Abstract
NudC is a highly conserved protein necessary for cytoplasmic dynein-mediated nuclear migration in Aspergillus nidulans. NudC interacts genetically with Aspergillus NudF and physically with its mammalian orthologue Lis1, which is crucial for nuclear and neuronal migration during brain development. To test for related roles for NudC, we performed in utero electroporation into embryonic rat brain of cDNAs encoding shRNAs as well as wild-type and mutant forms of NudC. We show here that NudC, like Lis1, is required for neuronal migration during neocorticogenesis and we identify a specific role in apical nuclear migration in radial glial progenitor cells. These results identify a novel neuronal migration gene with a specific role in interkinetic nuclear migration, consistent with cytoplasmic dynein regulation.
Keywords: NudC, Lis1, interkinetic nuclear migration, brain development, mitosis
Introduction
The ‘Nud’ (Nuclear distribution) genes (see Table 1) code for a family of proteins that are involved in the microtubule-based movement of nuclei in the germ tube of the filamentous fungus Aspergillus nidulans (Morris et al., 1998a). This process involves cytoplasmic dynein and dynein-related genes. NudF is homologous to the mammalian Lis1 (Xiang et al., 1995), mutations in which cause classical lissencephaly, a severe developmental disorder characterized by a smooth cerebral surface, cortical lamination defects, mental retardation and seizures (Table 1) (Lo Nigro et al., 1997; Reiner et al., 1993). Mutational and RNAi analysis of LIS1 in rodents supported a role in neuronal migration (Hirotsune et al., 1998; Shu et al., 2004; Tsai et al., 2005). We have addressed the cellular role of Lis1 by in utero electroporation of embryonic rat brain with Lis1 shRNAs combined with cDNAs encoding centrosome, nuclear, and microtubule markers. By live imaging we detected severe defects at several specific stages of neuronal migration (Tsai et al., 2005). As in Aspergillus, though via a different mechanism, LIS1 was essential for nucleokinesis during interkinetic nuclear migration (INM) in radial glial progenitor cells (RGPC) and in radially migrating postmitotic neurons (Tsai et al., 2007; Tsai et al., 2005; Tsai et al., 2010). LIS1 is also involved in mitosis directly (Faulkner et al., 2000; Siller et al., 2005; Yingling et al., 2008) and indirectly, by preventing nuclei in radial glial progenitor cells from reaching the ventricular surface (Tsai et al., 2005; Tsai et al., 2010). Cytoplasmic dynein heavy chain (HC) RNAi produces effects similar to those for LIS1, supporting a role as the target of LIS1 regulation (Tsai et al., 2007; Tsai et al., 2010).
Table 1.
listing of Aspergillus proteins and corresponding mammalian proteins and the consequences of their mutations.
| Aspergillus proteins |
Aspergillus mutation phenotype |
Aspergillus knock Out Phenotype |
Mammalian orthologs |
Consequences of Human mutation |
|---|---|---|---|---|
| NudF | Nuclear migration defect |
Nuclear migration defect |
Lis1 | Lissencephalya |
| NudE | Nuclear migration defect |
Nuclear migration defect |
NudE | Microcephaly with lissencephalyb |
| NudEL | unknown | |||
| NudC | Nuclear migration defect |
Lethal | NudC | unknown |
| NudCL | unknown | |||
| NudCL2 | unknown |
Another Aspergillus nuclear distribution gene, NudE, has mammalian orthologues, Nde1 (NudE) and Ndel1 (NudE-like, or NudEL), which also contribute to brain development. Depending on gene dosage NudE and NudEL mutant mice exhibit microcephaly as a result of mitotic defects (Feng and Walsh, 2004), or lissencephaly (Sasaki et al., 2005), also consistent with a role in the cytoplasmic dynein pathway. Human NudE mutations also cause microcephaly with lissencephaly (Alkuraya et al., 2011). Mammalian NudE and NudEL each interact with LIS1 and with cytoplasmic dynein (Feng et al., 2000; Niethammer et al., 2000; Sasaki et al., 2000). Recent biochemical and biophysical analysis has indicated that LIS1, NudE, and cytoplasmic dynein form a triple complex (McKenney et al., 2010). LIS1, positioned by NudE, was found to convert dynein to a persistent force producing state, likely required for transport of high load structures, such as nuclei.
NudC was identified as another Aspergillus gene in the cytoplasmic dynein pathway but about which much less is known. In Aspergillus, the mutant NudC3 leads to a nuclear migration defect similar to NudA (dynein heavy chain gene homologue) mutations and knock out (Chiu and Morris, 1995; Osmani et al., 1990). This mutation can be suppressed by overexpressing NudF, the LIS1 homologue (Xiang et al., 1995).
Three mammalian homologues have been identified, NudC (Axtell et al., 1995; Miller et al., 1999) and, more recently, NudC-like (Shu et al., 2006) and NudC-like protein 2 (Yang et al., 2010). Mammalian NudC has been reported to coimmunoprecipitate with dynein, dynactin and Lis1 (Aumais et al., 2001; Morris et al., 1998b; Zhou et al., 2003), supporting a role in dynein function. Direct interaction between mammalian NudC and Lis1 has also been observed in yeast two-hybrid and GST pull-down assays (Morris et al., 1998b). In addition, NudC has been implicated in cell proliferation (Miller et al., 1999). RNAi of NudC led to defects in cytokinesis and chromosome congression during karyokinesis in cultured non-neuronal cells (Aumais et al., 2003; Nishino et al., 2006; Zhang et al., 2002; Zhou et al., 2003). NudC contains a binding site for the mitotic kinase Plk1 (Zhou et al., 2003) and two Plk1 phosphorylation sites within its C-terminal domain (see Figure 2). This region of NudC is well conserved between Aspergillus and mammals (Morris et al., 1997; Zhang et al., 2002; Zhou et al., 2003), which, however, has an additional 133 a.a. N-terminal predicted coiled-coil extension of unknown function.
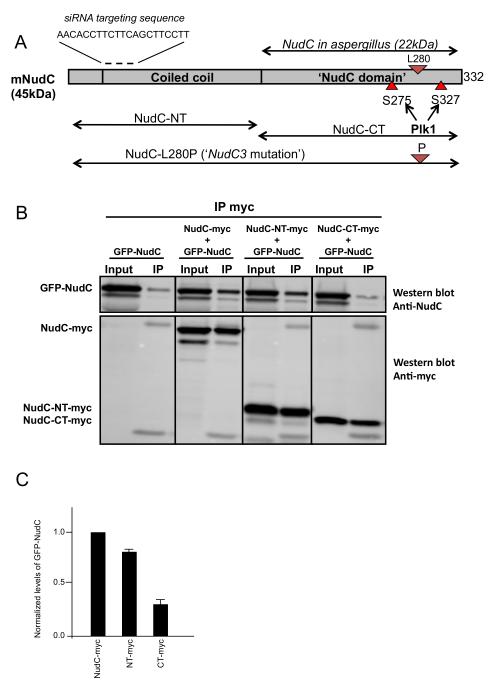
Figure 2. Domain organization of mammalian NudC.
(A) Schematic diagram showing NudC in A. Nidulans and mammals, NudC C-terminus, N-terminus and NudC-L280P and the phophorylation sites. (B) NudC dimerizes through the N-terminal domain: GFP NudC full length was coexpressed with NudC-myc full length, NudC-NT-myc or NudC-CT-myc in HeLa cells. Anti myc antibody was used to pull down myc proteins. CoIP of the GFP-NudC was detected using anti NudC antibody. (C) The amount of co-immunoprecipitated GFP-NudC protein was normalized to the corresponding amount of myc protein immunoprecipitated. The wild-type GFP-NudC/NudC-myc ratios were set to 1 and all other values were calculated relative to it, with n = 5 and *P < 0.0001 indicating significance (One-way ANOVA). Error bars represent SEM.
The mitotic effect of NudC inhibition is potentially consistent with a role in mammalian cytoplasmic dynein function. Human NudC mutations have not been identified (Table 1) (Matsumoto and Ledbetter, 1999). Nonetheless, we sought to test whether NudC plays a comparable role to LIS1, NudE, and NudEL in mammalian neuronal migration. This behavior is highly stereotypical in the neocortex, and provides an excellent system to test for roles in dynein and LIS1-mediated behavior. We find the NudC RNAi and overexpression phenotypes to be closely related to those for LIS1 and cytoplasmic dynein, identifying NudC as a novel brain developmental gene in the cytoplasmic dynein pathway.
Material and Methods
RNAi and cDNA constructs
For RNAi, we used pRNAT-U6.1/Neo (GenScript) which coexpress GFP with an shRNA for brain electroporation and oligos (Dharmacon) for transfection in cultured cells. The NudC targeting sequence, 5′-AACACCTTCTTCAGCTTCCTT-3′ has been previously described (Aumais et al., 2003) and targets a coding region which is identical in sequence in the human, rat and mouse genes. No related sequences were found in the genome of these species. The LIS1 targeting sequences, 5′-GGATGCTACAATTAAGGTGTG-3′ and the scrambled controls have been previously described (Tsai et al., 2005). Mouse NudC cDNA full length (pCDNA3.1-NudC-myc gift from Bruce Shaar) was cloned into the EcoRI and BamH1 site of the GFP vector PIC113 (Science’s stke). GFP and myc tagged NudC N-Terminus constructs were obtained by introducing a stop codon for the amino acid at position 160 by using site-directed mutagenesis in the full-length constructs. The different point mutations, GFP-NudC L280P, GFP-NudC EE and GFP-NudC AA were also obtained by site-directed mutagenesis (QuickChange kit, Stratagene). The C-terminus of NudC (160-332) was amplified by PCR and cloned into the EcoRI and BamH1 sites of the GFP vector pEGFP-C1 (BD Biosciences Clontech) and into the EcoRI and BamH1 sites of the pCDNA3.1 vector.
In utero electroporation
Plasmids were transfected using intraventricular injection followed by in utero electroporation (Saito and Nakatsuji, 2001; Tabata and Nakajima, 2001). In brief, pregnant Sprague Dawley rats (Hilltop) were used, and 1–2 μl cDNA (1–5 μg/μl) or 1 μg/μl siRNA were injected into the ventricle of embryonic brains at E16. A pair of copper alloy oval plates that were attached to the electroporation generator (Harvard Apparatus) transmitted five electric pulses at 50 V for 50 ms at 1-s intervals through the uterine wall. Animals were maintained according to protocols approved by the Institutional Animal Care and Use Committee at Columbia University. Each phenotypic analysis was done with at least 3 independent litters.
Western Blot
For western blotting HeLa cells and cortical cells were transiently transfected. HeLa Cells (40-50% confluent) cultivated in DMEM containing 10% FBS in 35mm dishes were transfected using effectene (Qiagen) for cDNA and oligofectamine (InVitrogen) for siRNA oligos as directed by the manufacturer. Primary cortical cells were obtained from E16 rat brains and cultured in Neurobasal/B27 medium. Briefly, cells were plated at a density of 1×105 on 24 well plates in Neurobasal medium containing B27, L-glutamine (0.5mM), and FCS (2%). Cortical E16 cells were transfected using the nucleofector kit from Amaxa Biosystems, Gaithersburg, MD, following manufacturer’s protocol. Briefly, cortical cells were spun down (80 rpm) and mixed with 100 μl of nucleofector solution and 2 μg of plasmids. The cells/plasmid mixture was then nucleofected using program A-033.
Transfected cells were then plated at a density of 5×106 per 60 mm culture dish. Cells were lysed 48 h after transfection in RIPA buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, and 1 mM EGTA) with protease inhibitor cocktail for mammalian tissues (Complete, Roche). Proteins were separated in 4-20% gradient SDS–PAGE and transferred to PVDF membranes (Immobilon-P; Millipore). NudC was immunodetected using an antibody to mouse NudC that was raised in chicken against the full-length protein with AvesLabs, Inc. Lis1 was immunodetected with a monoclonal anti-Lis1 antibody (clone LIS1-338, Sigma-Aldrich) and Dynein Intermediate Chain with a monoclonal anti-Dynein Intermediate Chain antibody (MAB1618, Chemicon).
Immunoprecipitation assays
HeLa cells, seeded on 6 well plates, were co-transfected with GFP-NudC and pCDNA3.1 vector for control, NudC-myc, NudC-NT-myc or NudC-CT-myc. Cells were lysed 24h after transfection, in 150μl RIPA buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, and 1 mM EGTA) containing protease inhibitor cocktail (Complete, Roche). Myc NudC was immunoprecipitated from 100 μl lysate using a mouse purified myc antibody (9E11) bound to protein-G sepharose beads (30 μl for each IP, GE Healthcare) for 2-4h at 4C. After IP, supernantants were saved and beads were washed 3 times in RIPA buffer and resuspended in 100 μl SDS sample buffer. Same volumes of Total, IP and supernatant were loaded on gel for western blotting. For quantification, the amount of co-immunoprecipitated GFP-NudC protein was normalized to the corresponding amount of myc protein immunoprecipitated. The wild-type GFP-NudC/NudC-myc ratios were set to 1 and all other values were calculated relative to it. Graph and statistical results were generated using GraphPad prism software.
Live cell imaging
Coronal slices were prepared 48–72 h after electroporation. Slices were placed on Millicell-CM inserts (Millipore) in culture medium containing 25% Hanks balanced salt solution, 47% basal MEM, 25% normal horse serum, 1× penicillin/streptomycin/glutamine (GIBCO BRL), and 0.66% glucose and were incubated at 37°C in 5% CO2. Multiple GFP-positive cells were imaged on an inverted microscope (Olympus) with a 40× objective. Time-lapse images were captured by camera (Hamamatsu Orca-ER) using MetaMorph software (Universal Imaging Corp.) at intervals of 10 min for 5–12 h.
Immunocytochemistry
Rat embryos were perfused transcardially with ice-chilled saline followed by 4% PFA (EMS) in 0.1 M PBS, pH7.4. Brains were postfixed in PFA overnight and sectioned on a Vibrotome (Leica). Slices were blocked at RT for 1 h with 10% serum, 0.5% Triton X-100 in PBS. Primary antibodies were applied overnight at 4° C in 10% serum, 0.5% Triton X-100 in PBS at the following concentrations: anti-PCNA (1:200, mouse, Dako); anti-TuJ1 (1:200, mouse, Covance); anti-Tbr2 (1:1000, Rabbit, Chemicon); anti-Phospho Histone 3 (1:200, Rabbit, Upstate). Sections were then washed with PBS and incubated for 1 hour at room temperature in 10% serum, 0.5% Triton X-100 in PBS with Cy3- or Cy5-conjugated secondary antibodies (1:200; Jackson ImmunoResearch Laboratories).
Confocal microscopy
Sections were imaged on an inverted laser-scanning confocal microscope (Zeiss LSM510 meta) with a 10, 20 and 40× water immersion objective (Zeiss). Excitation/emission wavelengths were 488/515 nm (GFP), 550/570 nm (Cy3), and 633/690 (Cy5). Z-series images were collected at 5 μm steps, and a projection of each stack was used for producing figures. In each brain slice, 100–500 cells could be found positive to GFP when electroporated with the constructs used. In experiments that involved colabeling or cell/process counting, images from individual optical sections were carefully examined.
In vivo cell analysis
Quantification of the number of VZ progenitors and SVZ precursors in vivo were performed on single optical sections by quantifying all PCNA-immunopositive cells and Tbr2-immunopositive cells and dividing by the number of transfected cells. Quantification of the number of mitotic cells was performed on single optical sections by quantifying all pH3–immunopositive cells and dividing by the number of transfected cells. The distribution of the transfected cells was quantified on single optical sections by counting all positive cells in a radial stripe comprising all cortical layers. The distance of the nuclei from the ventricular surface was calculated using the center of the soma, as indicated by cytoplasmic GFP staining, was used. We find this approach to provide accurate information when tested against CFP-histone H1.
Results
Effect of altered NudC expression on cellular redistribution during neocortical development
To explore the function of NudC in brain development and its potential involvement in lissencephaly, we used vector-based RNAi to downregulate NudC in the developing neocortex. A clear reduction in NudC levels (73%) was observed in HeLa cells and primary rat neocortical cells transfected with corresponding siRNAs to a common sequence in rat and human (Figure 1A,B and Supplemental Figure 1C). LIS1 levels were also reduced in some experiments as reported for the Aspergillus NudC3 mutant (Figure 1A,B and Supplemental Figure 1C) (Xiang et al., 1995). To investigate the function of NudC in the developing brain, NudC RNAi, scrambled RNAi control or empty vector were introduced into rat radial glial progenitor cells by in utero electroporation at E16. The distribution of electroporated cells was determined in fixed sections prepared at E19 and E21, corresponding to three and five days after electroporation, respectively. By E19 control progenitor cells could be observed in the ventricular zone (VZ), subventricular zone (SVZ), and intermediate zone (IZ), with a few cells having reached the cortical plate (CP) (Figure 1C and Supplemental Figure 1A). By E21 the majority of the electroporated cells had reached the CP (Figure 1D and Supplemental Figure 1B). Some transfected cells persisted within the VZ, presumably representing proliferating progenitor cells.
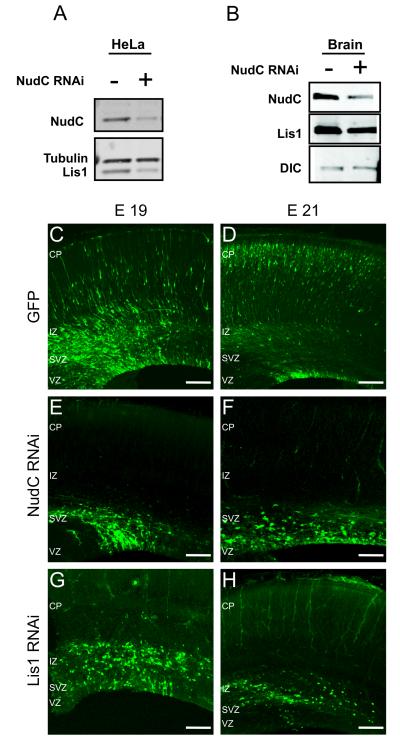
Figure 1. Effects of NudC RNAi on neuronal redistribution in the neocortex.
(A) Western blot of NudC and Lis1 in Hela cells 48 h after transfection with NudC RNAi.
(B) Western blot of NudC, Lis1 and Dynein intermediate chain (DIC) in cortical precursors isolated from E16 rat brains 48 h after transfection with NudC RNAi vector.
In Both systems, NudC levels in cells transfected with NudC RNAi were 73% lower than those in cells transfected with empty vector.
(C–H) Micrographs of coronal sections of the cerebral cortex at E19 and E21, electroporated as indicated. Note that GFP transfected cells are widely distributed in the entire neocortex, from the apical to the pial surface (C,D) while NudC RNAi or Lis1 RNAi transfected cells fail to migrate properly and reach the pial surface (E-H)
CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone. The apical and pial surfaces are at the boundaries of the VZ and CP, respectively. Scale bars: 100 μm.
In contrast, none of the NudC RNAi-electroporated cells reached the CP (Figure 1E,F and Supplemental Figure 1A,B). The cells showed evidence of accumulating in the VZ/SVZ by E19 (Figure 1E and Supplemental Figure 1A) and failed to progress any further by E21 (Figure 1F and Supplemental Figure 1B). The morphology of the cells gradually changed over this time period, and by E20/E21 the radial glial progenitor cells had lost their typical elongated shapes (see also Figure 4 E-H). However, these results do not imply that NudC must be involved in cell morphogenesis, but only note the effects of NudC downregulation on cell shape. Lis1 RNAi produced a similar redistribution phenotype, though RGPC morphology was preserved (Tsai et al., 2005; Tsai et al., 2010) (Figure 1 G,H, Figure 4 M-P and Supplemental Figure 1A,B).
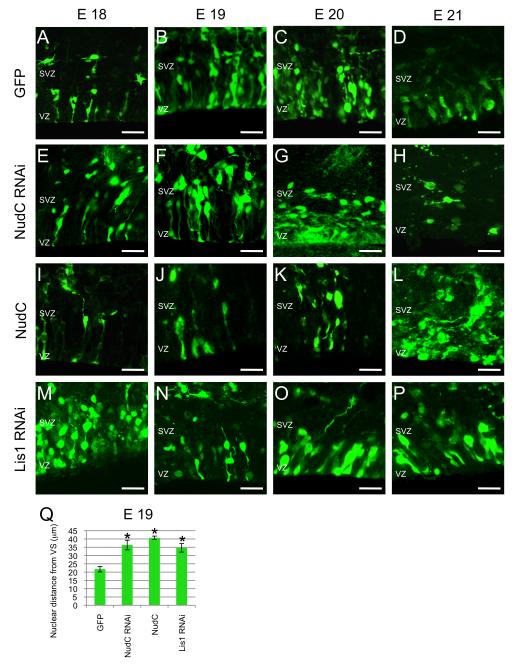
Figure 4. Effects of NudC RNAi and Dominant Negatives on Nuclear Distribution within the ventricular zone.
(A–P) Fluorescent micrographs of coronal sections of E18 (A,E,I,M), E19 (B,F,J,M), E20 (C,G,K,O) and E21 (D,H,L,P) from brains electroporated with GFP (A-D), NudC RNAi (E-H), NudC (I-L) and Lis1 RNAi (M-P) at E16.
(Q) Analysis of nuclear distance from ventricular surface of E19 brains (B,F,J,N). Note the increased average distance from ventricular surface of nuclei from NudC RNAi, NudC and Lis1 RNAi compared to GFP transfected cells.
SVZ, subventricular zone; VZ, ventricular zone; VS, ventricular surface. Error bars represent s.e.m. Scale bar: 20 μm. Student’s t-test, *P≤0.0001.
Contribution of N- and C-terminal NudC domains to neuronal migration
NudC is well conserved among eukaryotes. However, Aspergillus NudC (22kDa) is much shorter than NudC in Drosophila, mouse, human and rat, all of which contain an extra N-terminal domain (Figure 2A). The homology between Aspergillus NudC and the C-terminal domain of mNudC is 78%, with 68% identity (Morris et al., 1997). Moreover it has been shown that the mammalian C-terminal domain of NudC can complement the NudC3 mutation in Aspergillus, demonstrating that the full Aspergillus function is contained within the C-terminal domain of mNudC (Morris et al., 1997). The extra N-terminal region contains a predicted coiled-coil domain that seems to be a late evolutionary addition that might mediate self-association. Indeed, we find that GFP-NudC coimmunoprecipitated either with full-length myc-NudC or a myc-NudC-N-terminal fragment (Figure 2B,C; cf. (Zheng et al., 2011)). However, the C-terminal domain of NudC was not able to co-immunoprecipitate with GFP-NudC full length, demonstrating that NudC interacts with itself via the N-terminal domain.
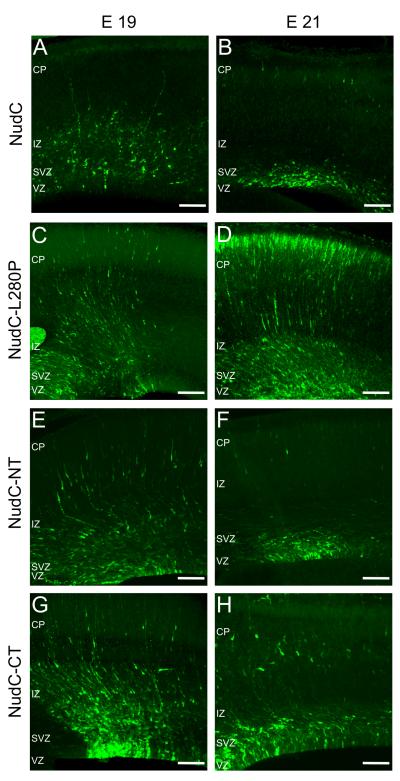
Overexpression of either full-length NudC or its fragments each affected neuronal migration in embryonic rat brain. The full-length protein produced a similar phenotype to that observed after NudC downregulation (Figure 3A,B and Supplemental Figure 1A,B). A reduced fraction of NudC-expressing progenitors reached the IZ by E19, none of which reached the CP (Figure 3A and Supplemental Figure 1A). By E21 NudC-expressing cells were mainly observed in the VZ/SVZ (Figure 3B and Supplemental Figure 1B), suggesting a loss of few further migrating cells. As observed with NudC RNAi, NudC overexpression resulted in a loss of RGPC processes by E21 (Figure 3B and 4I-L). We also tested a mutant form of NudC L280P, based on the original Aspergillus NudC3 mutation (L146P) (Chiu and Morris, 1995; Chiu and Morris, 1997). Three days after electroporation a very mild phenotype was observed (Figure 3C and Supplemental Figure 1A) and the cellular distribution was very similar to the control (Figure 1C and Supplemental Figure 1A). However, two days later, at E21, the cellular distribution and morphology was completely normal (Figure 3D and Supplemental Figure 1B). Thus NudC-L280P largely reversed the effects of NudC overexpression, thereby serving as a control for the specificity of the NudC overexpression phenotype.
Figure 3. Dominant Negative Effects of NudC constructs on neuronal redistribution in the neocortex.
(A–H) Micrographs of coronal sections of the cerebral cortex at E19 and E21, electroporated as indicated.
CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone. The apical and pial surfaces are at the boundaries of the VZ and CP, respectively. Scale bars: 100 μm.
Three days after electroporation of NudC N-terminus in the embryonic brain, a migration phenotype (Figure 3E and Supplemental Figure 1A) was observed compared to control (Figure 1A and Supplemental Figure 1A), which was more pronounced two days later (Figure 3F vs. 1D and Supplemental Figure 1B). Overexpression of the C-terminus of NudC resulted in a somewhat weaker phenotype (Figure 3G,H and Supplemental Figure 1A,B).
Effect of altered NudC expression on interkinetic nuclear migration in RGPCs
Recent interest in INM has led to several models for its underlying mechanism. Our own work has implicated cytoplasmic dynein in apical INM and kinesin-3 in basal INM (Tsai et al., 2007; Tsai et al., 2010). To test how NudC might participate in this process, we first examined the effect of NudC RNAi and overexpression on nuclear position in RGPCs.
Quantitation of nuclear distance from the ventricular surface was determined at E19 (Figure 4). Average distance with control plasmid was 21.8 ± 1.78 μm (Figure 4A-D). Similar to Lis1 downregulated cells (34.6 ± 2.6 μm, Figure 4M-P), the average distance for NudC RNAi electroporated cells was increased to 36.3 ± 2.9 μm (Figure 4E-H) and for cells overexpressing NudC to 40.7 ± 1.0 μm (Figure 4I-L), almost twice that of the control. These data suggested a role for NudC in regulating apically-directed nuclear movement in radial glial cells.
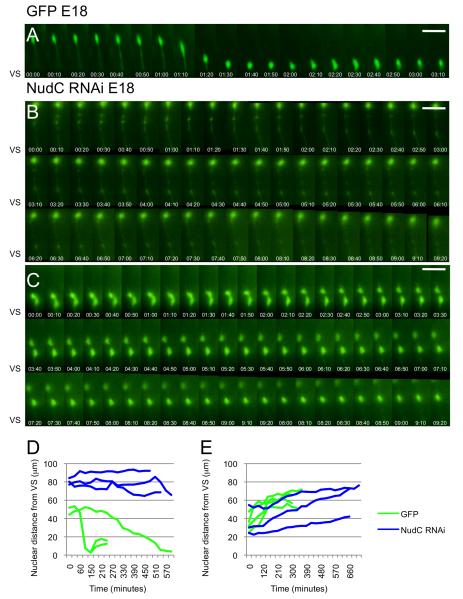
To test this possibility directly, control RNAi and NudC RNAi were introduced in E16 brains by in utero electroporation, and live imaging of E18 rat brain was performed two days later. Control cells (n=6 in 4 different slices) exhibited clear evidence of nuclear migration (Figure 5A) in both apical and basal directions (Figure 5D,E, green lines) at rates comparable to those in our previous studies (Tsai et al., 2005; Tsai et al., 2010). In contrast, cells electroporated with NudC RNAi showed no nuclear movement toward the ventricular surface (apically-directed movement) during the 8-9 h duration of observation (n=3; Figure 5B and 5D, blue lines). Basally-directed movement was, however, normal (n=3; Figure 5C, F, blue lines).
Figure 5. Live cell imaging of neural progenitor cell behavior within the VZ.
(A) Cell body of a control progenitor cell at the radial glial stage migrates away from and then toward the ventricular surface, where it divides. Tracings of cell body positions for six typical controls are shown in panels D,E (green lines). (B,C) Cell body cells transfected with NudC RNAi. Note that the nuclei migrate away from the ventricular surface (C) but they do not move toward the ventricular surface (B) over a 9-h time period. Tracings show these behaviors in D,E (blue lines). Times are shown in hours/minutes. VS, ventricular surface. Scale bar: 20 μm.
Effect on mitosis and differentiation
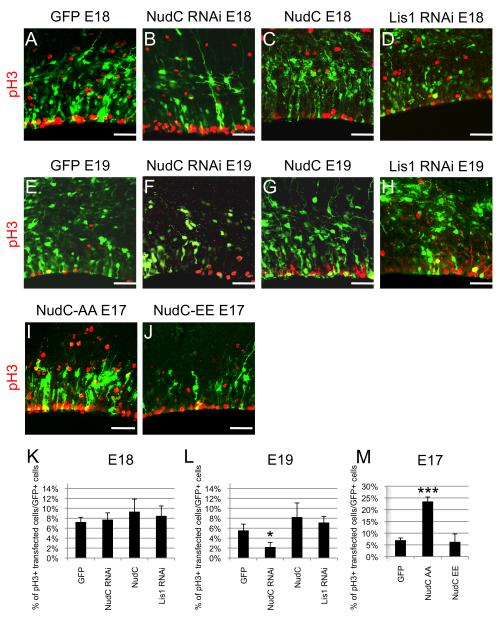
NudC as been reported to participate in mitosis in cultured non-neuronal cells (Aumais et al., 2003; Nishino et al., 2006; Zhou et al., 2003). To test whether altered mitotic behavior contributed to the NudC neuronal distribution defects we used antibodies to phosphohistone H3 (pH3) or phosphovimentin. By 3 days NudC RNAi resulted in a substantial decrease in mitotic cells (Figure 6A,B,E,F,K,L) whereas the other conditions examined showed little effect (Figure 6C,D,G,H,K,L).
Figure 6. Mitotic effects of NudC RNAi and Dominant Negatives.
(A–J) Fluorescent micrographs depict immunostainings with pH3 of E17, E18 and E19 coronal sections from brains electroporated with GFP (A,E), NudC RNAi (B,F), NudC (C,G), Lis1 RNAi (D,H), NudC-AA (I) and NudC-EE (J)
(K-M) Percentage of cells in mitosis at E17, E18 and E19 in the VZ/SVZ after electroporation with GFP, NudC RNAi, NudC, Lis1 RNAi, NudC-AA and NudC-EE. Error bars represent s.e.m. Scale bar: 20 μm. Student’s t-test, *P≤0.05, ***P≤0.0001.
NudC is phosphorylated by the mitotic kinase plk1 on two conserved sites (Ser275 and Ser327 in mouse). These phosphorylations have been shown to be important for mitosis and cytokinesis in tissue cultured cells (Aumais et al., 2003; Nishino et al., 2006; Zhou et al., 2003). We tested whether these phosphorylations were also important in our brain system. A phosphomutant form of NudC (NudC AA), in which the amino acids Ser 275 and Ser 327 are changed to alanine and a phosphomimetic form of NudC (NudCEE) in which the amino acids Ser 275 and Ser 327 are changed to glutamate were electroporated in E16 brains.
Almost no electroporated cells were detected after two days, due to either very weak expression of the constructs in brain or, as we suspect, to a loss of transfected cells. In favor of this hypothesis many transfected cells were observed at one day after electroporation. At this stage a very substantial increase in mitotic cells was observed in cells overexpressing the phosphomutant form of NudC (Figure 6I,M) while no effect was detected in cells overexpressing phosphomimetic NudC (Figure 6J,M).
NudC expression levels affect the proportion of VZ vs. SVZ cells
We also stained brain sections from electroporated rats using markers for proliferating cells (PCNA), for proliferating SVZ cells (Tbr2), and for newborn neurons (Tuj1) to investigate further effects of altered NudC expression on cortical development (Figure 7). At 3 days after electroporation control (Figure 7A and M) and NudC RNAi-expressing cells (Figure 7B and M) were mostly positive for PCNA, a marker of the proliferating state. To test whether the proliferating cells were also delayed in cell fate progression we double stained for Tbr2.
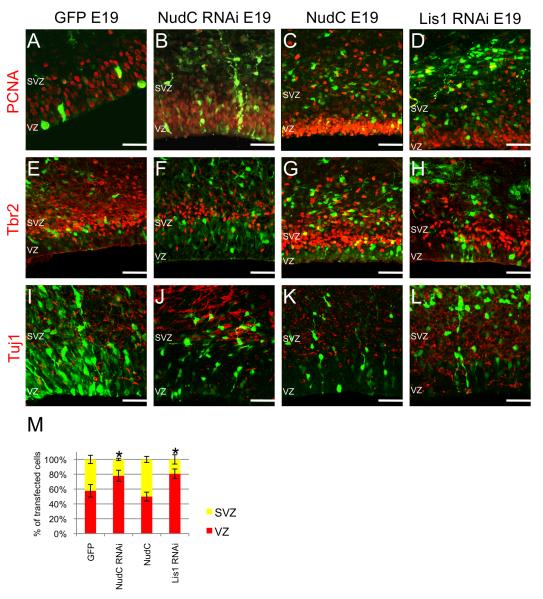
Figure 7. Effects of NudC RNAi on neuronal differentiation.
(A–L) Fluorescent micrographs depict immunostainings as indicated in the panels of E19 coronal sections from brains electroporated with GFP (A,E,I), NudC RNAi (B,F,J), NudC (C,G,K) and Lis1 RNAi (D,H,L) at E16.
(M) Percentage of ventricular zone progenitors (PCNA positive, Tbr2 negative cells, red bar) and subventricular zone progenitors (Tbr2 positive cells, yellow bar) of E19 brains electroporated with GFP, NudC RNAi, NudC and Lis1 RNAi. SVZ, subventricular zone; VZ, ventricular zone. Error bars represent s.e.m. Scale bars: 20 μm. Student’s t-test, *P≤0.01.
Surprisingly, NudC RNAi caused a marked decrease in Tbr2 staining and therefore in differentiation from the RGPC stage (Figure 7E,F and M) even though the average distance from the VZ was increased compared to control cells. This result indicates that cells arrested in apically-directed nuclear movement remained undifferentiated. On the contrary the fraction of NudC overexpressing cells positive for Tbr2 was about half the number of PCNA positive cells (Figure 7C,G and M) demonstrating that the varying levels of NudC had very different effects on cellular differentiation.
Discussion
NudC in brain development
Although a number of genes in the cytoplasmic dynein pathway have been implicated in neuronal migration and proliferation, the role of the less well understood NudC gene in brain development has not been addressed. Here we report that NudC is essential for the redistribution of neuronal precursors during neocortical development, with a particular role in INM.
We used a variety of NudC reagents to test its contribution to neocortical development. NudC RNAi produced a clear neuronal misdistribution phenotype, very similar to that seen for LIS1, dynein HC and NudEL RNAi (Shu et al., 2004; Tsai et al., 2007; Tsai et al., 2005). As further tests for the specificity of the NudC RNAi effect we overexpressed wild-type and mutant constructs. Full-length NudC also affected neuronal redistribution in developing rat neocortex, generating a pattern similar to that resulting from NudC RNAi. This result is similar to the effect of LIS1 overexpression on neuronal migration in embryonic rat brain (Tsai et al., 2005) and possibly to the phenotypic effects of human LIS1 gene duplications in humans (Bi et al., 2009). Although we do not understand the mechanism for the NudC overexpression phenotype, it was almost completely abolished by a single missense mutation at a.a. L280. The homologous site in Aspergillus NudC has been implicated in maintenance of NudF levels and in human NudC to its reported chaperonin-like role of in LIS1 stabilization (Zheng et al., 2011; Zhu et al., 2011). We also expressed N- and C-terminal NudC fragments, which each phenocopied the NudC RNAi and overexpression effects. The C-terminal construct is homologous to Aspergillus NudC, suggesting that it represents the functionally more important portion of NudC. We find the N-terminal domain to be involved in self-association and to heterooligomerize with full-length NudC. These results reveal that higher eukaryotic NudC self-associates and suggest that the N-terminal construct may act by producing nonfunctional heteromers. The effects on neuronal redistribution in the embryonic rat brain were similar in each case and clearly related to that observed for LIS1, cytoplasmic dynein and NudEL (Shu et al., 2004; Tsai et al., 2007; Tsai et al., 2005). These results strongly support a role for NudC in cytoplasmic dynein mediated neuronal migration.
NudC and interkinetic nuclear migration
We specifically tested for a role for NudC in INM, in which cytoplasmic dynein has recently been implicated (Del Bene et al., 2008; Tsai et al., 2005; Tsai et al., 2010). The centrosome in radial glial cells remains at the endfoot at the ventricular surface of the brain throughout INM (Tsai et al., 2010). Microtubules are oriented almost uniformly with minus ends apical and plus ends basal throughout INM (Tsai et al., 2010). We observed potent inhibition of apical migration, consistent with a role for NudC in dynein-mediated nuclear migration. As for dynein RNAi we observed no effect of NudC RNAi on basally directed migration (Tsai et al., 2010). These results argue against a general role in nuclear behavior, and for a very specific role in dynein mediated transport. We were unable to test the effects of NudC phosphorylation on INM directly. However, the accumulation of mitotic NudC-AA-expressing cells in mitosis at the ventricular surface suggests either that apical nuclear migration must persist in these cells.
NudC has also been implicated in mitosis in nonneuronal cells, but the contributions of NudC inhibition to mitosis in RGPCs seems complex. NudC RNAi led to a decrease in mitotic index in developing rat brain. This result likely results from the observed failure of the nuclei to reach the ventricular surface, rather than a direct effect on mitosis. Nonetheless, NudC phosphorylation is likely to affect mitosis directly, based on the known effects in nonneuronal cells in vitro. In contrast the dual phosphomutant form of NudC, NudC-AA caused a very striking and unusual increase in mitotic cells, most of which resided in ventricular zone. These results indicate that NudC and its phosphorylation by Plk1 are, in fact, required for mitosis in RGPCs in the developing brain. The specific enrichment of the NudC phosphomutant cells in mitosis at the ventricular surface either indicates a very early block in cell division in RGPCs, or the lack of a role for PLK1 phosphorlyation of NudC in apical INM. This suggests that NudC phosphorylation by Plk1 is not required for nuclear migration. NudC RNAi led to an increase in the pool of undifferentiated RGPCs. A similar result was obtained for kinesin-3 RNAi (Tsai et al., 2010). These results raise the possibility that interference with either apical or basal nuclear movement prevents RGPCs from generating neurons, an important conclusion that remains to be tested in detail.
Supplementary Material
Highlights.
NudC regulates neuronal migration
NudC downregulation abolishes apical-directed interkinetic nuclear migration
NudC downregulation alters cell morphology
Acknowledgments
We would like to thank Wei-Nan Lian, Shahrnaz Kemal and Garrett E. Seale for critical discussion and technical help, Tania Nayak and Daniel Hu for critical reading of the manuscript. This work was supported by NIH grant HD40182 to RBV, an EMBO Postdoctoral fellowship to SC, an American Heart Association Postdoctoral Fellowship to PM, and a March of Dimes grant to RBV.
Footnotes
S. Cappello’s present address: Institute of Stem Cell Research, Helmholtz Centre Munich, Germany
P. Monzo’s present address: Mechanobiology institute (MBI), 5A Engineering drive 1, Singapore 117411.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkuraya FS, et al. Human Mutations in NDE1 Cause Extreme Microcephaly with Lissencephaly. Am J Hum Genet. 2011;88:536–47. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumais JP, et al. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21:RC187. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumais JP, et al. Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci. 2003;116:1991–2003. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- Axtell SM, et al. Characterization of a prolactin-inducible gene, clone 15, in T cells. Mol Endocrinol. 1995;9:312–8. doi: 10.1210/mend.9.3.7776977. [DOI] [PubMed] [Google Scholar]
- Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–77. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Morris NR. Extragenic suppressors of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans. Genetics. 1995;141:453–64. doi: 10.1093/genetics/141.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Morris NR. Genetic and molecular analysis of a tRNA(Leu) missense suppressor of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans. Genetics. 1997;145:707–14. doi: 10.1093/genetics/145.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, et al. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–65. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Feng Y, et al. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–79. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by nde1 controls cerebral cortical size. Neuron. 2004;44:279–93. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nature Genetics. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Lo Nigro C, et al. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet. 1997;6:157–64. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Ledbetter DH. Molecular cloning and characterization of the human NUDC gene. Hum Genet. 1999;104:498–504. doi: 10.1007/s004390050994. [DOI] [PubMed] [Google Scholar]
- McKenney RJ, et al. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–14. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BA, et al. A homolog of the fungal nuclear migration gene nudC is involved in normal and malignant human hematopoiesis. Exp Hematol. 1999;27:742–50. doi: 10.1016/s0301-472x(98)00074-5. [DOI] [PubMed] [Google Scholar]
- Morris NR, et al. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998a;8:467–70. doi: 10.1016/s0962-8924(98)01389-0. [DOI] [PubMed] [Google Scholar]
- Morris SM, et al. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr Biol. 1998b;8:603–6. doi: 10.1016/s0960-9822(98)70232-5. [DOI] [PubMed] [Google Scholar]
- Morris SM, et al. A prolactin-inducible T cell gene product is structurally similar to the Aspergillus nidulans nuclear movement protein NUDC. Mol. Endocrin. 1997;11:229–236. doi: 10.1210/mend.11.2.9892. [DOI] [PubMed] [Google Scholar]
- Niethammer M, et al. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Nishino M, et al. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol. 2006;16:1414–21. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Osmani AH, et al. The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J. Cell Biol. 1990;111:543–551. doi: 10.1083/jcb.111.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–46. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Sasaki S, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–27. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, et al. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–96. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Shu T, et al. Ndel1 Operates in a Common Pathway with LIS1 and Cytoplasmic Dynein to Regulate Cortical Neuronal Positioning. Neuron. 2004;44:263–77. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Shu T, et al. Doublecortin-like kinase controls neurogenesis by regulating mitotic spindles and M phase progression. Neuron. 2006;49:25–39. doi: 10.1016/j.neuron.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Siller KH, et al. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell. 2005;16:5127–40. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–72. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Tsai JW, et al. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–9. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai JW, et al. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–45. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, et al. An Unconventional Kinesin and Cytoplasmic Dynein Are Responsible for Interkinetic Nuclear Migration in Neural Stem Cells. Nat Neurosci. 2010 doi: 10.1038/nn.2665. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, et al. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. NudC-like protein 2 regulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90. Proc Natl Acad Sci U S A. 2010;107:3499–504. doi: 10.1073/pnas.0914307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–86. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, et al. Involvement of the fungal nuclear migration gene nudC human homolog in cell proliferation and mitotic spindle formation. Exp Cell Res. 2002;273:73–84. doi: 10.1006/excr.2001.5414. [DOI] [PubMed] [Google Scholar]
- Zheng M, et al. Structural Features and Chaperone Activity of the NudC Protein Family. J Mol Biol. 2011;409:722–41. doi: 10.1016/j.jmb.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, et al. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5:127–38. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Zhu XJ, et al. The L279P mutation of nuclear distribution gene C (NudC) influences its chaperone activity and lissencephaly protein 1 (LIS1) stability. J Biol Chem. 2011;285:29903–10. doi: 10.1074/jbc.M110.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.