Abstract
Monoclonal B cell lymphocytosis (MBL) is a hematologic condition wherein small B cell clones can be detected in the blood of asymptomatic individuals. Most MBL have an immunophenotype similar to chronic lymphocytic leukemia (CLL), and “CLL-like” MBL is a precursor to CLL. We used flow cytometry to identify MBL from unaffected members of CLL kindreds. We identified 101 MBL cases from 622 study subjects; of these, 82 individuals with MBL were further characterized. Ninety-one unique MBL clones were detected: 73 CLL-like MBL (CD5+CD20dimsIgdim), 11 atypical MBL (CD5+CD20+sIg+), and 7 CD5neg MBL (CD5negCD20+sIgneg). Extended immunophenotypic characterization of these MBL subtypes was performed, and significant differences in cell surface expression of CD23, CD49d, CD79b, and FMC-7 were observed among the groups. Markers of risk in CLL such as CD38, ZAP70, and CD49d were infrequently expressed in CLL-like MBL, but were expressed in the majority of atypical MBL. Interphase cytogenetics was performed in 35 MBL cases, and del 13q14 was most common (22/30 CLL-like MBL cases). Gene expression analysis using oligonucleotide arrays was performed on 7 CLL-like MBL, and showed activation of B cell receptor associated pathways. Our findings underscore the diversity of MBL subtypes and further clarify the relationship between MBL and other lymphoproliferative disorders.
Introduction
Monoclonal B cell lymphocytosis (MBL) is an asymptomatic hematologic condition wherein small numbers (< 5.0 × 109 / L) of clonal B cells are detectable in blood (1). Most MBL have an immunophenotype similar to chronic lymphocytic leukemia (CLL): CD5+, CD19+, CD20dim, CD23+ and surface immunoglobulin (sIg)dim (2). Using four color flow cytometry, the population prevalence of MBL was reported to be approximately 3 - 5% among adults over age 50 in the general population (3-5), though recently the population prevalence of MBL was reported to be 12% when highly sensitive eight color flow cytometry was employed (6). MBL is a precursor state for CLL and to the best of our knowledge precedes essentially all cases of CLL (7). Longitudinal studies of subjects in which the majority of individuals were ascertained after identification of absolute lymphocytosis revealed an estimated rate of progression from MBL to CLL of approximately 1% per year (8, 9). These reports showed that the principal risk factor for progression from MBL to CLL is the absolute B lymphocyte count (B-ALC), which is proportional to the size of the abnormal B cell clone (8). Analysis of population-based screening studies of MBL showed that the great majority of MBL clones are quite small: 95% of all CLL-like MBL have a CLL-phenotype cell count < 56 × 106 cells / L. Therefore, the available population and clinical data indicate that the prevalence of MBL far exceeds CLL and predict that most MBL do not progress to CLL.
When identified in an individual with a normal B-ALC, MBL have little potential to progress to clinical disease
Therefore, the biologic differences between CLL and MBL likely define critical pathways required for MBL progression to CLL and thus clarify CLL pathogenesis. For example, investigation of immunoglobulin heavy chain variable region (IGHV) gene usage in high-count MBL clones showed predominant usage of IGHV genes that are common among CLL with a bias toward mutated and clinically favorable B-cell receptors (BCRs) (7, 8). In contrast, our previous investigations of low count MBL showed that MBL are frequently oligoclonal (10), and others have shown that low count MBL utilize an IGHV repertoire that, while restricted, is distinct from CLL (6, 11). Additionally, although larger MBL clones show the typical spectrum of acquired chromosomal defects observed in CLL (8, 9), deletion of 17p13 and 11q22 have not been reported in low count MBL (6, 10). These and other differences suggest that important biologic differences exist between low-count MBL, high-count MBL, and CLL.
Although most MBL have an immunophenotype similar to CLL, other less common phenotypes have also been described (5). In general, three subgroups of MBL have been defined: (1) CLL-like MBL; (2) atypical MBL, in which the immunophenotype is CD5+ and CD19+, but unlike CLL is CD20high/+, or sIghigh; and (3) CD5neg MBL, in which the immunophentype is CD5neg, CD19+, CD20+, and the ratio of κ:λ sIg is skewed to > 3:1 or < 1:3 (1, 2). The biology and clinical course of atypical and CD5neg MBL remain largely unexplored. We hypothesized that functional and biologic differences among MBL subtypes exist, and they would be reflected in the immunophenotypic characteristics of the MBL cases. To investigate our hypotheses and better define the biology of MBL, we performed extended immunophenotyping, interphase cytogenetics (FISH), and in a subset of CLL-like MBL, gene expression array analysis on a series of MBL cases ascertained through systematic screening of unaffected members of “high-risk” CLL kindreds (12).
Materials and Methods
Subjects
This study was performed by the Genetic Epidemiology of CLL (GEC) Consortium, a collaboration of researchers from seven institutions with the overall aim of investigating the genetic basis of CLL through analysis of CLL families (i.e., families with two or more relatives with CLL). CLL patients presenting to GEC consortium sites (Mayo Clinic, Duke University, MD Anderson Cancer Center, the National Cancer Institute (13), the University of California at San Diego, the University of Minnesota, and the University of Utah) were screened for a family history of CLL as part of routine clinical care. Those patients who reported two or more family members with CLL were invited to participate (12). Unaffected first or second degree relatives over age 30 from CLL families were enrolled and screened for MBL. Blood samples were obtained in-person or by mail. Whenever possible, a complete blood count (CBC) was obtained for each sample on the day of collection using an automated blood cell counter or, in some cases, a single platform determination using BD Trucount™ (Becton Dickinson, San Jose, CA, USA). PBMC were isolated by ficoll density centrifugation, and cells were cryopreserved by freezing in a commercial freezing chamber to −80°C in Hybridoma-SFM supplemented with FBS 20% v/v and DMSO 7% v/v. Extended immunophenotyping of cryopreserved cells was subsequently done at Duke University. Normal subjects, ascertained at Duke University, were unaffected siblings of MBL cases (n = 3) or healthy individuals (n = 7) not from CLL kindreds. The median age of controls was 51.4 years (range 22 – 78). The Institutional Review Boards at all GEC Consortium Sites approved the protocol, and all research participants provided written informed consent.
Flow Cytometry
MBL identification
This was reported elsewhere in greater detail (10). In brief, viably frozen mononuclear cells were rapidly thawed and diluted to a final concentration of 1.0 × 109 / L. Viability typically ranged from 70 – 95%. For fresh venous blood samples, PBMC were isolated using ficoll density centrifugation and diluted to the same final concentration. An aliquot of fresh blood was used for lymphocyte enumeration and determination of the B-ALC. 2.5 × 105 PBMC (either fresh or thawed) were incubated with the conjugated antibodies as described in Supplemental Table 1. Flow cytometry was performed on a Becton Dickinson ARIA flow cytometer. Analysis of flow cytometry results was performed using FlowJo software (Tree Star Inc; Ashland, OR). CLL-like MBL were defined as CD5+, CD19+, CD20dim, sIgdim B cells that comprised at least 50 events. Atypical MBL were defined as CD5+, CD19+ B cells with either CD20 surface density equal to that of normal B cells or sIghigh with a κ:λ ratio of >3.0 or <0.3. CD5neg MBL were defined as CD5−, CD19+ B cells with a κ:λ ratio of >3.0 or <0.3. The κ:λ ratio was also determined for both CLL-like and atypical MBL, but skewing of this ratio was not required for identification if the MBL population formed an immunophenotype that had a distinct cluster based upon expression of CD5 and CD20.
MBL immunophenotyping and fluorescence-activated cell sorting (FACS)
The frequency of cellular markers of clinical risk in CLL (14-20) was determined in MBL as follows: 2.5 × 105 PBMC were aliquoted and fluorescently stained as described above, except the following antibodies were substituted in different tubes for the FITC-, APC-, or PE-conjugated antibodies: CD10 PE, CD23 PE, CD27 FITC, CD38 FITC, CD49d APC, CD69 FITC, CD71 PE, CD79b PE, FMC-7 FITC, ZAP70 AF488, IgG APC, IgM FITC, and IgD PE. Intracellular staining for ZAP70 was performed after fixation and permeabilization with CytoFix/Cytoperm (BD Biosciences, Franklin Lakes, NJ) according to the manufacturer's instruction. Extracellular staining for membrane ROR1 was performed by incubating PBMC with polyclonal goat anti-human ROR1 IgG and subsequently stained with FITC-conjugated swine anti-goat IgG. Because the number of cells available for analysis was limited in some cases, complete immunophenotyping was not performed in all cases. A listing of antibody-tube combinations is provided in Supplemental Table 1, and flow cytometry reagents are listed in Supplemental Table 2.
Fluorescent in situ Hybridization (FISH)
If an adequate number of cells were available for study, 5 × 103 MBL cells were sorted in bulk directly onto two glass slides for FISH using the FACS procedure described above. FISH analysis of fixed interphase MBL clone cell chromosomes was performed using two panels of fluorescent probes to detect chromosomal abnormalities commonly identified in CLL. Panel 1: 13q14.3 (D13S319), 13q34 (LAMP1; for enumeration of chromosome 13), and a chromosomal enumeration probe (CEP; 12p11.1-q11) to detect trisomy 12. Panel 2: 17p13.1 (p53) and 11q22.3 (ATM) (Vysis; Des Plaines, IL). Following fixation of cells on slides, denaturation of MBL DNA and fluorescent probe labeling of MBL chromosomes, specimens were examined on a fluorescence microscope to detect chromosomal abnormalities. For each sample, 100 adequate cells were examined, and the percentage of MBL cells with each chromosomal abnormality was recorded. If there was limited sample adequate for only 1 FISH experiment, only panel 1 was performed. For panel 1, identification of del 13q14 or trisomy 12 in ≥5% of cells was considered positive; for panel 2, because a chromosomal enumeration control was not used for chromosomes 11 and 17 due to limited cell numbers, ≥14% of cells were required for determination of del 17p13 or del 11q22.
Gene Expression Array Analysis
mRNA expression analysis was performed to identify transcriptional differences between normal B cells, MBL and CLL. Seven CLL-like MBL ascertained from CLL kindreds, 6 control subjects, and 10 untreated sporadic CLL ascertained at Duke University were included to allow for immediate purification of target cell populations. Approximately 40 mL of heparinized blood was collected from CLL patients and controls; approximately 100 mL of heparinized venous blood was collected for MBL characterization. Blood was incubated with RosetteSep™ Human B Cell Enrichment Cocktail (Stem Cell Technologies, Vancouver, British Columbia, Canada) and purified by ficoll-Hypaque density centrifugation to enrich B cells prior to FACS. Enriched B cells were incubated as described in Supplemental Table 1; Tube 8. 1×105 CLL phenotype cells were sorted from both MBL and CLL using identical FACS procedures using a Becton Dickinson ARIA flow cytometer. Naïve (CD5–CD19+CD38+IgD+) and memory B (CD5–CD19+CD38dimIgD−) B lymphocytes were purified using FACS from normal controls without MBL. Cells were sorted into RNA lysis buffer, and mRNA was purified using a Gentra Versa-gene™ RNA preparation kit (Qiagen; Valencia, CA). RNA was labeled and hybridized to an Affymetrix Human 133 Plus 2.0 arrays (Affymetrix; Santa Clara, CA) using standard Affymetrix reagents and protocols and MAS5-normalized. Array experiments were performed by the microarray core facility at the Duke University Institute for Genome Science and Policy (IGSP).
Statistical Analysis
Analysis of variance was used to test differences in continuous values of immunophenotypic markers between the three MBL groups. Fisher's exact tests were used to assess the association of dichotomized immunophenotypic marker values and MBL group. All statistical tests were two-sided, p-values were considered significant at the 0.05 level, and analyses were performed using SAS v9.2 (SAS, Cary, NC).
Results
MBL Identification
To date, a total of 101 MBL cases from 63 families have been identified through systematic screening of 622 unaffected members of CLL families (families with at least 2 affected CLL cases). From these, either an additional sample was collected or a cryopreserved specimen was available from the 82 MBL cases for the analysis described herein. A total of 91 unique MBL clones were detected from the 82 study participants. Nine subjects (11%) had 2 distinct MBL clones: 1 with two CLL-like MBL clones (both sIg κ), 5 with one CLL-like and one atypical MBL (of these 5 biclonal cases, 3 showed sIg κ in 1 clone and sIg λ in the other); 2 with one CLL-like and one CD5neg MBL (both showed sIg κ in CD5neg clone and sIg λ in the CLL-like MBL clone); and 1 case with two atypical MBL clones (sIg κ in 1 clone and sIg λ in the other). These clones are considered separately in the following analyses. Among 91 total MBL clones analyzed, 73 MBL clones were CLL-like MBL, 11 were atypical MBL, and 7 were CD5neg MBL. Among the 11 atypical MBL cases, 10 showed a normal level of CD20 expression, and one showed bright sIg expression.
More than one person with MBL was identified in 16 different families, with a total of 51 unaffected individuals screened from these families. Eleven families were concordant with two or more individuals with CLL-like MBL, and five families had members who had discordant MBL subtypes. Of these, three families had at least one member with CLL-like MBL and one with atypical MBL, one family had one member with CLL-like MBL and one with CD5 neg MBL, and finally one family had a member with atypical MBL and one with CD5neg MBL.
A complete blood count with differential was obtained at the time of study enrollment in 53 of the 82 study subjects. The B cell absolute lymphocyte count (B-ALC) was < 5.0 × 109 cells / L in all of these subjects. The percentage of lymphocytes that were CD19+ (i.e., the proportion of the lymphocyte compartment that was comprised of B cells) was determined using flow cytometry in all cases. Among the 53 subjects with a CBC available, we observed that the ALC was < 3.3 × 109 cells / L and the B-ALC was < 1.4 × 109 cells / L in all cases in which the CD19+ cells comprised < 20% of the total number of lymphocytes. Among the 29 cases without a CBC, 23 of 29 had a CD19+ percentage < 20%, and are thus highly likely to be MBL. Of the remaining 6 with a CD19+ percentage ≥ 20%, all showed a CLL-like immunophenotype. Only two showed clear expansion of the B cell compartment with CD19+ percentages of 79% and 41%; among the other 4, the percentage of CD19+ cells was < 30%. We conclude that only 2 of 82 study subjects may be early stage CLL rather than MBL. Furthermore, extrapolating from those subjects in whom a CBC is available, over 80% of the MBL under investigation are “low count” MBL.
Immunophenotypic Characteristics
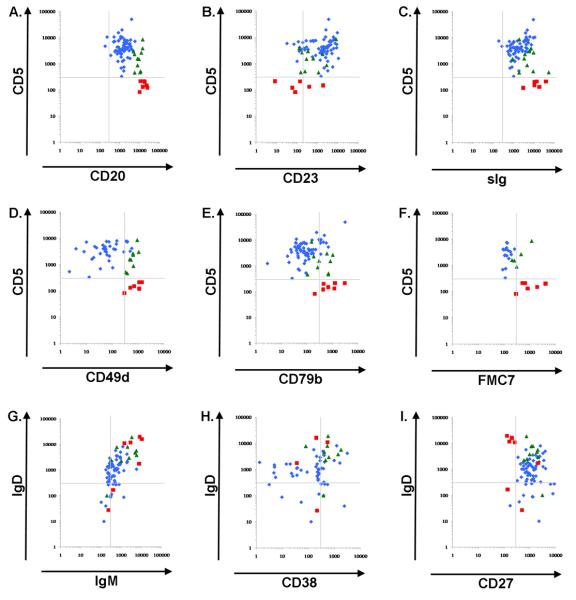
We performed flow cytometry to compare the immunophenotypic characteristics of the three different subtypes of MBL. Several cell surface markers were expressed at significantly different levels among the groups (Table 1 and Figure 1). Among all of the proteins included in this analysis, CD19 and CD27 were not different among the 3 groups. CD23, a cell surface receptor that is commonly used as a marker of B cell maturation, was expressed at higher levels in CLL-like MBL than in either atypical MBL or CD5neg MBL (CD23 MFI: 2684 in CLL-like,1468 in atypical, 365 in CD5neg,p=0.03). sIg was expressed at significantly lower levels in CLL-like MBL than in atypical MBL or CD5neg MBL (sIg MFI: 2671 in CLL-like, 9358 in atypical, 16100 in CD5 neg, p<0.0001). Similarly, CD79b, a co-factor associated with the BCR that is required for BCR-mediated signaling, was expressed at significantly lower levels in CLL-like MBL than in atypical MBL or CD5neg MBL (CD79b MFI: 178 in CLL-like, 488 in atypical,1046 in CD5neg, p<0.0001). The surface immunoglobulin isotype showed co-expression of IgM and IgD in all three groups, but again the surface expression was significantly lower among CLL-like MBL than either atypical or CD5neg MBL (IgD MFI: 1393 in CLL-like, 5593 in atypical, 8651 in CD5neg, p<0.0001; IgM MFI: 625 in CLL-like, 2672 in atypical, 4540 in CD5neg, p<0.0001). FMC7 had very low expression by CLL-like MBL, showed intermediate expression in atypical MBL, and was expressed among the majority of CD5neg MBL (FMC7 MFI: 135 in CLL-like, 489 in atypical, 1411 in CD5neg, p=0.002).
Table 1.
MBL Immunophenotypic characteristics
| Variablea | CLL-like MBL (n) |
Atypical MBL (n) |
CD5neg MBL (n) |
p-value |
|---|---|---|---|---|
| CD5 | 5547 ± 6148 (73) |
2346 ± 2346 (11) |
159 ± 51 (7) |
0.02 |
| CD20 | 1746 ± 1061 (73) |
9490 ± 3431 (11) |
16798 ± 5488 (7) |
<0.0001 |
| CD19 | 1714 ± 844 (73) |
1761 ± 935 (11) |
1890 ± 445 (7) |
0.86 |
| CD23 | 2684 ± 2573 (70) |
1468 ± 2421 (11) |
365 ± 716 (7) |
0.03 |
| CD27 | 1679 ± 1645 (65) |
1467 ± 786 (10) |
507 ± 737 (7) |
0.16 |
| CD49d | 91 ± 148 (42) |
625 ± 238 (9) |
718 ± 532 (7) |
<0.0001 |
| CD79b | 178 ± 405 (65) |
488 ± 328 (10) |
1046 ± 973 (7) |
<0.0001 |
| sIg | 2671 ± 2353 (71) |
9358 ± 15136 (11) |
16100 ± 12534 (6) |
<0.0001 |
| FMC7 | 135 ± 42 (17) |
489 ± 419 (5) |
1411 ± 1468 (6) |
0.002 |
| IgD | 1393 ± 1494 (64) |
5593 ± 5188 (10) |
8651 ± 8014 (7) |
<0.0001 |
| IgM | 625 ± 787 (64) |
2672 ± 2219 (10) |
4540 ± 4273 (7) |
<0.0001 |
| ROR1 | 2319 ± 1114 (23) |
1694 ± 1032 (8) |
796 ± 483 (4) |
0.03 |
For each cell surface marker, the mean MFI (mean fluorescence intensity) ± standard deviation and sample size (n) is reported. All statistical comparisons were performed using analysis of variance test.
Figure 1. Immunophentype profiling in MBL.
In frames A through I, the MFI for each MBL case is shown for each cell surface marker. CLL-like MBL are designated with blue diamonds, atypical MBL are green triangles, and CD5NEG MBL are shown as blue squares. A. CD5 by CD20; B. CD5 by CD23; C. CD5 by sIg, D. CD5 by CD49d, E. CD5 by CD79b, F. CD5 by FMC7, G. IgD by IgM. To characterize the B cell maturation state of different MBL subtypes, plots of IgD by CD38 (H) and IgD by CD27 (I) were displayed.
CLL prognosis is likely determined in large part by biological characteristics of the CLL cells. Prognosis is associated with the mutational status of immunoglobulin genes, specific chromosomal abnormalities, the expression of cell surface markers (CD38 and the ratio of CD69:CD71), and expression of ZAP70 (14, 15, 17, 21-27). As such, we compared the expression of various CLL prognostic markers measured in our MBL samples with those of a cohort of CLL patients followed at Duke University wherein identical flow cytometric methods were used for both MBL and CLL samples(19) (Table 2). CD38 was expressed by ≥30% of the cells in only 8 of 65 (12%) of the CLL-like MBL cases analyzed, significantly less frequent than the 27% observed in CLL (p=0.004) (19). Intracellular ZAP70 expression in ≥20% of cells was likewise observed at a lower frequency among CLL-like MBL than in CLL (26% vs. 54%, p=0.001). Cell surface expression of CD49d ≥45% was observed in CLL-like MBL at a similar frequency to a previously published CLL cohort (28). Both the ratio of CD69:CD71 ≥1.0 and high surface expression of IgM have previously been correlated to an unmutated IGHV status in CLL (15, 23). Only 28% of CLL-like MBL showed a CD69:CD71 ratio ≥1.0, and fewer CLL-like MBL expressed high levels (>40%) of sIgM when compared to CLL (IgM: 36% vs. 50%, p=0.06). These cell attributes suggest a biologically indolent, favorable risk phenotype among CLL-like MBL. In contrast, atypical MBL expressed CD38, ZAP70, and CD49d in most cases, and the increased expression of these risk stratification markers was significantly different than in CLL-like MBL and CD5neg MBL (Table 2).
Table 2.
CLL Risk stratification using immunophenotypic parameters in MBL
| CLL-like MBL |
Atypical MBL |
CD5neg MBL | p-valuea | CLLb | p-valuec | |
|---|---|---|---|---|---|---|
| CD38 > 30% | 8/65 (12%) | 9/10 (90%) | 1/7 (14%) | <0.0001 | 49/158 (27%) |
0.004 |
| Zap70 > 20% | 14/54 (26%) | 7/10 (70%) | 4/6 (67%) | 0.007 | 53/98 (54%) |
0.001 |
| CD49d > 45% | 8/42 (19%) | 9/9 (100%) | 6/7 (86%) | <0.0001 | d | n.d. |
| CD69:71 Ratio > 1.0 | 16/57 (28%) | 5/10 (50%) | 2/7 (29%) | 0.41 | e | n.d. |
| IgD > 40% | 47/64 (73%) | 9/10 (90%) | 6/7 (86%) | 0.56 | 162/187 (87%) |
0.02 |
| IgM > 40% | 23/64 (36%) | 8/10 (80%) | 6/7 (86%) | 0.002 | 93/187 (50%) |
0.06 |
Comparison among the 3 MBL subtypes
Analysis of longitudinal cohort of CLL patients followed at Duke University wherein identical flow cytometry methods were employed (19).
Comparison between CLL-like MBL and CLL; n.d.: not determined.
Previously reported frequency of CD49d expression > 45% in untreated CLL was 23% (37 of 146) (28)
Previously reported frequency of CD69:CD71 ratio > 1.0 was approximately 80% among IGHV unmutated CLL, and 20% in IGHV mutated CLL (15).
B cell maturation is a sequential process wherein discrete subgroups have unique biologic and immunophenotypic characteristics. We determined the maturation state of different MBL populations using standard flow cytometric criteria (Figure 1). Expression of IgD and CD38 was first determined. Among normal B cells, a CD19+CD38+IgD+ phenotype is characteristic of the naïve (pre-germinal center) subset, and most CD19+CD38dimIgD− B cells have a memory (post-germinal center, post-isotype switch) phenotype (29). This approach was uninformative for CLL-like MBL given the heterogeneity in CD38 expression. An alternative approach was then used based upon surface expression of IgD and CD27. Normal B cell subsets defined using this system include naïve (CD19+CD27-IgD+); post-germinal center, pre-isotype switch (CD19+CD27+IgD+); post-germinal center, post-isotype switch (CD19+CD27+IgD−); and plasma cells (CD19dim/−CD27hiIgD−) (30). Here, as shown in figure 1 both atypical and CLL-like MBL clustered in the region expected for post-germinal center, pre-isotype switch B cells.
MBL interphase cytogenetics analyses
Adequate numbers of MBL cells were available for FISH in 35 MBL cases including 30 CLL-like MBL, 4 atypical MBL, and 1 CD5neg MBL (Supplemental Table 3). Of these, adequate numbers of MBL cells were available for the full FISH panel in 29; in the remaining 6, only FISH for 13q14 and trisomy 12 was performed. Among the 30 CLL-like MBL, 22 subjects showed mono- or biallelic deletion of 13q14 as a sole abnormality (73%), 5 showed no abnormalities / normal FISH panel (17%), 2 showed trisomy 12 (7%), and 1 had del 17p13 (3%). The one case of del 17p13 was observed in a low-count MBL case, and 19% of cells showed this deletion. Among the 4 atypical MBL subjects, there were 2 MBL with trisomy 12, and one each with del 13q14 and del 17p13. In this del 17p13 subject, the absolute lymphocyte count was 8.8 × 109 / L and MBL phenotype cells constituted 81% of the B cell compartment. The single CD5neg MBL showed del 17p13 and also had a mild absolute lymphocytosis of 3.7 × 109 / L.
MBL Gene Expression Array Analyses
mRNA expression analysis was performed to identify transcriptional differences between normal memory B cells, CLL-like MBL, and CLL. Seven MBL and 10 untreated CLL were ascertained, and 1×105 CLL phenotype cells were sorted from both MBL and CLL blood using identical FACS procedures. Naïve (CD5−CD19+CD38+IgD+) and memory B (CD5−CD19+CD38−IgD−) lymphocytes were purified using FACS from normal subjects without MBL, and global gene expression patterns were determined using mRNA expression oligonucleotide arrays. We observed large transcriptional differences between MBL and normal memory cells; 238 genes were found to be differentially expressed at a significance level of p<0.0001, or 20-fold the number of probes expected to be found by chance alone. The genes that were most highly upregulated in MBL compared to memory B cells were CTLA4, LEF1, ROR1 and TCL1A. The genes that were downregulated in MBL compared to memory B cells are PAG1, EBF1, and SOX5. Transcriptional differences between MBL and CLL were also determined. Although the differences between CLL and MBL were less pronounced than between either CLL or MBL and memory B cells, we nonetheless identified 168 genes differentially expressed between MBL and CLL (p<0.0001). Interestingly, 151 of these 168 genes were expressed at a higher level in MBL than CLL. Genes upregulated in MBL as compared to CLL included DNA and RNA binding proteins (RBM22, ZC3H15), signal transduction proteins (PELI3, RGS2), and mediators of cellular differentiation (DNAJB9).
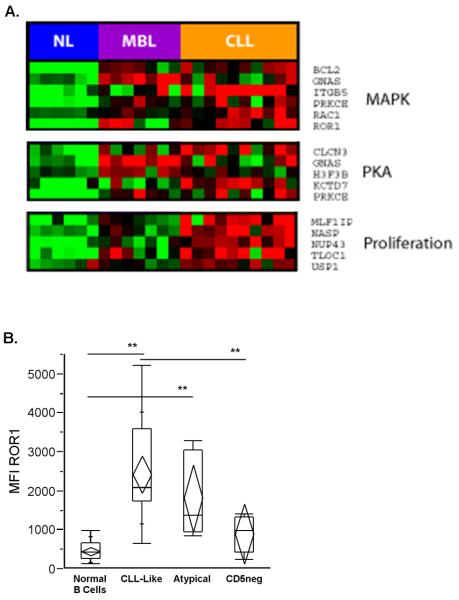
In order to identify the functional significance of altered gene expression in CLL and MBL, we selected genes that were differentially expressed (p<0.05) among CLL, MBL, and normal B cells. Genes that were highly expressed in CLL and MBL compared to controls were subjected to gene set enrichment analysis (31). The expression of pathways related to two gene sets reflecting activation of MAPKinase (MAPK) and activation of protein kinase A (PKA) were found to be enriched in MBL and CLL, whereas CLLs were found to have higher expression of genes regulating the cell cycle and proliferation when compared to both MBL and normal B cells (Figure 2A). Because both MAPK and PKA are downstream of the BCR (32), this pathway analysis highlights the potential role of the BCR in the pathogenesis of CLL and MBL.
Figure 2. Gene Expression Array Analysis and ROR1 Expression in MBL.
A. Gene set enrichment analysis of gene pathways comparing normal memory B cells (NL), CLL-like MBL cells, and CLL cells. Relative expression of differentially expressed genes is shown where green denotes lower expression and red shows increased expression. MAPKinase (MAPK), protein kinase A (PKA), and proliferation signatures show differential expression between groups. B. Quantile box plots of MFI ROR1 in MBL and residual normal CD19+ B lymphocytes. CLL-like MBL showed greater surface expression of ROR1 than either CD5neg MBL or normal B cells (p< 0.005, t-test; designated **). Atypical MBL showed greater surface expression than normal B cells.
ROR1 Expression in MBL
We chose to focus on further characterization of Receptor tyrosine kinase-like orphan receptor 1 (ROR1) which is an oncofetal cell surface antigen. ROR1 is constitutively expressed in CLL, provides pro-survival signals in CLL, and has been proposed as a target for both monoclonal antibody and immunotherapy in CLL(33, 34). ROR1 was highly upregulated in CLL-like MBL when compared to normal B cells (6.4 fold difference) in our gene expression array data. To confirm localization of ROR1 to the cell surface expression in CLL-like MBL, we used a polyclonal antibody to detect the extracellular domain of ROR1 using flow cytometry (Figure 2B). ROR1 was expressed in the majority of CLL-like MBL cases (19 of 23, 83%). Expression of ROR1 was statistically different (p=0.03) using MFI levels among the three MBL groups with higher values in CLL-like MBL than in atypical MBL, which was in turn higher than in CD5neg MBL.
Discussion
In this report, we have shown that that MBL are heterogeneous and have unique biologic characteristics. Although CLL-like MBL are phenotypically similar to CLL and are precursors to CLL (8, 9, 35), the majority of CLL-like MBL do not progress to CLL, and the identification of biologic differences between CLL-like MBL and CLL will likely illuminate critical aspects of leukemogenesis. In a large group of CLL-like MBL (n = 73) and consistent with prior reports (4-6), we found that the immunophenotype was very similar to that of CLL: CD5+ CD19+ CD20dim CD23+ CD27+ CD79bdim sIgdim. Gene expression array data discovered ROR1 over expression in CLL-like MBL, and this finding was confirmed by the identification of increased expression of ROR1 protein on the cell surface of CLL-like MBL. This finding suggests that cell surface expression of ROR1 may be important for maintenance of the CLL-like MBL, as well as CLL, clones (33, 34) . Nonetheless, phenotypic differences between CLL-like MBL and CLL were observed, and these differences may explain the apparent limited malignant potential of most CLL-like MBL. We found that both CD38 and ZAP70 were less frequently expressed among CLL-like MBL than in CLL. Only 28% of CLL-like MBL showed a ratio of CD69:CD71 > 1.0, and only 36% showed high expression of surface IgM. Both of these have been described to correlate with unmutated IGHV status (15, 23). This observation is concordant with prior work from our work and others showing that the majority of CLL-like MBL have a mutated IGHV (5, 8, 10).
Previous studies by Rawstron et al (36) and Rossi et al (37) comparing CLL-like MBL and CLL did not detect differences in expression of risk stratification parameters such as CD38 and CD49d between MBL and CLL. In both of these reports, the MBL populations were ascertained clinically: the median B-ALC in the Rawstron study was 3.2 × 109 / L and the median MBL cell count in the Rossi study was 2.8 × 109 / L. Because these CLL-like MBL were large clones, the biology was likely similar to CLL. In contrast, our cohort was comprised primarily of low count MBL (approximately 80%), suggesting that the low malignant potential of small CLL-like MBL clones is reflected in the reduced expression for markers of clinically higher risk in CLL.
Interphase cytogenetic analysis also provided important insights into the biology of CLL-like MBL. Del 13q14.3 as a sole abnormality, the most favorable cytogenetic subgroup in CLL, was observed in 22 of 30 (73%) cases of CLL-like MBL. This is higher than reported in prior CLL publications, including from our institution (19, 26). Concordant with our observations of low expression of CD38 and ZAP70 among this cohort of CLL-Like MBL, del 13q14 as a sole abnormality may be enriched within this apparently biologically indolent group when compared to clinical CLL. Alternatively, because highly purified sorted cells were analyzed, this may enhance detection sensitivity of low frequency chromosomal abnormalities. Additionally, we report, to our knowledge, the first case of del 17p13 in a low count CLL-like MBL. Because cases of del 17p CLL typically follow an aggressive disease course (26), we previously hypothesized that del 17p13 would rarely be observed in low-count MBL (10). However, a recent clinical report suggest that a subset of CLL with del 17p13 do follow a more indolent disease course, and one favorable risk factor is the observation of del 17p13 in <25% of cells, as was observed in this case (38). Overall, we stress that the FISH findings are consistent with an indolent disease biology with del 13q14 or normal cytogenetics observed in 27 of 30 (90%) of cases examined. Further, because those individuals with the smallest MBL clones typically did not have adequate sample for analysis, the reported results are biased toward analysis of the larger MBL clones.
The majority of MBL cases were CLL-like, but we also observed atypical MBL and CD5neg MBL (5). Our work provided more information on the extended immunophenotypic characterization of these MBL subtypes. The composite immunophenotype of atypical MBL was CD5+ CD19+ CD20+ CD23dim CD27+ sIg+. Among B lymphoid neoplasms, this immunophenotype is most consistent with CLL with an atypical immunophenotype or with mantle cell lymphoma (39). Although the total number analyzed was relatively low (n = 11), atypical MBL cells expressed markers for increased risk of progression as noted in CLL at high frequencies, with the majority of samples expressing CD38, CD49d, and ZAP70. This implies a more proliferative biology than CLL-like MBL. Paradoxically this phenotype is uncommonly encountered in clinical B LPDs. Because trisomy 12 is a relatively characteristic acquired chromosomal abnormality of CLL, the observation that 2 of 4 cases of atypical MBL had trisomy 12 suggests a shared biology between atypical MBL, CLL-like MBL, and CLL. We nonetheless believe that this subgroup merits future investigation as separate entity from “typical” CLL-like MBL because of the immunophenotypic differences and altered expression of risk stratification parameters. Regarding the CD5neg MBL, the composite immunophenotype was CD5neg CD10− CD19+ CD20+ CD23− CD27− FMC7+ sIg+. This immunophenotype is not specific to a single B-LPD, but it is consistent with marginal zone lymphoma, lymphoplasmacytic lymphoma (LPL), and hairy cell leukemia (HCL). Since familial clustering of CLL, Waldenstrom's/LPL, and HCL has been described, further characterization of this likely heterogenous group of MBLs is warranted (40).
We identified 9 individuals with multiple simultaneous, unique MBL clones. Of these, several showed different MBL subtypes and discordant expression of κ and λ, strongly suggesting a clonally distinct origin for each of the MBL clones. We have previously reported that oligoclonality is commonly observed in CLL-like MBL (10). In both this report and our prior work, the study subjects were ascertained from CLL kindreds. As such, these individuals may have an inherited susceptibility to the generation and maintenance of MBL clones; in support of this hypothesis is a recent report that showed an association between common SNPs and an inherited predisposition to MBL (41). Further, because CLL risk alleles may be enriched in CLL kindreds (42), family associated MBL potentially could have distinct biologic and clinical risk features as compared to “sporadic” MBL.
We utilized gene expression array methodology to further investigate the biology of CLL-like MBL ascertained from our familial kindreds. We explored transcriptional differences between CLL and MBL using gene set enrichment analysis, and this analysis highlighted the central role of BCR downstream signaling in both CLL and MBL with enhanced activation of PKA and MAPK pathways in these cells compared to normal B cells. Though not statistically significantly different, decreased expression of genes involved in cell-cycle and proliferation control were observed in CLL-like MBL as compared to CLL. BCR dependent signaling is quite complex, and the downstream response to signaling, whether proliferative or anergic, is dependent upon the strength and duration of BCR engagement as well as signaling through accessory pathways. Though CLL and CLL-like MBL share BCR dependent signaling, the consequences of this signaling could be quite divergent. Given the small number of samples studied, these findings require validation in independent datasets.
The strengths of this report include the large number of MBL cases studied and the extensive clinical, immunophenotypic, and molecular characterization, including the first examination of global gene expression array analyses in CLL-like MBL. One limitation of this study is that 29 of the 82 study subjects (35%) did not have a CBC with differential performed at the time of study enrollment. However, we used characteristics of the lymphocyte compartment to show that the majority of these cases did not have expansion of the B cell compartment, and thus are very likely to have MBL. Even if a small number of the MBL described herein are early stage CLL, this would not affect the overall findings because (1) the numbers are so small as to not change the conclusions, and (2) the inclusion of early stage CLL among the MBL group would decrease the likelihood of detecting a difference between the groups. Additional limitations include that the numbers of study subjects with atypical and CD5 neg MBL were small, thus limiting comparisons between groups, and that no atypical or CD5 neg MBL patients were available for gene expression analysis.
In summary, we report a large cohort of MBL cases derived from high-risk CLL kindreds. Three different MBL phenotypes were identified in this study, with CLL-like MBL accounting for approximately 80% of all MBL. CLL-like MBL mostly share molecular features with favorable risk CLL and appear indolent based upon this analysis. Ongoing exploration of the genetic and immunologic features of MBL will likely lead to important advances in the understanding of CLL and other B-LPDs.
Supplementary Material
Acknowledgements
MC Lanasa is supported by the Duke Clinical Oncology Research Clinical Development Program (K12) and is a fellow of the Leukemia and Lymphoma Society of America. This research is supported by the Leukemia and Lymphoma Society of America, The Harold Bernstein Family Fund, the VA Research Service, and grants from the National Institutes of Health (NCI R03 CA128030, NCI R01 CA95241, and NCI U01 CA118444). We thank the study research subjects for their willingness to participate in this study and the hematology-oncology nurses and physician assistants for their special help. Flow Cytometry was performed in the Duke Human Vaccine Institute Flow Cytometry Core Facility that is supported by the National Institutes of Health award AI-51445.
Footnotes
Conflicts of Interest
The authors declare no competing financial interest to disclose.
Authorship
All authors reviewed and approved the manuscript.
- Designed research: MCLa
- Performed research: MCLa, SDA
- Contributed vital analytical tools: SSD, BKG
- Contributed GEC samples and data: MCLa, SLS, GEM, NEK, NJC, CAH, LRG, VJC, CMV, LGS, LZR, JFL, SSS, TGC, JRC, MG
- Collected data: MCLa, SDA
- Analyzed and interpreted data: MCLa, SDA, SSD, MCLe, SLS, KGR, SJA, CL
- Wrote the manuscript: MCLa, JBW, NEC
References
- 1.Shanafelt TD, Ghia P, Lanasa MC, Landgren O, Rawstron AC. Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia. 2010 Mar;24(3):512–520. doi: 10.1038/leu.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marti GE, Rawstron AC, Ghia P, Hillmen P, Houlston RS, Kay N, et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol. 2005 Aug;130(3):325–332. doi: 10.1111/j.1365-2141.2005.05550.x. [DOI] [PubMed] [Google Scholar]
- 3.Shim YK, Middleton DC, Caporaso NE, Rachel JM, Landgren O, Abbasi F, et al. Prevalence of monoclonal B-cell lymphocytosis: a systematic review. Cytometry B Clin Cytom. 2010;78(Suppl 1):S10–18. doi: 10.1002/cyto.b.20538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawstron AC, Green MJ, Kuzmicki A, Kennedy B, Fenton JA, Evans PA, et al. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002 Jul 15;100(2):635–639. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 5.Ghia P, Prato G, Scielzo C, Stella S, Geuna M, Guida G, et al. Monoclonal CD5+ and CD5− B-lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 2004 Mar 15;103(6):2337–2342. doi: 10.1182/blood-2003-09-3277. [DOI] [PubMed] [Google Scholar]
- 6.Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF, et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009 Jul 2;114(1):33–37. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 7.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009 Feb 12;360(7):659–667. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008 Aug 7;359(6):575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 9.Shanafelt TD, Kay NE, Rabe KG, Call TG, Zent CS, Maddocks K, et al. Brief report: natural history of individuals with clinically recognized monoclonal B-cell lymphocytosis compared with patients with Rai 0 chronic lymphocytic leukemia. J Clin Oncol. 2009 Aug 20;27(24):3959–3963. doi: 10.1200/JCO.2008.21.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanasa MC, Allgood SD, Volkheimer AD, Gockerman JP, Whitesides JF, Goodman BK, et al. Single-cell analysis reveals oligoclonality among ‘low-count’ monoclonal B-cell lymphocytosis. Leukemia. 2010 Jan;24(1):133–140. doi: 10.1038/leu.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagklis A, Fazi C, Sala C, Cantarelli V, Scielzo C, Massacane R, et al. The immunoglobulin gene repertoire of low-count chronic lymphocytic leukemia (CLL)-like monoclonal B lymphocytosis is different from CLL: diagnostic implications for clinical monitoring. Blood. 2009 Jul 2;114(1):26–32. doi: 10.1182/blood-2008-09-176933. [DOI] [PubMed] [Google Scholar]
- 12.Goldin LR, Lanasa MC, Slager SL, Cerhan JR, Vachon CM, Strom SS, et al. Common occurrence of monoclonal B-cell lymphocytosis among members of high-risk CLL families. Br J Haematol. 2010 Oct;151(2):152–158. doi: 10.1111/j.1365-2141.2010.08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraumeni JF, Jr., Vogel CL, DeVita VT. Familial chronic lymphocytic leukemia. Ann Intern Med. 1969 Aug;71(2):279–284. doi: 10.7326/0003-4819-71-2-279. [DOI] [PubMed] [Google Scholar]
- 14.Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003 May 1;348(18):1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 15.Damle RN, Ghiotto F, Valetto A, Albesiano E, Fais F, Yan XJ, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 2002 Jun 1;99(11):4087–4093. doi: 10.1182/blood.v99.11.4087. [DOI] [PubMed] [Google Scholar]
- 16.Damle RN, Temburni S, Calissano C, Yancopoulos S, Banapour T, Sison C, et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood. 2007 Nov 1;110(9):3352–3359. doi: 10.1182/blood-2007-04-083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibrahim S, Keating M, Do KA, O'Brien S, Huh YO, Jilani I, et al. CD38 expression as an important prognostic factor in B-cell chronic lymphocytic leukemia. Blood. 2001 Jul 1;98(1):181–186. doi: 10.1182/blood.v98.1.181. [DOI] [PubMed] [Google Scholar]
- 18.Rassenti LZ, Jain S, Keating MJ, Wierda WG, Grever MR, Byrd JC, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008 Sep 1;112(5):1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinberg JB, Volkheimer AD, Chen Y, Beasley BE, Jiang N, Lanasa MC, et al. Clinical and molecular predictors of disease severity and survival in chronic lymphocytic leukemia. Am J Hematol. 2007 Dec;82(12):1063–1070. doi: 10.1002/ajh.20987. [DOI] [PubMed] [Google Scholar]
- 20.Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003 Jun 15;101(12):4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 21.Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995 Mar 15;85(6):1580–1589. [PubMed] [Google Scholar]
- 22.Dohner H, Stilgenbauer S, James MR, Benner A, Weilguni T, Bentz M, et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997 Apr 1;89(7):2516–2522. [PubMed] [Google Scholar]
- 23.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998 Oct 15;102(8):1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999 Sep 15;94(6):1840–1847. [PubMed] [Google Scholar]
- 25.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999 Sep 15;94(6):1848–1854. [PubMed] [Google Scholar]
- 26.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000 Dec 28;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 27.Hamblin TJ, Orchard JA, Ibbotson RE, Davis Z, Thomas PW, Stevenson FK, et al. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002 Feb 1;99(3):1023–1029. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]
- 28.Shanafelt TD, Geyer SM, Bone ND, Tschumper RC, Witzig TE, Nowakowski GS, et al. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: a prognostic parameter with therapeutic potential. Br J Haematol. 2008 Mar;140(5):537–546. doi: 10.1111/j.1365-2141.2007.06965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994 Jul 1;180(1):329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998 Nov 2;188(9):1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efremov DG, Gobessi S, Longo PG. Signaling pathways activated by antigen-receptor engagement in chronic lymphocytic leukemia B-cells. Autoimmun Rev. 2007 Dec;7(2):102–108. doi: 10.1016/j.autrev.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 33.Baskar S, Kwong KY, Hofer T, Levy JM, Kennedy MG, Lee E, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008 Jan 15;14(2):396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda T, Chen L, Endo T, Tang L, Lu D, Castro JE, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):3047–3052. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanafelt TD, Kay NE, Jenkins G, Call TG, Zent CS, Jelinek DF, et al. B-cell count and survival: differentiating chronic lymphocytic leukemia from monoclonal B-cell lymphocytosis based on clinical outcome. Blood. 2009 Apr 30;113(18):4188–4196. doi: 10.1182/blood-2008-09-176149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawstron AC, Shingles J, de Tute R, Bennett F, Jack AS, Hillmen P. Chronic lymphocytic leukaemia (CLL) and CLL-type monoclonal B-cell lymphocytosis (MBL) show differential expression of molecules involved in lymphoid tissue homing. Cytometry B Clin Cytom. 2010;78(Suppl 1):S42–46. doi: 10.1002/cyto.b.20534. [DOI] [PubMed] [Google Scholar]
- 37.Rossi D, Sozzi E, Puma A, De Paoli L, Rasi S, Spina V, et al. The prognosis of clinical monoclonal B cell lymphocytosis differs from prognosis of Rai 0 chronic lymphocytic leukaemia and is recapitulated by biological risk factors. Br J Haematol. 2009 Jun;146(1):64–75. doi: 10.1111/j.1365-2141.2009.07711.x. [DOI] [PubMed] [Google Scholar]
- 38.Tam CS, Shanafelt TD, Wierda WG, Abruzzo LV, Van Dyke DL, O'Brien S, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009 Jul 30;114(5):957–964. doi: 10.1182/blood-2009-03-210591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morice WG, Kurtin PJ, Hodnefield JM, Shanafelt TD, Hoyer JD, Remstein ED, et al. Predictive value of blood and bone marrow flow cytometry in B-cell lymphoma classification: comparative analysis of flow cytometry and tissue biopsy in 252 patients. Mayo Clin Proc. 2008 Jul;83(7):776–785. doi: 10.4065/83.7.776. [DOI] [PubMed] [Google Scholar]
- 40.Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica. 2009 May;94(5):647–653. doi: 10.3324/haematol.2008.003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowther-Swanepoel D, Corre T, Lloyd A, Gaidano G, Olver B, Bennett FL, et al. Inherited genetic susceptibility to monoclonal B-cell lymphocytosis. Blood. 2010 Dec 23;116(26):5957–5960. doi: 10.1182/blood-2010-07-294975. [DOI] [PubMed] [Google Scholar]
- 42.Slager SL, Goldin LR, Strom SS, Lanasa MC, Spector LG, Rassenti L, et al. Genetic susceptibility variants for chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev. 2010 Apr;19(4):1098–1102. doi: 10.1158/1055-9965.EPI-09-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.