Abstract
Trachealess (Trh) and Single-minded (Sim) are highly similar Drosophila bHLH/PAS transcription factors. They activate nonoverlapping target genes and induce diverse cell fates. A single Drosophila gene encoding a bHLH/PAS protein homologous to the vertebrate ARNT protein was isolated and may serve as a partner for both Trh and Sim. We show that Trh and Sim complexes recognize similar DNA-binding sites in the embryo. To examine the basis for their distinct target gene specificity, the activity of Trh–Sim chimeric proteins was monitored in embryos. Replacement of the Trh PAS domain by the analogous region of Sim was sufficient to convert it into a functional Sim protein. The PAS domain thus mediates all the features conferring specificity and the distinct recognition of target genes. The normal expression pattern of additional proteins essential for the activity of the Trh or Sim complexes can be inferred from the induction pattern of target genes and binding-site reporters, triggered by ubiquitous expression of Trh or Sim. We postulate that the capacity of bHLH/PAS heterodimers to associate, through the PAS domain, with additional distinct proteins that bind target-gene DNA, is essential to confer specificity.
Keywords: Gene expression, bHLH/PAS, Trachealess, Single minded, HIF1α, ARNT, trachea, midline
Specific induction of gene expression by transcription factors is key to all biological processes. Several central features are hallmarks of transcription regulation. First, specificity of transcription factor binding to DNA is crucial for the activation of the correct target genes. Second, the existence of heterodimeric complexes of transcription factors allows the generation of multiple combinations of assembly to accommodate the distinct specificities. Finally, many transcription factors are regulated by extracellular signaling pathways, for example, by phosphorylation, nuclear translocation, or degradation (Karin and Hunter 1995).
Many transcription factors bind DNA as dimers. The formation of heterodimeric complexes allows the extension of the DNA-binding site and increases possible combinations of distinct binding sites (Lamb and McKnight 1991). The family of basic helix–loop–helix (bHLH) transcription factors is a case in point. Central and diverse biological processes are induced by members of this family, including growth and apoptosis, myogenesis, neurogenesis, segmentation, and sex determination (Murre et al. 1994). In many instances, divergent biological outcomes of regulation by bHLH proteins have been attributed to associations with different heterodimeric partners (Kadesch 1993). For example, association of the Drosophila Daughterless protein with members of the Achaete-Scute class induces the formation of most neuronal precursors, whereas the Daughterless/Atonal heterodimer gives rise to the formation of different, nonoverlapping sense organs and photoreceptors (Jarman et al. 1993). The bHLH dimeric structure also confers, in some cases, the capacity to associate with transcriptional repressors, leading to inactivation of the complex (Murre et al. 1994).
Despite the enormous regulatory diversity provided by the heterodimeric structure of bHLH complexes, variations in the sequence of the DNA-binding site are not sufficient to account for the capacity to induce specific gene expression. For example, the heterodimeric MyoD/E2A complex induces muscle-specific gene expression while the E12 homodimer, which recognizes a similar DNA-binding site in vitro, activates transcription of immunoglobulin genes (Weintraub et al. 1994). Understanding the structural and functional basis for the recognition of different target genes by complexes that bind similar sites on the DNA remains a central challenge. It applies not only to bHLH proteins but also to other classes of transcription factors, including the AP1, CCAAT/enhancer-binding protein (C/EBP), activating transcription factor/cAMP response element-binding (ATF/CREB) and homeodomain proteins (Lamb and McKnight 1991).
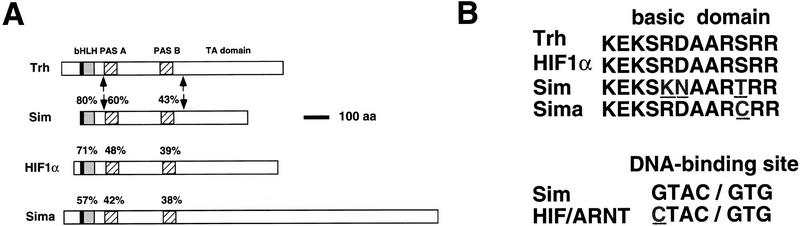
To address the issue of target-site specificity of heterodimeric transcription factor complexes during embryonic development of Drosophila, we focused on bHLH/PAS proteins, comprising a small bHLH subfamily which regulates central biological processes. Similar to all bHLH proteins, transcription factors comprising the bHLH/PAS class contain a basic DNA-binding domain of 12 residues, a helix–loop–helix dimerization motif, and a transcription-activation region. In addition, bHLH/PAS proteins contain a unique domain termed PAS (Fig. 1A). This region, comprised of two ∼50 amino acid repeats spaced by ∼150 residues, is critical for dimerization with other PAS-containing proteins (Huang et al. 1993; Lindebro et al. 1995).
Figure 1.
Structure of bHLH/PAS proteins and their DNA-binding sites. (A). A scheme of the structures of Drosophila Trh, Sim, and Sima and human HIF1α. Percentages of identical residues in each domain are shown, with respect to Trh. The arrows mark the borders of the Sim PAS domain that were used to replace the parallel domain of Trh, to generate the Trh–Sim PAS chimera. (B) Alignment of the basic domains of the four proteins, and the DNA-binding sites of the Sim complex and HIF1α/ARNT heterodimer. Nonidentical residues are underlined.
In vertebrates, several bHLH/PAS proteins have been identified. Depending on the association of the common partner aryl hydrocarbon receptor nuclear translocator (ARNT) (Hoffman et al. 1991) with two other bHLH/PAS proteins, diverse biological responses are induced. AhR/ARNT heterodimers are activated by toxic compounds to initiate the xenobiotic response, which involves the expression of detoxifying enzymes in the liver (Burbach et al. 1992). Hypoxia-inducible factor 1α (HIF1α)/ARNT heterodimers on the other hand, are stabilized by hypoxic conditions and induce the response to hypoxic stress, including the expression of glycolytic enzymes and the erythropoietin gene (Wang et al. 1995). A new bHLH/PAS protein encoded by the mouse circadian Clock gene has recently been isolated (King et al. 1997).
Three bHLH/PAS proteins have been identified in Drosophila. sima RNA is ubiquitously expressed during embryogenesis, but no biological functions have been assigned to the gene yet (Nambu et al. 1996). Single-minded (Sim) is expressed only in the embryonic midline, where the precursors of the glial cells of the central nervous system are formed. It was shown to be the central component in the induction of midline cell fates. In sim mutant embryos no midline is formed. Conversely, ectopic expression of Sim gives rise to the induction of midline cell fates in the entire ventral ectoderm (Nambu et al. 1991). The third member, Trachealess (Trh), is expressed in the tracheal pits and developing tracheal tree, which is a branched tubular structure supplying air to all tissues. It is also expressed in two other tubular structures: the posterior tracheal spiracles and the salivary duct (Isaac and Andrew 1996; Wilk et al. 1996). Again, Trh is a pivotal player in the induction of all three structures, as they do not form in trh mutant embryos. Ectopic Trh expression gives rise to extra tracheal pits in segments that normally do not form pits and to the expression of tracheal markers on the ectoderm (Wilk et al. 1996).
In spite of the sequence similarity between the Sim and Trh proteins, they activate nonoverlapping sets of target genes in the embryo and induce different cell fates. We show that in the embryo Trh and Sim complexes recognize similar DNA-binding sites. To examine the basis for their target specificity, the capacity of Trh–Sim chimeras to induce reporters and endogenous target genes was monitored. Although replacement of the basic, DNA-binding domain did not alter specificity, exchange of the Trh PAS domain with that of Sim had dramatic effects and was sufficient to convert it into a functional Sim protein. The PAS domain therefore mediates all the features conferring specificity to the complex and distinct recognition of target genes. The specificity of the Trh PAS domain is conserved in evolution, as the human HIF1α protein that is a key player in the response to hypoxic stress retained the capacity to activate Trh- but not Sim-target genes. The normal expression pattern of additional proteins essential for the activity of the Trh or Sim complexes can be inferred from the induction pattern of a binding-site reporter, as well as of endogenous target genes, triggered by ubiquitous expression of Trh or Sim. We postulate that the capacity of bHLH/PAS heterodimers to associate, through the PAS domain, with additional distinct proteins that bind target gene DNA, is essential to confer specificity.
Results
To study target gene specificity of bHLH/PAS proteins in vivo, we ectopically expressed Trh/Sim chimeric constructs driven by the heat shock promoter and monitored target gene induction. Because ectopic Trh or Sim gives rise to different outcomes, the biological activities of the chimeras could be easily discerned. Ubiquitous expression of Sim gave rise to expanded midline cell fates in the entire ventral ectoderm (Nambu et al. 1991). Conversely, ectopic Trh expression results in the formation of extra tracheal pits, in segments that normally do not form pits, and in the expression of tracheal markers on the ectoderm (Wilk et al. 1996). The constructs were induced by the heat shock promoter, which is expressed in all embryonic cells, and the synthesis of the chimeric proteins verified by the appropriate antibodies.
The basic DNA-binding domains of Trh and Sim do not confer specificity
The basic domains of Trh and Sim share a high degree of homology, where 9 out of the 12 amino acids are identical (Fig. 1B; Isaac and Andrew 1996; Wilk et al. 1996). To examine whether these subtle differences may contribute to the recognition of different target genes, site-directed mutagenesis was used to convert the basic domain of Trh to that of Sim. Early ubiquitous expression of the Trh–bSim construct in embryos, driven by the heat shock promoter, resulted in a phenotype identical to the one described previously for the overexpression of Trh (Wilk et al. 1996). In addition to the 10 tracheal pits of a wild-type embryo (Fig. 2A), two additional tracheal pits were observed, as well as the expression of tracheal lumen antigens on the ectoderm (Fig. 2B–D). The construct did not induce any of the Sim target genes (not shown). A chimera described below, in which other domains were replaced, demonstrated that the basic domain of Trh was capable of inducing midline target genes. We conclude that the basic domains of Trh and Sim bind the same site on the DNA and do not confer specificity.
Figure 2.
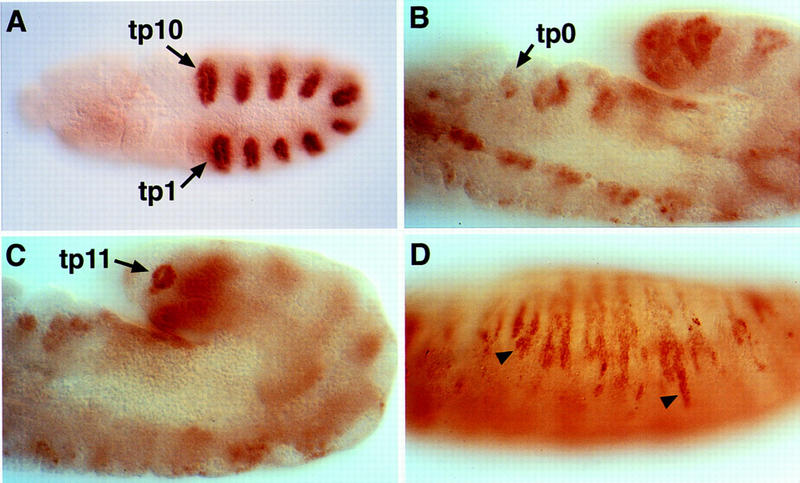
Induction of ectopic tracheal fates by the Trh–bSim construct. (A) Wild-type 1-eve-1 embryo shows staining of the trh enhancer trap in the 10 tracheal pits. (B,C) Ectopic expression of the Trh–bSim construct induced the formation of ectopic pits (tp0 and tp11), as monitored by the expression of the btl H82 enhancer trap. (D) The Trh–bSim construct also induced the expression of tracheal lumen antigens on the ectoderm at stage 15 (arrowheads), as monitored by anti-lumen antibodies (no. 84).
Isolation of the gene encoding Drosophila ARNT
Vertebrate bHLH/PAS proteins were shown to function as heterodimers. The DNA-binding site of these proteins is an asymmetric E-box-like element, where each half site is bound by a different subunit (Swanson et al. 1993; Wang and Semenza 1993; Wharton et al. 1994). The heterodimeric partners are typified by the ARNT protein. Although ARNT-related proteins also contain bHLH/PAS and transcription activation motifs, these sequences are highly conserved and distinct from those of AhR, HIF1α, Sim, or Trh.
To isolate Drosophila ARNT homologs, we used conserved sequences within the basic and HLH domains of ARNT as a basis for degenerate primers. Because the primers are spaced by only ∼150 bases that were not shown to be interrupted by introns, we used them to amplify by PCR the homologous gene(s) from Drosophila genomic DNA. A band of the expected size was generated. Sequence of multiple clones showed that only a single gene was amplified. Furthermore, a probe prepared from the fragment generated by PCR recognized only a single band in Southern blots of genomic DNA. These observations strongly suggest that there is a single ARNT homolog encoded in the Drosophila genome.
The cloned fragment was used to isolate clones from an embryonic cDNA library. Alignment of the cDNA sequence with human ARNT is presented in Figure 3A. The Drosophila ARNT (DARNT) protein shows complete identity to the human protein in the basic domain, 95% identity in the HLH region, and 56% identity in the region including PAS A, PAS B, and the spacer between them. This cDNA is ubiquitously expressed during embryogenesis (Fig. 3B). Elevated levels of the transcript can be seen in the tracheal placodes and pits.
Figure 3.

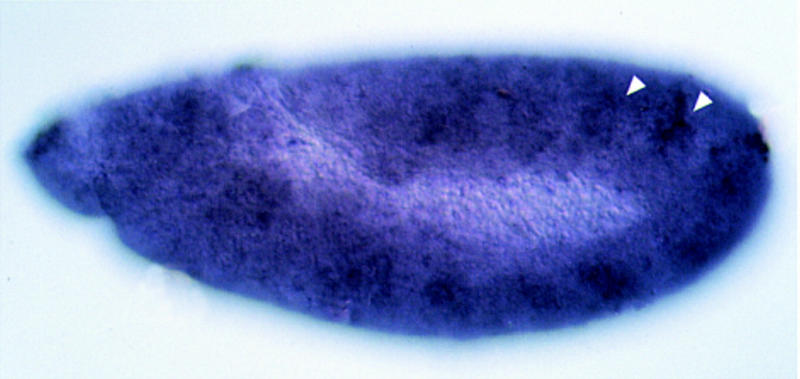
DARNT sequence and expression. (A) Alignment of DARNT (top line) with human ARNT. The first residue in DARNT represents the initiation codon. (B) Ubiquitous expression of DARNT in embryos. A stage 11 embryo is shown. Note the elevated levels of the transcript in the tracheal pits (arrowheads).
Trh and Sim complexes recognize similar DNA-binding sites
We demonstrated above that the basic domains of Trh and Sim recognize similar half-sites on the DNA. Isolation of only a single DARNT homolog suggested that this protein is a partner for both Trh and Sim. We would therefore expect the two heterodimeric complexes to bind similar sites on the DNA.
To check whether the entire site recognized by Trh or Sim and their partner is common, the expression pattern of a transgenic β-gal reporter, driven either by multimers of Sim or HIF1α/ARNT DNA-binding sites, was monitored during embryonic and larval development. The consensus DNA-binding site of Sim is highly similar to that of HIF1α/ARNT (Fig. 1B; Wang and Semenza 1993; Wharton et al. 1994). Because the reporter is present in all embryonic cells, we would expect to see expression in every tissue in which a functional bHLH/PAS complex recognizing the multimer sequence is normally available.
Expression of both reporters was detected in the tissues where Sim or Trh is known to be expressed and functional. Consistent with Sim function, the initial and most prominent expression is detected from embryonic stage 9 in the midline (Fig. 4A; Wharton et al. 1994). In addition, at stage 11, weak expression is detected in the tracheal pits, highlighting the domains where Trh is functional. Tracheal expression becomes more prominent during the formation of the tracheal tree and persists in all tracheal cells during the larval stages (Fig. 4C,D). Also consistent with Trh function, expression of the constructs is observed in the salivary duct at stage 15 (Fig. 4B; Isaac and Andrew 1996; Wilk et al. 1996). The precise correlation between the expression of the reporter and the sites of Sim or Trh expression demonstrates that both heterodimeric complexes can recognize the same DNA-binding site in embryos. Additional indications that expression of the reporter is driven directly by Trh or Sim are described below.
Figure 4.
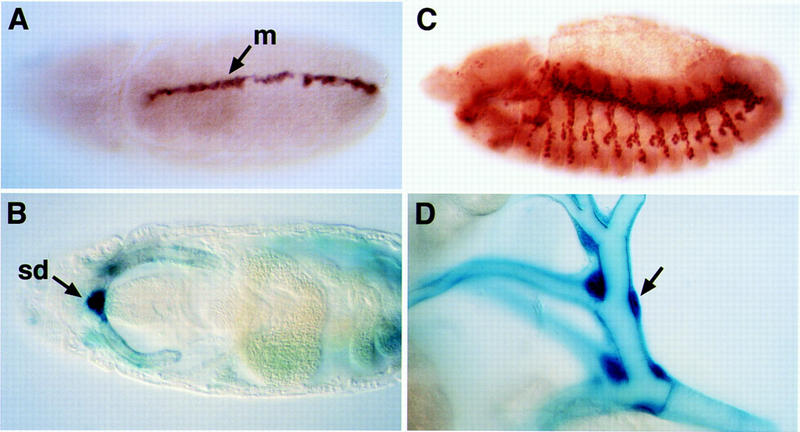
The DNA-binding sites of Sim or HIF1α are recognized by Sim or Trh complexes. The expression of lacZ driven by a pentamer of the HIF1α/ARNT binding site was monitored by X-gal or anti-β-gal staining. (A) Expression in the midline (m), from stage 9; (B) expression in the salivary gland duct (sd) at stage 15; (C) expression in the tracheal tree at stage 14; (D) expression in the tracheal tree of a third-instar larva. The arrow shows a tracheal cell. Note: A reporter driven by a tetramer of the Sim-binding site gave similar results.
The PAS domain of Sim provides midline specificity
The finding that the Sim and Trh basic domains are interchangeable suggested that other regions within these proteins are responsible for the distinct, nonoverlapping target specificities. To examine the role of the PAS domain, a chimera in which the PAS domain of Trh was replaced with that of Sim (termed Trh–Sim PAS) was generated.
Ubiquitous expression of this protein resulted in a phenotype identical to the one induced by ectopic Sim expression. Wide expansion of midline fates up to the ventral border of the tracheal pits, at the expense of the ventral ectoderm, was observed. This phenotype was examined with a variety of midline markers. Transcription of the sim gene itself is known to be autoregulated (Nambu et al. 1991; Wharton et al. 1994). Upon ubiquitous expression of Trh–Sim PAS, the expression of sim, which is normally confined to the midline, expands dramatically (Fig. 5A,B). Other genes that are known to be target genes of Sim, including breathless (btl), rhomboid (rho), and X55, were also expanded (Fig. 5C–E,G,H). Concomitant with the acquisition of Sim properties by the chimera, the capacity to activate Trh target genes was lost. Ectopic tracheal pits were not formed. In addition, in the ventral ectoderm, where the construct was capable of triggering midline-specific gene expression, no induction of tracheal markers was detected (Fig. 5I). All of these results are consistent with the expected behavior of a Sim protein.
Figure 5.
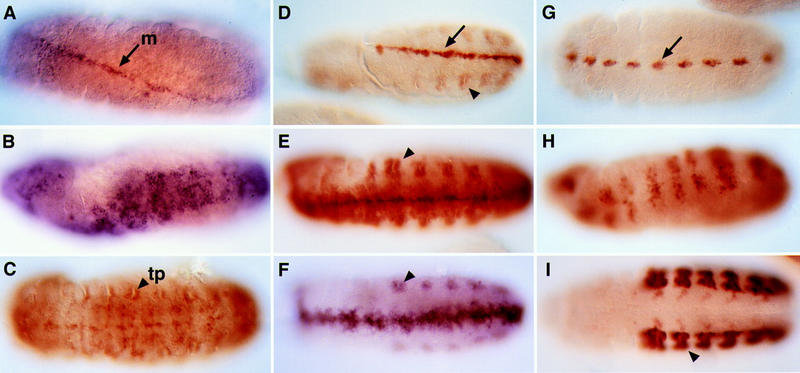
Induction of ectopic midline fates by the Trh–Sim PAS chimera. (A) Expression of sim RNA in the midline (m) of a wild-type embryo. (B) Ectopic Trh–Sim PAS induced the expression of sim in the entire ventral ectoderm. (C) The same construct also induced expansion of midline expression of the btl H82 enhancer trap. The tracheal pits (tp) are normal. (D) Expression of a rho enhancer trap in a wild-type embryo [(arrow) midline; (arrowhead) tracheal pits]. (E) Ectopic Trh–Sim PAS induces the expansion of rho midline expression. (F). A similar expansion of rho midline expression was obtained after inducing the chimera in homozygous simH9 mutant embryos. (G) Expression of the X55 enhancer trap in a subset of midline cells in a wild-type embryo. (H) X55 expression was expanded following induction of the chimera. (I) The Trh–Sim PAS protein did not induce tracheal markers in the ventral ectoderm or ectopic pits, as monitored by the expression of the trh enhancer trap 1-eve-1. All panels show ventral views.
In view of the expansion of sim transcription as a result of Trh–Sim PAS expression, it was possible that the induction of the other midline genes was driven by the endogenous Sim, rather than by the chimera. To examine this possibility, the Trh–Sim PAS chimera was induced in homozygous sim null mutant embryos, which do not normally express midline markers. Expanded expression of midline markers such as rho, was observed as in a wild-type background, demonstrating that the chimera is functionally indistinguishable from Sim (Fig. 5F). The Trh–Sim PAS protein, therefore, has the capacity to recognize a battery of midline genes and induced directly not only the expression of sim itself but also other Sim target genes. In conclusion, the ability to alter target specificities by exchanging the PAS domain demonstrates that this region is necessary and sufficient to confer distinct target specificities of the Trh and Sim proteins.
A conserved glutamine-rich region of ∼50 residues was identified, immediately carboxy-terminal to the PAS B domain of HIF1α, Sim, and Trh and termed HST (Isaac and Andrew 1996). This domain was included in the Sim fragment that was inserted into the Trh–Sim PAS chimera and may also contribute to specificity. Further dissection of the PAS domain was attempted, by inserting only the PAS A, or PAS B and HST domains of Sim into Trh. In both cases however, the constructs failed to produce a stable protein in embryos.
Different distributions of additional components in Trh and Sim complexes
The multimer reporter constructs contain only the binding site for Trh or Sim and their partner. Because the binding-site reporter constructs are expressed in all tissues where Sim and Trh are normally functional, binding of the Trh or Sim complex is sufficient to activate β-gal expression. DARNT is uniformly expressed in the embryo. Hence, we predict that in a situation where Trh or Sim is ubiquitously expressed, the expression pattern of the reporter should be dictated only by the distribution of other essential components that may be required for the activity of the Trh or Sim complexes.
Ubiquitous expression of Trh–Sim PAS (that was shown to mimic the activity of Sim) brought about induction of the reporter in all embryonic cells (Fig. 6A). On the other hand, uniform expression of Trh resulted in induction of the binding-site reporter in a broad yet defined domain of the embryo. All dorsal and dorsolateral ectodermal cells expressed β-gal. A clear demarcation of the expression border is observed, as an imaginary line running along the ventral edges of the tracheal pits (Fig. 6B). This result suggests that a factor restricted to this domain of the embryo may be essential for the activity of the Trh but not the Sim complex. This factor is likely to associate in trans with the Trh complex, as the binding-site reporter contains only the binding site for the bHLH/PAS heterodimer.
Figure 6.
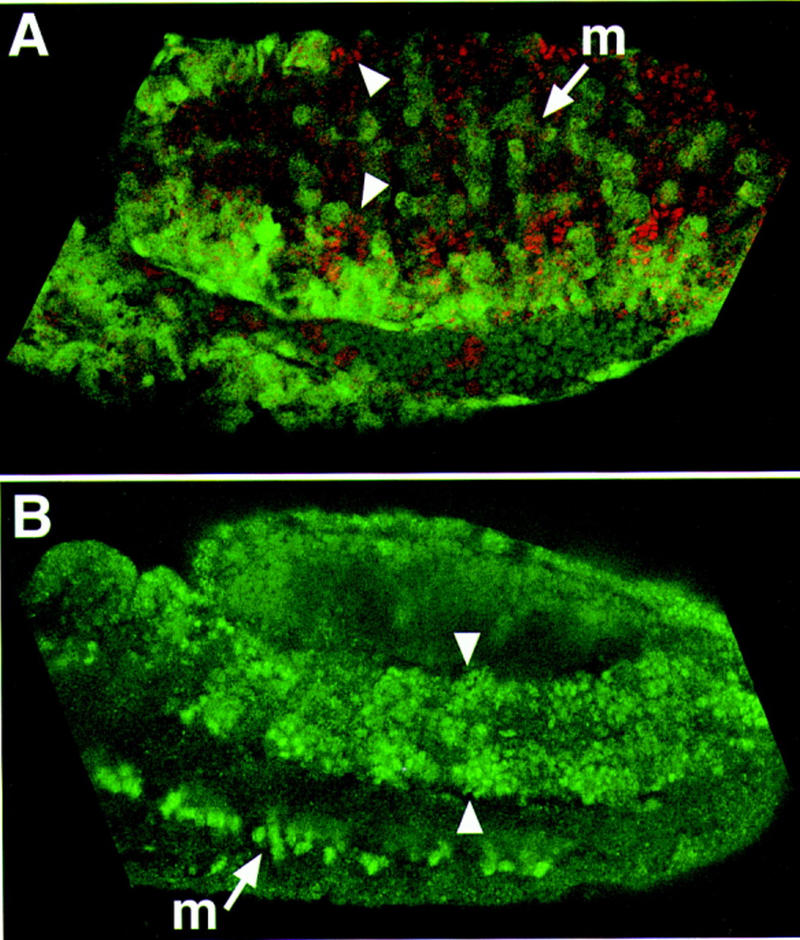
The distribution of Sim and Trh accessory factors. The HIF1α/ARNT pentamer reporter was used to follow the distribution of Sim or Trh accessory factors. (A) Following induction of Trh–Sim PAS (which we regard functionally as a Sim protein), ubiquitous expression of the reporter was observed (green). Arrowheads show the ventral borders of the tracheal pits. (B) The reporter is induced in all dorsal and dorsolateral ectodermal cells, after induction of Trh (borders shown by arrowheads). No expression was observed in the ventral ectoderm. Normal expression of the reporter in the midline (m) was also monitored. Red staining represents anti-Trh antibodies; these antibodies also recognize the Trh–Sim PAS chimera.
Activity of human HIF1α in Drosophila embryos
The similarity of human HIF1α protein to the Drosophila bHLH/PAS proteins and their similar DNA-binding sites suggested that HIF1α may be functional in Drosophila. This would depend on the capacity of HIF1α to form an active complex in Drosophila. We therefore examined the induction of the binding-site reporter after ubiquitous expression of HIF1α. Induction was monitored, and the pattern observed was similar to that induced by Trh, namely expression only in the dorsal and dorsolateral cells (Fig. 7A). In addition, a reduction in the expression of the reporter in the midline was detected, suggesting a dominant-negative effect on Sim target genes.
Figure 7.
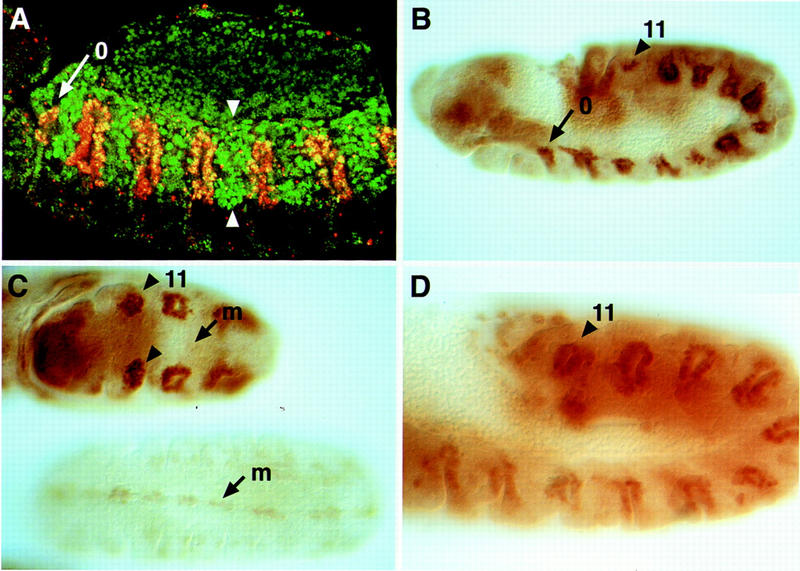
Trh-like activity of human HIF1α in embryos. (A) Ubiquitous HIF1α-induced expression of the pentamer reporter (green) in the dorsal and dorsolateral cells (borders shown by arrowheads), in a similar pattern to the one induced by Trh. Anti-Trh staining (red) showed the induction of extra tracheal pits (tp0). Green staining in the yolk represents autofluorescence. (B) The btl H82 enhancer trap also shows the induction of extra pits (tp0 and tp11) by HIF1α. (C) Comparison, in the same staining reaction, to H82 embryos which do not contain UAS-HIF1α, shows that HIF1α gave rise to an increased expression of btl in the placodes, and a decreased expression in the midline (m). (D) Ubiquitous expression of Trh in trh mutant embryos rescues the formation of tracheal pits in normal and ectopic segments, as followed by H82 expression. Note: HIF1α activates not only the transcription of trh itself but also appears to interact directly with Trh target genes. An elevated level of btl expression is observed after ectopic HIF1α induction in B and C. This is not an indirect consequence of Trh induction, as ectopic Trh expression does not give rise to higher btl levels in the tracheal pits, as seen in D.
The expression of the binding-site reporter indicated that HIF1α may form an active complex with the elements comprising the Trh complex, namely DARNT and the putative trans-acting factor implicated above. The activity of this complex was examined further under more stringent biological criteria. Induction of Trh target genes was monitored after ectopic HIF1α expression. Because trh expression is autoregulated (Wilk et al. 1996), it represents a Trh target gene. Staining with anti-Trh antibodies revealed the formation of extra tracheal pits and the induction of Trh expression in these pits (Fig. 7A). Examination of the btl marker, which is normally expressed in both midline and trachea, revealed several consequences: First, the formation of extra tracheal pits expressing btl is seen. The level of btl expression in all pits is significantly enhanced. On the other hand, elimination of btl expression in the midline is observed (Fig. 7B,C), confirming the dominant-negative effect of HIF1α and suggesting that HIF1α forms nonfunctional associations with proteins in the Sim complex.
Discussion
To address the issue of target gene specificity, we focused on bHLH/PAS proteins. These transcription factors are known to bind DNA as heterodimers of two different bHLH/PAS proteins. The binding sites for Sim and HIF1α, within their normal target genes, are similar (Wang and Semenza 1993; Wharton et al. 1994). The capacity of three different bHLH/PAS proteins (including Trh, Sim, and HIF1α) to recognize the same binding-site reporter in embryos was demonstrated, suggesting that all three proteins recognize similar sites within their normal target genes as well. Thus, alternative mechanisms must account for specificity. Replacement of the PAS domain of Trh with that of Sim was sufficient to convert it into a functional Sim-like protein. We suggest that the PAS domain, which is known to be involved in protein–protein interactions, confers specificity by dictating the assemblage of proteins in the complex.
Several bHLH/PAS proteins bind a common DNA site
The recognition of the DNA-binding site is mediated by the basic regions of the bHLH/PAS proteins, which form the contacts with the DNA. In the case of the bHLH/PAS proteins, however, the basic domain does not contribute to the specificity of Trh vs. Sim. The high degree of similarity between the basic domains of Trh and Sim suggested that they may recognize similar DNA-binding sites. This was indeed demonstrated experimentally by exchanging the basic domain without altering target specificity: The Trh protein containing the Sim basic sequence activated ectopic expression of tracheal-specific genes, whereas the Trh–Sim PAS chimera, containing the basic domain of Trh, induced midline target genes.
Another line of evidence that the binding site is common emerged from the expression pattern of the Sim or HIF1α binding-site reporter constructs. The reporter was detected in the embryo and larvae in the tissues where Trh and Sim are known to be expressed and functional and is therefore likely to be induced by the endogenous Trh or Sim complexes. Apart from these tissues, expression of the reporter was detected only in segmental clusters that may correspond to the chordotonal organs (not shown). Thus, only a restricted set of bHLH/PAS proteins may be functional during Drosophila embryogenesis.
A strong indication for the direct recognition of similar sites was provided by ectopic expression of Trh or Sim, which gave rise to an expanded expression pattern of the reporter. β-Gal appears to be induced directly by Trh or Sim, as its expression can also be observed in tissues where endogenous target genes of Trh or Sim are not induced (e.g., in the cells between the tracheal pits in the case of Trh, or in the dorsal and dorsolateral cells in the case of Trh–Sim PAS). Sima, a third Drosophila bHLH/PAS protein (Nambu et al. 1996) can also bind this site. Ectopic expression of Sima in embryos induced scattered expression of the binding-site reporter in all cells. Endogenous bHLH/PAS target genes such as rho and btl were similarly induced, whereas other genes (trh and X55) were induced only in the amnioserosa (not shown).
The endogenous target genes of Trh and Sim also appear to contain the same binding sites in their regulatory regions (Wharton et al. 1994). rho and btl are normally expressed in the midline and trachea. The transcription of these genes can be induced ectopically upon ubiquitous expression of Trh, Sim, Sima, or HIF1α. It is interesting to note that the btl promoter region contains two consensus bHLH/PAS DNA-binding sites (Murphy et al. 1995), and the rho promoter region also contains several such sites (T. Ip, pers. comm.). This raises the possibility that the same sites within these genes would be recognized by Trh or Sim in the respective tissues in which they are functional.
Another example of the common DNA-binding site for Drosophila bHLH/PAS proteins was provided recently (Nagao et al. 1996). Mobility-shift experiments, using extracts prepared from Drosophila Schneider cells that were maintained in hypoxic conditions, showed an increased association with the HIF1α/ARNT site. This experiment predicts the presence of a Drosophila protein that would be structurally and functionally similar to HIF1α, to induce transcriptional responses following hypoxic stress conditions. Finally, the conserved basic domains of two vertebrate Sim homologs (Fan et al. 1996) and the EPAS1 protein (Tian et al. 1997), suggest that they too will recognize the HIF1α/ARNT site.
The PAS domain determines target specificity
The PAS domain includes 250–300 amino acids, with two hydrophobic repeats of ∼50 amino acids each, known as PAS A and PAS B. Within the bHLH/PAS proteins, the PAS domain was suggested to be crucial for protein–protein interactions (Huang et al. 1993; Lindebro et al. 1995). Deletion of the Sim–PAS domain abolished the biological activity of Sim (Franks and Crews 1994). As described above, we demonstrated that specificity of bHLH/PAS proteins in Drosophila is not achieved at the level of different DNA-binding sites. Hence, the possibility that specificity is obtained by protein–protein interactions was examined.
The contribution of the PAS domain to the selection of target genes was examined by constructing the Trh–Sim PAS chimera, in which the PAS domain of Sim was used to replace that of Trh. The results showed that specificity is indeed determined by the PAS domain. By several criteria, the chimeric Trh protein bearing the Sim PAS domain was functionally indistinguishable from Sim. It was capable of triggering the transcription of sim itself, thus mimicking the Sim autoregulatory activity and also induced directly the transcription of Sim target genes. In parallel, as expected from a protein behaving like Sim, the chimeric protein completely lost the capacity to induce Trh target genes.
In conclusion, replacement of the PAS domain was sufficient to confer the specificity of target-gene induction. This was achieved without altering the DNA-binding site of the heterodimeric bHLH/PAS complex, thus raising the issue of the mechanism conferring specificity. Any model for such a mechanism will have to take into account that the PAS domain is crucial for protein–protein interactions and has not been reported to bind DNA directly.
Trh and Sim complexes contain common and distinct components
In bHLH/PAS proteins, the PAS domain is known to mediate the interaction with a second protein of the family, namely the partner, thus constituting a functional heterodimer. Degenerate oligonucleotides have isolated only a single, highly conserved, ARNT homolog, suggesting that this is a common partner for Trh, Sim, and Sima. The heterodimeric bHLH/PAS complex may not be sufficient for inducing target gene expression. DARNT is uniformly expressed in the embryo. If the bHLH/PAS heterodimer was sufficient, we would expect a uniform induction of the binding site reporter following ectopic expression of Trh. However, the reporter was induced only in the dorsolateral and dorsal ectodermal cells (Fig. 6B). This result highlights the spatial distribution of another component that restricts the activity of the Trh complex. This putative factor appears to be associated in trans with the complex without binding DNA, as the reporter contained only binding sites for the bHLH/PAS heterodimer. A similar experiment, following ubiquitous Trh–Sim PAS expression resulted in ubiquitous expression of the reporter (Fig. 6A). Thus, the Sim complex may not require additional factors or, alternatively, may interact with a different factor that is expressed uniformly.
Another indication for different associated components is provided by the ectopic expression of human HIF1α. Ubiquitous HIF1α expression resulted in opposite effects in the trachea and midline. While ectopic tracheal fates and target genes were induced, expression of midline markers was reduced or abolished. The most likely explanation for these opposing activities is that HIF1α forms a heterodimer with DARNT. This heterodimer is capable of associating with the accessory proteins of the Trh complex but not with those of the Sim complex, thus functioning as a dominant-negative construct in the midline.
HIF1α represents the cardinal factor in the response of vertebrate cells to hypoxic stress. It induces not only systemic responses like the transcription of erythropoietin and vascular endothelial growth factor (VEGF) but also cellular responses such as the transcriptional activation of glycolytic enzymes (Maxwell et al. 1993; Wang and Semenza 1993). On the other hand, the Trh protein is the primary regulator of the Drosophila respiratory system and is responsible not only for initiating but also for maintaining this tissue throughout development. The two proteins are functionally interchangeable, and the human HIF1α protein can induce tracheal structures. This suggests that the capacity to interact with DARNT, as well as with specific cofactors, has been conserved. The functional conservation of Trh and HIF1α highlights the common evolutionary role of their progenitor, which could be devoted to tubular patterning and systemic responses to hypoxic stress.
Recent identification of another vertebrate PAS protein termed [endothelial PAS1 (EPAS1)], which is highly similar to HIF1α, strengthens this notion further. EPAS1 is expressed in endothelial tissues during normal development, it appears to be induced by hypoxic stress, and recognizes the HIF1α-binding site (Tian et al. 1997). It is thus possible that EPAS1 is a central player in modeling the endothelial network after the initial patterning induced by VEGF has been completed. This modeling may include the recruitment of mesenchyme cells. EPAS1 was shown to induce the expression of Tie-2 (Tian et al. 1997), a receptor tyrosine kinase that is involved in endothelial–mesenchyme communication. bHLH/PAS proteins devoted to hypoxic responses may thus function in the target tissues like HIF1α, or in the tubular network itself like Trh and EPAS1.
A model for target specificity of bHLH/PAS proteins
The identification of similar binding sites for bHLH/PAS proteins in Drosophila poses a problem. In the midline Sim induces the expression of sim and its target genes, but not tracheal-specific genes, and vice versa for Trh in the trachea. Why, within the same tissue, are only some of the genes containing the bHLH/PAS-binding site selectively induced? Tissue-specific, non-DNA-binding proteins, which associate preferentially with the Trh or Sim complexes, cannot account for specificity. In addition, specificity cannot be explained by global, tissue-specific changes in chromatin structure or accessibility. The same gene, for example, btl, can be induced in extra tracheal pits by ectopic Trh but not by Trh–Sim PAS, indicating that although the btl regulatory region is accessible to transcription factors, specificity is maintained.
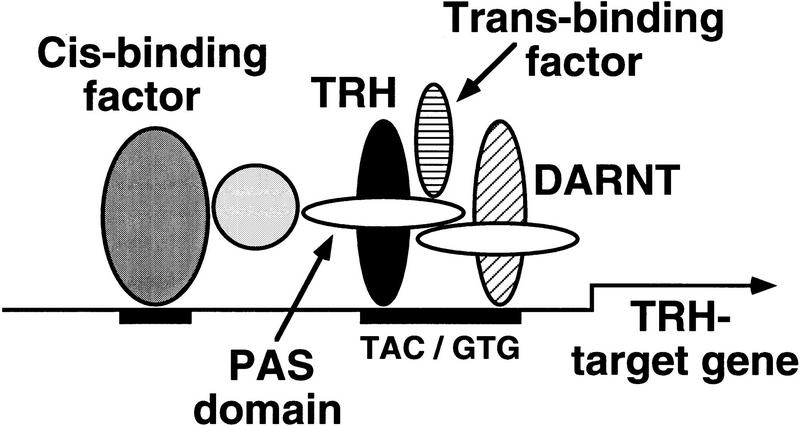
We suggest that target genes contain binding sites for tissue specific elements in their regulatory region, in addition to the canonical bHLH/PAS-binding site. These factors may interact with the bHLH/PAS heterodimer, possibly through mediation of additional proteins, to form a functional, higher order complex for transcriptional activation. A model that accounts for the results and provides a mechanism for tissue-specific expression of bHLH/PAS target genes is shown in Figure 8.
Figure 8.
A model for the capacity of Trh or Sim complexes to induce distinct target genes. In the tracheal cells Trh and its partner DARNT are expressed, and bind to the canonical binding site on the regulatory regions of tracheal, as well as midline target genes. The binding is not sufficient, however, to induce expression of these genes. Transcription will be induced only for genes that also have a binding site for an additional, tracheal-specific cis-binding factor that is capable of forming (directly or indirectly) protein–protein interactions with the Trh complex. Activity of the complex also requires the association with a trans-acting factor. The PAS domain of Trh or Sim plays a pivotal role in the complex, as it appears to mediate the interaction with the distinct cis- and trans-acting factors, as well as with DARNT.
The difference between the expression pattern of the binding-site reporter and the endogenous target genes, following ubiquitous expression of Trh or Trh–Sim PAS, provides evidence for the presence and distribution of tissue-specific elements that bind DNA and cooperate with the bHLH/PAS complex. The tissue-specific elements that associate with the Sim complex appear to be expressed in the entire ventral ectoderm while the element that cooperates with the Trh complex is restricted to the tracheal pit precursors. This was demonstrated, for example, by ectopic expression of Trh in trh mutant embryos, which was capable of rescuing the mutant and inducing tracheal pits in the correct positions and in segments that normally do not form pits (Fig. 7D). The combination of different regulatory DNA-binding sites in Trh versus Sim target genes on the one hand, and tissue-specific distribution of cis-binding accessory factors on the other, could provide a system to restrict the expression of tracheal or midline target genes, in spite of the common bHLH/PAS DNA-binding sites they posses.
In conclusion, this work has identified the PAS domain as the central player in nucleating the bHLH/PAS transcription complex. It is likely to associate not only with the DARNT protein but also with specific trans- and cis-acting factors. Exchanging the PAS domain alters target gene specificity by recruiting a different set of factors to the complex.
Materials and methods
DNA constructs
All constructs with a manipulated trh sequence are based on the full-length trh cDNA clone (Wilk et al. 1996). In the region between the trh PAS A and PAS B domains, two positions for alternative splicing were identified (Isaac and Andrew 1996; Wilk et al. 1996): Exon 4 has two possible donor sites, and exon 6 can be eliminated from the message. The trh cDNA used for the constructs has the smaller version of exon 4 and contains exon 6. The subsequent positions of amino acid residues relate to a trh cDNA sequence containing the longer exon 4 and exon 6, according to Wilk et al. (1996). All of the mutations and junctions generated in the chimeras were verified by sequencing.
To generate the Trh–bSim construct, three positions in the basic domain of Trh were mutated to the Sim sequence (R83K, D84N, S88T), using the uracil incorporation method of site-directed mutagenesis (Kunkel 1985). The Trh–bSim chimera was inserted into the pUAST vector (Brand and Perrimon 1993) at the EcoRI site.
As a prelude to generating the Trh–Sim PAS chimera, BglII sites were introduced by PCR (using Pwo DNA polymerase, Boehringer Mannheim) into the PAS junction regions of trh and sim. The BglII sites added the residues Arg and Ser to the encoded protein. The junctions in Trh were generated immediately after position Q166 and before L506. The junctions in Sim were introduced before L79 and after D363. The BglII fragment of sim, containing the PAS region, was used together with the two trh junction fragments to generate the Trh–Sim PAS construct, which was inserted into the EcoRI site of pUAST.
The 5XHIF binding-site construct was generated by oligonucleotides containing the 18-bp HIF1α/ARNT-binding site GCCCTACGTGCTGTCTCA (Wang and Semenza 1993), with a BglII site at the 5′ end and a BamHI site at the 3′ end. The oligonucleotides were treated with T4 polynucleotide kinase and ligated. After digestion with BglII and BamHI, the remaining head-to-tail multimers were separated on an acrylamide gel, and the pentamers purified and inserted into the BamHI site of pCaSpeR AUG β-gal (provided by C. Thummel, University of Utah, Salt Lake City). The number and orientation of the repeats were verified by sequencing.
The upstream activating sequence (UAS)–HIF1α construct was generated by excising the HIF1α cDNA (provided by D. Livingston, Harvard Medical School, Boston, MA) with EagI and KpnI and inserting it into the same sites in pUAST. The UAS–sima construct was generated by inserting an EcoRI–NotI fragment of the sima full-length cDNA (provided by S. Crews, University of North Carolina, Chapel Hill) into pUAST.
DARNT cloning
Degenerate oligonucleotides were synthesized according to the amino acid sequences conserved in all ARNT proteins. The amino-terminal sequence from the basic region (ENHSEIE) was used to generate the 5′ oligonucleotide. The sequence from the second helix region (MAVSHMK) was used to generate the 3′ oligonucleotide. These oligonucleotides were used to amplify the 147-bp fragment from Drosophila genomic DNA. Eight independent clones of the fragment exhibited the same sequence.
The amplified fragment was used to probe a Southern blot, as well as an embryonic 3- to 12-hr cDNA library in λgt10 (obtained from L. Kauvar and T. Kornberg, University of California, San Francisco). Sequence was obtained from the cDNA clones. A cDNA clone of 1.3 kb, including the bHLH and PAS domains, was used as a probe for in situ hybridization. The DARNT GenBank accession number is AF016053.
Fly lines
Transgenic lines were generated from the above-mentioned constructs by standard procedures. For Trh–Sim PAS, a construct inserted on the third chromosome was used. Similar patterns of induction were also observed with other insertions of the construct. For the UAS–Trh–bSim, UAS–HIF1α, UAS–sima, and 5XHIF constructs, a mixture of several independent insertions was used.
Other lines used include UAS–trhX (Wilk et al. 1996). For the trh rescue experiments, a line containing a chromosome with the trh null allele l(3)10512 and UAS–trh3, over TM3, was crossed to a line containing trh l(3)10512, the K25 sev HS–Gal4, and the btl enhancer trap H82, over TM3. The expression of Trh–Sim PAS in a sim mutant background, was induced by crossing w; 1.6 rho–lacZ; simH9, UAS–Trh–Sim PAS/TM3 flies to w; rho–Gal4; simH9/TM3 flies. The trh or sim homozygous mutant embryos were identified by absence of β-gal expression conferred by the balancer chromosomes.
As markers for the trachea or midline, the following enhancer traps were used: 1-eve-1 inserted upstream to the trh gene (Wilk et al. 1996), H82 inserted upstream to the btl gene (Klämbt et al. 1992), X55 inserted on the second chromosome (Klämbt et al. 1991), and 1.6 rho–lacZ inserted on the second chromosome (Ip et al. 1992). The 4Xsim line contains a tetramer of the Sim-binding site fused to lacZ and inserted on the third chromosome (Wharton et al. 1994). For heat shock inductions, the K25 sev HS–Gal4 line on the third chromosome was used.
Induction and staining procedures
To induce the different UAS constructs by HS–Gal4, embryos were incubated at 37°C for 20 min, 2.7 ± 0.5 hr after egg laying (AEL). They were then returned to 25°C and fixed at 7 or 11 hr AEL. For RNA in situ hybridization, the sim NruI–NotI fragment (which is not included in the Trh–Sim PAS construct and contains the sequences that are 3′ to the PAS), was used as a probe. Rabbit anti-β-gal antibodies (Cappel), guinea pig anti-tracheal lumen antibodies (no. 84) (Klämbt et al. 1992), and mouse or rat anti-Trh antibodies (which do not recognize other bHLH/PAS proteins including Sim, Sima, or HIF1α) were used (Wilk et al. 1996). Because these polyclonal antibodies also recognize a fragment after the PAS B domain, which is included in all chimeras, it was possible to verify the ubiquitous expression of the chimeras after heat shock induction. Standard procedures for RNA, antibody, and X-gal staining were used. The embryos were visualized by Nomarski optics or by a Bio-Rad 1024 confocal microscope.
Acknowledgments
We are grateful to S. Crews for providing the sim and sima clones and the 4Xsim reporter line, D. Livingston for the HIF1α cDNA, C. Thummel for the AUG β-gal clone, N. Perrimon for the 1-eve-1 line, C. Klämbt for the X55 line, M. Levine for the rho–lacZ line, E. Hafen for the K25 HS–Gal4 line, and L. Kauvar and T. Kornberg for the cDNA library. We thank R. Leiserowitz for excellent and devoted assistance in embryo injections and generating the anti-Trh antibodies, L. Glazer for help in library screens, the sequencing unit of the Weizmann Institute, and R. Schweitzer, Y. Shaul, C. Kahana, B. Cohen, I. Haviv, R. Agami, H. Greif, and all members of the Shilo laboratory for continuous advice and suggestions. We thank O. Gileadi, I. Haviv, Z. Paroush, Y. Shaul, and M. Walker for critical reading of the manuscript. This work was supported by a grant to B.S. from the Israel Academy of Sciences. P.W. was supported by a Campomar–Weizmann postdoctoral fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lvshilo@weizmann.weizmann.ac.il; FAX 972-8-9344108.
References
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C-M, Kuwana E, Bulfone A, Fletcher CF, Copeland NG, Jenkins NA, Crews S, Martinez S, Puelles L, Rubenstein JLR, Tessier-Lavigne M. Expression patterns of two murine homologs of Drosophila Single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol Cell Neurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- Franks RG, Crews ST. Transcriptional activation domains of the single-minded bHLH protein are required for CNS midline cell development. Mech Dev. 1994;45:269–277. doi: 10.1016/0925-4773(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Hoffman EC, Reyes H, Chu F-F, Sander F, Conley LH, Brooks BA, Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Isaac DD, Andrew D. Tubulogenesis in Drosophila: A requirement for the trachealess gene product. Genes & Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Kadesch T. Consequences of heterodimeric interactions among Helix-Loop-Helix proteins. Cell Growth Differ. 1993;4:49–55. [PubMed] [Google Scholar]
- Karin M, Hunter T. Transcriptional control by protein phosphorylation: Signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Vitaterna MH, Kornhauser JM, Lowery PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: A model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Klämbt C, Glazer L, Shilo BZ. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes & Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb P, McKnight SL. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- Lindebro MC, Poellinger L, Whitelaw ML. Protein-protein interaction via PAS domains: Role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Pugh CW, Ratcliffe PJ. Inducible operation of the erythropoietin 3′ enhancer in multiple cell lines: Evidence for a widespread oxygen-sensing mechanism. Proc Natl Acad Sci. 1993;90:2423–2427. doi: 10.1073/pnas.90.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AM, Lee T, Andrews CM, Shilo BZ, Montell DJ. The Breathless FGF receptor homolog, a downstream target of Drosophila C/EBP in the developmental control of cell migration. Development. 1995;121:2255–2263. doi: 10.1242/dev.121.8.2255. [DOI] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Nagao M, Ebert BL, Ratcliffe PJ, Pugh CW. Drosophila melanogaster SL2 cells contain a hypoxically inducible DNA binding complex which recognizes mammalian HIF-1 binding sites. FEBS Lett. 1996;387:161–166. doi: 10.1016/0014-5793(96)00484-x. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton JKA, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Chen W, Hu S, Crews ST. The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human hypoxia-inducible factor 1 alpha and Drosophila single-minded. Gene. 1996;172:249–254. doi: 10.1016/0378-1119(96)00060-1. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Tullis K, Denison MS. Binding of transformed AhR receptor complex to dioxin responsive transcriptional enhancer: evidence for two distinct heteromeric DNA-binding forms. Biochemistry. 1993;32:12841–12849. doi: 10.1021/bi00210a037. [DOI] [PubMed] [Google Scholar]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- Wang GL, Semenza G L. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang B-H, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: Determination of specificity by cis-acting repression elements. Genes & Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymetric E-box-like elements: Similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- Wilk R, Weizman I, Shilo BZ. trachealess encodes a bHLH–PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes & Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]