2 heterodimeric transcription factor. The ppi gene encoding peptidyl-prolyl cis/trans isomerase served as loading control. (C) Western blot analysis of Cdc24-GFP protein levels in strains AB31 and AB31Δrac1 before (0 h) and after induction (8 h) of the bE1/bW2 heterodimeric transcription factor. A monoclonal antibody against GFP was used for detection of Cdc24-GFP. Tubulin (Tub1) served as loading control. (D) The protein level of Cdc24 was determined at various time points after induction of bE1/bW2 overexpression in wild-type cells and in cells deleted for the endogenous copy of rac1. Cells were grown overnight in noninducing medium (0 h) and transferred to inducing medium. Proteins were isolated before induction and at 2, 4, 6, and 8 h after induction. (E) The protein level of Cdc24-3xHA expressed under control of its endogenous promoter was determined in strain AB31 and in conditional mutants of rac1 and cdc42 that express the GTPases under control of the crg promoter at the genomic loci. Cells were grown in liquid medium under noninducing (−) or inducing (+) conditions.
Next we asked which GEF is responsible for stimulation of Rac1 activity during polarized growth. Although in yeast the GEF Cdc24p is specific for Cdc42 (Zheng et al., 1994), its U. maydis orthologue (Cdc24) was proposed to act as Rac1-specific GEF (Mahlert et al., 2006). This view is supported by the observation that overexpression of Cdc24 induces filamentous growth in U. maydis (Castillo-Lluva et al., 2007) (Supplemental Figure 1A). Here we observed that induced expression of Cdc24 results in significantly increased levels of active Rac1-GTP but not Cdc42-GTP (Figure 1C). These results indicate that the b mating type–dependent morphogenetic transition involves stimulation of Rac1, most presumably by Cdc24.
Dimorphic switching is accompanied by down-regulation of Cdc24
To study the role of the GEF Cdc24 during dimorphic switching in more detail, we expressed a C-terminal Cdc24–green fluorescent protein (GFP) fusion protein under control of the constitutive tef promoter in AB31 cells. Before induction of the b mating type genes, Cdc24-GFP was evenly distributed in the cytoplasm and showed some accumulation in nuclei and at the bud tip (Figure 2A). Four hours after induction of the bE1/bW2 heterodimer, cells grow as filaments and Cdc24 accumulated at the growing tip (Figure 2A). Much to our surprise, the intensity of Cdc24-GFP fluorescence decreased dramatically upon prolonged induction (8 h) of the b mating type genes (Figure 2A). The mRNA level of Cdc24-GFP was not affected under these conditions (Figure 2B). Western analysis using anti-GFP antibodies confirmed that the level of Cdc24-GFP protein was strongly reduced upon induction of bE1/bW2 (Figure 2C). A time course experiment revealed that degradation of Cdc24 started within 2 h after induction of bE1/bW2 expression (Figure 2D). To rule out that the constitutive expression of Cdc24-GFP was responsible for the observed down-regulation of Cdc24, we tagged the genomic copy of Cdc24 at its C-terminus with a 3xHA epitope. It is reassuring that, also in these strains, significant degradation of Cdc24 was observed during mating type induced–filament formation (Figure 2E). Together, these data suggest that mating type–dependent dimorphic switching results in significant destabilization of Cdc24 protein.
Because induction of the bW2/bE1 heterodimer enhances the level of active Rac1 (Figure 1C), we determined whether degradation of Cdc24 depends on Rac1. If the rac1 gene is deleted in strain AB31Δrac1, down-regulation of Cdc24 upon expression of bE1/bW2 is completely abolished (Figure 2, A, C, and D). Of interest, Cdc24 was stabilized in the absence of Rac1 also in uninduced budding cells (Figure 2, A, C, and E). Furthermore, overexpression of Rac1 was sufficient to trigger destruction of Cdc24-3xHA (Figure 2E), whereas neither depletion nor overexpression of Cdc42 affected the protein level of Cdc24 (Figure 2E). This confirms our previous hypothesis that Cdc24 acts as Rac1-specific GEF and suggests that the Rac1-dependent destabilization of Cdc24 constitutes a negative feedback loop. Apparently, this feedback mechanism operates not only during filamentous growth, but also during budding.
The assumption that activation of Rac1 is required to trigger down-regulation of Cdc24 was corroborated by our observation that the nonactivatable variant Rac1T17N fails to destabilize Cdc24 (Figure 3A). In addition, a mutant version of Cdc24 (Cdc24T285A) that has no GEF activity (Aghazadeh et al., 1998) was significantly more stable than wild-type Cdc24 (Figure 3B). Surprisingly, expression of constitutive active Rac1Q61L did not result in degradation of Cdc24 (Figure 3A). Instead, we even observed significant stabilization of endogenously expressed Cdc24-3xHA (Figure 3A). Because overexpression of Rac1Q61L is deleterious in U. maydis (Mahlert et al., 2006), stabilization of Cdc24 might result from toxic side effects. This hypothesis could be ruled out since wild-type Rac1 was still able to trigger Cdc24 destruction even in the presence of Rac1Q61L (Supplemental Figure 1G). Thus it is not just the active form of Rac1 that destabilizes Cdc24. Obviously, negative feedback regulation of Cdc24 requires that Rac1 is able to cycle between its inactive, GDP-bound form and its active, GTP-bound form.
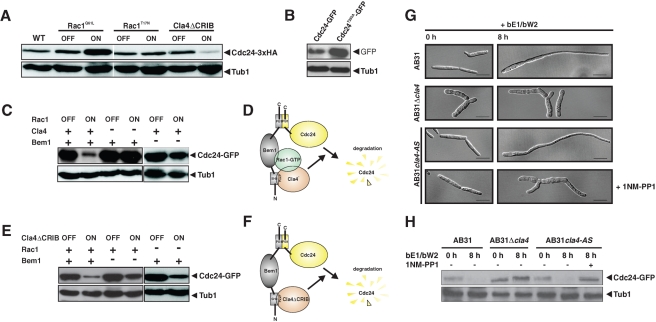
FIGURE 3:
Negative feedback regulation of Cdc24 depends on a functional Bem1-scaffolded signaling module. (A) The protein level of Cdc24-3xHA expressed under control of its endogenous promoter was determined in cells that overexpress constitutive active Rac1Q61L, dominant negative Rac1T17N, and constitutive active Cla4ΔCRIB kinase. Cells were grown in noninducing medium, and proteins were isolated before induction (OFF) and 8 h after induction (ON). (B) The protein level of constitutively expressed mutant Cdc24T285A-GFP was determined in comparison with the level of wild-type Cdc24-GFP expressed under the same conditions. (C) The effect of Rac1 overexpression on the protein level of Cdc24-GFP was determined in the presence (+) or absence (−) of either the Rac1 effector Cla4 or the scaffolding protein Bem1. Rac1 was ectopically expressed under control of the nitrate-inducible nar1-promoter in wild type, in Δcla4 mutants, and in a conditional bem1crg mutant strain that expresses the bem1 gene under control of the arabinose-inducible crg promoter. Cells were grown in liquid minimal medium containing glucose (−) or arabinose (+) as carbon source and ammonium (OFF) or nitrate (ON) as nitrogen source. (D) Rac1-GTP needs both the scaffold Bem1 and the effector Cla4 to target Cdc24 for degradation. (E) The protein level of Cdc24-GFP was determined before (OFF) and after induction of an ectopic copy of constitutive active Cla4ΔCRIB kinase in the wild type, in the rac1 deletion mutant, and in the conditional bem1 mutant. Growth conditions were the same as described in C. (F) Constitutively active Cla4ΔCRIB needs the bridging function of Bem1 to induce degradation of Cdc24. (G) Phenotypes of AB31, AB31Δcla4, and AB31cla4-AS before (0 h) and after induction of bE1/bW2 overexpression (8 h). Strain AB31cla4-AS expresses the analogue-sensitive mutant cla4M629A, which can be inhibited by 1NM-PP1. Scale bars, 10 μm. (H) Determination of endogenous Cdc24-3x-HA levels in the cells shown in G.
Feedback regulation of Cdc24 occurs within a ternary complex
Rho GEFs are often part of larger, multiprotein complexes (Marinissen and Gutkind, 2005; Garcia-Mata and Burridge, 2007). In the yeast Saccharomyces cerevisiae, Cdc24p interacts with the scaffold protein Bem1p, which also binds the GTPase Cdc42p and its downstream effector Cla4p (Gulli and Peter, 2001; Kozubowski et al., 2008). Homologues of both the p21-activated kinase Cla4p and the scaffold protein Bem1p exist in U. maydis (Leveleki et al., 2004; Alvarez-Tabares and Perez-Martin, 2008). To test for ternary complex formation, we performed pull-down assays using GST-Rac1 fusion proteins (Supplemental Figure 3C). Purified GST-Rac1 was preloaded with either GDP or nonhydrolyzable GTPγS and incubated with cell extracts from U. maydis strains overexpressing Cdc24-GFP, Cla4-GFP, or GFP-Bem1 either singly or in all possible pairwise and triple combinations. Rac1-GDP showed high affinity only for Cdc24 and for neither Bem1 nor Cla4 (Supplemental Figure 3D). In contrast, active Rac1-GTPγS was unable to precipitate Cdc24 but showed strong interactions with Cla4 and Bem1 (Supplemental Figure 3D). Further coprecipitation experiments revealed that Bem1 interacts directly with Cdc24 and Cla4 (Supplemental Figure 3E). Interaction between Cdc24 and Cla4 was observed only in the presence of Bem1 (Supplemental Figure 3, E–G), indicating that Bem1 acts as scaffold protein for Cdc24, Rac1, and Cla4.
To follow ternary complex formation in vivo, GFP fusion proteins of Cdc24, Cla4, and Bem1 were expressed in either wild-type cells or in cells depleted for either one of these proteins. In wild-type cells, all fusion proteins accumulated at the growing tips of filaments and buds (Supplemental Figure 2). Tip localization of Cdc24 and Cla4 required the presence of the scaffold protein Bem1 (Supplemental Figure 2), demonstrating that Bem1 is critical to recruiting Cdc24 and Cla4 to the growing tip.
Next we asked whether complex formation is required for the negative regulation of Cdc24. To avoid any side effects by other b-regulated genes (Scherer et al., 2006), we used Rac1-overexpression–induced destabilization of Cdc24-GFP as readout in the following experiments. First, we tested whether Cla4 kinase is required for negative autoregulation of Cdc24. Destruction of Cdc24 upon overexpression of Rac1 was abolished in Δcla4 mutants (Figure 3C) and in cells depleted for Bem1 (Figure 3C). Interaction between Cdc24 and Bem1 is mediated by their C-terminal PB1 domains (Supplemental Figure 3F; Bose et al., 2001). If we deleted the PB1 domain of Cdc24, the resulting protein Cdc24ΔPB1 became resistant to Rac1-induced destabilization (Figure 4C). These data strongly suggest that negative feedback regulation of Cdc24 requires ternary complex formation.
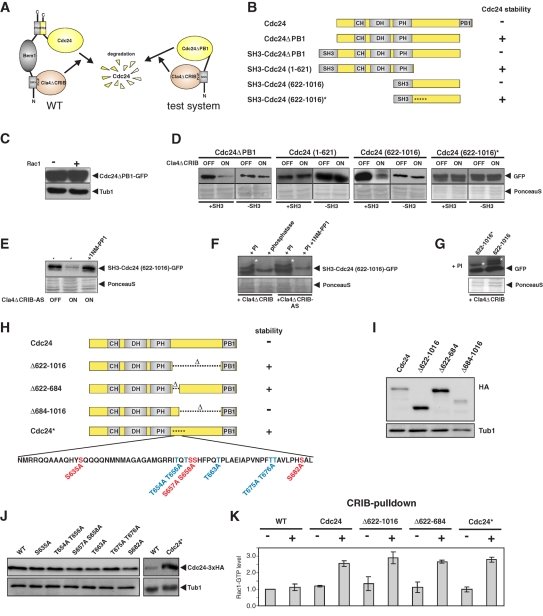
FIGURE 4:
Characterization of the region within Cdc24 that confers Cla4-dependent destabilization. (A) Schematic representation of the test system for Cdc24 fragments that are important for Cla4-induced Cdc24 destabilization. Bem1 normally interacts with Cdc24 via its PB1 domain and with Cla4 via an SH3 domain. The “bridging” function of Bem1 is mimicked by direct fusion of the SH3 domain of Bem1 to the Cdc24 fragments. (B) Schematic overview of the tested Cdc24 fragments. (C) Protein stability of ectopically expressed mutant Cdc24ΔPB1-GFP was determined in the presence (+) and absence (−) of Rac1. (D) Western blot analysis of C-terminal GFP fusions of the indicated Cdc24 fragments. Cdc24 fragments were expressed in Δrac1 mutant cells as N-terminal fusions to the SH3 domain of Bem1 (+SH3). Cdc24 fragments without the SH3 domain (−SH3) served as control. Probes were taken before (OFF) and after 4 h of Cla4ΔCRIB overexpression (ON). (E) Determination of the protein level of fragment SH3-Cdc24(622-1016)-GFP before (OFF) and 6 h after induction of the overexpression of the analogue-sensitive variant of Cla4ΔCRIB (Cla4ΔCRIB-AS), which carries the M629A mutation. Cultures were grown in the presence and absence of the inhibitor 1NM-PP1. (F) Detection of mobility shifts in SH3-Cdc24(622-1016)-GFP caused by expression of Cla4ΔCRIB or Cla4ΔCRIB-AS. Protein extracts of kinase-overproducing cultures were prepared in the presence of phosphatase inhibitors (PIs) or after addition of ALP. Cla4ΔCRIB-AS was expressed in the presence and absence of the inhibitor 1NM-PP1. Asterisks mark phosphorylated SH3-Cdc24(622-1016)-GFP. (G) Wild-type SH3-Cdc24(622-1016)-GFP and the stabilized protein SH3-Cdc24(622-1016)*-GFP were prepared in the presence of PIs shortly after induction of Cla4ΔCRIB overexpression. Mobility shifts are indicated by asterisks. (H) Schematic representation of internal Cdc24 deletions that were introduced at the genomic locus. In Cdc24* all serine (S) and threonine (T) residues of the indicated domain were exchanged for alanine (I). Protein levels of the indicated Cdc24 mutant proteins. For visualization, the proteins were C-terminally fused to a 3xHA tag. (J) Western blot analysis of the protein levels of Cdc24 mutants carrying the indicated mutations. (K) Quantification of active Rac1-GTP before (−) and after (+) 4 h of the overexpression of the indicated Cdc24 variants. The diagram shows the mean values of three independent determinations. SDs are indicated.
Cla4-dependent phosphorylation targets Cdc24 for destruction
The Rac1 downstream effector Cla4 kinase is required for filament formation, but overexpression of Cla4 is not sufficient to induce filamentous growth (Supplemental Figure 1) and had only a very minor effect on the protein level of Cdc24 (unpublished data). In contrast, expression of a mutant version of Cla4 that lacks the inhibitory CRIB domain (Cla4ΔCRIB; Leveleki et al., 2004) triggered destruction of Cdc24 (Figure 3E and Supplemental Figure 1A). Cla4ΔCRIB-induced degradation of Cdc24 was independent of Rac1 but still required the presence of the scaffold protein Bem1 (Figure 3E). These data strongly suggest that Cdc24 degradation is induced by active Cla4 kinase tethered to Cdc24 via Bem1 (Figure 3F).
To prove that Cla4 kinase activity is critical for degradation of Cdc24, we generated the analogue-sensitive variant Cla4M629A, which can be specifically inhibited by 4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1NM-PP1; Bishop et al., 1998; Liu et al., 1998; Weiss et al., 2000). If treated with 1NM-PP1, cells displayed a phenotype similar to that of Δcla4 mutants (Figure 3G; Leveleki et al., 2004). Inhibition of Cla4 kinase activity interfered with both b-induced filament formation and destabilization of endogenous Cdc24-3xHA (Figure 3, G and H). This indicates that the kinase activity of Cla4 is necessary to trigger Cdc24 degradation, most probably by direct phosphorylation.
To find out whether Cdc24 is a substrate for Cla4-dependent phosphorylation, we designed a test system. We reasoned that direct interaction between Cdc24 and active Cla4ΔCRIB should result in degradation of Cdc24, independent of Bem1 and Rac1 (Figure 4A). To this end, we fused the Cla4-interacting SH3 domain of Bem1 to C-terminally truncated Cdc24ΔPB1. Because Cdc24ΔPB1 is resistant to Cla4-dependent degradation (Figure 4C), we tested whether fusion of the SH3 domain restores Cla4-dependent degradation. Indeed, expression of Cla4ΔCRIB resulted in degradation of the SH3–Cdc24ΔPB1 fusion protein. This indicates that the bridging function of Bem1 during feedback regulation can be mimicked by direct interaction between Cla4 and SH3–Cdc24ΔPB1. We used this assay to delimit the region within Cdc24 that is sufficient to target Cdc24 for destruction and tested several fragments of Cdc24 for Cla4-induced destabilization (Figure 4B). As control, we expressed the respective Cdc24 fragments also without SH3 domain. These experiments revealed that a region comprising 394 amino acids between the pleckstrin homology domain and the C-terminal PB1 domain was sufficient to trigger Cla4-dependent degradation (Figure 4D). If analogue-sensitive Cla4ΔCRIBM629A was used, degradation could be inhibited by addition of 1NM-PP1 (Figure 4E). This confirms that kinase activity is essential for destabilization and suggests that Cdc24 is a direct substrate of Cla4 kinase.
To detect Cla4ΔCRIB-dependent phosphorylation of the 394–amino acid Cdc24 fragment, protein extracts were prepared shortly after induction of Cla4∆CRIB and in the presence of phosphatase inhibitors. When proteins were separated by SDS–gel electrophoresis, less mobile variants of the Cdc24 peptide were detected (Figure 4F). We confirmed that the observed mobility shift results from phosphorylation by treatment with alkaline phosphatase (ALP) (Figure 4F). If the analogue-sensitive variant Cla4ΔCRIBM629A was used for phosphorylation of the Cdc24 fragment, the mobility shift was observed only in the absence of inhibitor (Figure 4F). Together these data clearly indicate that phosphorylation of Cdc24 depends on Cla4 kinase activity.
Expression of a stabilized version of Cdc24 affects polarized growth
Next we addressed the biological function of negative autoregulation of Cdc24. We constructed a strain that expresses a mutant version of Cdc24 that lacks the region that was shown to be sufficient to trigger degradation (Figure 4H). The resulting protein, Cdc24Δ622-1016, displayed significantly enhanced stability compared with wild-type Cdc24 (Figure 4I). We were able to delimit the domain that is necessary to confer instability on a short region immediately adjacent to the PH domain (Figure 4, H and I). This domain consists of 63 amino acids and contains a total of nine serine or threonine residues, which might be phosphorylated by Cla4. We replaced these potential phosphorylation targets by alanine either singly or in groups of two. In neither of these mutants was the stability of the resulting protein significantly affected (Figure 4J). However, exchange of all serine and threonine residues within this region resulted in significant stabilization of the mutant protein Cdc24* (Figure 4J). When a fragment carrying these amino acid exchanges was assayed in our test system, we also observed significant stabilization (Figure 4, B and D). We noted that the stabilized fragment SH3–Cdc24(622-1016)* is still a substrate for Cla4 because it still displays a weak mobility shift in the SDS gel (Figure 4D). Apparently, this residual phosphorylation does not affect protein stability (Figure 4G). To exclude that the observed stabilization of mutant Cdc24* might result from loss of GEF activity, we tested whether Cdc24* was still able to activate Rac1. Significant increase of active Rac1-GTP was observed upon expression of Cdc24* indicating that GEF activity is not affected in this mutant (Figure 4K and Supplemental Figure 4).
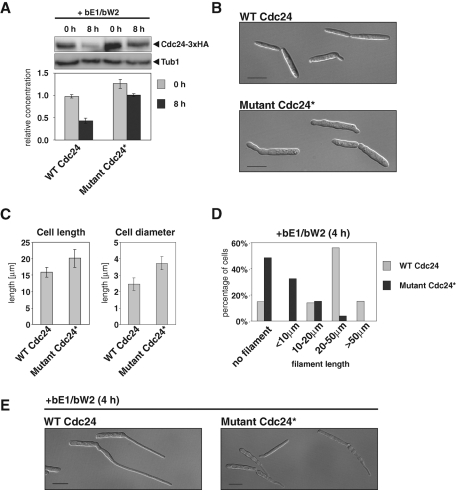
To determine whether removal of potential Cla4 phosphorylation sites affects the stability of Cdc24 during b-dependent dimorphic switching, we followed the level of endogenously expressed mutant Cdc24* protein in strain AB31. On expression of active bE1/bW2 heterodimer, the level of wild-type Cdc24 protein decreased by more than 50%, whereas mutant Cdc24* was only slightly affected (Figure 5A). Next we determined whether stabilization of Cdc24 interferes with its biological function. Cells expressing Cdc24* were significantly wider and longer (Figure 5, B and C). Remarkably, cells carrying Cdc24* were also affected in dimorphic switching. Both number and length of filaments were reduced in Cdc24* mutant cells (Figure 5, D and E). These data highlight the importance of negative autoregulation by Cla4-dependent degradation for the function of Cdc24 during both budding and hyphal tip growth.
FIGURE 5:
Expression of Cla4-resistant Cdc24* interferes with induction of filamentous growth. (A) The relative concentration of Cdc24-3xHA and Cdc24*-3xHA in which all serines and threonines within the critical region for destabilization were mutated into alanines was determined before (0 h) and after 8 h of bE1/bW2 overexpression. Shown are the mean values of three independent experiments. SDs are reflected by the bars. (B) Phenotype of U. maydis cells expressing either wild-type Cdc24 or Cla4-resistant Cdc24* at the endogenous locus. Scale bars, 10 μm. (C) Quantification of length and diameter of the cells shown in B (n = 50). (D) Quantification of the percentage of cells without filaments and cells with filaments of indicated length (shorter than 10 μm, between 10 and 20 μm, between 20 and 50 μm, and longer than 50 μm) 4 h after the induction of bE1/bW2 overexpression in strains expressing wild-type Cdc24 or mutated Cdc24* at the endogenous locus (n = 100). (E) Representative images of cells from the cultures quantified in D. Pictures were taken 4 h after bE1/bW2 induction. Bars, 10 μm.
DISCUSSION
In this study, we present evidence that polarized growth in U. maydis involves activation of Rac1 by the Rho-GEF Cdc24. We show that Cdc24 is part of a multiprotein complex that comprises the scaffold protein Bem1, the small GTPase Rac1, and the protein kinase Cla4. Activation of Rac1 by Cdc24 stimulates Cla4 kinase activity, which then triggers destruction of Cdc24. We identified a region within Cdc24 that confers Cla4-dependent destabilization. Expression of a Cla4-resistant variant of Cdc24 severely affected filament formation, demonstrating that negative feedback regulation of Cdc24 plays an important role during polarized growth.
We observed that in U. maydis the Rac1-specific GEF Cdc24 is subject to posttranslational control during both budding and filamentous growth. During both biological processes Cdc24 is destabilized by Cla4-dependent phosphorylation. Phosphorylation of Cdc24p by Cla4p also has been observed in budding yeast, but the function of this modification is a matter of debate (Gulli et al., 2000; Bose et al., 2001; Irazoqui et al., 2003). It was reported that Cla4p-dependent phosphorylation of Cdc24p triggers release from Bem1p and thus may be part of a negative feedback loop (Gulli et al., 2000). However, this view was challenged by conflicting data that suggested that phosphorylation of Cdc24p had no effects on Bem1p interaction but even enhanced Cdc24p GEF activity (Bose et al., 2001; Kozubowski et al., 2008). To distinguish between these competing models, phosphorylation sites have been mapped in yeast Cdc24p by mass spectrometry. Surprisingly, however, even removal of all mapped phosphorylation sites affected neither Cdc24p stability nor function (Wai et al., 2009).
Within U. maydis Cdc24 we could identify a short region of 63 amino acid residues that is both necessary and sufficient to trigger Cla4-dependent destruction. Mutation of all serine and threonine residues within this region abolished Cla4-dependent destabilization but did not affect the GEF activity. Comparison with the yeast Cdc24p sequence revealed that none of these putative phosphorylation sites is conserved between both organisms. Because exchange of single phosphorylation sites did not altered protein stability, we assume that phosphorylation of multiple residues within this region is required to trigger Cdc24 degradation. Multisite phosphorylation is a widespread phenomenon in protein regulation (Salazar and Hofer, 2009). In yeast, degradation of the CDK inhibitor Sic1p and the filamentous growth regulator Tec1p also requires multiple phosphorylation events (Nash et al., 2001; Bao et al., 2010). It has been shown that multisite phosphorylation of Sic1 and Tec1 triggers their recognition by the F-box adaptor protein Cdc4 of the SCF E3 ubiquitin ligase complex (Deshaies and Joazeiro, 2009). SCF-dependent degradation has also been shown for the mammalian Cdc42 GEFs FGD1 and FGD3 (Hayakawa et al., 2005, 2008). Thus we speculate that in U. maydis phosphorylated Cdc24 is recognized by a yet-uncharacterized ubiquitin ligase that targets Cdc24 for ubiquitin dependent degradation.
One of the most challenging aspects of cell polarization is to concentrate growth activity at a single, defined spot. In yeast, bud sites are normally formed at landmarks provided by the bud site selection machinery (Casamayor and Snyder, 2002). Of interest, cells defective in bud site selection nevertheless are able to form a single bud at a random position. This phenomenon has been termed “symmetry breaking” and has been used extensively to address the problem of how a gradient of active GTPase can be established and maintained (Wedlich-Söldner and Li, 2003). Two different models were proposed to explain spontaneous symmetry breaking. In the first model, active Cdc42p-GTP at the cell cortex promotes actin-driven transport of Cdc42p-containing vesicles, resulting in enhanced accumulation of Cdc42p. The growing cluster of Cdc42p attracts more actin cables, and this dynamic feedback loop results in stable and robust cell polarization (Wedlich-Söldner et al., 2003, 2004). This hypothesis has been challenged by mathematical modeling. Simulation of vesicle dynamics predicts that actin-directed traffic would perturb rather than reinforce polarization (Layton et al., 2011). As an alternative model, it was postulated that active Cdc42-GTP recruits cytoplasmic Cla4p–Bem1p–Cdc24p complexes to the membrane (Irazoqui et al., 2003). The membrane-associated complex activates additional Cdc42p molecules, resulting in highly focused concentration of active Cdc42p-GTP (Irazoqui et al., 2003; Goryachev and Pokhilko, 2008; Kozubowski et al., 2008). The growing foci of Cdc42p activity compete for the limiting pool of Cla4p–Bem1p–Cdc24 complexes, thus allowing only one of these foci to develop into a bud (Howell et al., 2009).
In U. maydis polarization depends strictly on formation of a ternary Cla4–Bem1–Rac1 complex. We assume that several mechanisms cooperate to generate a steep gradient of active Bem1–Rac1–Cla4 at the tip of growing buds and hyphae. We assume that the GEF Cdc24 interacts with Bem1–Cla4 and recruits active Rac1 into the transient Cdc24–Bem1–Rac1–Cla4 complex (for a model see Figure 6). The complex is anchored at the plasma membrane via interaction with the membrane-bound Rac1. Active Rac1-GTP interacts with and presumably stimulates its effector Cla4, which then triggers polarized growth by activation of yet-unknown downstream effector proteins. We could show that Cla4 concomitantly phosphorylates Cdc24, which is then removed from the complex and degraded. This regulatory scheme generates a constant flow of “active,” membrane-bound Bem1–Rac1–Cla4 complex, which then diffuses in lateral directions. We assume that activity of this complex is spatially restricted to the growing tip region by a subapical “zone of inhibition” presumably formed by Rac1 GAP proteins. A comparable zone of GAP activity was observed in budding yeast, where it prevents bud initiation within the previous bud site (Tong et al., 2007; Figure 6). We assume that destabilization of Cdc24 is required to prevent immediate reactivation of Rac1 and thus plays an important role in our model of polarized growth. We provide evidence that both inactivation of Rac1 and destabilization of Cdc24 are critical for polarized growth during budding and filament formation. First, mutants that express stabilized Cdc24* are characterized by wider bud tips and reduced filament formation. Second, expression of a constitutively active Rac1Q61L resulted in strong isotropic growth (Mahlert et al., 2006) and at the same time stabilized Cdc24. We assume that in both cases Rac1 activity is not sufficiently focused to the tip: in one case, Cdc24* is not removed from the ternary complex and therefore not able to reactivate Rac1 immediately; in the other case, it is the insensitivity of Rac1Q61L toward GAP inactivation that allows the GTPase to pass the supposed zone of inhibition. That overexpression of Rac1Q61L resulted in significant stabilization of Cdc24 appears unanticipated on first glance but can be reconciled with our model. Because Bem1 is continuously recycled during polarized growth, the observed stabilization of Cdc24 most likely results from sequestration of Bem1 in the Rac1Q61L–Bem1–Cla4 complex, which is insensitive to GAP inactivation. This was supported by the observation that overexpression of Bem1 suppresses the stabilizing effect of Rac1Q61L (Supplemental Figure 1I).
FIGURE 6:
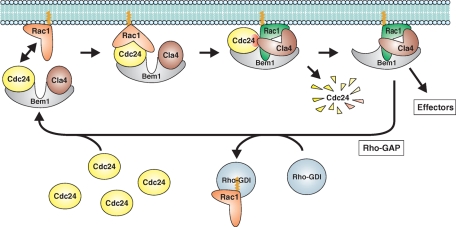
Schematic model of the negative feedback loop that triggers destruction of the Rac1-specifc Rho-GEF Cdc24. Cdc24 and Cla4 are assumed to be associated with the scaffold protein Bem1. Inactive GDP-loaded Rac1 (orange) is recruited into this complex by Cdc24. Within this transient quaternary complex Cdc24 activates Rac1 by stimulating GDP/GTP cycling. In its GTP-bound form, Rac1 (green) interacts with Cla4 and induces a conformational change in Cla4 (open shape). This relieves autoinhibition of the kinase domain and results in phosphorylation of Cdc24 (red dot). Phosphorylated Cdc24 is rapidly destroyed, most probably by proteasomal degradation. This results in the release of an active Bem1–Rac1–Cla4 complex, which triggers polar growth by interaction with yet-unknown effectors. Recycling of Bem1 and Rac1 is accomplished by interaction with Rac1-specific Rho-GAP and Rho-GDI proteins.
During the life cycle of U. maydis filament formation is controlled by the b mating type locus. We observed that induced expression of the active bW2/bE1 transcription factor results in a significant increase in the levels of active Rac1-GTP. There is no indication that expression of Rac1 or Cdc24 is significantly enhanced in the presence of the active bW2/bE1 transcription factor. This implies that b mating type induction either stimulates the activity of the Rac1-specific GEF Cdc24 or promotes its relocalization to the tip. In budding cells significant nuclear localization of Cdc24 is observed, which decreases after b induction. This may indicate that active bW2/bE1 transcription factor triggers translocation of Cdc24 rather than activation. A similar nucleocytoplasmic shuttling was described for budding yeast, where Cdc24p is sequestered in the nucleus by interaction with Far1 (Nern and Arkowitz, 1999; Toenjes et al., 1999; Shimada et al., 2000). Cell cycle–dependent release of Cdc24p is triggered by Cdc28-dependent degradation of Far1 (Nern and Arkowitz, 2000). Because U. maydis does not contain an obvious homologue of Far1, this mechanism of nucleocytoplasmic shuttling appears to be not conserved. Therefore we assume that if comparable nucleocytoplasmic shuttling of Cdc24 occurs in U. maydis, it must operate by a different mechanism.
Negative feedback regulation of Cdc24 appears to be critical for induction of filamentous growth during dimorphic switching in the phytopathogenic fungus U. maydis. Similar morphogenetic transitions are features of many plant and human pathogenic fungi. Thus it will be interesting to elucidate whether in other fungal systems similar regulatory feedback loops operate during hyphal tip growth.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions
The haploid U. maydis strains Bub8 and AB31 were used (Schulz et al., 1990; Brachmann et al., 2001). Cells were grown as described (Leveleki et al., 2004; Mahlert et al., 2006). Deletion mutants for rac1, cdc42, and cla4 were generated by replacing the open reading frame with a hygromycin or nourseothricin resistance cassette using the method described in Brachmann et al. (2004). Conditional mutants of rac1, cdc42, and bem1 were obtained by replacing the endogenous promoter with the arabinose-inducible crg promoter (Bottin et al., 1996). To generate a strain expressing the Cdc24-3xHA fusion protein, the epitope sequence was C-terminally fused to the cdc24 open reading frame. Strain AB31/3xHA-Rac1 was obtained by replacing the wild-type rac1 allele by 3xHA-rac1, which codes for an N-terminally extended version, followed by the hygromycin resistance cassette. For transformation the construct was flanked by 1000 base pairs upstream and downstream of the rac1 open reading frame. Correct integration of all constructs was verified by Southern analysis. For overexpression of Rac1, Cdc42, Cla4, Cdc24, and Bem1, the open reading frames were cloned into the U. maydis vectors pRU2 and pRU11 (Brachmann et al., 2001). Single amino acid substitutions (Rac1Q61L, Rac1T17N, Cdc24S635A, Cdc24T654AT656A, Cdc24S657AS658A, Cdc24T663A, Cdc24T675AT676A, Cdc24S682A) were introduced into the open reading frames by PCR-directed mutagenesis and verified by DNA sequencing. GFP fusions were constructed in plasmid p123, which carries the constitutive otef promoter (Spellig et al., 1996). For RFP and cMyc fusions derivatives of p123 were used in which GFP was replaced by the respective epitopes. All constructs were targeted to the cbx locus by homologous recombination, and single-copy integration was verified by PCR (Brachmann et al., 2001). cdc24-GFP under control of the otef promoter was integrated into the nar1 locus by homologous recombination. Correct integration was verified by Southern analysis. Cla4-RFP was expressed from a plasmid that was randomly integrated into the genome. Construction of plasmids was performed using standard procedures (Sambrook and Russell, 2001). Detailed cloning procedures can be requested from the authors.
Microscopy and staining
U. maydis cells from logarithmically growing cultures were visualized by differential interference contrast (DIC) and epifluorescence microscopy using a Zeiss (Jena, Germany) Axiovert 200 microscope. Images were taken using a cooled CCD camera (Hamamatsu Orca-ER, Hamamatsu, Japan) with an exposure time of 100–1000 ms. Image acquisition was performed using Volocity software (Improvision, PerkinElmer, Waltham, MA). Image analysis and processing was performed using ImageJ (National Institutes of Health, Bethesda, MD; Abramoff et al., 2004).
Protein expression and purification
The DH-PH domain of Cdc24 (Cdc24DH-PH) and full-length Rho-family GTPases were expressed in Escherichia coli as glutathione-S-transferase fusion proteins using the pGEX vector system (GE Healthcare, Piscataway, NJ) or as NusA-6His fusion proteins using pETM-60 (Dümmler et al., 2005). Purification was performed in accordance with published procedures (Self and Hall, 1995). Cells were resuspended in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, and 5 mM dithiothreitol (DTT) or 2.5 mM 2-mercaptoethanol, supplemented with protease inhibitors (Roche, Mannheim, Germany), and disrupted using a French press. After centrifugation, the clarified supernatant was loaded on glutathione agarose (Macherey-Nagel, Düren, Germany) or Ni-NTA agarose (Qiagen, Valencia, CA), respectively. Proteins bound to glutathione beads were washed with lysis buffer and eluted with 10 mM reduced glutathione at pH 8.8. Ni-NTA–bound proteins were washed with lysis buffer containing 50 mM imidazole and eluted in the presence of 1 M imidazole. Proteins were stored at –80°C.
Pull-down assays
Purified GTPases were bound to glutathione agarose (Macherey-Nagel) or Ni-NTA agarose (Qiagen). For nucleotide loading, bound GTPases were incubated in lysis buffer containing 20 mM EDTA and either 10 mM GDP or 1 mM GTPγS for 1 h at 4°C. The reaction was stopped by adding MgCl2 to a final concentration of 20 mM. Nucleotide-loaded GTPases were washed twice with lysis buffer containing 10 mM MgCl2. For preparation of whole-cell extracts, U. maydis cells were resuspended in lysis buffer containing protease inhibitor cocktail (Sigma), 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, and 5 mM DTT or 2.5 mM 2-mercaptoethanol and disrupted with glass beads. A total of 10 μl of matrix bound GTPases was added to 100 μl of cleared extract and incubated for 1 h at 4°C. To maintain the loading status, 10 mM GDP or 1 mM GTPγS was added. Beads were washed five times with lysis buffer and loaded on a SDS–polyacrylamide gel, and proteins were visualized by Western blotting. For CRIB pull-down assays, the glutathione agarose was loaded with purified GST-CRIB protein. Extracts from U. maydis strains overexpressing either the bE1/bW2 heterodimeric transcription factor or the various Cdc24 versions were prepared as described earlier, but the lysis buffer was supplemented with 1% (vol/vol) Triton X-100. Cleared extracts with equalized protein concentrations were incubated with equal amounts of GST-CRIB–loaded beads for 30 min at 4°C. Beads were washed three times with lysis buffer, and proteins were visualized by Western blotting.
Supplementary Material
Acknowledgments
This work was supported by the Loewe Center for Synthetic Microbiology, the International Max Planck Research School for Environmental and Cellular Microbiology Marburg, and the Deutsche Forschungsgemeinschaft Research Unit 1334.
Abbreviations used:
- 1NM-PP1
4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine
- ALP
alkaline phosphatase
- CRIB
Cdc42 and Rac1 interaction and binding
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GSH
glutathione SH
- GST
glutathione S transferase
- PAK
p21 activated kinase
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0314) on July 14, 2011.
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Aghazadeh B, Zhu K, Kubiseski TJ, Liu GA, Pawson T, Zheng Y, Rosen MK. Structure and mutagenesis of the Dbl homology domain. Nat Struct Biol. 1998;5:1098–1107. doi: 10.1038/4209. [DOI] [PubMed] [Google Scholar]
- Alvarez-Tabares I, Perez-Martin J. Cdk5 kinase regulates the association between adaptor protein Bem1 and GEF Cdc24 in the fungus Ustilago maydis. J Cell Sci. 2008;121:2824–2832. doi: 10.1242/jcs.026286. [DOI] [PubMed] [Google Scholar]
- Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- Bao MZ, Shock TR, Madhani HD. Multisite phosphorylation of the Saccharomyces cerevisiae filamentous growth regulator Tec1 is required for its recognition by the E3 ubiquitin ligase adaptor Cdc4 and its subsequent destruction in vivo. Eukaryot Cell. 2010;9:31–36. doi: 10.1128/EC.00250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- Bölker M. Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology. 2001;147:1395–1401. doi: 10.1099/00221287-147-6-1395. [DOI] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Bottin A, Kämper J, Kahmann R. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol Gen Genet. 1996;253:342–352. doi: 10.1007/pl00008601. [DOI] [PubMed] [Google Scholar]
- Brachmann A, König J, Julius C, Feldbrügge M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics. 2004;272:216–226. doi: 10.1007/s00438-004-1047-z. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Weinzierl G, Kämper J, Kahmann R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol. 2001;42:1047–1063. doi: 10.1046/j.1365-2958.2001.02699.x. [DOI] [PubMed] [Google Scholar]
- Brefort T, Doehlemann G, Mendoza-Mendoza A, Reissmann S, Djamei A, Kahmann R. Ustilago maydis as a pathogen. Annu Rev Phytopathol. 2009;47:423–445. doi: 10.1146/annurev-phyto-080508-081923. [DOI] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva S, Alvarez-Tabares I, Weber I, Steinberg G, Perez-Martin J. Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J Cell Sci. 2007;120:1584–1595. doi: 10.1242/jcs.005314. [DOI] [PubMed] [Google Scholar]
- Cole KC, Barbour JE, Midkiff JF, Marble BM, Johnson DI. Multiple proteins and phosphorylations regulate Saccharomyces cerevisiae Cdc24p localization. FEBS Lett. 2009;583:3339–3343. doi: 10.1016/j.febslet.2009.09.033. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dümmler A, Lawrence AM, de Marco A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb Cell Fact. 2005;4:34. doi: 10.1186/1475-2859-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42–the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Burridge K. Catching a GEF by its tail. Trends Cell Biol. 2007;17:36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Gulli M, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M. Phosphorylation of the cdc42 exchange factor cdc24 by the PAK-like kinase cla4 may regulate polarized growth in yeast. Mol Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Gulli MP, Peter M. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 2001;15:365–379. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Kitagawa H, Miyazawa K, Kitagawa M, Kikugawa K. The FWD1/beta-TrCP-mediated degradation pathway establishes a “turning off switch” of a Cdc42 guanine nucleotide exchange factor, FGD1. Genes Cells. 2005;10:241–251. doi: 10.1111/j.1365-2443.2005.00834.x. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Matsushima M, Hagiwara H, Oshima T, Fujino T, Ando K, Kikugawa K, Tanaka H, Miyazawa K, Kitagawa M. Novel insights into FGD3, a putative GEF for Cdc42, that undergoes SCF(FWD1/beta-TrCP)-mediated proteasomal degradation analogous to that of its homologue FGD1 but regulates cell morphology and motility differently from FGD1. Genes Cells. 2008;13:329–342. doi: 10.1111/j.1365-2443.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- Heimel K, Scherer M, Vranes M, Wahl R, Pothiratana C, Schuler D, Vincon V, Finkernagel F, Flor-Parra I, Kämper J. The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog. 2010;6:e1001035. doi: 10.1371/journal.ppat.1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J, Reichmann M, Romeis T, Bölker M, Kahmann R. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveleki L, Mahlert M, Sandrock B, Bölker M. The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol Microbiol. 2004;54:396–406. doi: 10.1111/j.1365-2958.2004.04296.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shah K, Yang F, Witucki L, Shokat KM. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bölker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2006;59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- Marinissen MJ, Gutkind JS. Scaffold proteins dictate Rho GTPase-signaling specificity. Trends Biochem Sci. 2005;30:423–426. doi: 10.1016/j.tibs.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Mionnet C, Bogliolo S, Arkowitz RA. Oligomerization regulates the localization of Cdc24, the Cdc42 activator in Saccharomyces cerevisiae. J Biol Chem. 2008;283:17515–17530. doi: 10.1074/jbc.M800305200. [DOI] [PubMed] [Google Scholar]
- Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J Cell Biol. 1999;144:1187–1202. doi: 10.1083/jcb.144.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J Cell Biol. 2000;148:1115–1122. doi: 10.1083/jcb.148.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho- type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Salazar C, Hofer T. Multisite protein phosphorylation–from molecular mechanisms to kinetic models. FEBS J. 2009;276:3177–3198. doi: 10.1111/j.1742-4658.2009.07027.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Scherer M, Heimel K, Starke V, Kämper J. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell. 2006;18:2388–2401. doi: 10.1105/tpc.106.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 1990;60:295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- Self AJ, Hall A. Purification of recombinant Rho/Rac/G25K from Escherichia coli. Methods Enzymol. 1995;256:3–10. doi: 10.1016/0076-6879(95)56003-3. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Gulli MP, Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat Cell Biol. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Garcia-Pedrajas MD, Hong W, Yu Z, Gold SE, Perlin MH. An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot Cell. 2004;3:180–189. doi: 10.1128/EC.3.1.180-189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T, Bottin A, Kahmann R. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet. 1996;252:503–509. doi: 10.1007/BF02172396. [DOI] [PubMed] [Google Scholar]
- Steinberg G, Perez-Martin J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Toenjes KA, Sawyer MM, Johnson DI. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr Biol. 1999;9:1183–1186. doi: 10.1016/S0960-9822(00)80022-6. [DOI] [PubMed] [Google Scholar]
- Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R, Zahiri A, Kämper J. The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol Microbiol. 2010;75:208–220. doi: 10.1111/j.1365-2958.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- Wai SC, Gerber SA, Li R. Multisite phosphorylation of the guanine nucleotide exchange factor Cdc24 during yeast cell polarization. PLoS One. 2009;4:e6563. doi: 10.1371/journal.pone.0006563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Li R. Spontaneous cell polarization: undermining determinism. Nat Cell Biol. 2003;5:267–270. doi: 10.1038/ncb0403-267. [DOI] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL, Bishop AC, Shokat KM, Drubin DG. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.