Abstract
CXCR7 is a receptor for chemokines including CXCL12 (SDF-1), a molecule that promotes tumor growth and metastasis in breast cancer and other malignancies. Building upon the recent observation that CXCR7 sequesters CXCL12, we investigated mechanisms for CXCR7-dependent uptake of chemokines. Breast cancer cells expressing CXCR7 accumulated chemokines CXCL12 and CXC11 present at concentrations < 1 ng/ml, unlike cells expressing CXCR4. CXCR7-dependent accumulation of chemokines was reduced by inhibitors of clathrin-mediated endocytosis. Following CXCR7-mediated internalization, CXCL12 trafficked to lysosomes and was degraded, although levels of CXCR7 remained stable. CXCR7 reduced CXCL12 in the extracellular space, limiting amounts of chemokine available to acutely stimulate signaling through CXCR4. CXCR7 constitutively internalized and recycled to the cell membrane even in the absence of ligand, and addition of chemokines did not significantly enhance receptor internalization. Chemokines at concentrations less than the Kd for ligand-receptor binding did not alter levels of CXCR7 at the cell surface. Higher concentrations of chemokine ligands reduced total cell surface expression of CXCR7 without affecting receptor internalization, indicating that receptor recycling was inhibited. CXCR7-dependent uptake of chemokines and receptor trafficking were regulated by β-arrestin 2. These studies establish mechanisms through which CXCR7 regulates availability of chemokine ligands in the extracellular space.
Keywords: breast cancer, chemokines, chemokine receptors, bioluminescence imaging, luciferase, protein fragment complementation
Introduction
Chemokine CXCL12 (SDF-1) signaling through receptor CXCR4 has been implicated in tumor progression and metastasis in breast cancer and more than 20 other malignancies (Balkwill, 2004; Kollmar et al., 2007; Luker and Luker, 2006; Muller et al., 2001; Orimo et al., 2005; Vandercappellen et al., 2008; Zhang et al., 2007). CXCL12 secreted by stromal fibroblasts in primary breast tumors increases proliferation of tumor cells and angiogenesis (Allinen et al., 2004; Orimo et al., 2005). Gradients of CXCL12 are proposed to enhance cell migration, intravasation of cancer cells into the vasculature, and organ-specific metastases (Kang et al., 2003; Muller et al., 2001; Smith et al., 2004). More recently, chemokine CXCL11 (I-TAC) has been implicated in cancer, although functions of this molecule in malignancy are less defined. Some studies suggest that CXCL11 accelerates progression of malignancies such as ovarian cancer (Atanackovic et al., 2008; Furuya et al., 2007), while other research indicates that this chemokine limits tumor growth by inhibiting angiogenesis or directly reducing cell proliferation and survival (Yang et al., 2005) (Strieter et al., 2006). Given multiple functions of CXCL12 and CXCL11 in cancer, molecules that control availability of these chemokines will substantially impact tumor progression and represent potential therapeutic targets.
Bioavailability of chemokines is controlled by several different mechanisms. CXCL12 and CXCL11 bind to heparan sulfate glycosaminoglycans on cell membranes and extracellular proteins, increasing local concentrations of chemokine available for signaling (Fermas et al., 2008; Laquri et al., 2007). Binding to glycosaminoglycans also protects chemokines from degradation by extracellular enzymes (Hoogewerf et al., 1997). For both CXCL12 and CXCL11, extracellular proteases remove amino acids required for signaling through CXCR4 and CXCR3, respectively, and generate receptor antagonists (Cox et al., 2008; Proost et al., 2001; Sadir et al., 2004). Proteolysis has been regarded as the primary mechanism for regulating signaling by secreted CXCL12 and CXCL11.
Chemokines also may be controlled by decoy receptors, which sequester chemokines without activating signaling pathways (Hansell et al., 2006). Decoy receptors generally act as tumor suppressors. For example, decoy receptor D6 in breast cancer limits proliferation of malignant cells, tumor angiogenesis, and metastasis in mice. D6 expression correlates positively with disease-free survival in patients (Wu et al., 2008). Such studies further emphasize effects of chemokines on tumor progression and metastasis.
Recent studies suggest that CXCR7 functions as a decoy receptor for CXCL12 (Boldajipour et al., 2008; Naumann et al., 2010). CXCR7 removes CXCL12 from the extracellular space and is essential for migration of primordial germ cells in zebrafish, potentially by controlling gradients of CXCL12 to maintain signaling through CXCR4. However, mechanisms of action for CXCR7 in sequestering chemokines and the fate of internalized ligands remain undetermined, the latter of which is important because decoy receptors may degrade or release chemokines (Pruenster et al., 2009).
In this study, we showed that CXCR7 internalized chemokine ligands and degraded these molecules in lysosomes, controlling amounts of chemokines in the extracellular space for signaling through other receptors. Accumulation of chemokines was mediated by constitutive internalization and recycling of CXCR7 to the cell membrane. Elevated concentrations of chemokines did not significantly enhance CXCR7 internalization, but ligands more substantially reduced total CXCR7 on the cell surface. CXCR7-dependent uptake of chemokines and receptor internalization proceeded through clathrin-mediated endocytosis and the cytosolic scaffolding protein β-arrestin 2. Association of CXCR7 with β-arrestin 2 also regulated ligand-dependent depletion of CXCR7 from the cell surface. These studies establish mechanisms through which CXCR7 regulates availability of chemokines in the tumor microenvironment.
Results
CXCR7 promotes accumulation of chemokine ligands CXCL12 and CXCL11
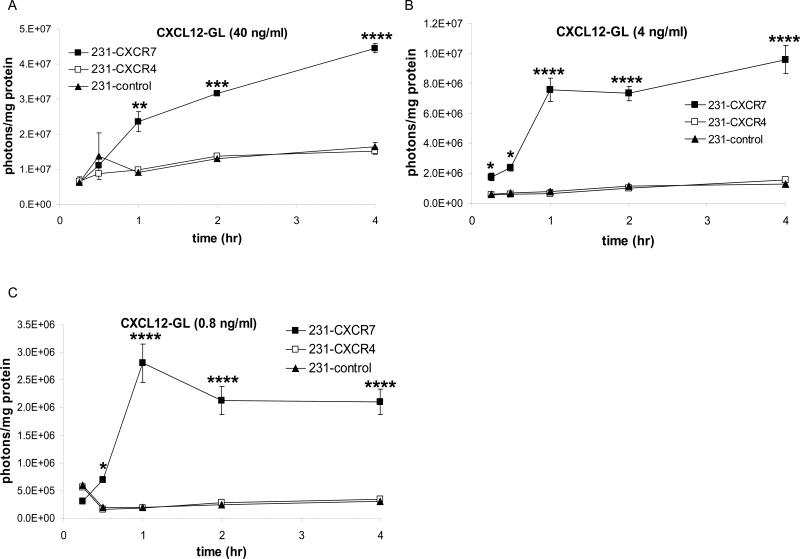
To quantify CXCR7-dependent accumulation of chemokines, we used MDA-MB-231 human breast cancer cells stably transduced with CXCR7, CXCR4, or GFP (231-CXCR7, 231-CXCR4, and 231-control, respectively. MDA-MB-231 cells do not express endogenous CXCR7 (Luker et al., 2009a). We incubated cells with ≈ 40, 4, or 0.8 ng/ml of CXCL12 fused to Gaussia luciferase (CXCL12-GL). CXCL12-GL retains normal signaling functions of unfused CXCL12 and allows quantification by bioluminescence imaging (Luker et al., 2009a). For all concentrations of chemokine, 231-CXCR7 cells accumulated significantly more CXCL12-GL than 231-CXCR4 or 231-control cells within 1 hour (p < 0.01) (Fig 1A-C). When incubated with 40 ng/ml CXCL12-GL, 231-CXCR7 cells accumulated increasing amounts of chemokine (Fig 1A). At lower concentrations near the ≈ 4 ng/ml Kd for CXCL12-CXCR7 binding (Balabanian et al., 2005; Burns et al., 2006), CXCR7-mediated uptake of chemokine either reached steady-state (4 ng/ml) or declined (0.8 ng/ml) after 1 hour (Fig 1B, C). Accumulation of CXCL12-GL did not differ between 231-CXCR4 and 231-control cells. 231-CXCR7 cells also accumulated CXCL11, the other ligand for this receptor, when incubated with CXCL11-GL at concentrations ranging from 0.8 – 30 ng/ml (Fig S1A, B). These data show that CXCR7 promotes uptake and accumulation of even low concentrations of chemokine ligands.
Figure 1. CXCR7-dependent accumulation of CXCL12.
MDA-MB-231 cells stably expressing CXCR4 (231-CXCR4), CXCR7 (231-CXCR7), or GFP (231-control) were incubated with ≈ 40 (A), 4 (B), or 0.8 (C) ng/ml CXCL12-GL. Cells were acid washed to remove extracellular chemokine prior to bioluminescence imaging. Data were expressed as mean values of photons/cell protein per well ± SEM (n = 4 each). *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001.
CXCR7 degrades CXCL12 and limits acute CXCR4 activation
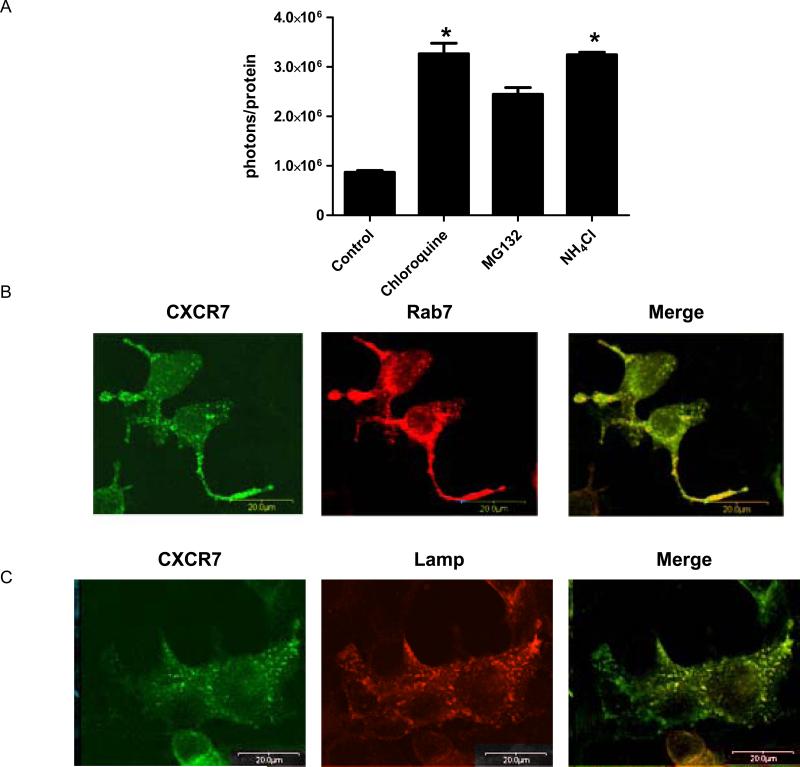
Internalized chemokines typically are degraded, although receptors including CXCR4 and Darc may resecrete chemokines into the extracellular space (Dar et al., 2005; Pruenster et al., 2009). To investigate outcomes for internalized CXCL12, we pulse labeled 231-CXCR7 cells with ≈ 40 ng/ml CXCL12-GL and then switched cells to chemokine-free medium. Approximately 50% of CXCL12-GL signal was lost after 4 hours as quantified by bioluminescence in live cells and lysates (Fig 2A and data not shown). Loss of CXCL12-GL was blocked by lysosomotropic agents chloroquine and NH4Cl (p < 0.05), while treatment with proteasome inhibitor MG132 did not significantly increase levels of intracellular chemokine as determined by post-hoc testing for multiple comparisons. Differences in levels of internalized CXCL12-GL were confirmed by Western blotting (Fig S2). Degradation of CXCL11-GL in 231-CXCR7 cells also was inhibited by chloroquine (Fig S3), but CXCR7 itself remained stable as quantified with a CXCR7-Gaussia luciferase fusion protein (Fig S4). Culture media from 231-CXCR7 cells during the 4-hour chase period did not contain detectable CXCL12-GL (data not shown), showing that internalized chemokines were not released.
Figure 2. CXCR7-dependent degradation of chemokines in lysosomes.
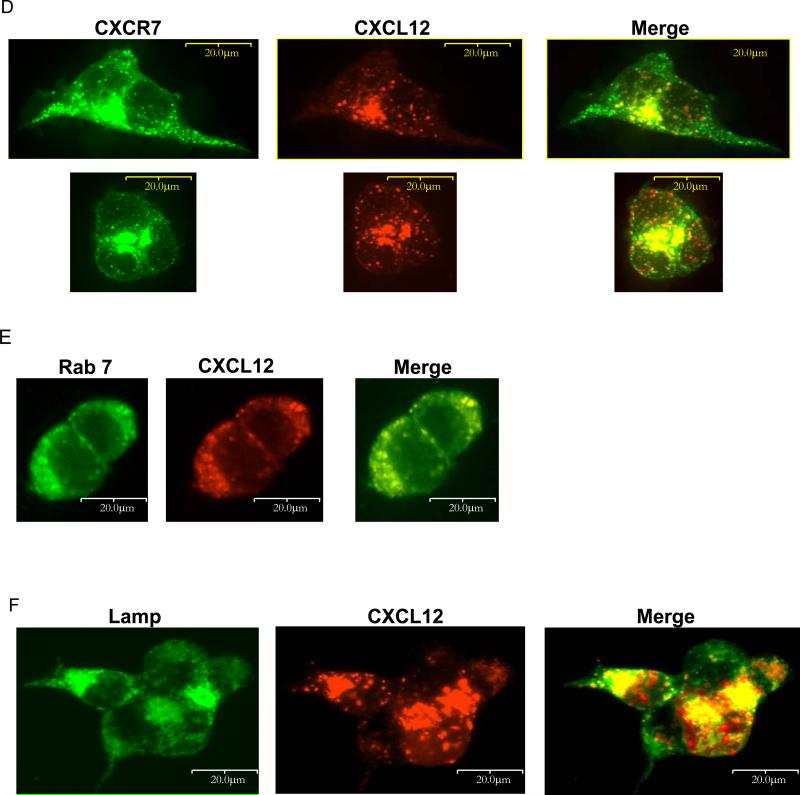
(A) 231-CXCR7 cells were incubated with ≈ 40 ng/ml CXCL12-GL for 15 minutes, washed, and then incubated without bioluminescent chemokine for 4 hours in medium containing 50 μM chloroquine, 50 mM NH4CL, 25 μM MG132, or vehicle. Cells were acid washed to remove extracellular chemokine before bioluminescence imaging. Photons were normalized to total cellular protein in each sample, and data were expressed as mean values + SEM for per cent initial uptake of CXCL12-GL after the 15 minute pulse (n = 4 each). *, p < 0.05. (B, C) 293T cells were co-transfected with CXCR7-citrine (depicted as green) and either Rab7-CFP (B) or Lamp-CFP (depicted as red) (C), cultured in normal growth medium, and imaged by fluorescence microscopy two days after transfection using a 40X objective. (D – F) 293T cells were transfected with CXCR7-GFP (D) or co-transfected with unfused CXCR7 and Rab7-CFP (E) or Lamp-CFP (F). Two days after transfection, cells were incubated for 30 minutes with ≈ 8 ng/ml CXCL12-cherry. Cells were fixed for fluorescence microscopy and viewed with a 40X objective.
We used fluorescence microscopy to monitor intracellular localization of CXCR7 and internalized CXCL12. Under baseline conditions, intracellular CXCR7 partially co-localized with markers of late endosomes (Rab7) and lysosomes (Lamp) (Fig 2B, C). After incubation for 30 or 60 minutes with ≈ 8 ng/ml CXCL12-cherry fusion protein, some internalized chemokine localized with CXCR7-GFP in intracellular vesicles (Fig 2D and data not shown). Similar to CXCR7, CXCL12-cherry was present in Rab7-positive late endosomes and Lamp-positive lysosomes (Fig 2E, F), indicating that CXCR7 trafficks chemokines through late endosomes to lysosomes for degradation. Similar co-localization of CXCR7 with endogenous Rab7 and lysosomes was demonstrated by immunostaining (Fig S5).
To quantify effects of CXCR7 on extracellular CXCL12, we measured depletion of CXCL12-GL by 231-CXCR7 or 231-CXCR4 cells. From a starting concentration of ≈ 40 ng/ml CXCL12, there was ≈ 50% loss of chemokine in medium incubated for a total of 1 hour with 231-CXCR7 cells, while < 20% of CXCL12-GL was depleted during incubation with 231-CXCR4 cells (Figure 3A) (p < 0.05). Using a comparable starting concentration of Gaussia luciferase, losses of unfused enzyme did not differ between cell types and were indistinguishable from effects of 231-CXCR4 cells on CXCL12-GL.
Figure 3. CXCR7 depletes CXCL12 and limits CXCR4 signaling.
(A, B) 293T cells were transfected with CXCR7. Localization of CXCR7 and Rab7 (A) 231-CXCR7 or 231-CXCR4 cells were incubated with ≈ 40 ng/ml CXCL12-GL or a comparable amount of unfused GL for 30 minutes and then transferred to the same cell type for an additional 30 minutes. Following two rounds of depletion, bioluminescence from CXCL12-GL and GL was quantified in 5 μl of supernatants. Data were expressed as mean values + SEM for per cent of initial bioluminescence (n = 4 each). (B) 231-CXCR4 cells were incubated with supernatants from B for 5 minutes and then analyzed by Western blot for phosphorylated AKT. Membranes were reprobed for total AKT as a loading control. (C) Supernatants from B were incubated with 293T cells stably expressing a firefly luciferase complementation reporter for activation of CXCR4 (CXCR4-NLuc and β-arrestin 2-CLuc). CXCL12-GL and GL that had not been incubated with 231-CXCR4 or 231-CXCR7 cells were used as controls. Bioluminescence was quantified in live cells, normalized to total protein in each sample, and graphed as mean values + SEM (n = 4 each). *, p < 0.05.
Having established that CXCR7 decreases extracellular CXCL12, we measured effects on CXCR4 signaling upon acute exposure to ligand. Following incubation with 231-CXCR7 or 231-CXCR4 cells, we transferred medium containing the remaining CXCL12-GL to new 231-CXCR4 cells and analyzed phosphorylation of AKT, a protein activated by CXCR4 signaling through Gαi in 231 cells (Zhao et al., 2008). There was reduced activation of AKT by CXCL12-GL in medium transferred from 231-CXCR7 cells relative to medium from 231-CXCR4 cells (Fig 3B). We also measured CXCL12-dependent recruitment of β-arrestin 2 to CXCR4, another early step in signaling, using firefly luciferase complementation (Luker et al., 2008). CXCL12-GL from 231-CXCR4 medium and CXCL12-GL that had not been pre-incubated with cells produced comparable complementation of β-arrestin 2 and CXCR4 (Fig 3C). Medium incubated with 231-CXCR7 cells produced significantly less activation of CXCR4, consistent with partial depletion of CXCL12-GL (p < 0.05). Unfused GL had no effect on bioluminescence. These data establish that CXCR7-dependent depletion of CXCL12 from the extracellular space reduces acute CXCR4 signaling.
Chemokine regulation of CXCR7 internalization and cell surface expression
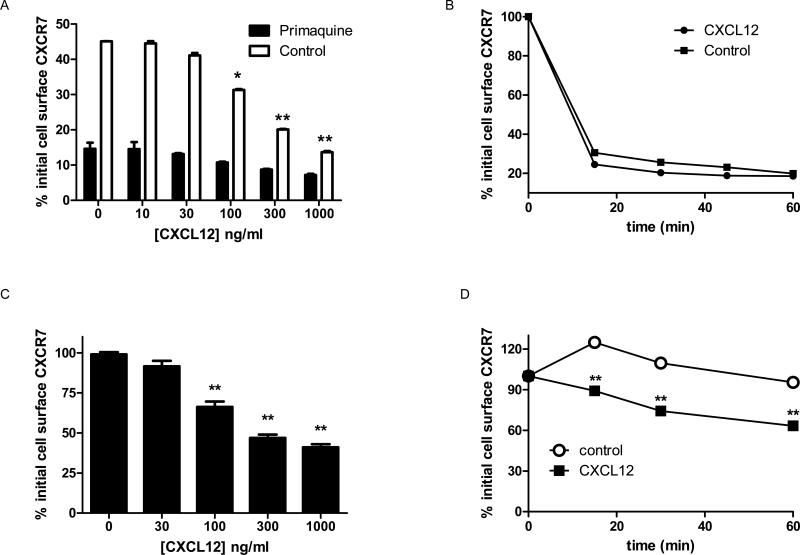
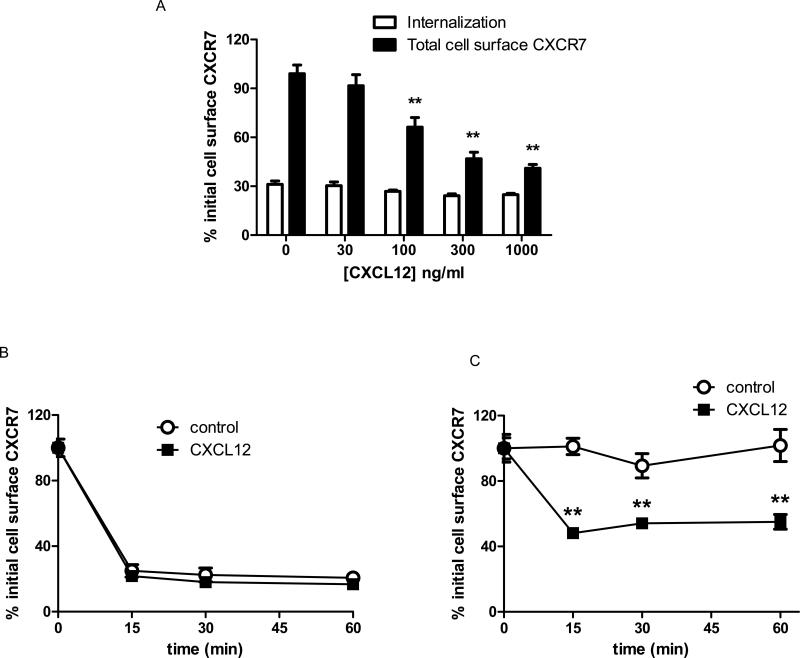
While signaling chemokine receptors typically remain at the cell surface until stimulated by ligand (Galliera et al., 2004; Weber et al., 2004), decoy receptors may internalize constitutively. We used flow cytometry to quantify internalization of CXCR7 in the presence or absence of primaquine, an agent known to block recycling endosomes (van Weert et al., 2000). In the absence of ligand, > 85% of CXCR7 internalized from the surface of primaquine-treated cells within 30 minutes. Incubation with 100 ng/ml or more CXCL12 minimally increased receptor internalization from the cell surface to ≈ 90% (Fig 4A). Without primaquine, cell surface levels of CXCR7 decreased by only ≈ 55% after 30 minutes of incubation without ligand, showing that ≈ 30% of antibody-labeled receptors internalized and recycled to the cell membrane during this time. At 100 ng/ml or higher concentrations of CXCL12, differences between cells incubated without or with primaquine progressively decreased. Net internalization of CXCR7 was ≈ 85% in cells incubated with 1 μg/ml CXCL12 and no primaquine, indicating that high concentrations of ligand limited CXCR7 recycling to the cell membrane. Approximately 75% of CXCR7 internalization occurred within 15 minutes, and treatment with 100 ng/ml CXCL12 only minimally increased kinetics of receptor internalization (Fig 4B). CXCL11 at 100 ng/ml or higher also modestly enhanced net internalization of CXCR7 (p < 0.05 relative to untreated cells), suggesting that high concentrations of this chemokine also may limit recycling of internalized CXCR7 to the cell membrane (Fig S6).
Figure 4. Internalization and cell membrane levels of overexpressed CXCR7.
(A) 231-CXCR7 cells were labeled with anti-CXCR7 primary antibody before treatment with increasing concentrations of CXCL12 for 30 minutes to determine receptor internalization in the presence or absence of primaquine to block recycling endosomes. (B) Cells were labeled as in A and then incubated without or with 100 ng/ml CXCL12 in the presence of primaquine. (C) 231-CXCR7 cells were incubated for 30 minutes with increasing concentrations of CXCL12, and then total levels of cell surface CXCR7 were quantified by flow cytometry. (D) Total cell surface CXCR7 was quantified at various times after incubation without or with 100 ng/ml CXCL12. Mean fluorescence intensity for cell surface CXCR7 was measured by flow cytometry, and data were expressed as the percentage of initial cell surface CXCR7. Error bars denote SEM. *, p < 0.05; **, p < 0.01.
We measured effects of ligands on total cell surface CXCR7. Over 30 minutes, levels of CXCR7 on the cell membrane remained relatively constant in untreated cells and cells treated with up to 30 ng/ml CXCL12 (Fig 4C). Higher concentrations of CXCL12 produced dose-dependent reductions in CXCR7 on the cell membrane, resulting in ≈ 65% less receptor at the surface following treatment with 1 μg/ml CXCL12 (Fig 4C). Incubation with 100 ng/ml CXCL12 significantly decreased total cell surface CXCR7 by 15 minutes with progressively greater reductions through 60 minutes (p < 0.05 relative to untreated cells at comparable times) (Fig 4D). Within the 60 minute period of these assays, blocking protein synthesis with cycloheximide did not affect results for total cell surface CXCR7 (data not shown). CXCL11 also produced dose-dependent reductions in total cell surface CXCR7 (Fig S6).
We investigated internalization of endogenous CXCR7 in human MCF-7 breast cancer cells, which show CXCR7-dependent accumulation of CXCL12 (Luker et al., 2009a). Approximately 70% of cell surface CXCR7 internalized in 30 minutes without ligand, and incubation with CXCL12 did not alter internalization significantly (Fig 5A). Similar to 231-CXCR7 cells, almost all receptor internalization occurred by 15 minutes (Fig 5B). However, primaquine did not increase net internalization of CXCR7 in MCF-7 cells (data not shown), indicating that kinetics of receptor recycling in these cells were slower than 231-CXCR7 cells. MCF-7 cells treated with 100 ng/ml CXCL12 or higher had significantly reduced total CXCR7 on the cell membrane within 15 minutes of incubation, which persisted through 60 minutes (p < 0.01) (Fig 5A, C). These data establish that CXCR7 undergoes ligand-independent internalization with the extent and kinetics of recycling varying between breast cancer cell lines. Chemokines minimally alter receptor internalization while having more pronounced effects to reduce total levels of cells surface CXCR7.
Fig 5. Internalization and cell membrane levels of endogenous CXCR7.
(A) MCF-7 cells were labeled with anti-CXCR7 antibody and then incubated with various concentrations of CXCL12 for 30 minutes. Receptor internalization and total cell surface levels of CXCR7 at the end of the incubation period were measured by flow cytometry. (B, C) Cells were treated with 100 ng/ml CXCL12 for increasing periods of time before assaying receptor internalization (B) or total cell surface CXCR7 (C) by flow cytometry. Data were expressed as mean values for per cent of initial cell surface CXCR7. *, p < 0.05; **, p < 0.01.
β-arrestin 2-dependent trafficking of CXCR7 and accumulation of CXCL12
We and othershave shown that CXCR7 interacts with β-arrestin 2, and ligand-dependent association of these proteins increases over time (Kalatskaya et al., 2009; Luker et al., 2009c; Zabel et al., 2009). Since β-arrestins mediate clathrin-mediated endocytosis and trafficking of several other chemokine receptors (Shenoy and Lefkowitz, 2003), we hypothesized that uptake of chemokines and internalization of CXCR7 require β-arrestins. We initially incubated 231-CXCR7 cells with ≈ 4 ng/ml CXCL12-GL in the presence or absence of agents that disrupt clathrin-mediated endocytosis by blocking clathrin (hypertonic sucrose) or dynamin (dynasore) (Heuser and Anderson, 1989) (Macia et al., 2006). Each inhibitor significantly reduced intracellular uptake of chemokine (p < 0.05) (Fig 6A), showing that CXCR7-dependent accumulation of ligands proceeds via clathrin-mediated endocytosis.
Figure 6. Clathrin-mediated endocytosis and β-arrestin 2 regulate CXCR7-dependent accumulation of chemokines, receptor internalization, and total cell membrane CXCR7.
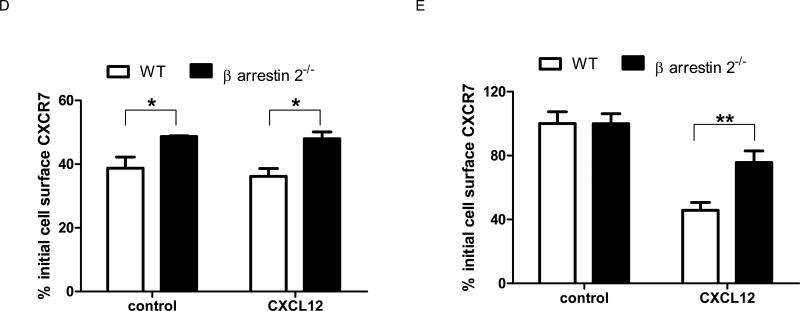
(A) 231-CXCR7 (CXCR7) and 231-control (231) cells were incubated with 0.4M sucrose, 80 μM dynasore, or vehicle control for 30 minutes prior to incubation with ≈ 4 ng/ml CXCL12-GL for various periods of time. Amounts of intracellular chemokine were normalized to total protein per well and graphed as mean ± SEM at each time point (n = 4 per condition). *, p < 0.05 for 231-CXCR7 control samples relative to 231-CXCR7 cells with sucrose or dynasore. (B) β-arrestin 1-/-, β-arrestin 2-/-, and matched wild-type mouse embryonic fibroblasts (MEFs) were transduced with lentiviruses for CXCR7-GFP. Expression of CXCR7-GFP was measured by flow cytometry for GFP. Filled symbols are untransduced cells, while open symbols denote MEFs transduced with CXCR7-GFP. (C) Stably transduced wild-type (WT), β-arrestin 1-/-, or β-arrestin 2-/- MEFs were incubated with ≈ 4 ng/ml CXCL12-GL for 30 minutes in medium containing 100 nM of the CXCR7-specific inhibitor CCX733 or vehicle. Cells were acid washed to remove extracellular chemokine prior to quantifying CXCL12-GL bioluminescence in living cells. Data were graphed as mean values for photons/mg protein + SEM (n = 4 each). (D) Wild-type and β-arrestin 2-/- MEFs were incubated for 30 minutes with 1 mM primaquine in the presence or absence of 100 ng/ml CXCL12 for 30 minutes before quantifying internalization of cell surface CXCR7 by flow cytometry. (E) MEFs were incubated for 30 minutes with or without 100 ng/ml CXCL12, and total mean fluorescence intensity for cell surface CXCR7 at the end of the incubation period was quantified by flow cytometry. Data in D and E were expressed as per cent initial cell surface CXCR7 as in Figure 2. *, p < 0.05; **, p < 0.0.
In addition to β-arrestin 2, the related adapter protein β-arrestin 1 may regulate endocytosis of chemokine and other seven transmembrane receptors (Kohout et al., 2001). We expressed CXCR7-GFP in β-arrestin 1-/-, β-arrestin 2-/-, and matched wild-type mouse embryonic fibroblasts (MEFs). Matched pairs of MEFs had comparable expression of CXCR7-GFP (Fig 6B). Accumulation of CXCL12-GL did not differ between WT and β-arrestin 1-/- MEFs, while accumulation was significantly lower in β-arrestin 2-/- MEFs (p < 0.01) (Fig 6C). Uptake was of CXCL12-GL was blocked with CCX733, a specific inhibitor of CXCR7. These studies show that β-arrestin 2 is required for CXCR7-dependent uptake of chemokines.
Using primaquine to block recycling endosomes, we showed that ligand-independent internalization of cell surface CXCR7 over 30 minutes was significantly greater in wild-type MEFs relative to β-arrestin 2-/- MEFs (Fig 6D) (p < 0.05). Hypertonic sucrose also reduced ligand-dependent internalization of CXCR7 in 231-CXCR7 cells, providing further evidence for trafficking through clathrin-mediated endocytosis (Fig S7). While loss of β-arrestin 2 did not alter total cell surface CXCR7 under basal conditions, CXCL12 reduced total cell surface CXCR7 to a greater extent in wild-type cells (p < 0.01) (Fig 6E). These results demonstrate that β-arrestin 2 regulates internalization and ligand-dependent depletion of CXCR7 from the cell membrane.
Removing CXCL12 increases receptor recycling
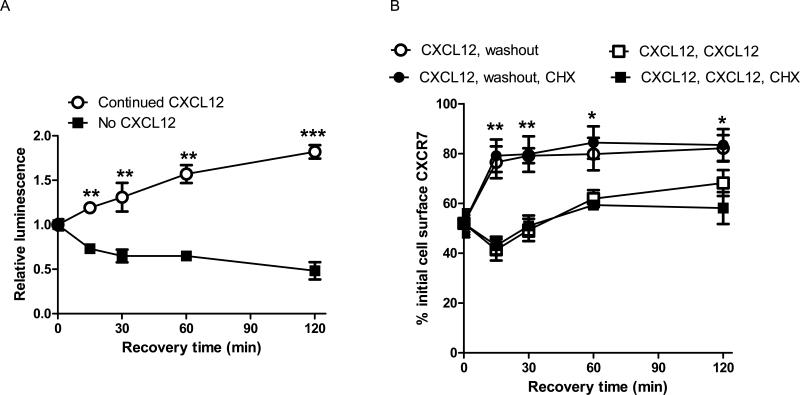
We hypothesized that CXCR7 and β-arrestin 2 would dissociate upon removal of CXCL12, increasing recovery of receptors at the cell membrane. We quantified association of CXCR7 and β-arrestin 2 using a firefly luciferase complementation assay (Luker et al., 2009c). MDA-MB-231 breast cancer cells stably transduced with the CXCR7/β-arrestin complementation reporter were treated with 300 ng/ml CXCL12 for 30 minutes to promote association of CXCR7 and β-arrestin 2. Following removal of chemokine, bioluminescence decreased by ≈ 20% within 15 minutes and then declined gradually at subsequent time points, showing dissociation of CXCR7 and β-arrestin 2 (Fig 7A). Consistent with our previous work, bioluminescence from association of CXCR7 and β-arrestin 2 increased over time when cells were maintained with CXCL12 (p < 0.005) (Luker et al., 2009c).
Figure 7. Removing CXCL12 dissociates CXCR7 from β-arrestin 2 and enhances recovery of cell surface receptors.
(A) 231 CXCR7/β-arrestin complementation reporter cells were incubated with 300 ng/ml CXCL12 for 30 minutes (time 0). Parallel samples of cells were washed and then incubated in medium without or with 300 ng/ml CXCL12 for various periods of time. Firefly luciferase activity produced by association of CXCR7 with β-arrestin 2 was quantified by bioluminescence imaging of living cells. Bioluminescence was normalized to total cell protein per well and expressed as changes in luminescence relative to cells at time 0 with no recovery period. Data points are mean values ± SEM (n = 4 per sample). (B) 231-CXCR7 cells were incubated for 30 minutes with 300 ng/ml CXCL12. Cells then were either maintained in CXCL12 or washed to remove extracellular CXCL12 and transferred to fresh medium. Parallel samples of cells were incubated with 100 μg/ml cycloheximide to block protein synthesis during the recovery time. Cell surface CXCR7 was determined using mean values for fluorescence measured by flow cytometry. Error bars denote SEM. *, p < 0.05; **, p < 0.01, ***, p < 0.005.
By flow cytometry, levels of CXCR7 at the cell surface increased by ≈ 25% within 15 minutes after removing CXCL12 with small additional increases through 2 hours (Fig 7B). Blocking protein synthesis with cycloheximide did not alter recovery of cell surface CXCR7. Increases in cell membrane CXCR7 had similar kinetics to dissociation of CXCR7 and β-arrestin 2, suggesting that β-arrestin 2 contributes to intracellular retention of CXCR7. Cells incubated continuously with CXCL12 had minimal recovery of cell surface CXCR7, mediated largely by synthesis of new receptors. These data demonstrate that suppression of CXCR7 recycling by CXCL12 reverses rapidly following removal of ligand.
Discussion
CXCR7 is expressed in malignant cells and vascular endothelium in many human cancers (Miao et al., 2007; Raggo et al., 2005) (Wang et al., 2008), but mechanisms of action for this receptor remain poorly defined. CXCL11 and CXCL12, the known chemokine ligands for CXCR7, both regulate primary tumor growth and progression to metastatic disease in breast cancer and other common malignancies (Atanackovic et al., 2008; Furuya et al., 2007; Luker and Luker, 2006; Yang et al., 2005). Therefore, functions of CXCR7 to control availability of these chemokines may affect multiple steps in cancer progression. Establishing mechanisms of action for CXCR7 and its effects on homeostasis of these chemokines are essential for understanding chemokine signaling in cancer and targeting these pathways for therapy.
We established that breast cancer cells expressing CXCR7 accumulated CXCL12 over a large range of concentrations in the extracellular space, unlike CXCR4, the other receptor for this chemokine. We also showed that CXCR7 promotes uptake of CXCL11. Following CXCR7-dependent internalization, chemokines trafficked through Rab7-positive late endosomes to lysosomes for degradation and were not released back into the extracellular space. These results build upon recent work showing that CXCR7 scavenges and degrades chemokine ligands (Boldajipour et al., 2008; Naumann et al., 2010). Although internalized chemokines were degraded in cells expressing CXCR7, we established that the receptor remained stable. Stability of CXCR7 during internalization and degradation of chemokines likely contributes to its function as a decoy receptor, unlike CXCR4 and other receptors that may be degraded following ligand binding (Marchese and Benovic, 2001).
CXCR7 constitutively internalized and recycled to the cell surface in the absence of chemokine ligands, maintaining relatively constant levels of receptor at the cell membrane. Chemokines did not significantly increase receptor internalization. However, ligands at 100 ng/ml or higher concentrations reduced total levels of CXCR7 at the cell membrane, likely by inhibiting receptor recycling. This concentration is considerably higher than Kd values for CXCL11 and CXCL12 binding to CXCR7 (≈ 30 and ≈ 4 ng/ml for CXCL11 and CXCL12, respectively) (Balabanian et al., 2005) (Burns et al., 2006). Receptor internalization and accumulation of chemokines were partially dependent on β-arrestin 2, indicating that constitutive internalization of CXCR7 is necessary for uptake of chemokines from the extracellular space. Recycling of CXCR7 to the cell surface following incubation with CXCL12 had comparable kinetics to dissociation of CXCR7 and β-arrestin 2, further indicating that β-arrestin 2 regulates trafficking of CXCR7. Trafficking of CXCR7 most closely parallels decoy receptor D6, although chemokine ligands for D6 increase levels of receptor at the cell membrane (Galliera et al., 2004; Weber et al., 2004). Further studies are needed to establish how different concentrations of chemokine ligands regulate the extent and kinetics of CXCR7 recycling.
By removing chemokines from the extracellular environment, CXCR7 regulates signaling through CXCR4. CXCR7-mediated depletion of CXCL12 reduces acute activation of CXCR4 and its downstream signaling pathways. Similar results for effects of CXCR7 on immediate responses of CXCR4 to CXCL12 have been described, although the prior study reported that cells expressing CXCR7 completely abrogated CXCR4 signaling rather than decreasing it as we observed (Boldajipour et al., 2008). While we used the same experimental protocol as the prior study, differing results may be due to discordant concentrations of CXCL12 and/or expression levels of CXCR4 and/or CXCR7. Nevertheless, both studies show that CXCR7 limits CXCL12-CXCR4 signaling in response to short-term stimulation.
CXCR7-dependent depletion of CXCL12 and inhibition of short-term CXCR4 signaling might be expected to oppose effects of CXCL12-CXCR4 on growth and metastasis of cancer cells. However, CXCR7 increases tumor progression in animal models of several different cancers (Miao et al., 2007; Raggo et al., 2005; Wang et al., 2008). Although some effects of CXCR7 in cancer are independent of CXCR4, CXCR7 is known to heterodimerize with CXCR4 and modify CXCR4 signaling, suggesting that CXCR7 may combine with CXCR4 to promote tumor growth and/or metatasis (Levoye et al., 2009; Luker et al., 2009b; Sierro et al., 2007). Integrated functions of CXCR7 on somatic cells and CXCR4 on germ cells are necessary for proper chemotaxis of the latter cell type during zebrafish development, where CXCR7 is proposed to maintain gradients of CXCL12 (Boldajipour et al., 2008). Since fundamental processes in development frequently are recapitulated in cancer, CXCR7 may control levels and distribution of CXCL12 in the tumor microenvironment to potentiate CXCR4 signaling in malignant and/or stromal cells. We currently are investigating to what extent CXCR7 regulates availability of CXCL12 and functions of CXCR4 in breast cancer in vivo. In summary, we established that CXCR7 undergoes constitutive internalization and ligand-regulated recycling regulated by β-arrestin 2 to accumulate and degrade chemokines. These results provide a mechanism through which CXCR7 may modify gradients of chemokines and chemokine receptor signaling in cancer. Functions of CXCR7 as a decoy receptor do not preclude alternative mechanisms of action, such as direct activation of signaling pathways or cellular responses (Wang et al., 2008; Zabel et al., 2009). Understanding effects of CXCR7 on chemokine homeostasis in the tumor microenvironment will advance ongoing efforts to target chemokine receptors for therapy in breast cancer and other malignancies.
Materials and Methods
Cells
MDA-MB-231 and MCF-7 human breast cancer cells (ATCC, Manassas, VA, USA), HEK293T cells (Open Biosystems, Huntsville, AL, USA), wild-type, β-arrestin 1-/-, and β-arrestin 2-/- mouse embryonic fibroblasts (MEFs) (Robert Lefkowitz, Duke University) were cultured in DMEM (Invitrogen, Carlsbad, CA, USA), 10% fetal bovine serum, 1% glutamine, and 0.1% penicillin/streptomycin/gentamicin. We used MDA-MB-231 cells stably transduced with CXCR4-GFP (231-CXCR4), CXCR7-GFP (231-CXCR7), or GFP alone (231-control) (Luker et al., 2009a). MDA-MB-231 cells stably expressing firefly luciferase complementation reporters for CXCR7 and β-arrestin 2 (CXCR7-NLuc and β-arrestin 2-CLuc) and 293T cells with CXCR4-NLuc and β-arrestin 2-CLuc have been described (Luker et al., 2008; Luker et al., 2009c).
Plasmids
To fuse CXCR7 to Gaussia luciferase (CXCR7-GL), we digested CXCR7-GFP with EcoRI and NotI and replaced GFP with GL from CXCL12-GL (Luker et al., 2009a). GL used in fusion constructs lacks the native secretion signal. We fused CXCL12 to mCherry (Roger Tsien, UCSD) by digesting the CXCL12-GL plasmid with EcoRI and NotI and replacing GL with mCherry. To fuse CXCR7 to fluorescent protein citrine (Griesbeck et al., 2001), we digested CXCR7-GFP with AgeI and NotI to remove GFP and insert citrine (Luker et al., 2009c). Rab7-CFP and Lamp-CFP fusion proteins were provided by Joel Swanson (University of Michigan).
Lentiviruses
Lentiviruses were prepared by transient transfection of 293T cells and used to stably express CXCR7-GFP in MEFs (Lois et al., 2002) (Luker et al., 2009c).
Chemokine supernatants
CXCL11 or CXCL12 fused to Gaussia luciferase (CXCL11-GL or CXCL12-GL) was prepared from 293T cells (Luker et al., 2009a). Concentrations of chemokines were determined using standard curves of bioluminescence relative to amounts of chemokine determined by ELISA (R&D Systems, Minneapolis, MN, USA) (Luker et al., 2009a).
Accumulation of bioluminescent chemokines
Cells were plated into black wall 96 well plates (1.5 × 104 cells per well) and used the subsequent day. Cells were incubated with CXCL11-GL or CXCL12-GL diluted in DMEM with 0.2% BSA (Probumin, Millipore, Billerica, MA, USA). In selected experiments, cells were incubated with 0.4M sucrose (Sigma, St. Louis, MO, USA), 80 μM dynasore (Sigma,), or vehicle for 30 minutes before adding bioluminescent chemokine. To measure chemokine degradation, cells were incubated with CXCL11- or CXCL12-GL for 15 minutes, washed twice with PBS, and then incubated for 4 hours in DMEM-0.2% BSA containing 50 μM chloroquine, 50 mM NH4Cl, 25 μM MG132 (Sigma), or vehicle. After incubations, cells were washed twice with acidic solution (0.2M acetic acid, 0.5M NaCl) at 4°C followed by PBS to remove extracellular chemokine (Kelly et al., 2006; Luker et al., 2009c).
Bioluminescence imaging in live cells was performed on an IVIS 100 (Caliper, Hopkinton, MA, USA) (Luker et al., 2008; Luker et al., 2009a). Data were quantified as photons and normalized to cellular protein determined by sulforhodamine B (Sharma et al., 1996).
Fluorescence microscopy
293T cells were transfected with CXCR7-citrine or unfused CXCR7 in combination with Rab7-CFP or Lamp-CFP. Alternatively, cells were transfected with CXCR7-GFP. Two days after transfection, cells were analyzed under baseline conditions or following incubation with incubated with ≈ 8 ng/ml CXCL12-Cherry in DMEM with 0.2% BSA for 30 or 60 minutes. Cells were washed once with PBS, fixed in 2% formaldehyde solution in PBS for 5 minutes, and then mounted with fluorescent mounting medium (Prolong Gold, Invitrogen). Slides were viewed by epifluorescence microscopy using a 40X objective.
Chemokine depletion
231-CXCR4 or 231-CXCR7 cells were plated in 96 well plates as described above. CXCL12-GL or unfused GL supernatants were incubated with 231 cells for 30 minutes, transferred to cells expressing the same receptor, and incubated for an additional 30 minutes. Supernatants then were transferred to 231-CXCR4 cells that had been cultured overnight in DMEM medium with 0.2% BSA. Cells were lysed after 5 minutes and probed by Western blotting for phospho-473 and total AKT (Cell Signaling, Danvers, MA, USA) (Luker et al., 2009a). Alternatively, CXCL12-GL supernatants were transferred to 293T cells expressing firefly luciferase complementation reporters for CXCR4/β-arrestin 2 and incubated for 10 minutes before quantifying bioluminescence. We also measured bioluminescence from CXCL12-GL or unfused GL in 5 μl of supernatants.
Flow cytometry
Internalization of CXCR7 from the cell surface was measured by flow cytometry (Galliera et al., 2004). Cells were washed with DMEM containing 1% BSA and 25 mM Hepes, pH 7.4 (DMEM-1% BSA). Cells were incubated with mAb 11G8 to CXCR7 (gift of ChemoCentryx, Mountain View, CA, USA) for 1 hour on ice, washed twice with DMEM-1% BSA, resuspended in DMEM-1% BSA without or with CXCL12 or 1 mM primaquine (Sigma) as indicated, and shifted to 37°C. In selected experiments, cells were incubated with 0.4M sucrose or vehicle for 30 minutes on ice before shifting to 37°C. Control cells were maintained on ice. All samples were washed twice with cold DMEM-1% BSA and incubated for 30 minutes on ice with 1:200 dilution of APC-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA, USA). To determine total cell surface CXCR7 after incubation at 37°C, cells were incubated again with anti-CXCR7 antibody, washed twice with DMEM-1% BSA, and then stained with secondary antibody.
To analyze effects of protein synthesis on recovery of cell surface CXCR7, cells were incubated at 37°C for 30 minutes in DMEM-1% BSA without or with 300 ng/ml CXCL12. Cells incubated with CXCL12 were either maintained with CXCL12 or washed twice with DMEM-1% BSA and transferred to medium without CXCL12. Cycloheximide (100 μg/ml) (Sigma) was added to subsets of cells to block protein synthesis during 2 hour incubation at 37°C (Luker et al., 2003). Staining for cell surface CXCR7 was performed as described.
Cells were analyzed by flow cytometry using a FACSAria (BD Biosciences, San Jose, CA, USA), collecting ≈ 1 × 104 events per sample. CXCR7 internalization was calculated using mean fluorescence intensity (MFI) of APC for cells incubated at 37°C and those maintained on ice (Galliera et al., 2004). Total receptor on the cell surface following incubation at 37°C was assessed by MFI of APC for cells restained with primary antibody.
Firefly luciferase complementation for interactions between CXCR7 and β-arrestin 2
231 breast cancer cells stably expressing CXCR7-NLuc and β-arrestin 2-CLuc were incubated at 37°C with 300 ng/ml CXCL12. Cells either were maintained in medium containing CXCL12 or washed twice with DMEM-1% BSA and incubated without CXCL12 for various times through 2 hours. Firefly luciferase activity was quantified as described (Luker et al., 2008).
Statistics
Data were plotted as mean values with SEM. Cell culture experiments were performed 2-5 times each, and representative data were presented. Statistically significant differences (p < 0.05) were determined by Mann-Whitney U test (GraphPad Prism). Dunn's multiple comparison test was used for post-hoc analysis of groups of data.
Supplementary Material
Acknowledgments
Research was supported by NIH grants R01CA136553, R01CA136829, and P50CA093990. We thank ChemoCentryx for mAb 11G8 and CCX733.
Footnotes
Conflict of Interest Statement
The authors declare no competing financial interests.
References
- Allinen M, R B, Cai L, Brennan C, Lahti-Domerci J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Atanackovic D, Cao Y, Kim J, Brandl S, Thorn I, Faltz C, et al. The local cytokine and chemokine milieu within malignant effusions. Tumour Biol. 2008;29:93–104. doi: 10.1159/000135689. [DOI] [PubMed] [Google Scholar]
- Balabanian K, Lagane B, Infantino S, Chow K, Harriague J, Moepps B, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- Boldajipour B, Mahabaleshwar S, Kardash E, Reichman-Fried M, Blaser H, Minina S, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Burns J, Summers B, Wang Y, Melikian A, Berahovich R, Miao Z, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Dean R, Roberts C, Overall C. Matrix metalloproteinase processing of CXCL11/I-TAC results in loss of chemoattractant activity and altered glycosaminoglycan binding. J Biol Chem. 2008;283:19389–19399. doi: 10.1074/jbc.M800266200. [DOI] [PubMed] [Google Scholar]
- Dar A, Goichberg P, Shinder V, Kalinkovich A, Kollet O, Netzer N, et al. Chemokine receptor CXCR4-dependent internalization and resecretion of functional chemokine SDF-1 by bone marrow endothelial and stromal cells. Nat Immunol. 2005;6:1038–1046. doi: 10.1038/ni1251. [DOI] [PubMed] [Google Scholar]
- Fermas S, Gonnet F, Sutton A, Charnaux N, Mulloy B, Du Y, et al. Sulfated oligosaccharides (heparin and fucoidan) binding and dimerization of stromal cell-derived factor-1 (SDF-1/CXCL 12) are coupled as evidenced by affinity CE-MS analysis. Glycobiology. 2008;18:1054–1064. doi: 10.1093/glycob/cwn088. [DOI] [PubMed] [Google Scholar]
- Furuya M, Suyama T, Usui H, Kasuya Y, Nishiyama M, Tanaka N, et al. Up-regulation of CXC chemokines and their receptors: implications for proinflammatory microenvironments of ovarian carcinomas and endometriosis. Hum Pathol. 2007;37:1676–1687. doi: 10.1016/j.humpath.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Galliera E, Venkatakrishna R, Trent J, Bonecchi R, Signorelli P, Lefkowitz R, et al. b-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–25597. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- Griesbeck O, Barid G, Campbell R, Zacharias D, Tsien R. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Hansell C, Simpson C, Nibbs R. Chemokine sequestration by atypical chemokine receptors. Biochem Soc Trans. 2006;34:1009–1013. doi: 10.1042/BST0341009. [DOI] [PubMed] [Google Scholar]
- Heuser J, Anderson R. Hypertonic media inhibit receptor-mediated but not fluid-phase endocytosis by blocking clathrin-coated pit formation. J Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewerf A, Kuschert G, Proudfoot A, Borlat F, Clark-Lewis I, Power C, et al. Glycosaminoglycans mediate cell surface oligomerization of chemokine. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- Kalatskaya I, Berchiche Y, Gravel S, Limberg B, Rosenbaum J, Heveker N. AMD3100 is a CXCR7 ligand with allosteric agonist properties. Mol Pharmacol. 2009;75:1240–1247. doi: 10.1124/mol.108.053389. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel P, Shu W, Drobnjak M, Kakonen S, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kelly K, Narhendorf M, Yu A, Reynolds F, Weissleder R. In Vivo Phage Display Selection Yields Atherosclerotic Plaque Targeted Peptides for Imaging. Mol Imaging Biol. 2006;8:1536–1632. doi: 10.1007/s11307-006-0043-6. [DOI] [PubMed] [Google Scholar]
- Kohout T, Lin F-T, Perry S, Conner D, Lefkowitz R. β-Arrestin 1 and 2 differentially regulate heptahelical receptor signaling and trafficking. Proc Natl Acad Sci U S A. 2001;98:1601–1606. doi: 10.1073/pnas.041608198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar O, Rupertus K, Scheuer C, Junker B, Tilton B, Schilling M, et al. Stromal cell-derived factor-1 promotes cell migration and tumor growth of colorectal metastasis. Neoplasia. 2007;9:862–870. doi: 10.1593/neo.07559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laquri C, Sadir R, Rueda P, Baleux F, Gans P, Arenzana-Seisdedos F, et al. The novel CXCL12gamma isoform encodes an unstructured cationic domain which regulates bioactivity and interaction with both glycosaminoglycans and CXCR4. PLoS One. 2007;2:e1110. doi: 10.1371/journal.pone.0001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong E, Pease S, Brown E, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- Luker K, Gupta M, Luker G. Imaging CXCR4 signaling with firefly luciferase complementation. Anal Chem. 2008;80:5565–5573. doi: 10.1021/ac8005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Gupta M, Luker G. Bioluminescent CXCL12 fusion protein for cellular studies of CXCR4 and CXCR7. Biotechniques. 2009a;47:625–632. doi: 10.2144/000113126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Gupta M, Luker G. Imaging chemokine receptor dimerization with firefly luciferase complementation. FASEB J. 2009b;23:823–834. doi: 10.1096/fj.08-116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Gupta M, Steele J, Foerster B, Luker G. Imaging Ligand-dependent Activation of CXCR7. Neoplasia. 2009c;11:1022–1035. doi: 10.1593/neo.09724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Luker G. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238:30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Marchese A, Benovic J. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J Biol Chem. 2001;276:45509–45512. doi: 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- Miao Z, Luker K, Summers B, Berahovich R, Bhojani M, Rehemtulla A, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanon M, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Naumann U, Cameroni E, Pruenster M, Mahabaleshwar S, Raz E, Zerwes H, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Proost P, Schutyser E, Menten P, Struyf S, Wuyts A, Opdenakker G, et al. Amino-terminal truncation of CXCR3 agonists impairs receptor signaling and lymphocyte chemotaxis, while preserving antiangiogenic properties. Blood. 2001;98:3554–3561. doi: 10.1182/blood.v98.13.3554. [DOI] [PubMed] [Google Scholar]
- Pruenster M, Mudde L, Bombosi P, Dimitrova S, Zsak M, Middleton J, et al. The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nat Immunol. 2009;10:101–108. doi: 10.1038/ni.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggo C, Ruhl R, McAllister S, Koon H, Dezube B, Fruh K, et al. Novel cellular genes essential for transformation of endothelial cells by Kaposi's sarcoma-associated herpesvirus. Cancer Res. 2005;65:5084–5095. doi: 10.1158/0008-5472.CAN-04-2822. [DOI] [PubMed] [Google Scholar]
- Sadir R, Imberty A, Baleux F, Lortat-Jacob H. Heparan sulfate/heparin oligosaccharides protect stromal cell-derived factor-1 (SDF-1)/CXCL12 against proteolysis induced by CD26/dipeptidyl peptidase IV. J Biol Chem. 2004;279:43865–43860. doi: 10.1074/jbc.M405392200. [DOI] [PubMed] [Google Scholar]
- Sharma V, Crankshaw C, Piwnica-Worms D. Effects of multidrug resistance (MDR1) P-glycoprotein expression levels and coordination metal on the cytotoxic potency of multidentate (N4O2) (ethylenediamine)bis[propyl(R-benzylimino)]metal(III) cations. J Med Chem. 1996;39:3483–3490. doi: 10.1021/jm950823c. [DOI] [PubMed] [Google Scholar]
- Shenoy S, Lefkowitz R. Multifaceted roles of beta-arrestins in the regulation of seven-membrane-spanning receptor trafficking and signalling. Biochem J. 2003;375:503–515. doi: 10.1042/BJ20031076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro F, Biben C, Martinez-Munoz L, Mellado M, Rashohoff R, Li M, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, et al. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- Strieter R, Burdick M, Mestas J, Gomperts B, Keane M, Belperio J. Cancer CXC chemokine networks and tumour angiogenesis. Eur J Cancer. 2006;42:768–778. doi: 10.1016/j.ejca.2006.01.006. [DOI] [PubMed] [Google Scholar]
- van Weert A, Geuze H, Groothuis B, Stoorvogel W. Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direction intearction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. Eur J Cell Biol. 2000;79:394–399. doi: 10.1078/0171-9335-00062. [DOI] [PubMed] [Google Scholar]
- Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- Wang J, Shiozawa Y, Wang J, Wang Y, Jung Y, Pienta K, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- Weber M, Blair E, Simpson C, O'Hara M, Blackburn P, Rot A, et al. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–2508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Ou Z, Feng L, Luo J, Wang L, Shen Z, et al. Chemokine decoy receptor d6 plays a negative role in human breast cancer. Mol Cancer Res. 2008;6:1276–1288. doi: 10.1158/1541-7786.MCR-07-2108. [DOI] [PubMed] [Google Scholar]
- Yang X, Chu Y, Wang Y, Guo Q, Xiong S. Vaccination with IFN-inducible T cell alpha chemoattractant (ITAC) gene-modified tumor cell attenuates disseminated metastases of circulating tumor cells. Vaccine. 2005;24:2966–2974. doi: 10.1016/j.vaccine.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Zabel B, Wang Y, Lewen S, Berahovich R, Penfold M, Zhang P, et al. Elucidation of CXCR7-mediated signaling events and inhibition of CXCR4-mediated tumor cell transendothelial migration by CXCR7 ligands. J Immunol. 2009;183:3204–3211. doi: 10.4049/jimmunol.0900269. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yeger H, Das B, Irwin M, Baruchel S. Tissue microenvironment modulates CXCR4 expression and tumor metastasis in neuroblastoma. Neoplasia. 2007;9:36–46. doi: 10.1593/neo.06670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Mueller B, DiScipio R, Schraufstatter I. Akt plays an important role in breast cancer cell chemotaxis to CXCL12. Breast Cancer Res Treat. 2008;110:211–222. doi: 10.1007/s10549-007-9712-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.