Abstract
The efficient mechanism by which double stranded DNA bacteriophages deliver their chromosome across the outer membrane, cell wall, and inner membrane of Gram-negative bacteria remains obscure. Advances in single particle electron cryo-microscopy have recently revealed details of the organization of the DNA injection apparatus within the mature virion for various bacteriophages, including epsilon15 (ε15) and P-SSP7. We have used electron cryo-tomography and 3D subvolume averaging to capture snapshots of ε15 infecting its host Salmonella anatum. These structures suggest the following stages of infection. In the first stage, the tailspikes of ε15 attach to the surface of the host cell. Next ε15's tail hub attaches to a putative cell receptor and establishes a tunnel through which the injection core proteins behind the portal exit the virion. A tube spanning the periplasmic space is formed for viral DNA passage, presumably from the rearrangement of core proteins or from cellular components. This tube would direct the DNA into the cytoplasm and protect it from periplasmic nucleases. Once the DNA has been injected into the cell, the tube and portal seals, and the empty bacteriophage remains at the cell surface.
Keywords: virus, infection, Salmonella, electron, cryo-tomography
Introduction
Salmonella bacteria are the leading source of food-borne and waterborne gastrointestinal illnesses. According to the Centers for Disease Control (www.cdc.gov), about 40,000 cases are reported each year in the U.S. alone, resulting in about 400 deaths, but it is estimated that at least 30 times more cases are not reported. In 2008, the U.S. Department of Agriculture Economic Research Service (http://www.ers.usda.gov/data/foodborneillness/) estimated the cost of Salmonella illness to be over $2.6 billion. The Gram-negative Salmonella anatum is the host cell for bacteriophage epsilon15 (ε15). Double stranded DNA (dsDNA) bacteriophages such as ε15, P22, and related viruses are important vectors for gene transfer between Salmonella populations, including genes for virulence, antibiotic resistance, and other determinants of pathogenity 1. One feature of bacteriophage physiology contributing to these processes is their efficient DNA injection mechanisms.
For Escherichia coli bacteriophage T4 and other viruses with contractile tails, the contraction process drives the tail tube through the cell envelope into the cytoplasm, forming a tunnel for DNA passage 2-5. This process requires initial interaction of the long tail fibers with cell surface receptors. Activation of the baseplate results in extension of the short tail fibers, triggering of sheath contraction, and release of the tail tube tip from the baseplate 6-8. The tip of the tail tube incorporates a lysozyme, presumably to aid passage of the tail tube through the cell wall9.
For dsDNA bacteriophages lacking a contractile tail and tail tube, the DNA transport mechanism has been obscure. The infection process is under active investigation for E. coli siphophage lambda 10,11 and Bacillus subtilis siphophage SPP112. For most of the well-studied bacteriophages, two stages have been identified: initial binding to a cell surface primary receptor such as lipopolysaccharride (LPS) by bacteriophage tailspikes or tail fibers; and secondary interaction with a host receptor protein integral to the outer membrane 13.
Another bacteriophage lacking a contractile tail is the E. coli podophage T7. Electron cryo-microscopy (cryo-EM) studies revealed T7 to have an external tail (composed of gp11 and gp12) attached to one vertex of its icosahedral capsid (gp10a) 14. Thin, flexible tail fibers (gp17) extend away from this tail 15. Coaxial with the tail is an internal core composed of gp14, gp15 and gp16, as well as two other virion proteins (gp6.7 and gp7.3) of unidentified function 14,16,17. Infection begins with the tail fibers attaching to the LPS on the E. coli cell surface 18. A signal is transmitted by the tail fibers through gp7.3 to release the viral DNA, leading to the loss of gp6.7 from the particle 17. The three internal core proteins are injected into the cell, resulting in gp14 residing in the outer membrane, while gp15 and gp16 (which can hydrolyze peptidoglycan) are found in the soluble fraction16,19. DNA ejection begins with the transportation (translocation) of the first “left” 850 base pairs through the cell membrane. This initial viral genome entry into the cell is mediated by gp15 and gp16, which act as a molecular motor that uses the membrane potential of the cell to translocate the DNA at ~70 bp/s20. The host cell's RNA polymerase internalizes the next ~7,000 base pairs of genome at ~40 bp/s, and the T7 RNA polymerase completes the process at 200-300 bp/s16. Unlike viruses with contractile tails that can mechanically puncture the host cell, T7 carries internal enzymes that can integrate into the host cell membranes and hydrolyze peptidoglycan and relies on polymerases to finish pulling its genome into the host cell.
Bacteriophage ε15 is also a podophage like T7. Single particle reconstruction from cryo-EM data reveals ε15 to have an icosahedral protein capsid ~70 nm in diameter, and the fold of its major capsid protein is similar to those of other HK97-like viruses, including bacteriophage P-SSP7 and Herpesviruses 21-23. At the virion's tail vertex, 6 tailspikes attach to a central 6-fold-symmetric tail hub. This hub may be equivalent to Salmonella typhimurium bacteriophage P22's hub (Table 1), which assembles onto the capsid during virion assembly to close the portal channel after termination of DNA packaging 24-27. In the Prochlorococcus marinus bacteriophage P-SSP7, the hub is made up of two proteins known as adaptor and nozzle 23. In ε15, the ~17 nm long hub is connected to the 12-fold-symmetric portal ring inside the capsid. Contiguous with the portal and projecting into the virion center is a protein core. This core is likely to be an analogue of the T7 core and P22 pilot proteins (Table 1). In P22, it has been shown that the pilot proteins are assembled into the virion and are needed subsequently for productive infection 28-30. The ε15 genome winds around the core, with a short segment of terminal DNA passing through the axis of the core and portal 22. A recent study of P-SSP7 alone and in association with its host cell showed drastic structural changes in the portal vertex upon viral genome release 23. The ability to resolve all the virion proteins needed for DNA injection, together with advances in electron cryo-tomography (cryo-ET), enables an attempt at visualizing the structural transformations occurring during infection 31,32.
Table 1.
Structural components of selected bacteriophages with short, non-contractile tails.
| ε15 22 | P22 23,26 | T7 14,15 | P-SSP7 51,52 | |
|---|---|---|---|---|
| Major capsid protein | gp7 | gp5 | gp10a | gp10 |
| Tailspike, tail fiber | gp20 | gp9 | gp17 | gp17 |
| Tail hub, tail tube, needle | gp15, gp16 | gp4, gp10, gp26 | gp7.3, gp11, gp12 | gp11, gp12 |
| Internal core, pilot protein | gp17 | gp7, gp16, gp20 | gp14, gp15, gp16 | gp15, gp16 |
| Portal, connector | gp4 | gp1 | gp8 | gp8 |
Results
Electron Cryo-Tomography
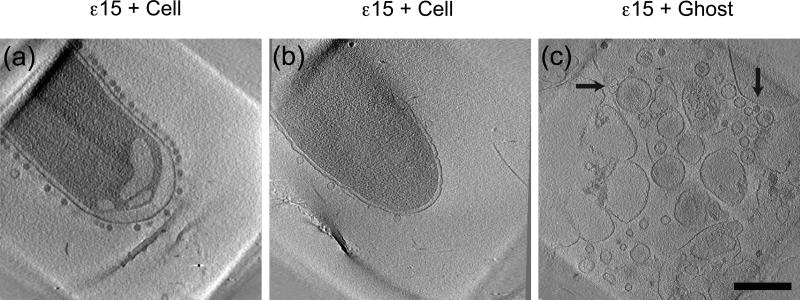
Infection of S. anatum by ε15 initiates with adsorption. Bacteriophage ε15's tailspikes come into contact with and digest the LPS covering the bacterium surface 33-35. Presumably, the bacteriophage “walks” or “chews” down to the cell surface and orients itself so that the tail axis is perpendicular to the surface. Subsequently, in a poorly understood process, a tunnel forms through the cell's outer membrane, peptidoglycan layer, and inner membrane, leading to translocation of the viral DNA into the cell 36. To visualize the infection of Salmonella by ε15, we initially mixed host cells in resting state with high-titer, infectious bacteriophages and imaged this sample by cryo-ET 31,32. A slice through the 3D reconstruction (Fig. 1a) shows bacteriophages attached around the periphery of the cell. The dark capsids indicate they had not yet injected their DNA. The inner membrane appears to have receded from the outer membrane. Next we examined ε15 incubated with Salmonella cells in mid-log phase. Figure 1b shows a cell with empty bacteriophages attached (white capsids). In contrast to the resting cell, the outer membrane appears to be wrinkled, while the inner membrane abuts the outer membrane. In order to trap the bacteriophages in an intermediate infection state due to insufficient or missing cellular components, they were incubated with ghost cells lacking most of the cytoplasm and presumably some factors necessary for efficient infection37. As seen in Figure 1c, the result is a mixture of full and empty bacteriophages attached to bacterial membranes, as well as free particles not attached.
Figure 1. Electron cryo-tomography.
(a) A slice through the 3D reconstruction of bacteriophage ε15 incubated with resting cells shows full capsids attached to the periphery of the cell. (b) A slice through the 3D of ε15 incubated with cells at log phase shows empty capsids around the cell. (c) A slice through the 3D shows full (vertical arrow) and empty capsids (horizontal arrow) attached to ghost cells (bacterial membranes). Bar is 500 nm.
Subvolume average control
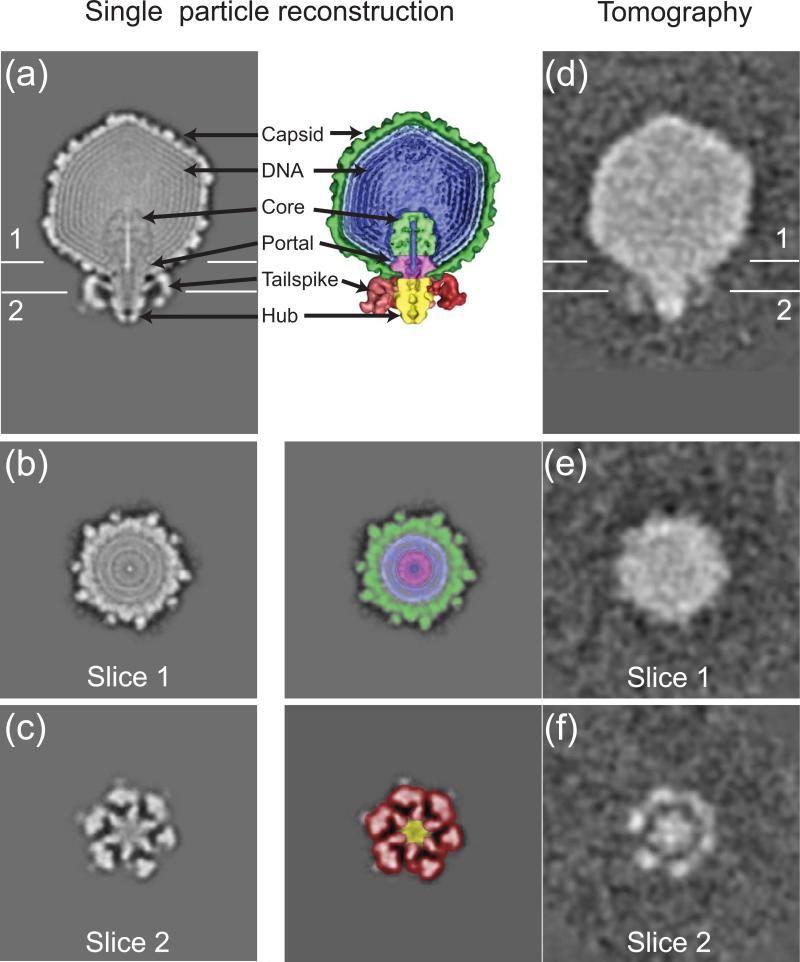
To compensate for the missing wedge and increase the signal, subvolumes containing a single bacteriophage were computationally cut out from the 3D reconstructions, aligned, and averaged together 38,39. As a control, Figure 2a (left panel) is a longitudinal slice through the map previously obtained by single particle reconstruction with no symmetry enforced 22. On the right panel, the bacteriophage components are annotated in color. Slicing the map laterally at a different position along the tail axis (as indicated in Figure 2a) shows portal, DNA, and capsid (Fig. 2b, left panel). These components have been pseudo-colored in the right panel. A lower slice shows the tail hub (Fig. 2c, left panel) surrounded by 6 tailspikes. The right panel annotates these components in color.
Figure 2. Bacteriophage ε15 in isolation.
(a) On the left, a slice through the center of the single-particle map shows the tailspikes, tail hub, portal, core, capsid, DNA shells, and DNA segment 22. On the right, these components are annotated in color (reproduced from Jiang et al 22). (b) On the left, a slice through the capsid position labeled in Figure 2a shows the DNA segment (bright dot in the center), surrounding portal, DNA shells around the portal, and capsid. On the right, these components are pseudo-colored. (c) On the left, a slice through the labeled tail position shows the closed tail hub as a 6-sided star surrounded by 6 tailspikes. On the right, these components are pseudo-colored. (d) A slice through the center of the map derived from subvolume averaging shows the tailspikes, tail hub, and full capsid, but the portal, core, and DNA can not be resolved. (e) A slice through the labeled capsid position shows a filled capsid. (f) A slice through the labeled tail position shows the tail hub as a solid density with 6-fold characteristic and surrounded by 6 tailspikes.
Figure 2d is a slice through the averaged map of 80 free bacteriophages reconstructed by cryo-ET; no cells or bacterial membranes were added. After processing, the tailspikes, tail hub, and capsid can be distinguished. However, compared to the single particle map (Fig. 2a), the capsid is filled with a punctate density, and the portal, core, and DNA cannot be resolved, probably due to the limit of the resolution. Figure 2e slices through the capsid at the portal position, and it also shows a full capsid. A lower slice (Fig. 2f) shows a filled tail hub with 6-fold symmetry surrounded by 6 tailspikes. Even though detailed features cannot be resolved, the similarity between the maps from the single particle reconstruction (Fig. 2a) and subvolume averaging of cryo-ET data (Fig. 2d) validates our computational approach.
Subvolume average of bacteriophage ε15 attachment
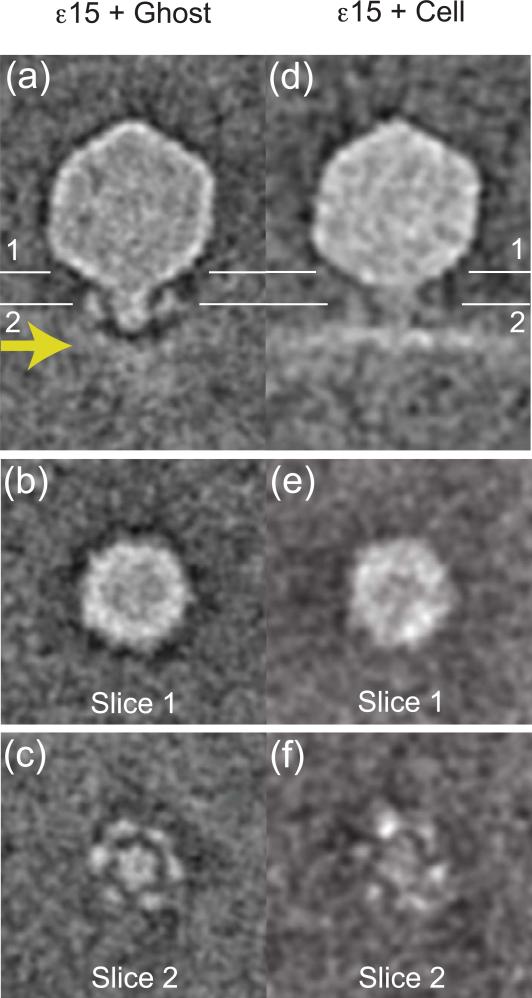
The tailspike adhesins of ε15 bind LPS, a component of the outer leaflet of the bacterial outer membrane. After the bacteriophage has adsorbed to the cell, it must find a secondary receptor for docking and injection of DNA 13. In bacteriophage lambda, this second receptor is the maltose binding protein LamB 10, while T5 binds the ferrichrome transporter FhuA 40,41. Figure 3a is a longitudinal slice through the average map of full bacteriophages attached to ghost cells (Fig. 1c). The ghost cell infection was intended to block some infection processes from completion because of the limited supply of necessary cellular factors, for example, membrane potential42-44. Although the bacteriophage map (Fig. 3a) is similar to the previous map (Fig. 2d), a bowl-shaped density beneath the tail hub (Fig. 3a, arrow) suggests the bacteriophage indents the host outer membrane at the beginning of infection to search for a secondary receptor or to puncture the membrane. It is also possible that the tailspikes have attached to LPS that are far apart, thereby causing the membrane to sag under the tail hub. Another possibility is that the membranes from ghost cells are not under osmotic tension so they may sag (Fig. 1c), while those from cells are rigid because they are osmotically intact (Fig. 1a). Yet another explanation of the bowl-shaped density is that not all tailspikes have attached equally and fully, resulting in the membrane not being perpendicular to the tail axis. However, the tail hub does not connect to the membrane (dark gap between the bowl and tail hub), suggesting some cellular factors may be missing. Slicing laterally through the capsid (Fig. 3b) at the indicated plane shows the viral DNA is still inside the capsid. A slice through the tail hub (Fig. 3c) suggests a hint of an opening in the center (for example, compare to the tail hub in Fig. 2f). The interactions between the tailspikes and LPS may cause the tail hub to change conformation in preparation for the next step, such as releasing the core or binding to a secondary receptor. In a similar way, structural changes occur in the portal vertex of P-SSP7 when the bacteriophage interacts with its host cell 23.
Figure 3. Subvolume averaging of full bacteriophages.
(a) A slice through the center of the map of bacteriophages attached to ghost cells shows the tailspikes, tail hub, full capsid, and indented cellular membrane (the bowl below the tail, arrow), but the tail hub is not connected to the cell membrane. (b) A slice through the capsid position labeled in Figure 3a shows a punctate but mostly solid density at the portal location. (c) A slice through the labeled tail position shows the tail hub having 6-fold characteristics and surrounded by 6 tailspikes. (d) A slice through the center of the map of bacteriophages attached to resting cells shows the tailspikes, tail hub, full capsid, and cell surface (horizontal density beneath). The tail hub density is continuous with and attached to the cell. (e) A slice through the capsid position labeled in Figure 3d shows the portal and DNA as punctate density. (f) A slice through the labeled tail position shows the tail hub surrounded by tailspikes.
Following attachment to the putative secondary receptor, a pore or tunnel for DNA passage must be formed. Figure 3d shows the subvolume average of full bacteriophages attached to resting cells. From the side, the presence of higher density in the cell membrane just beneath the tail hub (Fig. 3d) may be attributed to the putative secondary receptor or even the bacteriophage core. This putative membrane protein appears to be a closed pore. Slicing through the capsid (Fig. 3e) shows the presence of DNA. The reason for the difference in DNA density (compare Figures 3a and 3d) may be due to the difference in the overall contrast between the two data sets. The vitreous ice embedding the ghost cell preparation (Fig. 3a) is thinner than the ice embedding the intact cells (Fig. 3d). As a result, Figure 3a shows more contrast than Figure 3d. Alternatively, some of the bacteriophages that were manually classified as full may be partially empty, thereby leading to a lower DNA density in the average. Slicing through the tail hub (Fig. 3f) shows fainter densities than before. The fainter density may be the result of averaging tails that are not rigid or structurally homogeneous. For example, the tails may have been in different conformational states or in motion at the time of vitrification because they were searching for a putative secondary receptor in the proper orientation and state (such as one from a cell in log phase). For instance, the tail hub may reversibly attach to the putative secondary receptor, and this process may require several sequential intermediate steps. In a similar way, bacteriophage lambda binds reversibly with its receptor lamB, so the lambda-lamB complex may also be structurally heterogeneous 45.
Subvolume average after DNA has been ejected from bacteriophage ε15
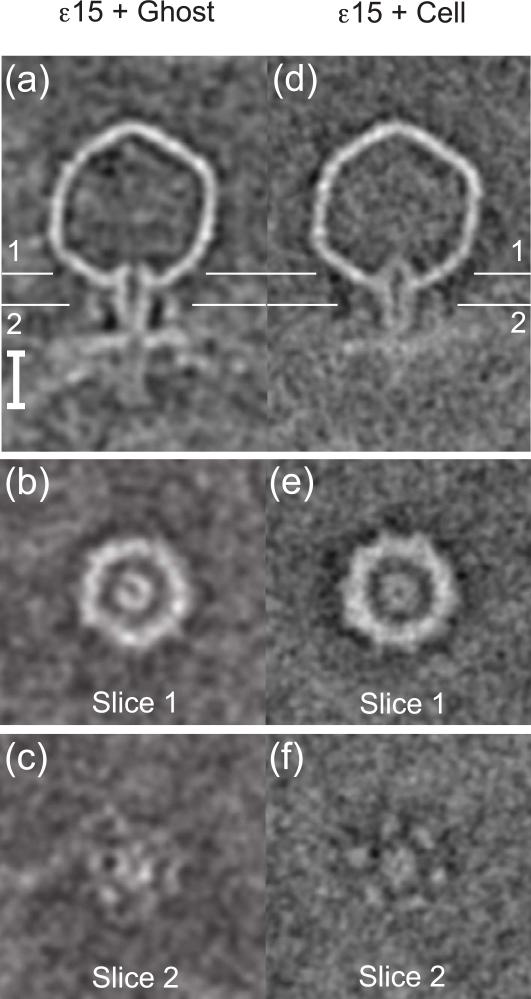
A tunnel spanning the outer membrane, peptidoglycan, and inner membrane is required for viral DNA to reach the cytoplasm. Figure 4a is the average of empty bacteriophages attached to ghost cells; the DNA and core has been ejected from the capsid. A ~21 nm tall tubular density extends from the outer membrane into the periplasmic space (Fig. 4a, vertical bar), which is 21-24 nm wide in other rod-shaped, Gram-negative cells 46. This length is just sufficient to reach the inner membrane, but the missing putative inner membrane pore probably dissociated after DNA passage to prevent leakage of cytoplasmic contents. Presumably the bacteriophage core plays a role in forming this tubular structure and may recruit cellular factors, such as an inner membrane pore. Using this tube is one way to protect the viral DNA from the cell's periplasmic nucleases 47,48. The portal region (Fig. 4b) has a ring density, indicating that it remains open after DNA ejection. The tail hub (Fig. 4c) has 6-fold symmetry and is surrounded by tailspikes.
Figure 4. Subvolume averaging of empty bacteriophages.
(a) A slice through the center of the map of bacteriophages attached to ghost cells shows tailspikes, tail hub with hollow interior, empty capsid (absence of core and DNA), cellular membrane, and a tubular density in the periplasmic region. Vertical bar is 21 nm tall. (b) A slice through the capsid position labeled in Figure 4a shows the portal as a hollow ring and absence of DNA. (c) A slice through the labeled tail position shows faint densities corresponding to a hollow tail hub and tailspikes. (d) A slice through the center of the map of bacteriophages attached to cells shows tailspikes, tail hub with hollow interior, empty capsid, cell surface, and tapered density in the periplasmic region. (e) A slice through the capsid position labeled in Figure 4d shows the portal as a hollow ring. (f) A slice through the labeled tail position shows a hollow tail hub surrounded by 6 tailspikes.
Once the viral DNA has been injected into the cell, the pores would need to be closed or remain impermeable to the surrounding medium in order to maintain the integrity of the cell 49. The empty capsid that stays on the cell surface (Fig. 1b) until the end of the infection cannot serve as a plug because it is permeable, as demonstrated by the observation that EDTA can chelate magnesium from the viral DNA and cause the capsid to rupture 50. Thus EDTA or magnesium can diffuse through the capsid. Figure 4d shows the average of empty bacteriophages attached to cells. The periplasmic tube is shorter and tapered (Fig. 4d), perhaps providing a seal after viral DNA injection. It is possible that the components that make up this tube may have dissociated, or the length of the tube may be obscured by the presence of other macromolecules in this thicker region. The portal is still a ring (Fig. 4e), but it also tapers and closes. The tail hub (Fig. 4f) is open and surrounded by 6 tailspikes. Thus the tail hub is capped at both ends by tapered densities, and this may consequently serve as an impermeable block.
Discussions
Adsorption
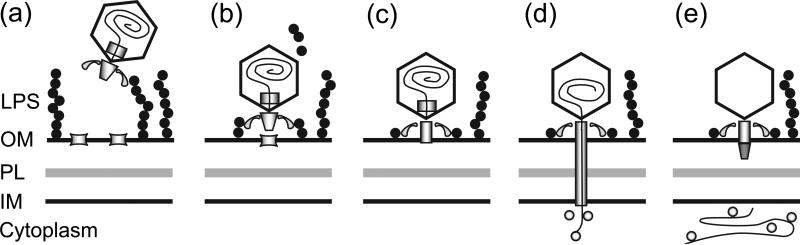
These experimental snapshots of different stages of bacteriophage/host interaction can be incorporated into an integrated model of the infection process (Fig. 5). Adsorption is the first step in the infection process. The bacteriophage randomly collides with a suitable cell, and the tailspikes or tail fibers recognize the LPS on the cell surface (Fig. 5a) 13. In ε15, the tailspike gp20 has endorhamnosidase activity and digests the LPS (Fig. 5b) 33,35. In P22, the same function is present in gp9, and digestion of the LPS brings the capsid near the cell outer membrane 51. Presumably a similar function for recognizing, attaching, and approaching the cell surface is present in the T7 tail fiber gp17 15,18. In P-SSP7, interactions with the host cell cause the tail fibers gp17 to change from inclined to horizontal before injection of DNA 23. The experimental snapshots presented here do not show gross changes in the structure of ε15's tailspikes like those of P-SSP7, even with a putative hinge 22. However, the resolution does not permit visualization of fine details, and the snapshots do not cover the complete infection sequence. For example, it is possible that the tailspikes may change conformations as they digest the LPS to reach the cell surface. Additional experiments are required to make a more definitive conclusion about the hinge in the tailspikes.
Figure 5. Infection model.
(a) Free-floating bacteriophage tailspikes come into contact with and recognize an LPS on the cell surface. (b) The virus digests the LPS to presumably reach the cell surface and (c) binds to a receptor on the cell outer membrane, causing the tail hub to open. (d) The core exits the capsids and forms a tube across the periplasmic space, thereby directing the viral DNA into the host cell cytoplasm. (e) After the viral genome has been completely internalized, the perisplamic tube seals to prevent leakage of cytoplasm, while the empty capsid remains on the cell surface. Key: lipopolysaccharides (LPS), outer membrane (OM), peptidoglycan layer (PL), inner membrane (IM).
Secondary receptor
Once the bacteriophage reaches the cell outer membrane, it interacts with a putative secondary cell receptor (Fig. 5c). The secondary receptor for ε15, P22, T7, and P-SSP7 has not been identified. However, the siphovirus T5 tail protein pb5 binds the ferrichrome transporter FhuA, SPP1 recognizes YueB, and lambda tail protein gpJ attaches to the maltoporin LamB 10,41,52.
Changes in the tail vertex
Interactions with the secondary receptor result in conformational changes in the bacteriophage tail vertex (Fig. 5c). In ε15, the portal and tail hub forms a channel to allow passage of the capsid contents (Fig. 4). Similarly, P22's internal proteins gp7, gp16, and gp20 are ejected at the initiation of the infection 28-30,53,54. Interestingly, the tail hub gp4 can hydrolyze peptidoglycan, so it must penetrate the outer membrane to reach the peptidoglycan layer in an unknown manner 19. In T7, the interaction with the cell transmits a signal through gp7.3, leading to the loss of gp6.7 17. In addition, 3 interior core proteins are ejected from the capsid. In P-SSP7, the tail nozzle changes in order to accommodate DNA and core passage 23. In SPP1, the flexible tail tip with a structure analogous to P22's tailspike gp9 is no longer visible after interaction with YueB, while the dome-shaped tail cap changes into an open 6-sided star shape55. The inner wall of the helical tail tube becomes narrower to presumably signal the release of the tape measure protein.
Pathway into the cytoplasm
The changes in the bacteriophage tail vertex lead to the formation of a pathway to the cytoplasm (Fig. 5d). In ε15, the core exits the capsid through the open tail complex and presumably forms a periplasmic tube with host proteins, such as a putative inner membrane pore (Fig. 4). The high-energy state of the packaged DNA would result in spontaneous transport once a tunnel has opened to the cell cytoplasm 56-58. Cytoplasmic proteins may bind to the invading DNA. In P22, gp16 partitions with the inner membrane and can mediate the active transport of DNA into liposomes 44. In T7, the internal proteins that are ejected are proposed to transform into a 40-55 nm long “injection complex”, sufficient to cross the ~24 nm with of the cell envelope 59. This “injection complex” is reminiscent of ε15's periplasmic tube as observed in this study. Furthermore, T7's gp15 and gp16 starts the translocation of DNA into the cell, and RNA polymerases complete the translocation 16,20. The initial translocation by gp15 and gp16 requires energy, like P22's gp16. Similarly, the core proteins in ε15 may use energy for DNA translocation, but resting cells may lack a membrane potential so that DNA is not injected into these cells (Fig. 1a). Less puncturing of the cell envelope may permit more bacteriophages to bind without causing lysis from without 60. In T5, the pb2 tail fiber can penetrate both single and double bilayered proteoliposomes, which mimics the host cell outer and inner membranes 40. Subsequently, pb2 changes conformation, becoming shorter and wider, and opens a channel that can accommodate dsDNA passage. In 2008, Boulanger showed that pb2 can fuse liposomes and hydrolyze peptidoglycan, and suggested that pb2 may degrade the peptidoglycan layer and fuse the inner and outer membrane to form the DNA pore, which would protect the viral DNA from periplasmic nucleases 61.
It is not known whether ε15 also requires a polymerase to complete DNA translocation like T716,20. The fact that ε15 capsids attached to ghost cells can become empty (Figs. 1c and 4a), which suggest that a polymerase is not needed, is similar to previous observations. For example, T5 can inject its DNA into liposomes without polymerases, and those empty capsids appear similar to those presented here 40. Likewise, P-SSP7 released its DNA into the buffer in the absence of cells, but its tail fibers were horizontally oriented like that observed in the presence of cells 23. One possibility is that a small number of receptors co-purified with the virus. Images of SPP1 incubated with purified YueB also show empty capsids 52,55. These observations suggest that a polymerase is not required for these systems, but the situation may be different in vivo.
After injecting the viral DNA
Once the viral DNA has been injected into the cell, the pore would need to be actively closed or remain impermeable to the surrounding medium in order to maintain the integrity of the cell so that new bacteriophages may form (Fig. 5e) 49. In ε15, the putative inner membrane pore detaches from the periplasmic tube to prevent cytoplasmic leakage, thereby causing the periplasmic tube to collapse and seal (Fig. 4d). The empty particle remains at the cell surface until cell lysis at the end of the infection (Fig. 1b) 62.
A common theme is emerging whereby bacteriophages carry most (if not all) of the machinery necessary to puncture or span the host cell envelope and use a bacterial receptor for recognition and docking. Although the receptor and a peptidoglycan hydrolase for ε15 have not been identified, we show here that ε15 can form a tube across the host cell's periplasmic space for viral DNA injection. In the future, identifying and characterizing these bacteriophage or host proteins will reveal more details about the infection mechanism. While T4 and other bacteriophages with contractile tails may use a mechanical syringe to physically puncture the cell envelope, bacteriophages with short or non-contractile tails have adopted alternative strategies.
Materials and methods
Bacteriophages were prepared as described previously by cesium chloride (CsCl) density centrifugation22,63,64. This method separates particles containing both DNA and protein from complexes and aggregates of protein alone. The samples thus produced were of high titer (~2.5×1012 pfu/ml) and high purity, as judged by SDS-PAGE and negative stain TEM (data not shown).
Purified ε15 was mixed with 15 nm gold, applied to a copper grid (Quantifoil Micro Tools GmbH) in a Vitrobot (FEI Company), and vitrified in liquid ethane65. The grids were imaged at -180°C with SerialEM software66 in a JEM3200FSC electron microscope (JEOL, Tokyo) operated at 20,000×, 300KeV, in-column energy filter with 20eV slit centered at the zero loss peak. The tilt series were collected on a Gatan 4k CCD (Gatan, Pleasanton) at 2° steps over a range of at least 120°, 6-9 μm defocus, and 35-65 electrons/Å2 total dose. The 3D reconstructions were performed with IMOD software using the gold as fiducial markers 67. Subvolumes, each containing a single bacteriophage, were computationally extracted from the reconstructions, aligned to the single particle map22, and averaged together as previously described 38,39. The averaged map has 80 subvolumes, and its resolution is estimated to be ~4 nm by comparing the icosahedrally-symmetrized capsid against the single particle map using 0.5 Fourier shell correlation as a criterion 22,68. The map was filtered to 5 nm for display.
For the full capsids attached to cells, Salmonella anatum were grown in LB media to log phase, pelleted, and resuspended in buffer at 4°C. The resting cells were mixed with ε15 (approximate MOI=400) and gold, incubated for 10 minutes at 25°C, vitrified, imaged, and processed as before. The averaged map has 83 subvolumes and was filtered to 7 nm.
For empty capsids attached to cells, the cells were grown to log phase, mixed with ε15 (approximate MOI=40) and gold, incubated for 30 minutes at 37°C, vitrified, imaged, and processed as before. The averaged map has 85 subvolumes and was filtered to 5 nm.
Ghost cells (bacterial membranes) were prepared37, incubated with ε15 for 50 minutes at 37°C (more than enough time to empty all the capsids in the previous experiment), mixed with gold, vitrified, imaged, and processed as before. However, the full and empty capsids attached to ghost cells were split into 2 groups by visual inspection and separately processed, resulting in 44 and 10 subvolumes in the averaged maps, respectively. They were filtered to 5 nm and 7 nm.
The averaged maps have been deposited in the electron microscopy data bank (www.emdatabank.org) with accession numbers: EMD-5216 (full ε15 attached to cell); EMD-5217 (empty ε15 attached to cell); EMD-5218 (full ε15 attached to ghost); EMD-5219 (empty ε15 attached to ghost).
Acknowledgments
We thank Irina Serysheva, Que Ngo, Theodore Wensel, Jared Gilliam, and Vera Moiseenkova-Bell for assistance with cell and ghost cell preparation; Steven Ludtke for helpful discussions on image processing. This work was supported by the Robert Welch Foundation (Q1242) and NIH grants (R01AI0175208 and P41RR002250).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daubin V, Lerat E, Perrière G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon LD, Anderson TF. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology. 1967;32:279–297. doi: 10.1016/0042-6822(67)90277-2. [DOI] [PubMed] [Google Scholar]
- 3.Simon LD, Anderson TF. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology. 1967;32:298–305. doi: 10.1016/0042-6822(67)90278-4. [DOI] [PubMed] [Google Scholar]
- 4.Simon LD. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. 3. Membrane-associated intracellular bacteriophages. Virology. 1969;38:285–296. doi: 10.1016/0042-6822(69)90370-5. [DOI] [PubMed] [Google Scholar]
- 5.Simon LD. Infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope: T4 head morphogenesis. Proc Natl Acad Sci U S A. 1972;69:907–911. doi: 10.1073/pnas.69.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aksyuk AA, Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure. 2009;17:800–808. doi: 10.1016/j.str.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 10.Berkane E, Orlik F, Stegmeier JF, Charbit A, Winterhalter M, Benz R. Interaction of bacteriophage lambda with its cell surface receptor: an in vitro study of binding of the viral tail protein gpJ to LamB (Maltoporin). Biochemistry. 2006;45:2708–2720. doi: 10.1021/bi051800v. [DOI] [PubMed] [Google Scholar]
- 11.Roessner CA, Ihler GM. Formation of transmembrane channels in liposomes during injection of lambda DNA. J Biol Chem. 1986;261:386–390. [PubMed] [Google Scholar]
- 12.Lhuillier S, Gallopin M, Gilquin B, Brasilès S, Lancelot N, Letellier G, Gilles M, Dethan G, Orlova EV, Couprie J, Tavares P, Zinn-Justin S. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci U S A. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinga I, Sao-Jose C, Tavares P, Santos MA. Bacteriophage entry in the host cell. In: Wegrzyn G, editor. Modern Bacteriophage Biology and Biotechnology. Research Signpost; Kerala, India: 2006. pp. 165–205. [Google Scholar]
- 14.Agirrezabala X, Martín-Benito J, Castón JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steven AC, Trus BL, Maizel JV, Unser M, Parry DA, Wall JS, Hainfeld JF, Studier FW. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 16.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 17.Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–317. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 18.Krüger DH, Schroeder C. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol Rev. 1981;45:9–51. doi: 10.1128/mr.45.1.9-51.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moak M, Molineux IJ. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol. 2004;51:1169–1183. doi: 10.1046/j.1365-2958.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- 20.Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1823. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- 26.Casjens S, King J. Virus assembly. Annu Rev Biochem. 1975;44:555–611. doi: 10.1146/annurev.bi.44.070175.003011. [DOI] [PubMed] [Google Scholar]
- 27.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 28.Bryant JL, King J. DNA injection proteins are targets of acridinesensitized photoinactivation of bacteriophage P22. J Mol Biol. 1984;180:837–863. doi: 10.1016/0022-2836(84)90260-2. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman B, Levine M. Bacteriophage P22 virion protein which performs an essential early function. I. Analysis of 16-ts mutants. J Virol. 1975;16:1536–1546. doi: 10.1128/jvi.16.6.1536-1546.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman B, Levine M. Bacteriophage P22 virion protein which performs an essential early function. II. Characterization of the gene 16 function. J Virol. 1975;16:1547–1559. doi: 10.1128/jvi.16.6.1547-1559.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumeister W, Grimm R, Walz J. Electron tomography of molecules and cells. Trends Cell Biol. 1999;9:81–85. doi: 10.1016/s0962-8924(98)01423-8. [DOI] [PubMed] [Google Scholar]
- 32.Lucić V, Förster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 33.Kanegasaki S, Wright A. Studies on the mechanism of phage adsorption: interaction between phage epsilon15 and its cellular receptor. Virology. 1973;52:160–173. doi: 10.1016/0042-6822(73)90406-6. [DOI] [PubMed] [Google Scholar]
- 34.McConnell M, Wright A. Variation in the structure and bacteriophage-inactivating capacity of Salmonella anatum lipopolysaccharide as a function of growth temperature. J Bacteriol. 1979;137:746–751. doi: 10.1128/jb.137.2.746-751.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda K, Uetake H. In vitro interaction between phage and receptor lipopolysaccharide: a novel glycosidase associated with Salmonella phage 15 . Virology. 1973;52:148–159. [PubMed] [Google Scholar]
- 36.Dreiseikelmann B. Translocation of DNA across bacterial membranes. Microbiol Rev. 1994;58:293–316. doi: 10.1128/mr.58.3.293-316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaback HR. Bacterial Membranes. In: Jakoby WB, editor. Methods in Enzymology, Volume 22. Academic Press; San Diego, California, USA: 1971. pp. 99–120. [Google Scholar]
- 38.Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid MF, Booth CR. Methods for aligning and for averaging 3D volumes with missing data. J Struct Biol. 2008;161:243–248. doi: 10.1016/j.jsb.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Böhm J, Lambert O, Frangakis AS, Letellier L, Baumeister W, Rigaud JL. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr Biol. 2001;11:1168–1175. doi: 10.1016/s0960-9822(01)00349-9. [DOI] [PubMed] [Google Scholar]
- 41.Bonhivers M, Ghazi A, Boulanger P, Letellier L. FhuA, a transporter of the Escherichia coli outer membrane, is converted into a channel upon binding of bacteriophage T5. EMBO J. 1996;15:1850–1856. [PMC free article] [PubMed] [Google Scholar]
- 42.Labedan B, Goldberg EB. Requirement for membrane potential in injection of phage T4 DNA. Proc Natl Acad Sci U S A. 1979;76:4669–4673. doi: 10.1073/pnas.76.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalasauskaite EV, Kadisaite DL, Daugelavicius RJ, Grinius LL, Jasaitis AA. Studies on energy supply for genetic processes. Requirement for membrane potential in Escherichia coli infection by phage T4. Eur J Biochem. 1983;130:123–130. [PubMed] [Google Scholar]
- 44.Perez GL, Huynh B, Slater M, Maloy S. Transport of phage P22 DNA across the cytoplasmic membrane. J Bacteriol. 2009;191:135–140. doi: 10.1128/JB.00778-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz M. Reversible interaction between coliphage lambda and its receptor protein. J Mol Biol. 1975;99:185–201. doi: 10.1016/s0022-2836(75)80167-7. [DOI] [PubMed] [Google Scholar]
- 46.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dürwald H, Hoffmann-Berling H. Endonuclease-I-deficient and ribonuclease I-deficient Escherichia coli mutants. J Mol Biol. 1968;34:331–346. doi: 10.1016/0022-2836(68)90257-x. [DOI] [PubMed] [Google Scholar]
- 48.Wu SI, Lo SK, Shao CP, Tsai HW, Hor LI. Cloning and characterization of a periplasmic nuclease of Vibrio vulnificus and its role in preventing uptake of foreign DNA. Appl Environ Microbiol. 2001;67:82–88. doi: 10.1128/AEM.67.1.82-88.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci U S A. 2010;107:2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartwieg E, Bazinet C, King J. The DNA injection apparatus of phage p22. Biophys J. 1986;49:24–26. doi: 10.1016/S0006-3495(86)83579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Israel V, Rosen H, Levine M. Binding of bacteriophage P22 tail parts to cells. J Virol. 1972;10:1152–1158. doi: 10.1128/jvi.10.6.1152-1158.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.São-José C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J Biol Chem. 2006;281:11464–11470. doi: 10.1074/jbc.M513625200. [DOI] [PubMed] [Google Scholar]
- 53.Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J Mol Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- 54.Poteete AR, King J. Functions of two new genes in Salmonella phage P22 assembly. Virology. 1977;76:725–739. doi: 10.1016/0042-6822(77)90254-9. [DOI] [PubMed] [Google Scholar]
- 55.Plisson C, White HE, Auzat I, Zafarani A, São-José C, Lhuillier S, Tavares P, Orlova EV. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007;26:3720–3728. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Locker CR, Fuller SD, Harvey SC. DNA organization and thermodynamics during viral packing. Biophys J. 2007;93:2861–2869. doi: 10.1529/biophysj.106.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evilevitch A, Fang LT, Yoffe AM, Castelnovo M, Rau DC, Parsegian VA, Gelbart WM, Knobler CM. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophys J. 2008;94:1110–1120. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Löf D, Schillén K, Jönsson B, Evilevitch A. Forces controlling the rate of DNA ejection from phage lambda. J Mol Biol. 2007;368:55–65. doi: 10.1016/j.jmb.2007.01.076. [DOI] [PubMed] [Google Scholar]
- 59.Serwer P, Wright ET, Hakala KW, Weintraub ST. Evidence for bacteriophage T7 tail extension during DNA injection. BMC Res Notes. 2008;1:36. doi: 10.1186/1756-0500-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delbrück M. THE GROWTH OF BACTERIOPHAGE AND LYSIS OF THE HOST. J Gen Physiol. 1940;23:643–660. doi: 10.1085/jgp.23.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boulanger P, Jacquot P, Plançon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem. 2008;283:13556–13564. doi: 10.1074/jbc.M800052200. [DOI] [PubMed] [Google Scholar]
- 62.HERSHEY AD, CHASE M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- 64.Meselson M, Stahl FW, Vinograd J. EQUILIBRIUM SEDIMENTATION OF MACROMOLECULES IN DENSITY GRADIENTS. Proc Natl Acad Sci U S A. 1957;43:581–588. doi: 10.1073/pnas.43.7.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 66.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 68.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 69.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]