Abstract
Src homology 2 (SH2) domains mediate protein-protein interactions by recognizing phosphotyrosine (pY)-containing sequences of target proteins. In all of the SH2 domain-pY peptide interactions described to date, the SH2 domain binds to a single pY peptide. Here, determination of the co-crystal structure of the N-terminal SH2 domain of phosphatase SHP-2 bound to a class IV peptide (VIpYFVP) revealed a noncanonical 1:2 (protein/peptide) complex. The first peptide binds in the canonical manner with its pY side chain inserted in the usual binding pocket, while the second pairs up with the first to form two anti-parallel β-strands that extend the central β sheet of the SH2 domain. This unprecedented binding mode was confirmed in solution phase by NMR experiments and shown to be adopted by pY peptides derived from cellular proteins. Site-directed mutagenesis and surface plasmon resonance studies revealed that the binding of the first peptide is pY dependent, but phosphorylation is not required for the second peptide. Our findings suggest a potential new function for the SH2 domain as a molecular clamp to promote dimerization of signaling proteins.
Src homology-2 (SH2) domain is a small protein module consisting of ~100 amino acids. It was first identified in the N-terminal regions of Src family protein tyrosine kinases and subsequently found in >100 human proteins, many of which are involved in intracellular signaling processes.1 SH2 domains recognize specific phosphotyrosyl (pY) residues in their partner proteins, promoting protein-protein interactions. The sequence specificity of an SH2 domain-pY protein interaction is primarily determined by the pY residue and the three to four residues immediately C-terminal to pY,2,3 although some SH2 domains require a more extended region of the pY peptide for high affinity and specificity of the interactions.4–8 The structural basis for the specific interaction has been very well studied.9–14 A key interaction, which is common to all SH2 domains, is the insertion of the pY side chain into a deep pocket in the SH2 domain, where an invariant arginine residue (Arg βB5) forms a bidentate interaction with the pY phosphate group. Additional binding energy is provided by interactions between amino acids adjacent to pY, particularly the three residues immediately C-terminal to pY, and the less conserved surface of the SH2 domain. This latter interaction also governs the selectivity of a given SH2 domain in binding to a specific pY partner.
SHP-2 is a non-transmembrane protein tyrosine phosphatase (PTP), which contains two N-terminal SH2 domains and a C-terminal catalytic domain.15 In the absence of any pY ligand, SHP-2 exists in an inactive conformation, with its N-terminal SH2 domain occluding the substrate-binding pocket of the PTP domain.16 Occupation of the N-SH2 domain with a pY-containing ligand leads to release of the inhibitory N-SH2-PTP interaction and subsequent activation of the enzyme.17, 18 The N-terminal SH2 domain of SHP-2 thus acts as a molecular switch that controls both the specificity and the activity of the enzyme. The sequence specificity of both SH2 domains has previously been determined using combinatorial libraries.6,7 Unlike the classical SH2 domains, SHP-2 SH2 domains require more extended sequences both N- and C-terminal to the pY residue (positions pY-2 to pY+5) for high-affinity binding. The C-SH2 domain binds pY peptides of a single consensus sequence (T/V/I/y)XpY(A/s/t)X(I/V/L) (X = any amino acid and the lowercase letters represent less preferred residues). On the other hand, the N-SH2 domain of SHP-2 can recognize four distinct classes of sequences (class I–IV), with consensus sequences of (I/L/V/m)XpY(T/V/A)X(I/V/L/f), W(M/T/v)pY(y/r)(I/L)X, (I/V)XpY(L/M/T)Y(A/P/T/S/g), and (I/V/L)XpY(F/M)XP, respectively.6 X-ray crystal structures of SHP-2 N-SH2 domain in complex with the class I peptides have been reported.12 To further investigate the structural basis for recognition of the four different classes of peptide ligands, we attempted to determine the structures of SHP-2 N-SH2 domain bound to peptides from Class I (RLNpYAQLWHR), Class III (RIHpYLYALNR), and Class IV (RVIpYFVPLNR) by X-ray crystallography. We show that the binding mode of SHP-2 N-SH2 domain with class I and III ligands closely matches that of a high-affinity complex between the N-SH2 domain and a class I peptide derived from the platelet derived growth factor receptor (VLpY1009TAV).12 In contrast, the class IV ligand bound to the SH2 domain as an antiparallel dimer, which was confirmed by solution-phase NMR studies. To our knowledge, this is the first example of a single SH2 domain binding to two pY ligands, thus expanding the functional implications of SH2-peptide interactions.
EXPERIMENTAL PROCEDURES
Materials
Antibiotics, Sephadex G-25 resin, organic solvents were obtained from Sigma-Aldrich (St. Louis, MO). Talon resin for IMAC purification was purchased from Clontech (Mountain View, CA). Q-Sepharose Fast-Flow column and Thrombin was purchased from Amersham (Pittsburgh, PA). Reagents for peptide synthesis were from Advanced ChemTech (Louisville, KY), Peptides International (Louisville, KY), and NovaBiochem (La Jolla, CA). Protein concentration was determined by the Bradford method using bovine serum albumin as standard.
Expression and Purification of SHP-2 N-SH2 Domain
Plasmid pET28a-SHP2-NSH2, which codes for the N-SH2 domain of SHP-2 (amino acids 1–106) plus an N-terminal six-histidine tag, had previously been described.6 Escherichia coli BL21(DE3) cells harboring the above plasmid were grown in LB medium supplemented with 0.1 mg/mL ampicillin at 37 °C. When the OD600 value reached 0.6, the cells were induced by the addition of 0.1 mM isopropyl β-D-thiogalactoside and allowed to grow for 4 additional hours at 30 °C. The cells were harvested by centrifugation and resuspended in 20 mL (for 1 L of cell culture) of Talon column buffer (50 mM sodium phosphate, pH 7.8, 500 mM NaCl, and 5 mM imidazole) containing protease inhibitors (1 mM PMSF, 10 µg/mL trypsin inhibitor, 1 µM pepstatin A), lysozyme (1 mg/mL), and protamine sulfate (5 mg/mL). After incubation for 30 min at 4 °C, the mixture was sonicated briefly. The crude lysate was centrifuged at 13,000 g for 20 min and the supernatant was loaded onto a Talon column (2 cm × 10 cm containing 20 ml of resin) that had been equilibrated in Talon column buffer. The column was washed with Talon column wash buffer (50 mM sodium phosphate, pH 7.8, 500 mM NaCl, and 25 mM imidazole) and eluted with a buffer containing 50 mM sodium phosphate, pH 7.8, 500 mM NaCl, and 125 mM imidazole. The purified SH2 protein was concentrated to ~20 ml by Amicon Ultra-15 centrifugal filter units (Millipore). Buffer exchange and removal of the six-histidine tag were achieved simultaneously by adding 200 units of thrombin (GE healthcare) into the dialysis bag and dialyzing overnight against 2 L of buffer A (25 mM Tris, pH 8.0, 20 mM NaCl) at room temperature. The resulting digested protein sample was passed through the Talon column again to remove any uncleaved protein. The flow-through fraction was collected and loaded onto Q-Sepharose column (2 cm × 10 cm) equilibrated in buffer A. The column was eluted with buffer A containing a linear gradient of 0–500 mM NaCl. The fractions containing the pure SHP-2 N-SH2 protein (as judged by SDS-PAGE) were pooled and concentrated to >10 mg/mL by Amicon Ultra-15 centrifugal filter units. The concentrated protein was dialyzed against 10 mM Mes (pH 5.5) and 50 mM KCl. After the addition of 0.5 mM tris(carboxyethyl)phosphine, the protein solution was flash frozen in an isopropanol/dry ice bath and stored at −80 °C.
Preparation of [15N]-SHP-2 N-SH2
E. coli BL21(DE3) cells harboring the above DNA construct were cultured in M9 minimal medium [prepared with (15NH4)2SO4] supplemented with 0.4 % (w/v) glucose, 5 mM MgSO4·7H2O, 10 µM FeSO4·7H2O, and 0.01% thiamine at 37 °C to an OD600 of 0.7. The cells were induced with 0.1 mM isopropyl β-D-thiogalactoside at 20 °C for 15 h and harvested by centrifugation. Cell lysis and purification by metal affinity chromatography on the Talon column were performed as described above. The elution buffer was exchanged into an appropriate NMR buffer by dialysis against 20 mM sodium phosphate, pH 7.4, 150 mM NaCl, and 1 mM dithiothreitol [for 2D 1H-15N heteronuclear single-quantum correlation (HSQC) and 1H-13C HSQC experiments] or 50 mM HEPES, pH 7.4, and 150 mM NaCl (for 31P NMR spectroscopy). The protein solution was lyophilized overnight and stored at −80 °C. Protein concentration was determined by both the Bradford method using bovine serum albumin as standard and measuring its absorbance at 280 nm (the two methods gave values that were within 10% difference).
Peptide Synthesis
Peptides were synthesized on CLEAR-amide resin (0.4 mmol/g) using standard Fmoc/HBTU chemistry. After cleavage and side-chain deprotection with a modified reagent K (6.5% phenol, 5% water, 5% thioanisole, 2.5% ethanedithiol, 1% anisole in trifluoroacetic acid), the peptides were precipitated by cold ether and purified by reversed-phase HPLC on a C18 column (Varian 120 Ǻ, 4.6 × 250 mm). The identities of the peptides were confirmed by MALDI-TOF mass spectrometric analyses. The peptides used for 13C-1H HSQC experiments were acetylated with (13CH3CO)2O at the N-termini. Peptides used for surface plasmon resonance (SPR) studies were biotinylated at their N-termini by treatment with N-hydroxysuccinimido-biotin (Sigma). The concentrations of the peptides were determined by quantitative ninhydrin test or their absorbance at 280 nm (for peptides that contained tyrosine in their sequences). For the peptides used in 15N-1H and 13C-1H HSQC experiments, the concentration was confirmed by various methods including direct weighing, ninhydrin tests, absorbance increase at 280 nm upon phosphatase treatment, and 1D proton NMR (Figure S1 in Supporting Information).
Crystallization and Data Collection
The purified N-terminal SH2 domain of SHP-2 was co-crystallized with peptides RVIpYFVPLNR (referred to as pYFVP hereafter), RLNpYAQLWHR (referred to as pYAQL), and RIHpYLYALNR (referred to as pYLYA) at room temperature by hanging drop vapor diffusion. The SH2 domain (10 mg/ml) in 0.1 M Mes (pH 5.5), 50 mM KCl, was mixed with a slight molar excess of peptide (1.1 equiv) dissolved in water. The hanging drops were prepared by mixing 2 µL of the complex and 2 µL of the reservoir solution, which consisted of 20% PEG 3350, 0.1 M bis-Tris (pH 5.5), 0.2 M Li2SO4. Large tetragonal crystals grew within approximately three days. Crystals were transferred to a cryo-protectant solution containing the reservoir solution supplemented with 20% glycerol and frozen in liquid nitrogen. The diffraction data were collected at −180 °C at beamline 19BM of the Advanced Photon Source. Date were integrated and scaled with DENZO/SCALEPACK.19
Structure Determination
The structure was determined by molecular replacement with MOLREP of the CCP4 suite20 using the structure of free SHP-2 N-SH2 domain (PDB code 1AYD)12 as the search model. The structures with the pYFVP and pYAQL peptides were refined to 1.80 Å and 2.05 Å, respectively, using Refmac of the CCP4 suite.20 After the first round of refinement, the electron density maps revealed strong density for two molecules of the pYFVP peptide and one molecule of the pYAQL peptide, which were built into the models. For pYLYA, no density corresponding to the peptide was observed. Several rounds of refinement with Refmac and model building using the program COOT21 led to final models. The final refined structure of SH2-pYFVP consists of residues 4–103 of the SH2 domain (chain A), residues (−2)-(+4) of the first peptide (peptide A) and residues (−2)-(+3) of the second peptide (peptide B). The final SH2-pYAQLW structure consists of residues 5–103 of the protein and residues (−2)-(+4) of the peptide (chain B). Side chains of residues, Arg-4, Lys-BC1 and Lys-BG7 in the complex with pYFVP, and Arg-5, Lys-BC1, Lys-BG5 and Lys-BG7 in the complex with pYAQL, were not resolved in the electron density and were truncated to alanine. The C-terminal peptide residues, Arg(+6), Asn(+5) of each peptide and Leu(+4) of peptide B in the complex with pYFVP, and His(+5), Arg(+6) of the peptide in the pYAQLW complex were not observed in the electron density and were not included in the final structures. Data collection and refinement statistics are presented in Table 1. Structural figures were prepared using PYMOL (PyMOL Molecular Graphics System, Schrödinger, LLC). The secondary structural elements are defined using Dali server22 and named according to the nomenclature established previously for SH2 domains.11, 12 Solvent accessible surface area calculation was performed using the AREAIMOL feature of CCP4 with a probe radius of 1.4 Å.20
Table 1.
Crystallographic and Refinement Statistics.
| NSH2-pYFVP | NSH2-pYAQL | |
|---|---|---|
| X-ray Diffraction Data | ||
| Space group | P43212 | P43212 |
| Unit Cell dimensions | ||
| a = b (Å) | 62.9 | 62.7 |
| c (Å) | 75.3 | 75.3 |
| Resolution (Å) | 48.3-1.8 (1.89-1.80) | 48.2-2.05 (2.13-2.05) |
| No. unique reflections | 15,827 | 9,943 |
| Redundancy | 11.2 (11.6) | 10.9 (11.3) |
| Completeness (%) | 99.3 (100) | 99.0 (100) |
| I/σ | 61.5 (5.0) | 46.5 (6.7) |
| Rmerge | 0.030 (0.382) | 0.064 (0.364) |
| Refinement Statistics | ||
| Resolution (Å) | 48.3-1.80 | 48.2-2.05 |
| No. of reflections | 13,727 | 9,354 |
| Rwork/Rfree | 0.197/0.216 | 0.193/0.234 |
| No. of non-hydrogen atoms | 992 | 860 |
| Mean B factor (Å2) | 25.4 | 25.9 |
| R.M.S.D bond length (Å) | 0.021 | 0.025 |
| R.M.S.D bond angle (°) | 1.96 | 2.10 |
| Ramachandran plot | ||
| Favored regions | 98 (97.0%) | 95 (96.0%) |
| Allowed regions | 3 (3.0%) | 4 (4.0%) |
NMR Spectroscopy
Unless stated otherwise, data were recorded at room temperature on a Bruker DMX-600 spectrometer equipped with a TXI probe and three-axis gradients, and standard Bruker pulse sequences were used. 15N-Lablelled SHP-2 N-SH2 domain was prepared in 500 µl of NMR buffer (20 mM sodium phosphate, pH 7.4, 150 mM NaCl, and 1 mM dithiothreitol) to give a final concentration of 0.46 mM in 90% H2O/10% D2O. For two-dimensional 1H-15N HSQC experiments, the protein sample was titrated with peptides to reach the protein/peptide ratios of 1:0.5, 1:1, 1:1.5, 1:2, and 1:2.5. Three-dimensional (3D) 15N-edited NOESY-HSQC (200 ms NOE mixing time) and 3D 15N-edited TOCSY-HSQC spectra (60 ms DIPSI-2 mixing time) were recorded prior to as well as at the end of peptide titration. Two-dimensional 1H-13C HSQC experiments were carried out by titrating 200 µM of 13C-labeled peptide with 50, 100, 150 µM SHP-2 N-SH2 domain to reach protein/peptide ratios of 0.25:1, 0.5:1, 0.75:1. 31P NMR experiments were conducted on the Bruker DMX-600 spectrometer equipped with a BBO probe by titrating 300 µM peptide in Hepes buffer (50 mM Hepes, 150 mM NaCl, pH 7.4) with various amounts of SHP-2 N-SH2 domain to reach targeted protein/peptide ratios.
Affinity Measurement by SPR
All measurements were made at room temperature on a BIAcore 3000 instrument, using the N-terminal histidine-tagged SHP-2 N-SH2 domain. Peptide biotin-(miniPEG)2-RVIYFVPLNR-NH2 was immobilized onto a streptavidin-coated sensorchip. The peptide was dissolved in HBS-EP buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20, pH 7.4) to a final concentration of 17 µM, and flowed over the streptavidin chip until no further response unit increase could be observed. The immobilized peptide was exposed to HBS-EP solutions containing a fixed concentration of the SH2 domain (10 µM) and increasing concentrations of the pY peptide H2N-(miniPEG)2-VIpYFVPLNYRR-NH2 (0–60 µM). Each solution was passed over the sensorchip for 2 min at a flow rate of 15 µL/min. The equilibrium response unit (RUeq) for each pY peptide concentration was obtained by subtracting the response of the blank flow cell (which had no immobilized peptide) from that of the sample cell. To prepare the surface for the next run, the sensorchip was washed with a solution containing 0.05% SDS, 200 mM NaCl, and 10 mM NaOH for 30 s at a flow rate of 100 µL/min. The apparent dissociation constant (KDapp) was determined by nonlinear regression fitting of the data to the following equation:
where RUmax is the maximum response unit.
For competition studies, peptide NH2-RLNpYAQLWHR-NH2 was immobilized onto a CM5 chip at the N-terminal amine. Briefly, 50 µL of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide in ddH2O was mixed with 50 µL of 0.1 M N-hydroxysuccinimide in ddH2O and passed through the desired channel at a flow rate of 10 µL/min. Next, the pY peptide (30 µg/mL) in the HBS-EP buffer was passed through the channel at a rate of 5 µL/min until a response unit increase of 200 RU was reached. Any excessive reactive groups on the surface was quenched by flowing 50 µL of 1 M ethanolamine (pH 8.5) at a rate of 5 µL/min. Competing peptides of concentrations ranging from 0.25 to 80 µM were incubated with 0.4 µM SHP-2 N-SH2 domain for 10 min and passed over the chip at 15 µL/min. The RUeq values were plotted against the competing peptide concentration and the IC50 values (concentration of competing peptide at which the RUeq value was reduced by 50%) were estimated from the curves.
RESULTS
Crystal Structures of SHP-2 N-SH2 in Complex with Class I and IV Peptides
To understand the structural basis for the ability of SHP-2 N-SH2 domain to recognize multiple classes of pY ligands, we attempted to determine the crystal structures of the SH2 domain in complex with representative class I, III, and IV ligands previously selected from a peptide library [peptides pYAQL, pYLYA, and pYFVP, respectively]. In all three structures, the protein exhibits a typical SH2 domain fold consisting of a central anti-parallel β sheet flanked by two α helices, one on either side. The electron density map of the SH2-pYLYA complex showed density for a phosphate ion in the expected pY-binding pocket, but no density for the rest of the peptide. The structure of the SH2 domain is nearly identical to the unbound SH2 domain structure previously reported (PDB code 1AYD).12 The electron density map for the SH2-pYAQL complex allowed tracing of residues LNpYAQLW of the peptide, but the side chains of residues Gln (pY+2) and Trp (pY+4), which face out into solvent, were not resolved. The complex with the pYAQL peptide is very similar to the previous SH2 domian-peptide complexes (PDB code 1AYA, 1AYB, and 1AYC), and will not be discussed in detail here.
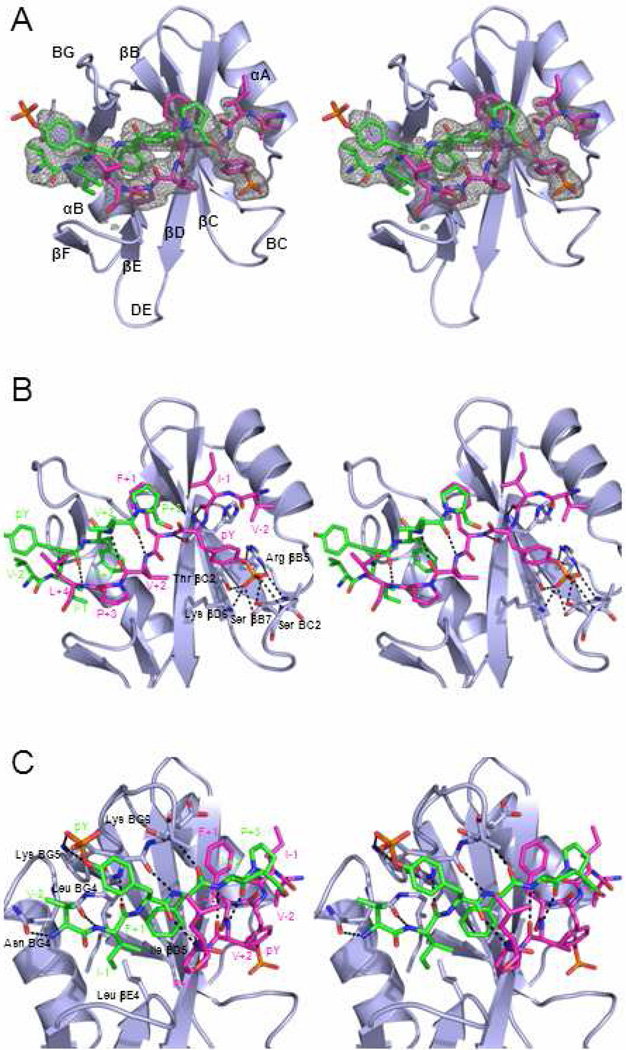
The electron density map for the complex with the pYFVP peptide, however, is surprisingly different (Figure 1A). In contrast to all previous SH2 domain-peptide complexes, which contain a single peptide per SH2 domain, we observe two copies of the pYFVP peptide bound to the peptide-binding surface on the SHP-2 N-SH2 domain. This was the case for all three of the data sets that we collected on three different crystals. The 1:2 binding mode was observed consistently, despite the fact that only a slight molar excess of the peptide (1.1 equiv) was present during crystallization. The first peptide, peptide A, binds in the canonical manner, as an extended β-strand that pairs with strand βD of the SH2-domain and inserts its pY residue into a positively charged pocket formed in part by Arg-βB5 and Lys-βD6 (Figure 1B). The second peptide, peptide B, binds in an extended conformation running anti-parallel to peptide A (Figure 1C). Residues IpYF of peptide B pair up with residues 88–90 of the BG loop of the SH2 domain, which form a short β-strand. Overall, the two peptides and the two short strands of the SH2 domain form an antiparallel four-stranded β-sheet, which adds onto the central four-stranded β-sheet (βA, βB, βC, and βD) of the SH2 domain to form a twisted eight-stranded β-sheet that almost forms a closed barrel.
Figure 1.
Crystal structure of SHP-2 N-SH2 domain in complex with two pYFVP peptides. (A) Stereo ribbon diagram of the complex with the peptides shown in stick form. Peptide A, which binds in the canonical manner, is in magenta, and peptide B is in green. The final 2Fobs − Fcalc electron density map calculated with model phases is contoured around the peptides. (B) Close-up stereo view of interactions of peptide A. Hydrogen bonds are shown as dashed lines. Peptides A and B pair up in an anti-parallel manner to form three β-sheet hydrogen bonds. (C) Close-up stereo view of interactions involving peptide B, which forms four parallel β-sheet hydrogen bonds with the BG loop. The Phe+2 residue of peptide B is buried in a hydrophobic pocket formed by Leu βE4, Leu BG4, Ile βD5, and Pro+3 of peptide A. The pY residue of peptide B is mostly solvent exposed, but forms a loose ion pair with Lys BG5.
The pY residue of peptide B is exposed on the surface of the structure, but forms a potentially favorable electrostatic interaction with Lys-BG5, whose ε-amino group is located 2.8 Å from the phosphate in the refined structure. The side chain of Lys-BG5, which was not at all resolved in the structures of the unbound SH2 domain or the complex with peptide pYAQL, is partially ordered in this structure. However, the electron density for the phosphate of peptide B, which is predominantly exposed, is considerably weaker than that for the buried phosphate of peptide A, suggesting that the former is likely to be flexible (Figure 1A). Accordingly, the average temperature factors for the phosphate atoms of peptides A and B are 47.8 and 68.2 Å2, respectively. Thus, although there is the potential for a weak ion pair, the absence of tight interactions with the phosphate of peptide B suggests that peptide B needs not to be phosphorylated to form the 1:2 complex.
In addition to the hydrogen bonding interactions, binding of the two peptides is also stabilized by their hydrophobic interactions with the SH2 domain and with one another (Figure 1B, C). In particular, the side chain of Phe (pY+1) of peptide B is anchored into a hydrophobic pocket on the SH2 domain formed by the side chains of Ile-βD5, Leu-βE4, Leu-BG4, and Ile-BG12 of the SH2 domain (Figure 1C). In the canonical SH2 domain-pY peptide structures, this hydrophobic pocket typically interacts with the nonpolar side chain of the pY+3 residue. The side chains of Ile (pY−1) of peptide B and Phe (pY+1) residue of peptide A are also partially buried, although their interactions appear to be less optimal since they are less tightly packed (Figure 1C). Due to the anti-parallel configuration, the Pro at the pY+3 position of peptide A stacks closely over the pY+1 Phe side chain of peptide B, and vice versa, to form additional hydrophobic interactions that appear to stabilize the complex. The side chains of the pY+2 valine residues of both peptides are exposed at the surface of the complex, but could form stabilizing hydrophobic interactions with each other, although they are not tightly packed (Figure 1B). Based on the structure, it appears that hydrophobic interactions between the FVP residues of the two peptides with one another and with the SH2 domain are in large part responsible for the specificity of the FVP sequence for the SHP-2 N-SH2 domain.
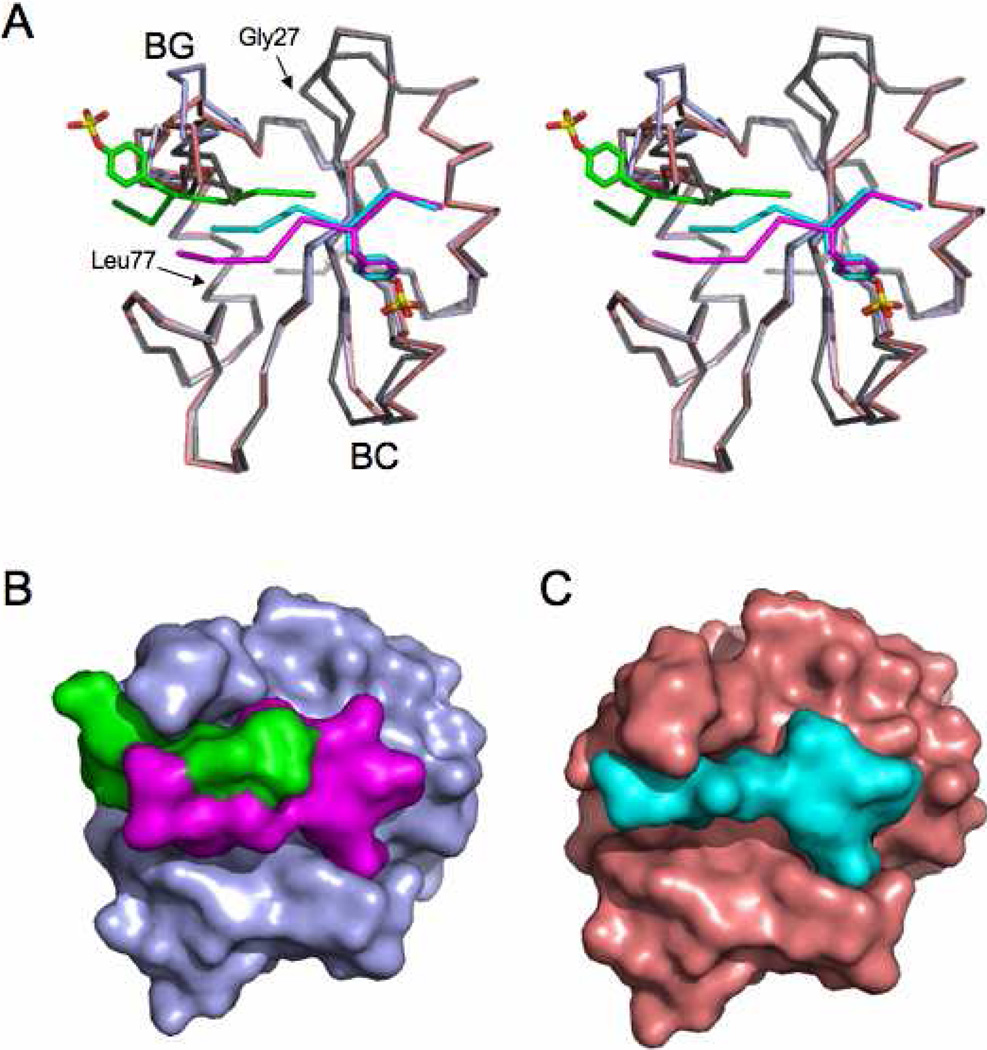
As seen previously in structures of N-SH2 domain of SHP-2,12 the peptide-binding surface appears as a wide groove that extends perpendicular to the plane of the central β-sheet of the SH2 domain. In order to accommodate peptide B of the pYFVP complex, the BG loop of the SH2-domain moves up and away from the peptide-binding site by ~4 Å, as compared to the structure of this domain alone (Figure 2). In addition, the residues C-terminal to the pY of peptide A are pushed down to create room for the C-terminal residues of peptide B to bind. Consistent with the wider peptide-binding groove, these movements result in a significantly increased binding surface in the pYFVP structure as compared to the pYAQL or previously published SH2 domain structures. A total of 670.6 Å2 solvent-accessible surface area of the SH2 domain is buried at the SH2-pYFVP interface, which is contributed approximately equally by the two peptides, while a total of 417.7 Å2 surface area of the protein is buried at the SH2-pYAQL interface.
Figure 2.
Conformational changes in the SHP-2 N-SH2 domain upon peptide binding. (A) Superposition of the unbound SH2 domain (grey), complex with pYFVP peptides (light blue, cyan, and green), and complex with pYAQL peptide (salmon and cyan). The BG loop moves up to accommodate the AQLW peptide and even further up to accommodate the two pYFVP peptides. Smaller conformational changes occur for the BC loop, which is the same for both complexes. (B) Surface view of the SHP-2-pYFVP complex. (C) Surface view of the SHP-2-pYAQL complex. Comparison of panels B and C shows how the binding groove on the SH2 domain opens up to accommodate the two pYFVP peptides.
Observation of the 1:2 SH2 Domain/Peptide Complex in Solution by NMR
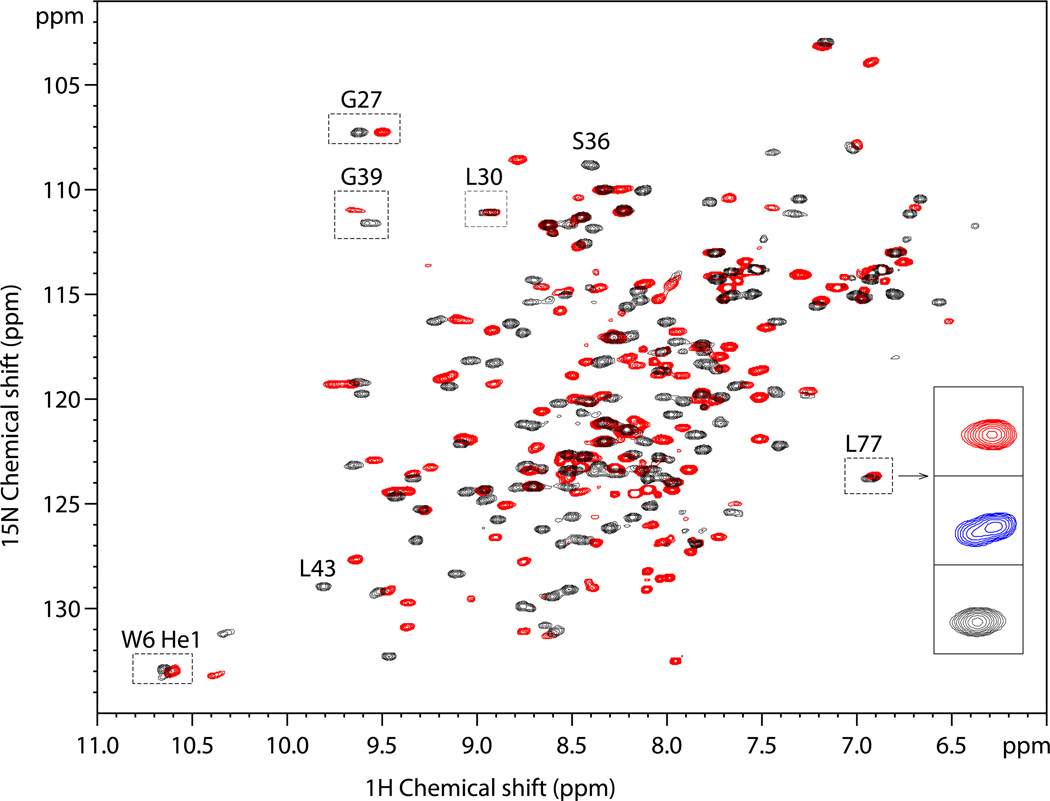
To determine if the 1:2 complex observed crystallographically also exists in solution, we initially attempted to determine the stoichiometry of the SH2 domain-pY peptide complex by isothermal titration calorimetry. The experiment was unsuccessful due to the very small amount of heat released, consistent with the above observation that hydrophobic interactions are largely responsible for the complex formation. We therefore examined the SH2 domain-pYFVP complex by NMR spectroscopy. 2D 1H-15N HSQC NMR spectroscopy of the uniformly 15N-labeled SHP-2 N-SH2 domain revealed good chemical shift dispersion of the backbone amides (Figure 3), indicative of a well-folded conformation in the free form. A few well-separated cross peaks were tentatively assigned on the basis of sequence alignment and spectral similarity to that of the Grb2 SH2 domain.23 The assignments were supported by the 3D 15N-edited NOESY-HSQC and TOCSY-HSQC studies. For instance, in the TOCSY spectrum, Trp-6 Hε1 displays a cross-peak at 7.38 ppm that could be assigned to Trp-6 Hδ1; whereas Gly-27 HN exhibits two cross-peaks at 4.01 and 3.45 ppm, respectively, presumably attributed to its Hα1/α2. Furthermore, Gly-27 HN in the NOESY spectrum shows medium-sized NOEs to the resonances of 3.03 and 2.69 ppm, which naturally fall into the chemical shift range expected for Hβ1/β2 of the preceding residue Asp-26.
Figure 3.
The overlap of 2D 1H-15N HSQC NMR spectra of the free 15N-labeled SH2 domain (black) and the protein/peptide mixture at the 1:2.5 ratio (red). Inset, the close-up view of L77 signals of the free SH2 protein (black), the 1:1 (mol/mol) protein/peptide mixture (blue), and the 1:2.5 protein/peptide mixture (red). Two peaks of approximately equal intensity are clearly discernable for L77 at the 1:1 protein/peptide ration despite the small chemical shift difference in 1H dimension (~0.025 ppm).
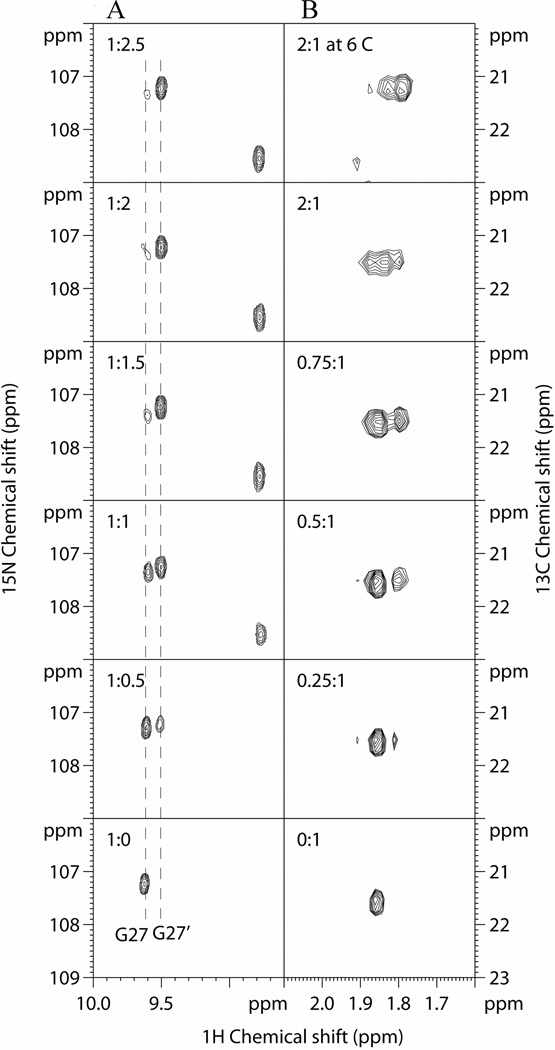
The 15N-labelled SH2 protein was then titrated with increasing concentrations of the pYFVP peptide and the 2D 1H-15N HSQC spectra were recorded at protein/peptide ratios of 1:0.5, 1:1, 1:1.5, 1:2, and 1:2.5. Addition of the peptide shifted a significant fraction of the cross-peaks including those of Trp-6 Hδ1 and Leu-77 HN, which are remote from the peptide binding site (Figure 2A). This suggests that binding to the pYFVP peptide causes substantial global conformational changes of the SH2 domain (Figure 3). The binding interaction has apparently high affinity (and/or slow exchange rate), as the signals underwent intensity changes rather than gradual shifting during the course of titration. For example, Leu-77 experienced only a minor perturbation whose magnitude was comparable to the resonance linewidth. However, its signal still split into two cross-peaks before reaching saturation (Figure 3). Most importantly, the binding interaction seems to involve a 1:2 protein/peptide ratio. This is evident from the data on Leu-77, whose signal split into two peaks of roughly equal intensity at the protein/peptide ratio of 1:1. Residue Gly-27, which is located in a β-turn region (Figure 2A) and experiences a larger perturbation upon peptide binding, was subsequently analyzed to determine the exact binding ratio. In the absence of peptide ligand, it has a cross peak at 9.6/107.3 ppm (Figure 4A). Upon addition of the pYFVP peptide, a new peak appeared at 9.5/107.2 ppm, with the concomitant decrease in the intensity of the Gly-27 signal of the free protein. The cross-peak at 9.5/107.2 ppm is assigned to Gly-27 in the protein-peptide complex (denoted as Gly-27’ hereafter) on the basis of the parallel NOESY and TOCSY data sets recorded at the end of peptide titration. Quantitative evaluation of their relative peak volumes successfully led to a 1:2 ratio of protein to peptide binding in support of the crystal structure (Supplementary Table S1).
Figure 4.
(A) 1H-15N HSQC spectra of 15N-labeled SHP-2 N-SH2 domain (460 µM) in the absence (1:0 protein/peptide) and presence of increasing concentrations of the pYFVP peptide (at the protein/peptide molar ratios of 1:0.5, 1:1, 1:1.5, 1:2, and 1:2.5). The intensity changes of G27 (and G27´) and an unassigned new cross-peak are shown. (B) 1H-13C HSQC spectra of the N-terminally 13C-labeled pYFVP peptide (200 µM) in the absence and presence of increasing concentrations of SHP-2 N-SH2 domain (at protein/peptide ratios of 0:1, 0.25:1, 0.5:1, 0.75:1, and 2:1). All spectra were recorded at room temperature except for that of the top panel in (B), which was recorded at 6 °C.
To gain additional evidence for the two differently bound peptides on a single SH2 domain, we performed a reverse titration experiment in which a specifically 13C-labeled peptide (13CH3CO-VIpYFVPLNYRR-NH2) was titrated with increasing concentrations of the SH2 protein. The free peptide showed a single 1H-13C cross-peak at 1.86/21.6 ppm (Figure 4B). Addition of the SH2 protein resulted in a new signal at 1.80/21.5 ppm, whose intensity increased with the protein concentration. Unfortunately, the interpretation of the results was somewhat complicated by the observation that the signal for one of the bound N-acetyl groups overlapped with that of the free peptide and that at high SH2 protein concentration (2:1 protein/peptide), all of the cross-peaks (those of the free peptide and the two differently bound peptides) appeared to collapse together, likely due to increased exchange rates. Cooling of the sample to 6 °C improved the quality of the spectrum, which clearly showed two cross-peaks of equal integral at 1.82/21.3 and 1.78/21.3 ppm, respectively. We assign these two cross-peaks to the N-terminal acetyl groups of the two differentially bound pY peptides. The lack of significant signal shift of the N-terminal acetyl groups is not unexpected, since the pY-3 residue was not resolved in the crystal structure, presumably due to a lack of interactions with the protein (Figure 1).
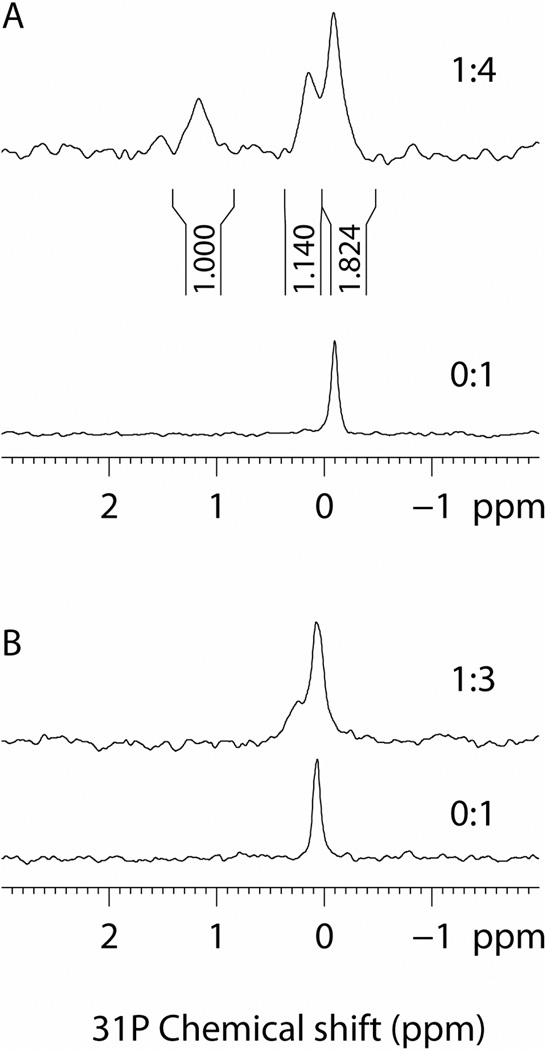
Finally, the SH2 domain-pYFVP interaction was examined by 1D 31P NMR spectroscopy. The free peptide showed a single resonance at −0.1 ppm (Figure 5A). Upon the addition of 0.25 equiv of the N-SH2 domain (1:4 protein/peptide ratio), two new peaks emerged at 0.2 and 1.1 ppm. Integration of the two new peaks as well as the free peptide peak gave a ratio of 1.0 : 1.1 : 1.8, in agreement with the 1:2 protein/peptide binding mode. Further addition of the SH2 protein to a 1:1 protein/peptide ratio resulted in the disappearance of the free peptide signal and increased intensities for the peaks at 0.2 and 1.1 ppm (data not shown). As a comparison, binding of a class I ligand (NH2-VIpYIDVQSYRR-NH2) to the N-SH2 domain gave only a single resonance at 0.2 ppm, consistent with the formation of a classical 1:1 SH2 domain-pY peptide complex (Figure 5B). The crystal structure shows that the pY side chain of peptide A is engaged in canonical SH2 domain-pY interactions including electrostatic interactions with Arg-βB5 and Lys-βD6, whereas the pY residue of peptide B is engaged in comparatively fewer interactions with the SH2 domain (e.g., Lys-BG5). Therefore, the two pY residues are clearly in different chemical environments, giving rise to different 31P NMR signals.
Figure 5.
(A) 1D 31P NMR spectra of class IV peptide NH2-VIpYFVPLNYRR-NH2 (300 µM) in the presence (1:4 protein/peptide) and absence of 75 µM SHP-2 N-SH2 domain (0:1) in 50 mM Hepes (pH 7.4) and 150 mM NaCl. The integrated peak area of each peak is shown under the peak. (B) 1D 31P NMR spectra of class I peptide NH2-VIpYIDVQSYRR-NH2 (300 µM) in the presence (1:3) and absence of 100 µM SHP-2 N-SH2 domain (0:1) in the same buffer as in (A).
Binding of Peptide B Does Not Require Tyrosine Phosphorylation
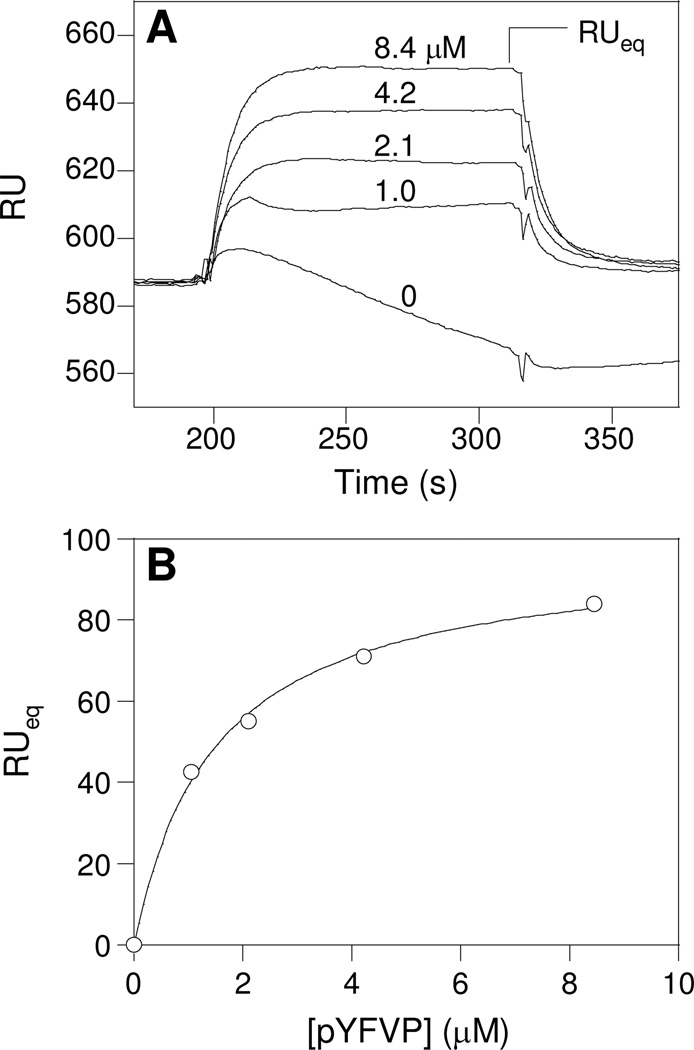
Since the pY side chain of peptide B is exposed to the solvent and appears to make only weak electrostatic interaction with the side chain of Lys-BG5, we suspected that the phosphate group might not be required for the formation of the 1:2 complex. To test this notion, we immobilized an unphosphorylated peptide [biotin-(miniPEG)2-RVIYFVPLNR-NH2 (YFVP peptide)] onto a streptavidin-coated SPR sensorchip and examined its ability to bind the N-SH2 domain in the absence and presence of the pYFVP peptide. As expected, neither the SH2 domain nor the pYFVP peptide alone showed any binding to the surface (Figures 6A and S2). However, when solutions containing both the SH2 domain (at a fixed concentration of 10 µM) and the pYFVP peptide (1.0–8.4 µM) were flowed over the surface, protein binding to the surface was observed. Moreover, the amount of binding to the surface increased with the pYFVP concentration. A plot of the RUeq values as a function of pYFVP concentration and curve fitting by nonlinear regression analysis gave an apparent KD value of 1.5 ± 0.2 µM (Figure 6B). Interestingly, the amount of SH2 domain binding plateaued at ~10 µM pYFVP peptide and then decreased upon further increase of the peptide concentration (i.e., when [pYFVP] > [SH2]); at 60 µM pYFVP peptide, binding by the SH2 domain to the SPR surface was completely abolished (Figure S2). Finally, mutation of Lys-BG5 of the SH2 domain into an alanine had little effect on its binding affinity for the immobilized pYFVP peptide (data not shown). The simplest explanation for these observations is that the SHP-2 N-SH2 domain simultaneously bound the pYFVP peptide at the canonical pY peptide-binding site and the YFVP peptide at the binding site for peptide B to form a 1:1:1 heterotrimeric complex. Since peptide B may be either a pY peptide or an unphosphorylated peptide, the presence of excess pYFVP peptide (relative to the SH2 domain) would compete with the immobilized peptide B for binding to the second site on the SH2 domain.
Figure 6.
SPR analysis of the binding of SHP-2 N-SH2 domain (10 µM) to immobilized peptide biotin-(miniPEG)2-RVIYFVPLNR-NH2 in the presence of varying concentrations of the pYFVP peptide (0, 1.0, 2.1, 4.2, and 8.4 µM). (A) Overlaid sensograms at indicated pYFVP concentrations. (B) Plot of RUeq as a function of pYFVP peptide concentration. The curve was fitted to the data according to the equation: RUeq = RUmax[pY]/(KDapp + [pY]).
Dimerization of Peptide pYFVP Increases Binding Affinity
The antiparallel relationship of peptides A and B in the SH2 domain-pY peptide complex suggests that joining the C-terminus of peptide A with the N-terminus of peptide B (or vice versa) may facilitate the formation of the 1:2 complex and enhance its stability. To test this notion, we designed two tandem pY peptides, RVIpYFVPGGGGGVIpYFVPYR-NH2 (bis-pYFVPG5) and RVIpYFVPGGGGGGVIpYFVPYR-NH2 (bis-pYFVPG6), in which the two pYFVP motifs are joined by flexible pentaglycine and hexaglycine linkers, respectively. SPR binding analysis of the N-SH2 domain to the immobilized bis-pYFVPG5 and bis-pYFVPG6 peptides gave apparent KD values of 0.18 and 0.33 µM, respectively (Figure S3). Under the same conditions, the SH2 domain bound to the immobilized pYFVP peptide with an apparent KD value of 0.044 µM.6 Since immobilization of the peptides to a surface may facilitate (or interfere with) their dimerization and complicate the quantitative analysis (see Discussion), we next performed a competition assay to determine the binding affinities of these peptides to the N-SH2 domain in solution. We immobilized a class I peptide ligand (pYAQL) onto an SPR sensorchip via its N-terminus and examined the ability of bis-pYFVPG5 and other peptides to compete with the immobilized pY ligand for binding to the SHP-2 N-SH2 domain. An IC50 value (concentration at which the amount of SH2 binding is reduced to 50% relative to the no-inhibitor control) of 0.65 µM was obtained for bis-pYFVPG5 (Figure S4 and Table 2). As a comparison, the pYAQL peptide and the monomeric pYFVP peptide had IC50 values of 0.60 and 2.8 µM, respectively. A peptide derived from protein CTLA-4, Ac-QPpYFIPINYRR-NH2, had an IC50 value of >80 µM. On the basis of the previously reported KD value (0.26 µM) for the interaction between the SHP-2 N-SH2 domain and the pYAQL peptide,7 we estimate that peptides pYFVP, bis-pYFVPG5, and QPpYFIP have KD (or apparent KD) values of 1.2, 0.28, and >35 µM, respectively. Note that the apparent KD value of 1.2 µM for the pYFVP peptide is similar to the KD value (1.5 µM) derived from the binding of N-SH2/pYFVP to the YFVP peptide (Figure 6). Thus, the bis-pYFVPG5 peptide binds to the SH2 domain with ~4-fold higher affinity than the monomeric pYFVP peptide (or ~2-fold enhancement after correction for the higher pY content of the bis-pYFVPG5 peptide).
Table 2.
Binding Affinity of SHP-2 N-SH2 Domain for pY Peptides
| peptide | IC50 (µM) | KD (µM) |
|---|---|---|
| pYAQL | 0.60 ± 0.10 | 0.26 ± 0.02 |
| pYFVP | 2.8 ± 0.8 | 1.2 ± 0.3 |
| bis-pYFVPG5 | 0.65 ± 0.10 | 0.28 ± 0.04 |
| pYFIP | >80 | >35 |
DISCUSSION
Among the SH2 domains whose ligand specificities have been well characterized, the N-SH2 domain of SHP-2 is unique in that it recognizes four distinct classes of pY ligands, all of which apparently bind to the same pY-binding site.6 To understand how the same SH2 domain recognizes vastly different peptide sequences, we investigated the binding mode of SHP-2 N-SH2 domain bound to three different classes of peptide ligands by X-ray crystallography. Both class I and III ligands bind to the SH2 domain in a manner very similar to those of numerous other SH2 domain-pY peptide complexes.9–14 In contrast, the class IV ligand (pYFVP) binds to the SH2 domain as a homodimer. NMR and SPR studies confirmed that the unusual binding mode exists in the solution phase. Further, a database search revealed 32 human proteins that contain one or more putative class IV SHP-2 N-SH2 ligands (Table S2). NMR analysis of five of these putative ligands showed that at least one of them (e.g., pYFLP of TEX10) also bound to the SHP-2 SH2 domain to form a 1:2 protein/peptide complex (Figure S5). We hypothesize that formation of the 1:2 complex with the SHP-2 SH2 domain may be a general property of all class IV ligands. There has been a previous report that the C-terminal SH2 domain of PLCγ binds to a doubly phosphorylated peptide from Syk kinase and forms a 1:1 SH2/pY peptide complex, in which the first pY occupies the canonical pY-binding pocket and the second pY makes additional contacts with the SH2 domain surface.24 The 1:2 complex in this study is distinct from the previous system in that the two pY peptides (or a pY peptide plus a non-pY peptide) in our system may come from two different proteins.
Dimerization of the class IV peptides appears to be required for binding to the SHP-2 N-SH2 domain. For peptides pYFVP and pYFLP, which bind to the SH2 domain tightly, the dissociation rate of the SH2 domain-pY peptide complex is slow enough to allow the NMR signals of the free peptide (or protein) to be resolved from that of the complex. Under all of the conditions tested (with either the peptide or the SH2 domain in excess), only the NMR signals corresponding to the free peptide (or the free protein) and the 1:2 protein/peptide complex were observed. This indicates that the canonical 1:1 peptide/SH2 domain complex is either not formed at all or is present at very low concentrations (i.e., below the detection limit of NMR spectroscopy). This in turn suggests that the 1:1 complex, if formed at all, has substantially lower stability than the 1:2 complex. Peptide dimerization is apparently induced by SH2 domain binding, since in the absence of the SH2 domain the pYFVP peptide showed no detectable binding to the immobilized YFVP peptide (KD ≫ 60 µM). Interestingly, the apparent KD value of peptide pYFVP as measured by the competition assay in this work (1.2 µM) is significantly higher than the previously reported KD value (0.044 µM), which was determined with the pYFVP peptide immobilized onto an SPR sensorchip.6 Clearly, immobilization of the pYFVP peptide onto a surface facilitated dimerization of the peptide (due to high surface density), thereby increasing the stability of the 1:2 SH2 domain/pY peptide complex. The propensity of the pYFVP peptide to dimerize in the presence of the SH2 domain is likely the reason why it was selected from the peptide library at the first place. Consistent with this notion, covalent crosslinking of two pYFVP peptides (e.g., bis-pYFVPG5) resulted in 2–4-fold higher affinity. It is possible that a more optimal linker than pentaglycine may result in even greater binding affinity.
The different interactions between the two bound peptides and the protein may have important implications. The pY side chain of peptide A is engaged in canonical interactions with the pY-binding pocket of the SH2 domain and binding of peptide A should therefore be phosphorylation-dependent. The pY side chain of peptide B is exposed to the solvent and makes only weak electrostatic interaction with the side chain of Lys-BG5, suggesting that binding of peptide B should not require tyrosine phosphorylation. Indeed, our results show that the SHP-2 N-SH2 domain can simultaneously bind a pY peptide (e.g., pYFVP) and an unphosphorylated peptide (e.g., YFVP) to form a 1:1:1 heterotrimeric complex. Further, it appears that the two peptides do not need to have the same sequence to form the 1:2 complex (RLH and DP, unpublished results). In fact, the discovery of the 1:2 complex in which the two peptide ligands are identical is rather fortuitous, due to fact that all of the peptides on each peptide library bead have the same sequence. In the context of proteins, formation of the 1:2 complexes may have two different manifestations. It may form intermolecularly, resulting a trimeric complex involving the SH2 domain, a pY protein, and a third protein (which may or may not be tyrosine phosphorylated), or intramolecularly, by forming a heterodimer between SHP-2 and a pY protein in which the pY peptide folds into a hairpin structure and binds to the SH2 domain. Obviously, further study is needed to test this hypothesis. It will also be important to determine whether the class I, II, and III ligands are capable of binding to the SHP-2 SH2 domain as dimers (either homo- or heterodimers), or whether other SH2 domains are capable of recognizing peptide dimers.
Supplementary Material
ABBREVIATIONS
- HSQC
heteronuclear single-quantum correlation
- PTP
protein-tyrosine phosphatase
- pY
phosphotyrosine
- SH2
Src homology 2
- SPR
surface plasmon resonance
Footnotes
This work was supported by grants from the National Institutes of Health (GM062820 and CA132855). R.L.H. was supported by an NIH predoctoral training grant (GM08512). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Supporting Information. Additional NMR spectra and SPR data showing the SH2 domain-pY peptide binding stoichiometry. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins – establishing the boundaries of phosphotyrosine signaling. Mol. Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 3.Huang H, Li L, Wu C, Schlibli D, Colwill K, Ma S, Li C, Roy P, Ho K, Songyang Z, Pawson T, Gao Y, Li SS-C. Defining the specificity space of the human Src homology 2 domain. Mol. Cell. Proteomics. 2008;7:768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Burshtyn DN, Yang W, Yi T, Long EO. A novel phosphotyrosine motif with a critical amino acid at position -2 for the SH3 domain-mediated activation of tyrosine phosphatase SHP-1. J. Biol. Chem. 1997;272:13066–13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 5.Poy F, Yaffe MB, Sayos J, Saxna K, Morra M, Sumegi J, Eck MJ. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol. Cell. 1999;4:555–561. doi: 10.1016/s1097-2765(00)80206-3. [DOI] [PubMed] [Google Scholar]
- 6.Sweeney MC, Wavreille A-S, Park J, Butchar J, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: Sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 7.Imhof D, Wavreille A-S, May A, Zacharias M, Tridandapani S, Pei D. Sequence specificity of SHP-1 and SHP-2 Src Homology 2 domains: Critical roles of residues beyond the pY+3 position. J. Biol. Chem. 2006;281:20271–20282. doi: 10.1074/jbc.M601047200. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko T, Huang H, Zhao B, Li L, Liu H, Voss CK, Wu C, Schiller MR, Li SS-C. Loops govern SH2 domain specificity by controlling access to binding pockets. Sci. Signal. 2010;3:ra34. doi: 10.1126/scisignal.2000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waksman G, Kominos D, Robertson SC, Pant N, Baltimore D, Birge RB, Cowburn D, Hanafusa H, Mayer BJ, Overduin M, Resh MD, Rios CB, Silverman L, Kuriyan J. Crystal structure of the phosphotyrosine recognition domain SH2 of v-Src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 10.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high-affinity phosphotyrosyl peptide to the Src SH2 domain - crystal-structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 11.Eck M, Shoelson SE, Harrison SC. Recognition of a high affinity phosphotyrosyl peptide by the Src homology 2 domain of p56lck. Nature. 1993;362:87–91. doi: 10.1038/362087a0. [DOI] [PubMed] [Google Scholar]
- 12.Lee C, Kominos D, Jacques S, Margolis B, Schlessinger J, Shoelson SE, Kuriyan J. Crystal structures of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure. 1994;2:423–438. doi: 10.1016/s0969-2126(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 13.Pascal SM, Singer AU, Gish G, Yamazaki T, Shoelson SE, Pawson T, Kay LE, Forman-Kay JD. Nuclear-magnetic-resonance structure of an SH2 domain of phospholipase c-gamma-1 complexed with a high-affinity binding peptide. Cell. 1994;77:461–472. doi: 10.1016/0092-8674(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 14.Xu RX, Word JM, Davis DG, Rink MJ, Willard DH, Gampe RT. Solution structure of the human pp60(c-Src) SH2 domain complexed with a phosphorylated tyrosine pentapeptide. Biochemistry. 1995;34:2107–2121. doi: 10.1021/bi00007a003. [DOI] [PubMed] [Google Scholar]
- 15.Neel BG, Gu H, Pao L. The “Shp”ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 16.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal Structure of the Tyrosine Phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 17.Lechleider RJ, Sugimoto S, Bennett AM, Kashishian AS, Cooper JA, Shoelson SE, Walsh CT, Neel BG. Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding-site, phosphotyrosine-1009, on the human platelet-derived growth-factor receptor-beta. J. Biol. Chem. 1993;268:21478–21481. [PubMed] [Google Scholar]
- 18.Pluskey S, Wandless TJ, Walsh CT, Shoelson SE. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J. Biol. Chem. 1995;270:2897–2900. doi: 10.1074/jbc.270.7.2897. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology 276, Macromolecular Crystallography, part A. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucl. Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornton KH, Mueller WT, McConnell P, Zhu G, Saltiel AR, Thanabal V. Nuclear magnetic resonance solution structure of the growth factor receptor-bound protein 2 Src homology 2 domain. Biochemistry. 1996;35:11852–11864. doi: 10.1021/bi952615s. [DOI] [PubMed] [Google Scholar]
- 24.Groesch TD, Zhou F, Mattila S, Geahlen RL, Post CB. Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J. Mol. Biol. 2006;356:1222–1236. doi: 10.1016/j.jmb.2005.11.095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.