Summary
Alphaviruses, including several emerging human pathogens, are a large family of mosquito-borne viruses with Sindbis virus being a prototypical member of the genus. The host factor requirements and receptors for entry of for this class of viruses remain obscure. Using a Drosophila system, we identified the divalent metal ion transporter Natural Resistance-Associated Macrophage Protein (NRAMP), as a host cell surface molecule required for Sindbis virus binding and entry into Drosophila cells. Consequently, flies mutant for dNRAMP were protected from virus infection. NRAMP2, the ubiquitously expressed vertebrate homolog, mediated binding and infection of Sindbis virus into mammalian cells, and murine cells deficient for NRAMP2 were non-permissive to infection. Alphavirus glycoprotein chimeras demonstrated that the requirement for NRAMP2 is at the level of Sindbis virus entry. Given the conserved structure of alphavirus glycoproteins, and the widespread use of transporters for viral entry, other alphaviruses may use conserved multi-pass membrane proteins for infection.
Introduction
Alphaviruses are important emerging pathogens causing human disease worldwide (Gould et al., 2009). These viruses circulate among a vertebrate reservoir in the wild, and many cause disease after spillover transmission to humans and agriculturally important domestic animals such as horses (Gould et al., 2009; Greene et al., 2005; Ryman and Klimstra, 2008; Weaver and Reisen, 2010). Sindbis virus is considered the prototypical virus of this genus, and has been used as a model to define key viral and host determinants of alphavirus-induced disease. In humans, Sindbis virus infection generally leads to non-life-threatening but debilitating, chronic illness, including polyarthritis, arthralgia, and Pogosta Disease (Kurkela et al., 2005; Sane et al.). However, there are other alphaviruses that lead to more severe disease and are considered serious human pathogens, including Ross River virus, Chikungunya virus, and Venezuelan Equine Encephalitis virus (Gould et al., 2009). Even though alphaviruses have been extensively studied for many years, there are still no effective antivirals or vaccines available. This reflects an incomplete understanding in alphavirus pathogenesis and limited knowledge of cellular factors involved in viral replication and pathogenesis.
Alphaviruses contain a nonsegmented single-stranded, positive-sense RNA genome with a 5’-cap and a 3’ poly(A) tail (Strauss and Strauss, 1994). The lipid membranes of these viruses have 80 glycoprotein spikes comprised of trimeric E1 and E2 heterodimers which are required for viral entry (Lescar et al., 2001; Zhang et al., 2002). E2 is a single transmembrane glycoprotein that contains the receptor binding site at the N terminus (Voss et al.), while E1 is a single transmembrane glycoprotein responsible for mediating fusion between the virus and host cell membranes (Omar and Koblet, 1988; Wahlberg and Garoff, 1992). While previous studies have established a requirement for a protein receptor for viral entry, the specific cell surface receptor required for entry has not yet been identified for infection of either the insect or vertebrate host (Strauss et al., 1994). In contrast, attachment factors including laminin receptor and heparin sulfate are not required for virus entry, though their presence has been shown to enhance viral infection (Klimstra et al., 1998; Wang et al., 1992).
A putative Sindbis virus receptor should be ubiquitously expressed given the exceptionally broad host range of the virus, and receptor engagement should lead to internalization via clathrin-mediated endocytosis (Strauss et al., 1994). The clathrin-coated vesicle fuses with endosomes where a low pH triggers conformational changes in E1 leading to a cholesterol and sphingolipid-dependent fusion of the viral envelope with the endosomal membrane (Ng et al., 2008; Strauss and Strauss, 1994). Importantly, Sindbis virus and other alphaviruses can fuse with artificial lipid membranes at low pH (Smit et al., 1999). Thus, the presence of a viral receptor per se is not required for the membrane fusion reaction, but rather for the efficient delivery of the virus to an appropriate endocytic organelle.
Using a Drosophila system, we identified the divalent metal ion tranporter Natural Resistance-associated Macrophage Protein (NRAMP), dNRAMP (Mvl), as required for binding and infection of Drosophila cells and that adult flies mutant for dNRAMP were protected from infection. We also found that NRAMP2, the ubiquitously expressed vertebrate homolog, mediated binding and entry into mammalian cells, and that murine cells deficient for NRAMP2 were non-permissive to Sindbis virus infection. The requirement for NRAMP is specific to Sindbis virus, as infection by another alphavirus, Ross River virus (RRV), was NRAMP-independent. Taken together, our findings that NRAMP is a conserved receptor used by Sindbis across disparate hosts informs our understanding of virus tropism and pathogenesis.
Results
dNRAMP is required for Sindbis virus infection
Previous studies have established an entry requirement for an as yet unidentified protein receptor for Sindbis virus (Strauss et al., 1994). We recently completed a genome-wide RNAi screen in Drosophila cells and identified nine transmembrane genes required for Sindbis virus infection, and dNRAMP (Mvl) was the only conserved, ubiquitously expressed, plasma membrane-associated protein identified (Rose, et al, unpublished). dNRAMP is the single Drosophila homologue of the mammalian natural resistance-associated macrophage proteins (NRAMPs), which are a family of highly conserved and broadly expressed twelve transmembrane proteins that mediate transport of heavy metals including iron (Figure S1a,b) (Andrews, 1999; Nevo and Nelson, 2006). Interestingly, plasma membrane-associated transporters are a common class of viral cellular receptors (Manel et al., 2005; Weiss and Tailor, 1995). Due to its broad conservation and ubiquitous expression, we set out to further characterize the role of dNRAMP in Sindbis infection. In Drosophila only one gene represents the NRAMP family, while in mammals there are two, NRAMP1 and NRAMP2 (Nevo and Nelson, 2006). dNRAMP is highly expressed in macrophages and neuronal cells similar to NRAMP1 (Southon et al., 2008), and in the intestine, with a low level of ubiquitous expression similar to NRAMP2 (Folwell et al., 2006). At the cellular level, dNRAMP is present at the plasma membrane similar to NRAMP2, and in endosomal compartments similar to NRAMP1 (Nevo and Nelson, 2006; Southon et al., 2008).
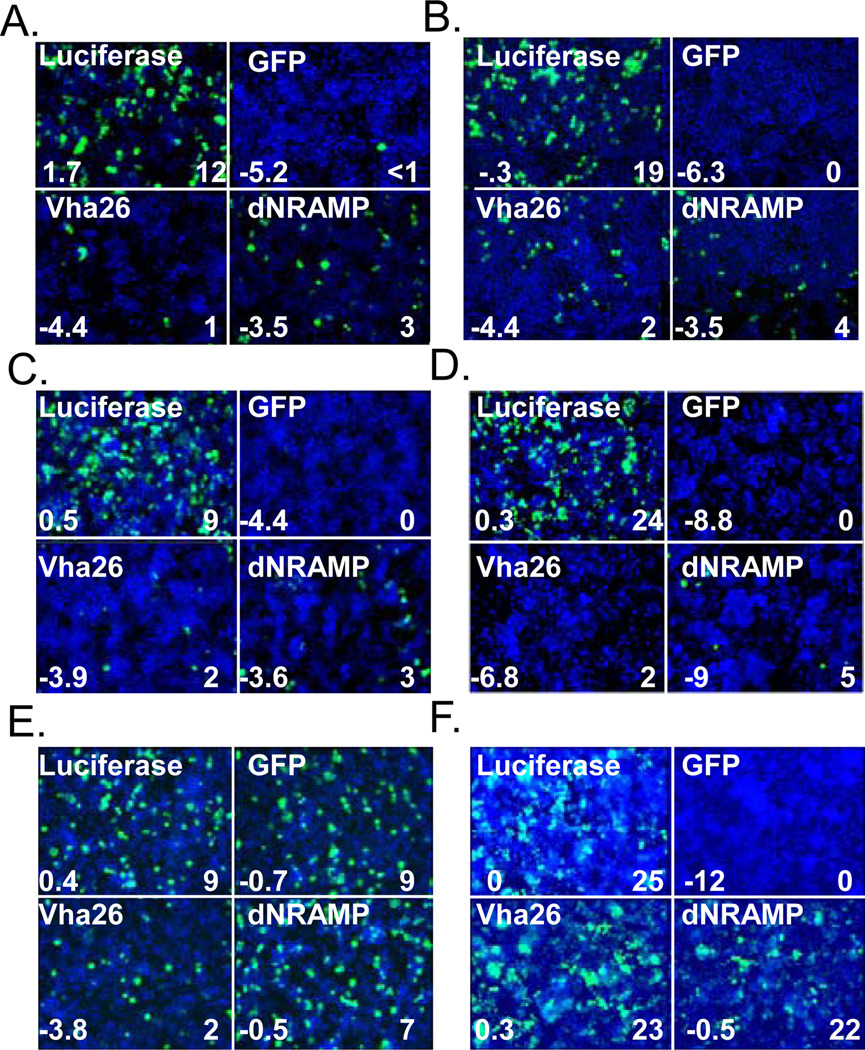
To characterize the requirement for dNRAMP during Sindbis virus infection, we tested independent dsRNAs in both the original Drosophila cell line used for screening (Figure 1a) as well as Kc167, another Drosophila cell line (Figure 1b), and found that infection with Sindbis virus HRsp expressing a GFP reporter gene was significantly decreased in both cases compared to negative controls (luciferase dsRNA). We treated cells with dsRNA against GFP and vha26 (required for endosomal acidification) as positive controls against viral or cellular genes, respectively (Figure 1a–d). In addition, we found that another, more pathogenic Sindbis virus strain, dsTE12H, was also dependent on dNRAMP for infection (Figure 1c) (Johnston et al., 2001). Since Sindbis virus strains HRsp and dsTE12H are lab-adapted, we also tested whether the wild type strain S.A.AR86 (Heise et al., 2000) was also dependent upon dNRAMP for infection. Indeed, we found that this strain was also dependent upon dNRAMP (Figure 1d). The Sindbis S.A.AR86 strain was a replicon which is unable to spread demonstrating that the requirement is during the first round of infection. Like Sindbis virus, West Nile Virus is an arbovirus (of the Flavivirus family) which is known to enter cells via clathrin-mediated endocytosis and traffics to an acidified compartment for fusion (Chu et al., 2006). Using wild type West Nile virus we found that while many of the canonical components of the clathrin-mediated endocytic pathway and the vATPase acidification machinery were required for both Sindbis virus and West Nile Virus infection of Drosophila cells, dNRAMP was completely dispensable for West Nile virus infection (Figure 1e). In addition, we found that dNRAMP was dispensable for infection with Vesicular Stomatitis Virus (VSV) an additional virus that is dependent upon clathrin-mediated endocytosis for entry (Figure 1f) (Cherry and Perrimon, 2004; Sun et al., 2005). These data suggest that dNRAMP was not ubiquitously involved in the clathrin-dependent uptake of viruses in general but rather specifically required for Sindbis virus infection.
Figure 1.
Drosophila NRAMP is required for Sindbis virus infection. dsRNA against dNRAMP, the vATPase component vha26, the SINV reporter GFP and the negative control luciferase were challenged with a. Sindbis virus-GFP (HRsp) in DL1 cells b. Sindbis virus-GFP (HRsp) in Kc167 cells c. Sindbis virus-GFP (dsTE12H) in DL1 cells. d. Sindbis-GFP (S.A.AR86) in DL1 cells. e. Wild type WNV detected using anti-E in DL1 cells. F. VSV GFP in DL1 cells. a–f Number on bottom left is robust Z-score and bottom right percent infection (virus, green; nuclei, blue).
dNRAMP is required for binding and entry
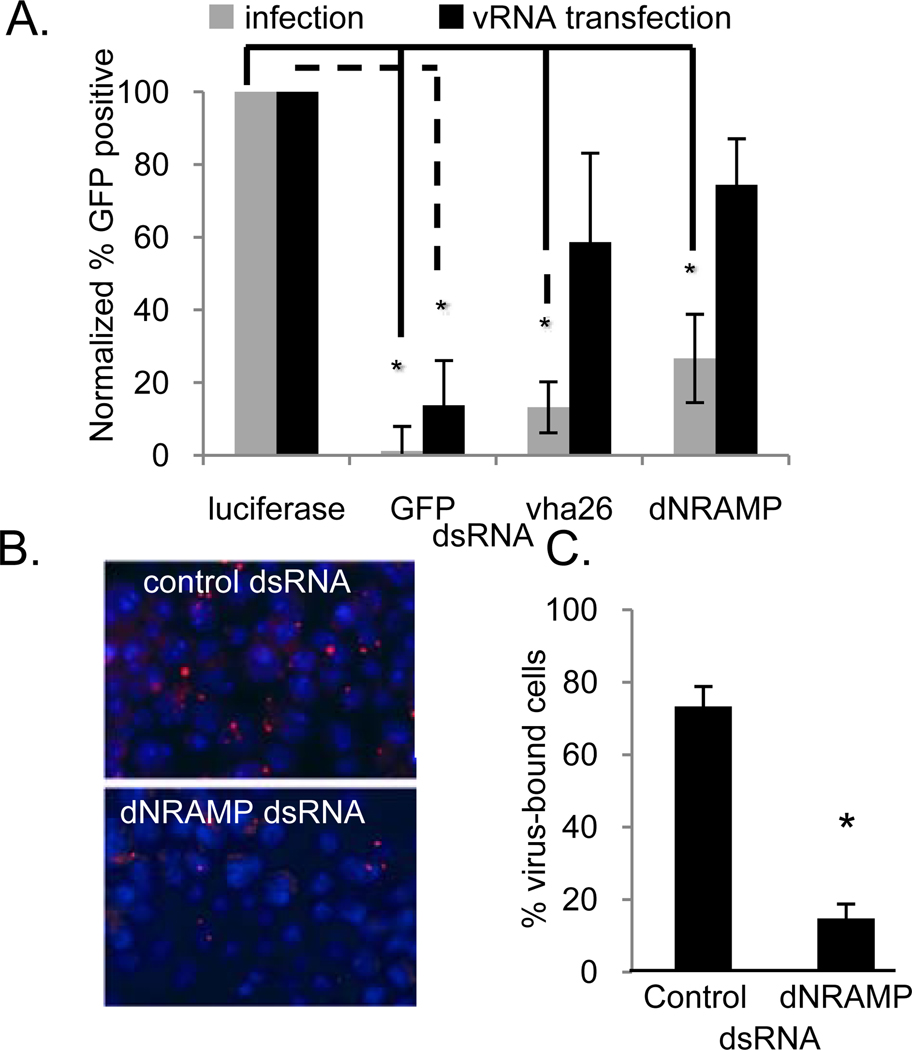
We next investigated the step in the Sindbis virus lifecycle that is dependent upon dNRAMP. To determine if the requirement for dNRAMP was at the level of entry, we performed a viral RNA-bypass assay. When transfected into cells, genomic viral RNA is directly translated and can launch the infection bypassing any entry requirements. We compared the production of virally-expressed GFP between infection (entry-dependent) and viral RNA transfection (entry-independent) in cells depleted for the control luciferase compared to the vATPase or dNRAMP. Using this assay, we found that the requirement for dNRAMP, as well as the vATPase, was largely bypassed by viral RNA transfection suggesting a role for dNRAMP in entry (Figure 2a). In contrast, treatment of cells with dsRNA against the virally expressed reporter GFP could not be bypassed, since infection levels were decreased in the RNA bypass assay. This also demonstrates that dNRAMP is not required for a step downstream of entry in Drosophila cells.
Figure 2.
dNRAMP is required for binding and entry in Drosophila cells. a. Cells pre-treated with the indicated dsRNA were either infected (gray) or transfected with vRNA (black) and triplicate Mean± S.D. shown; *p<0.05. b. Cells were pre-treated with dsRNA against dNRAMP or control dsRNA and bound with biotinylated Sindbis virus (virus, red; nuclei, blue). c. The percentage of cells that had at least one viral particle bound is graphed with Mean± S.D. for triplicate experiments; *p<0.05.
Next, we assayed whether dNRAMP was required for Sindbis virus binding to cells since dNRAMP is a plasma membrane-localized transporter (Nevo and Nelson, 2006). For these experiments, cells were pre-treated with dsRNA targeting dNRAMP or the negative control luciferase, and then challenged with biotinylated infectious Sindbis virus at 4°C. Free virus was removed by extensive washing, and bound virus was visualized with Streptavidin-Texas Red (Figure 2b). Quantification revealed that cells depleted of dNRAMP showed a ~5-fold reduction in binding compared to control dsRNA treated cells (Figure 2c). Taken together, these data suggest that dNRAMP is an entry receptor for Sindbis virus in Drosophila cells.
dNRAMP is required for Sindbis infection of adult flies
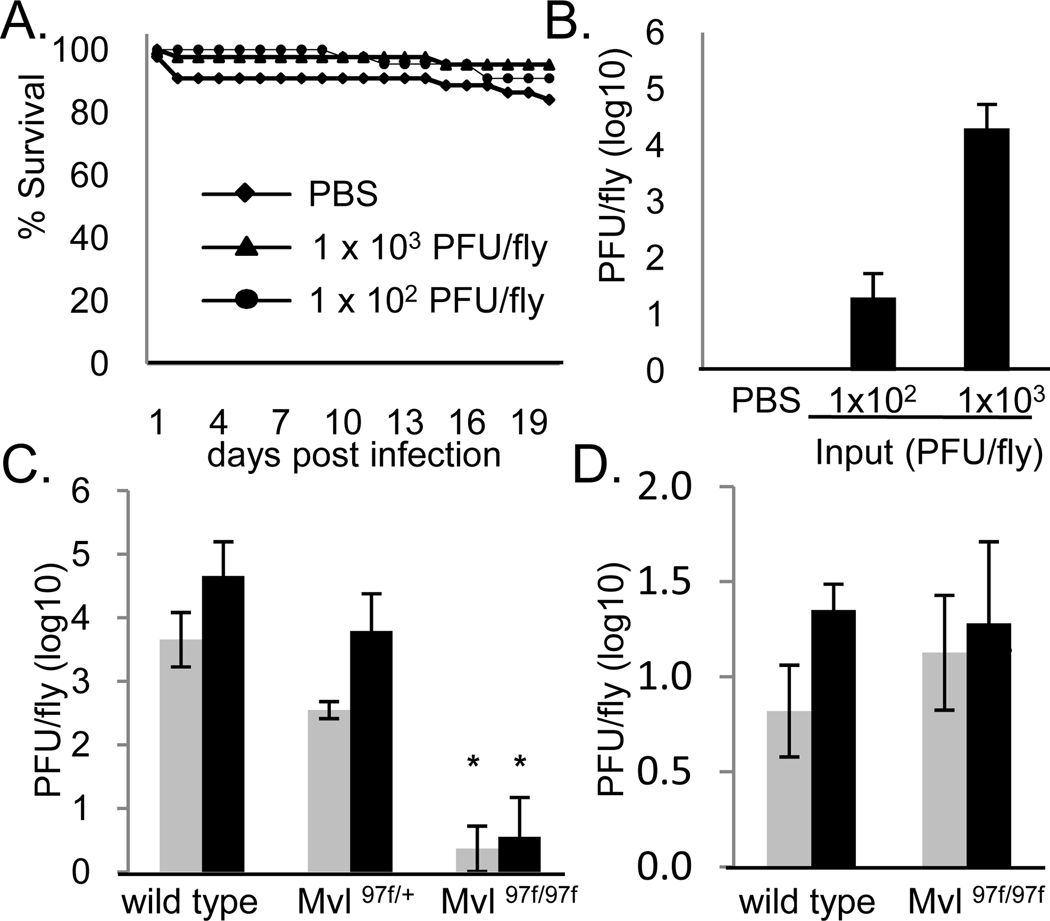
We next sought to determine if dNRAMP was required for infection not only of tissue culture cells, but also of adult flies. Sindbis virus infection of both the natural mosquito host and adult flies is non-lethal (Figure 3a) (Cirimotich et al., 2009; Galiana-Arnoux et al., 2006) and the generation of infectious virus in vivo was dose-dependent (Figure 3b). A transposon insertion allele of dNRAMP (Mvl97f) has been previously characterized as strong loss-of-function allele that behaves as a null allele in some behavioral assays (Rodrigues et al., 1995). We found that there was a 4-log reduction in viral titers of flies that were infected with Sindbis virus but deficient for dNRAMP (Mvl97f) compared to heterozygous or wild-type controls while VSV infection was unaffected (Figure 3c and d, respectively). Altogether, these data illustrate that dNRAMP is required for infection both at the cellular and organismal level.
Figure 3.
dNRAMP is required for Sindbis infection of adult flies. a. Survival curve of SINV challenged flies compared to control. b. Viral titers of flies challenged with the indicated SINV concentrations or control at 6 days post infection. Mean± S.D. of three experiments. c. Sindbis virus titers from flies of the indicated genotypes at either 3 (gray) or 6 (black) days post infection. Mean± S.D. of three experiments; *p<0.05. d. VSV virus titers from flies of the indicated genotypes at either 3 (gray) or 6 (black) days post infection; Mean± S.D. of three experiments.
Posttranslational regulation of NRAMP by iron inhibits Sindbis virus infection in insects
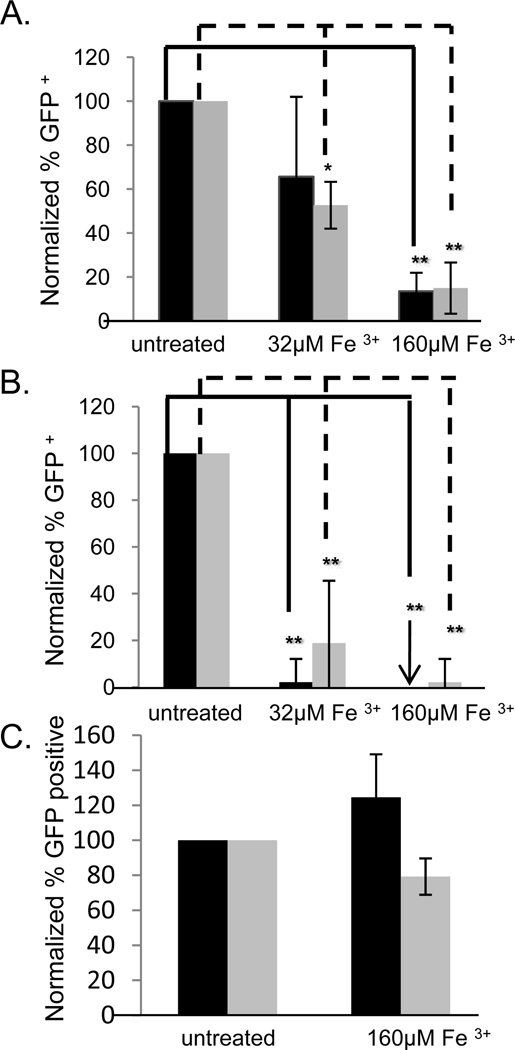
Since Drosophila is not the natural host of Sindbis virus, we were interested in determining if NRAMP was also required for Sindbis virus infection of one of the natural mosquito hosts, Aedes aegypti (Strauss et al., 1994). For these studies we took advantage of the fact that NRAMP is a metal ion transporter sensitive to intracellular iron concentrations whereby high concentrations of iron lead to a reduction in NRAMP expression (Martini et al., 2002). NRAMP downregulation can occur on two levels. First, NRAMP2 is regulated through effects on mRNA stability; when iron levels are high NRAMP2 mRNA is degraded (Martini et al., 2002). Second, NRAMP is posttranscriptionally degraded; in the presence of high iron NRAMP interacts with recycling adaptor complexes and is removed from the cell surface for degradation (Foot et al., 2008; Liu and Culotta, 1999). These mechanisms of regulation synergistically deplete NRAMP2 from cells. If NRAMP is a Sindbis virus receptor, high iron treatment would be predicted to attenuate infection. Indeed, iron treatment of Drosophila cells attenuated infection with both lab-adapted (Figure 4a) and wild type (Figure 4b) Sindbis virus while having no effect on infection with VSV (Figure 4c). Likewise, treatment of Aedes Aegypti Aag-2 cells with iron also significantly impeded Sindbis virus infection of lab-adapted (HRsp) and wild type (S.A.AR86) Sindbis virus strains in a dose-dependent manner but not VSV (Figure 4a–c). Again, the use of the S.A.AR86 replicon that cannot spread, demonstrates that this requirement is during the first round of infection. Taken altogether the data strongly suggested that dNRAMP is required for Sindbis virus entry into insects including vector mosquitoes.
Figure 4.
Sindbis virus infection is sensitive to iron treatment in both Drosophila and mosquito cells. a–c. Iron treatment of Drosophila DL1 cells (black) or Aedes aegypti Aag-2 cells (gray) challenged with (a) Sindbis virus (HRsp), (b) Sindbis virus (S.A.AR86), or (c) VSV. Mean± S.D. of three experiments; * p<0.05.
Sindbis virus infection of mammalian cells is iron-regulated
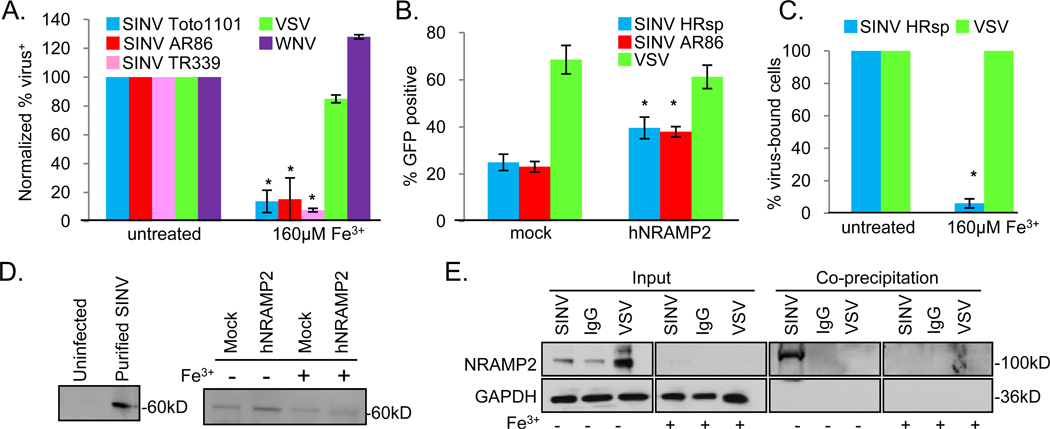
Due to the conservation between NRAMPs (Figure S1a, b) (Pinner et al., 1997), we tested whether NRAMP played a role in Sindbis virus infection of mammalian cells. NRAMP1 has a restricted expression pattern, while NRAMP2 is more ubiquitously expressed (Nevo and Nelson, 2006). In addition, NRAMP1 is found predominantly in intracellular compartments, while NRAMP2 is expressed more ubiquitously at the plasma membrane and in recycling endosomes (Nevo and Nelson, 2006). Since Sindbis virus can infect many cell types and is thought to initially interact with its entry receptor(s) at the plasma membrane, we reasoned that NRAMP2 was more likely to function in entry. First, we set out to determine if Sindbis virus infection was sensitive to iron as NRAMP2 expression is decreased by treatment with high iron (Figure S2a). Indeed, treatment of mammalian cells including human U2OS with iron significantly attenuated lab-adapted (Toto 1101) and wild type (S.A.AR86, TR339) Sindbis virus infection, and did so during the first round of infection because the two wild type strains are replicons unable to spread (Figure 5a, Figure S2b). Furthermore, iron treatment had no effect on West Nile virus or VSV infection (Figure 5a) demonstrating a specific dependency for Sindbis virus. We also found that iron-treated human 293T cells and mouse embryonic fibroblasts (MEFs) were refractory to Sindbis virus infection (Figure S2c and S2d, respectively).
Figure 5.
NRAMP2 facilitates binding and infection of Sindbis virus in mammalian cells. a. U2OS cells challenged with the indicated virus in the presence or absence of iron. Mean± S.D. of three experiments; * p<0.05. b. U2OS cells transfected with hNRAMP2 or the control vector (mock) were challenged with the indicated virus. Mean± S.D. of three experiments; *p<0.05. c. The percentage of cells that had at least one viral particle bound is graphed for the indicated virus with Mean± S.D. for triplicate experiments shown; *p<0.05. d. Mock or hNRAMP2-transfected U2OS cells were either untreated or iron treated and bound with biotinylated Sindbis virus, precipitated with steptavidin beads and visualized by immunoblot. Left panel shows that cells that were uninfected do not have a band while input labeled virus is visualized as a 60kD band. Representative of 3 experiments. e. U2OS cells were either untreated or iron treated and bound with biotinylated Sindbis virus, biotinylated VSV or biotinylated IgG, precipitated with streptavidin beads and visualized by immunoblot. GAPDH and NRAMP2 are observed in the input, whereas only Sindbis virus precipitates with NRAMP2; and only in the absence of iron treatment which reduces NRAMP2 levels. Representative of 5 experiments.
NRAMP2 facilitates infection and binding of Sindbis virus in mammalian cells
As many receptors are rate-limiting for viral infection (Breiner and Schaller, 2000; Cheng et al., 2007), we tested whether NRAMP2 over-expression impacted Sindbis virus infection. Indeed, we found that over-expression of NRAMP2 led to a 2-fold increase in the percentage of infected cells compared to controls for both wild type (S.A.AR86) and lab-adapted (HRsp) Sindbis virus strains but had no affect on VSV infection (Figure 5b). Again, this was during the first round of infection because the wild type strain is a replicon unable to spread. When NRAMP2 over-expressing cells were treated with iron, we observed a decrease in NRAMP2 expression (Figure S2a). We used these cells to determine if NRAMP2 mediated binding of Sindbis virus to mammalian cells. Using biotinylated Sindbis virus, we observed a ~17-fold reduction in virus binding to iron-treated cells compared to controls when we visualized bound virus using the microscopy-based assay (Figure 5c, Figure S2e). Iron-treatment had no affect on VSV binding using the same assay (Figure 5c, Figure S2e). Furthermore, we biochemically monitored binding by immunoprecipitating bound virus using streptavidin beads and could specifically detect Sindbis virus (Figure 5d, left panel). We observed increased binding to NRAMP2-overexpressing cells, and this binding was attenuated by iron treatment of both wild type cells and cells over-expressing NRAMP2 (Figure 5d, right panel). Next, we bound cells with either biotinylated Sindbis virus, biotinylated VSV or biotinylated IgG and precipitated the bound proteins using streptavidin beads and performed an immunoblot. We found that NRAMP2, but not GAPDH, specifically co-precipitated with Sindbis virus while the VSV and IgG did not (Figure 5e). Furthermore, this interaction is lost under high iron concentrations which significantly deplete NRAMP2 from cells (Figure 5e). Taken together, these data suggest that Sindbis virus binding to mammalian cells is mediated by NRAMP2.
NRAMP2-deficient cells are non-permissive for Sindbis virus infection
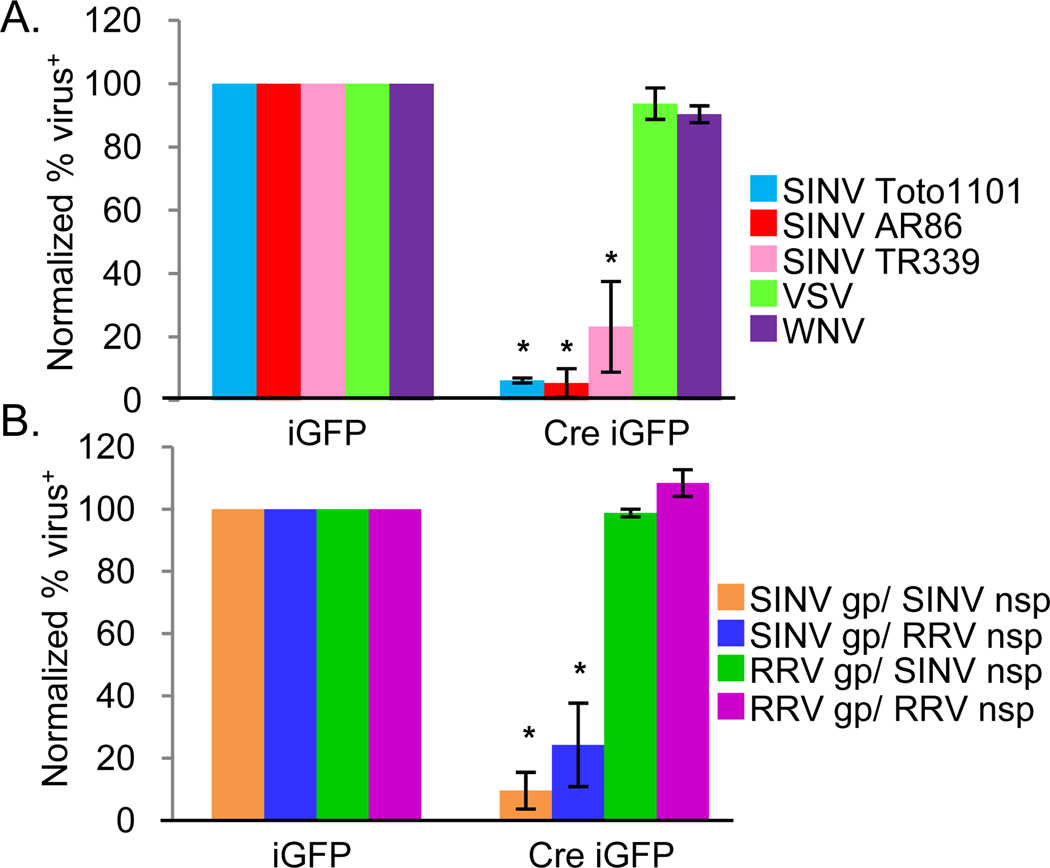
Next, we set out to determine if NRAMP2 is required for Sindbis virus infection of mammalian cells. As there are no known cell lines refractive to Sindbis virus infection, and NRAMP2 knockout mice are lethal, we generated SV40 T-antigen transformed MEFs from mice carrying floxed NRAMP2 alleles (NRAMP2fl/fl) (Gunshin et al., 2005). These cells express NRAMP2 but not NRAMP1 (Figure S3a). Next, the NRAMP2fl/fl cells were transduced with either a control retrovirus that expresses GFP (MSCV iGFP) or a retrovirus that co-expresses Cre and GFP (MSCV Cre iGFP) (Cherry et al., 2000). We performed PCR to monitor the status of the NRAMP2 locus and found that Cre transduction led to deletion of the floxed exons in the NRAMP2 locus (Figure S3b). When NRAMP2 deleted MEFs were challenged with Sindbis virus (Toto 1101), there was a 50-fold decrease in infection compared to Cre negative transduced MEFs (Figure 6a). Wild type Sindbis virus (A.R.AR86, TR339) infection using these replicons was also dependent upon NRAMP2 demonstrating the requirement in the first round of infection (Figure 6a). In contrast, loss of NRAMP2 had no effect on West Nile virus or VSV infection (Figure 6a).
Figure 6.
NRAMP2 is required for Sindbis virus infection and entry. a. NRAMP2fl/fl cells were transduced with MSCV-Cre-ires-GFP or MSCV-ires-GFP retroviruses and challenged with the indicated virus. The percent infection of retrovirally transduced cells (GFP+) is shown for triplicate experiments Mean± S.D.; *p<0.05. b. Sindbis and Ross River chimeric viruses were used to challenge NRAMP2fl/fl cells transduced with MSCV-Cre-ires-GFP or MSCV-ires-GFP retroviruses as indicated. Mean± S.D. of three experiments; * p<0.05. (gp, glycoproteins; nsp, nonstructural proteins)
NRAMP2 is required for Sindbis virus entry
To underscore the specificity of NRAMP2 as a receptor for Sindbis virus, we tested whether Ross River virus, another alphavirus, is dependent upon NRAMP for infection. We found that Ross River virus infection is both insensitive to iron treatment (not shown) and NRAMP2 deletion (Figure 6b). This allowed us to take advantage of Sindbis virus/Ross River virus chimeras (Kuhn et al., 1996; Kuhn et al., 1991), in which the structural and nonstructural proteins are swapped, to demonstrate that NRAMP2 is specifically required for entry and not downstream events in the Sindbis virus life cycle. Viruses containing Sindbis virus structural proteins with either the Sindbis virus or Ross River virus nonstructural proteins were NRAMP2-dependent while viruses containing the Ross River virus structural proteins with either the Sindbis virus or Ross River virus nonstructural proteins were NRAMP2-independent (Figure 6b). Taken together, these data show that NRAMP2 is required for Sindbis virus binding, entry and infection in both mammalian and insect cells.
Discussion
Using a Drosophila system with robust RNAi technology we identified NRAMP as a receptor for the prototypical alphavirus Sindbis virus in cells. Molecular and biochemical assays revealed a requirement for dNRAMP for binding and entry. In addition, the ease of organismal genetics also allowed us to demonstrate that dNRAMP is the only receptor present in flies, as mutant flies were non-permissive to infection. Importantly, we demonstrated that mosquito cells could be rendered non-permissive under high iron conditions, suggesting that they too use NRAMP as a receptor. Whether additional cellular receptors can be used by Sindbis virus in mosquitoes remains unknown.
We extended our findings to mammalian systems where we hypothesized that the plasma membrane-bound homolog NRAMP2 may also be used by Sindbis virus for entry. A variety of molecular and biochemical assays revealed that NRAMP2 indeed binds and facilitates entry of Sindbis virus in mammalian cells. The identification of NRAMP as a cellular receptor for Sindbis virus in both the insect and vertebrate host sheds light on the mechanism of entry for this virus. The ubiquitous plasma membrane expression of NRAMP, and its trafficking via the clathrin-dependent endocytic pathway for delivery into acidified compartments, fulfills two requirements for a potential Sindbis virus receptor (Strauss et al., 1994). However, the residual infection observed in mammalian cells (2-log reduction) can be explained by at least two different models. First, residual infection may be due to low affinity interactions, which traffic the virus to a low pH compartment thereby allowing for low levels of infection. Since the fly mutants do not support infection (>4-log reduction) it would suggest that these low affinity interaction are less efficient in Drosophila. A second model is that the residual infectivity in mammalian cells is due to the presence of another receptor which plays a less important role in the cell types we tested. Future studies will clarify these issues.
Since we tested both natural hosts (humans) as well as artificial hosts (Drosophila) we suggest that the lack of non-permissive cells for Sindbis virus infection is due to conserved residues in NRAMP. While the overall similarity of dNRAMP with human NRAMP2 is only 54%, we found that many of the extracellular loops have much higher homology which suggests that Sindbis virus relies on the conservation of NRAMP across species to infect diverse hosts. In particular, the most conserved extracellular loop contains the iron binding site, and iron uptake is essential for all organisms, therefore we hypothesize that Sindbis virus binds to this region making it difficult for organisms to escape from Sindbis virus interactions.
Furthermore, by demonstrating that Sindbis virus strains obtained from infected humans and mosquitoes are all NRAMP-dependent, we hypothesize that NRAMP is likely a receptor for all Sindbis virus isolates globally. Since the portion of the viral E2 glycoprotein used for host recognition is highly conserved among different alphaviruses, all of which exhibit broad species tropisms (Li et al.; Voss et al.), it is possible that conserved, ubiquitously expressed multi-pass membrane proteins may support entry of other alphaviruses (Kielian; Li et al.; Voss et al.). Lastly, our data suggest that this loss-of-function screening approach in Drosophila may allow for the identification of ubiquitously expressed cellular receptors for arthropod-borne viruses or other viruses with broad species tropism.
Experimental Procedures
Cells and virus and reagents
DL1, KC167, and Aag-2 cells were maintained in Schneider’s Medium (Invitrogen-GIBCO, Carlsbad, CA), supplemented with 10% heat inactivated Fetal Bovine Serum (Sigma), 100 U/ml of penicillin, 100 mg/ml of streptomycin (Invitrogen-GIBCO, Carlsbad, CA), and 100 mg/ml Hepes (Invitrogen-GIBCO, Carlsbad, CA). U2OS, 293T, and MEFs were maintained in DMEM (Invitrogen-GIBCO, Carlsbad, CA), supplemented with 10% heat inactivated Fetal Bovine Serum (Invitrogen-GIBCO, Carlsbad, CA), 100 U/ml of penicillin, 100 mg/ml of streptomycin (Invitrogen-GIBCO, Carlsbad, CA). Other chemicals were purchased from Sigma. Texas Red conjugated secondary antibodies were obtained from Jackson Immunochemicals. Streptavidin conjugated Texas Red or -HRP were obtained from Thermo Scientific. Anti-NRAMP2 was obtained from Santa Cruz, and anti-GAPDH was obtained from Calbiochem. Sindbis virus (HRsp and dsTE12H) were generated in BHK cells and then propagated in C6/36 cells (Burnham et al., 2007). VSV and VSV-GFP were propagated in BHK cells (Ramsburg et al., 2005). WNV (WNV lineage I strain 3000.0259 NY 2000) virus was propagated in C6/36 cells (Hanna et al., 2005). Wild type Sindbis virus-GFP (S.A.AR86) (Heise et al., 2000), Sindbis virus-GFP (TR339) (McKnight et al., 1996), Ross River virus-GFP (T48) replicons were packaged using wild type coats for their corresponding viruses as previously described (Pushko et al., 1997). Sindbis virus and Ross River virus chimeras were propagated as previously described (Kuhn et al., 1996; Kuhn et al., 1991). MSCV-iGFP and MSCVCreiGFP retroviral supernatants were generated as described (Cherry et al., 2000).
Drosophila RNAi
18,000 cells were seeded into 384-well plates pre-arrayed with 250ng/well dsRNA (Ambion) in 10μL of serum free media using automated liquid handling (Wellmate). One hour later 20μL of complete media was added, and the plates were incubated for three days and then infected with Sindbis virus (MOI=10, determined on BHK) for 36 hours. The plates were fixed in PBS/4% Formaldehyde for 10 minutes and washed twice in PBS. Cells were then stained with the indicated antibody and 5μg/mL Hoechst 33432 and washed twice with PBS. 3 sites per well per wavelength were imaged at 20X (ImageXpress Micro, Molecular Devices). Automated image analysis segmented the images and was used to determine the number of DAPI+ cells and the number of virus+ cells. The percentage of infected cells was calculated, averaged for the three sites. These metrics were used to calculate a robust Z score. The percent infection was calculated for each well, log-transformed, and the interquartile range (IQR) and plate median was used to calculate a robust Z score for each well using the following equation: [(log10(%infection)-log10(median)/(IQR*0.74)] (Zhang et al., 2006).
Insect cell infections
Insect cells were infected with Sindbis virus HRsp (MOI=10), dsTE12H (MOI=10), or S.A.AR86 (MOI=10), West Nile Virus (MOI=1), or VSV (MOI=2) by spin-infection for 2 hours at 1200rpm, room temperature in serum-free media. Afterwards serum was increased to 7%. Infected insect cells were fixed between 30 and 36 hours post infection.
RNAi vRNA Bypass
34,000 cells were seeded into 96-well plates pre-arrayed with 750ng/well dsRNA in 30μL of serum free media. One hour later 70μL of complete media was added and the plates were incubated for three days. Next, the cells were infected with SINV virus (MOI=10), or transfected with 0.1μg purified Sindbis virus genomic RNA using Effectene (Qiagen) following the manufacturer’s protocol. 36 hours later the cells were fixed and processed for microscopy.
Virus Biotinylation and Immunoprecipiations
Supernatant containing Sindbis virus (HRsp) harvested from C6/36 cells or VSV harvested from BHK cells was pre-cleared (1200rpm for 4 min), and pelleted through a 25% sucrose cushion (SW32 @ 21,000rpm, 4°C) for 2hrs and resuspended in 100μL PBS. This purified virus was biotinylated with 3.5μg/μL EZ-link Sulfo-NHS-LC-biotin (Thermo Scientific, Rockford, IL) for 30 minutes at room temperature and quenched with 1mM Tris-Cl (pH 7.5). Biotinylated Sindbis virus, biotinylated VSV or IgG (Sigma) was bound to pre-chilled cells for 45 minutes at 4°C. Unbound virus or IgG was removed and cells were processed for microscopy using Streptavidin-Texas Red. For biochemical analysis cells were lysed in1% Triton in PBS containing 1X Protease Inhibitor cocktail (Roche) for 30 min at 4°C and insoluble fractions were removed by a 30 min spin at 10,000 × g. Samples were pre-cleared and precipitated with Streptavidin-agarose. Washed beads were boiled in SDS-Lysis buffer and immunobloted.
Adult Fly Infections
Flies of the indicated genotypes were obtained from the Bloomington stock center and maintained on standard medium at room temperature. 4- to 7-day-old adult female flies were inoculated with Sindbis virus or VSV as previously described (Cherry and Perrimon, 2004). Groups of at least 20 flies were challenged for mortality studies. Flies were processed for plaque assay by crushing and tittering on BHK cells.
Mammalian Transfections and infections
U2OS cells were transfected with using Fugene per manufacturer’s protocol. Mammalian cells were infected with Sindbis virus (HRsp, MOI=0.1), wild type Sindbis virus (S.A.AR86, MOI=0.5), West Nile virus (MOI=1), VSV-GFP (MOI=0.1), Ross River virus (MOI=0.5) and fixed at 12 hours post infection.
Iron Treatment
Cells were treated with 160μM fresh Ammonium Iron(III) Citrate (Sigma) daily for three days prior to and throughout the infection.
mNRAMP2fl/fl Assays
mNRAMP2fl/fl MEFs were generated and immortalized with SV40 T-antigen. Cells were infected with either MSCViGFP or MSCVCre-iGFP as described (Cherry et al., 2000) and infected with Sindbis virus (Toto1101) (MOI=0.1), Sindbis virus (S.A.AR86, TR339) (MOI=0.5), VSV (MOI=0.1), WNV (MOI=1), chimeras (MOI=1) and visualized using anti-Sindbis virus E1 (gift from D.E. Griffin) for Toto1101, anti-Sindbis ascites (gift from R. Tesh) for S.A.AR86,TR339 and chimeric virus series with Sindbis virus glycoproteins, anti-VSV G (gift from R. Doms), anti-WNV E (gift from R. Doms), anti-RRV ascites for chimeric viruses with Ross River virus glycoproteins, (gift from R. Tesh) 14hr post infection.
Highlights.
Drosophila NRAMP, a metal ion transporter is required for Sindbis virus infection
dNRAMP is required for binding, entry and infection of Drosophila cells and adult flies
Sindbis virus infection is sensitive to iron treatment
NRAMP2 is required for Sindbis virus entry and infection of mammalian cells
Supplementary Material
Acknowledgements
The authors would like to thank RW. Doms and P. Bates for critically reading the manuscript; members of the Cherry, Ross, and Doms laboratory for technical advice and helpful discussions; W.B. Klimstra for Sindbis virus (TR339); R. Kuhn for chimeric viruses; J. Rose for VSV-eGFP; P. Gros for pCB6-NRAMP2; N. Andrews for NRAMP2-floxed mice; The Bloomington Stock center for fly stocks; anti-SINV E1 from D. Griffin; anti-VSV G and anti-WNV E from R. Doms; anti-RRV and anti-Sindbis ascites from R. Tesh and the NIAID and the World Reference Center for Emerging Viruses and Arboviruses; and Aag2 cells from A. Raikel. This work was supported by NIH training grant T32-AI055400 and NIH F32-AI078666 to PPR and NIH grants RO1AI079451 and U54AI057168 to SC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
PPR and SC designed the project, analyzed results and wrote the manuscript. PPR, SLH, SAB performed experimental work. PPR, AS, NW, and SC were involved in bioinformatics analysis. RH, DB KM, DW, MH and SR generated reagents and assisted in experimental implementation.
References
- Andrews NC. The iron transporter DMT1. Int J Biochem Cell Biol. 1999;31:991–994. doi: 10.1016/s1357-2725(99)00065-5. [DOI] [PubMed] [Google Scholar]
- Breiner KM, Schaller H. Cellular receptor traffic is essential for productive duck hepatitis B virus infection. J Virol. 2000;74:2203–2209. doi: 10.1128/jvi.74.5.2203-2209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007 doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Cheng HH, Anderson MM, Overbaugh J. Feline leukemia virus T entry is dependent on both expression levels and specific interactions between cofactor and receptor. Virology. 2007;359:170–178. doi: 10.1016/j.virol.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5:81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SR, Beard C, Jaenisch R, Baltimore D. V(D)J recombination is not activated by demethylation of the kappa locus. Proc Natl Acad Sci U S A. 2000;97:8467–8472. doi: 10.1073/pnas.150218497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JJ, Leong PW, Ng ML. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus mosquito (C6/36) cells. Virology. 2006;349:463–475. doi: 10.1016/j.virol.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009;9:49. doi: 10.1186/1471-2180-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folwell JL, Barton CH, Shepherd D. Immunolocalisation of the D. melanogaster Nramp homologue Malvolio to gut and Malpighian tubules provides evidence that Malvolio and Nramp2 are orthologous. J Exp Biol. 2006;209:1988–1995. doi: 10.1242/jeb.02193. [DOI] [PubMed] [Google Scholar]
- Foot NJ, Dalton HE, Shearwin-Whyatt LM, Dorstyn L, Tan SS, Yang B, Kumar S. Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood. 2008;112:4268–4275. doi: 10.1182/blood-2008-04-150953. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Gould EA, Coutard B, Malet H, Morin B, Jamal S, Weaver S, Gorbalenya A, Moureau G, Baronti C, Delogu I, et al. Understanding the alphaviruses: Recent research on important emerging pathogens and progress towards their control. Antiviral Res. 2009 doi: 10.1016/j.antiviral.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene IP, Paessler S, Austgen L, Anishchenko M, Brault AC, Bowen RA, Weaver SC. Envelope glycoprotein mutations mediate equine amplification and virulence of epizootic venezuelan equine encephalitis virus. J Virol. 2005;79:9128–9133. doi: 10.1128/JVI.79.14.9128-9133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin H, Fujiwara Y, Custodio AO, Direnzo C, Robine S, Andrews NC. Slc11a2 is required for intestinal iron absorption and erythropoiesis but dispensable in placenta and liver. J Clin Invest. 2005;115:1258–1266. doi: 10.1172/JCI24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise MT, Simpson DA, Johnston RE. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J Virol. 2000;74:4207–4213. doi: 10.1128/jvi.74.9.4207-4213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Jiang W, Chu T, Levine B. Identification of genes involved in the host response to neurovirulent alphavirus infection. J Virol. 2001;75:10431–10445. doi: 10.1128/JVI.75.21.10431-10445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M. Structural biology: An alphavirus puzzle solved. Nature. 468:645–646. doi: 10.1038/468645a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra WB, Ryman KD, Johnston RE. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Griffin DE, Owen KE, Niesters HG, Strauss JH. Chimeric Sindbis-Ross River viruses to study interactions between alphavirus nonstructural and structural regions. J Virol. 1996;70:7900–7909. doi: 10.1128/jvi.70.11.7900-7909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Niesters HG, Hong Z, Strauss JH. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- Kurkela S, Manni T, Myllynen J, Vaheri A, Vapalahti O. Clinical and laboratory manifestations of Sindbis virus infection: prospective study, Finland, 2002–2003. J Infect Dis. 2005;191:1820–1829. doi: 10.1086/430007. [DOI] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Culotta VC. Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J Biol Chem. 1999;274:4863–4868. doi: 10.1074/jbc.274.8.4863. [DOI] [PubMed] [Google Scholar]
- Manel N, Battini JL, Taylor N, Sitbon M. HTLV-1 tropism and envelope receptor. Oncogene. 2005;24:6016–6025. doi: 10.1038/sj.onc.1208972. [DOI] [PubMed] [Google Scholar]
- Martini LA, Tchack L, Wood RJ. Iron treatment downregulates DMT1 and IREG1 mRNA expression in Caco-2 cells. J Nutr. 2002;132:693–696. doi: 10.1093/jn/132.4.693. [DOI] [PubMed] [Google Scholar]
- McKnight KL, Simpson DA, Lin SC, Knott TA, Polo JM, Pence DF, Johannsen DB, Heidner HW, Davis NL, Johnston RE. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strains which affect cell culture and in vivo phenotypes. J Virol. 1996;70:1981–1989. doi: 10.1128/jvi.70.3.1981-1989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo Y, Nelson N. The NRAMP family of metal-ion transporters. Biochim Biophys Acta. 2006;1763:609–620. doi: 10.1016/j.bbamcr.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Ng CG, Coppens I, Govindarajan D, Pisciotta J, Shulaev V, Griffin DE. Effect of host cell lipid metabolism on alphavirus replication, virion morphogenesis, and infectivity. Proc Natl Acad Sci U S A. 2008;105:16326–16331. doi: 10.1073/pnas.0808720105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar A, Koblet H. Semliki Forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology. 1988;166:17–23. doi: 10.1016/0042-6822(88)90141-9. [DOI] [PubMed] [Google Scholar]
- Pinner E, Gruenheid S, Raymond M, Gros P. Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a ember of the mammalian natural resistance-associated macrophage protein family. J Biol Chem. 1997;272:28933–28938. doi: 10.1074/jbc.272.46.28933. [DOI] [PubMed] [Google Scholar]
- Pushko P, Parker M, Ludwig GV, Davis NL, Johnston RE, Smith JF. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- Ramsburg E, Publicover J, Buonocore L, Poholek A, Robek M, Palin A, Rose JK. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J Virol. 2005;79:15043–15053. doi: 10.1128/JVI.79.24.15043-15053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues V, Cheah PY, Ray K, Chia W. malvolio, the Drosophila homologue of mouse NRAMP-1 (Bcg), is expressed in macrophages and in the nervous system and is required for normal taste behaviour. Embo J. 1995;14:3007–3020. doi: 10.1002/j.1460-2075.1995.tb07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman KD, Klimstra WB. Host responses to alphavirus infection. Immunol Rev. 2008;225:27–45. doi: 10.1111/j.1600-065X.2008.00670.x. [DOI] [PubMed] [Google Scholar]
- Sane J, Guedes S, Kurkela S, Lyytikainen O, Vapalahti O. Epidemiological analysis of mosquito-borne Pogosta disease in Finland, 2009. Euro Surveill. 15 doi: 10.2807/ese.15.02.19462-en. [DOI] [PubMed] [Google Scholar]
- Smit JM, Bittman R, Wilschut J. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol. 1999;73:8476–8484. doi: 10.1128/jvi.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southon A, Farlow A, Norgate M, Burke R, Camakaris J. Malvolio is a copper transporter in Drosophila melanogaster. J Exp Biol. 2008;211:709–716. doi: 10.1242/jeb.014159. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Wang KS, Schmaljohn AL, Kuhn RJ, Strauss EG. Host-cell receptors for Sindbis virus. Arch Virol Suppl. 1994;9:473–484. doi: 10.1007/978-3-7091-9326-6_46. [DOI] [PubMed] [Google Scholar]
- Sun X, Yau VK, Briggs BJ, Whittaker GR. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Garoff H. Membrane fusion process of Semliki Forest virus. I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RA, Tailor CS. Retrovirus receptors. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, Holder DJ, Ferrer M, Espeseth AS. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics. 2006;7:299–309. doi: 10.2217/14622416.7.3.299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.