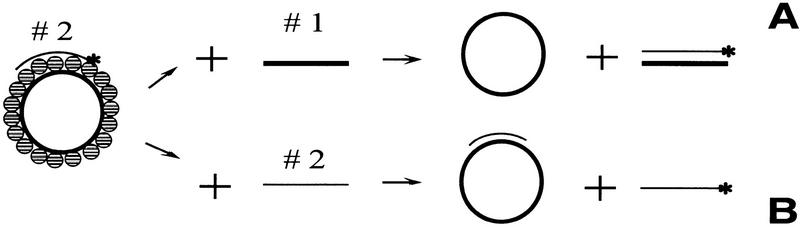Abstract
Traditionally, recombination reactions promoted by RecA-like proteins initiate by forming a nucleoprotein filament on a single-stranded DNA (ssDNA), which then pairs with homologous double-stranded DNA (dsDNA). In this paper, we describe a novel pairing process that occurs in an unconventional manner: RecA protein polymerizes along dsDNA to form an active nucleoprotein filament that can pair and exchange strands with homologous ssDNA. Our results demonstrate that this “inverse” reaction is a unique, highly efficient DNA strand exchange reaction that is not due to redistribution of RecA protein from dsDNA to the homologous ssDNA partner. Finally, we demonstrate that the RecA protein–dsDNA filament can also pair and promote strand exchange with ssRNA. This inverse RNA strand exchange reaction is likely responsible for R-loop formation that is required for recombination-dependent DNA replication.
Keywords: DNA pairing, RecA protein, DNA strand exchange, recombination, R-loop
Genetic recombination is a complex biochemical pathway that proceeds through a series of reactions, which include DNA pairing, exchange of DNA strands, and DNA heteroduplex extension. Detailed analysis of Escherichia coli RecA protein has provided considerable insight into this process (Clark and Sandler 1994; Kowalczykowski et al. 1994; Roca and Cox 1997; Bianco et al. 1998). The canonical mechanism involves the following steps: RecA protein polymerizes on single-stranded DNA (ssDNA) to form an active presynaptic complex that is responsible for homologous recognition of double-stranded DNA (dsDNA). This nucleoprotein filament binds the dsDNA, homologously aligns it with the ssDNA, and exchanges DNA strands to form an intermediate joint molecule. Finally, DNA heteroduplex extension permits the complete exchange of DNA strands.
RecA protein-mediated strand exchange between two duplex DNA molecules has a strict requirement for a region of ssDNA in one of the duplexes (Conley and West 1990; Lindsley and Cox 1990). This ssDNA region is used both for loading RecA protein onto the DNA (Shaner et al. 1987) and for initiation of the recombinational process. After loading onto the ssDNA region, RecA protein continues to polymerize into dsDNA, resulting in a nucleoprotein filament assembled on dsDNA; however, DNA strand exchange always initiates in the single-stranded region. The fact that RecA protein can assemble into a filament on dsDNA raises the question of whether the dsDNA within the filament is actually competent for the initial steps of DNA strand exchange. It has been shown that contacts between a dsDNA nucleoprotein filament and homologous duplex DNA contribute to the efficiency of synaptic pairing (Conley and West 1989; Chiu et al. 1990). It has also been shown that, once initiated in a ssDNA region, the DNA heteroduplex extension can occur between duplex regions (West and Howard-Flanders 1984; Conley and West 1990). But, despite the many experiments arguing that the RecA–dsDNA complex has the capacity to recognize homologous DNA, this issue remains controversial. Instead, it was suggested that the experiments which measure duplex–duplex interactions by topological changes in dsDNA (Conley and West 1989; Chiu et al. 1990) involve a signal that is potentially too ambiguous to study these weak interactions (Cox 1995). Also, the other argument for homologous recognition of dsDNA by a RecA protein–dsDNA complex, which is its ability to effect the bypass of a double strand break in the dsDNA partner (West and Howard-Flanders 1984), was recently discounted in favor of an alternative view that proposed that ATP-mediated DNA torsional stress produced by RecA protein was responsible for duplex–duplex exchange (Shan and Cox 1998). And finally, DNA strand exchange initiating from a fully duplex RecA nucleoprotein filament has never been demonstrated, implying that the dsDNA nucleoprotein filament lacks the capacity to recognize and pair with homologous DNA.
However, recent characterization of the dsDNA–RecA protein filament revealed that the bound RecA protein is in the high-affinity DNA-binding state and suggested to us that these dsDNA nucleoprotein filaments might be active for DNA strand exchange with either ssDNA or dsDNA (Zaitsev and Kowalczykowski 1998). The possibility that this dsDNA RecA nucleoprotein filament can promote DNA strand exchange with ssDNA is examined here. To facilitate discussion, we refer to the canonical DNA strand exchange reaction between the ssDNA–RecA protein filament and free dsDNA as either “conventional” or “forward” DNA strand exchange. We refer to the DNA strand exchange reaction between the dsDNA–RecA protein filament and free ssDNA as the “inverse” DNA strand exchange reaction, as proposed by Stasiak and colleagues (Muller et al. 1990b). This nomenclature also avoids confusion with the term reverse DNA strand exchange that was used to define a reaction in which DNA strand exchange proceeded in the “reverse” direction by virtue of an exonuclease I contamination (Bedale et al. 1991, 1993).
Here, we examine whether RecA protein is capable of inverse DNA strand exchange. Our results demonstrate that (1) highly efficient DNA strand exchange can indeed initiate from a RecA–dsDNA nucleoprotein filament; (2) the inverse DNA strand exchange reaction is not simply reversal of the conventional forward reaction, but is a novel DNA strand exchange reaction promoted by RecA protein; (3) DNA strand exchange is not due to redistribution of RecA protein from dsDNA to the ssDNA; (4) free ssDNA is not produced from the dsDNA as an intermediate of this reaction; and (5) the inverse reaction can use ssRNA as nucleic acid substrate instead of ssDNA, a finding that parallels results from the Kogoma laboratory (Kasahara et al. 2000). We also discuss the biological relevance of this previously unreported nucleic acid strand exchange process.
Results
Inverse DNA strand exchange: homologous pairing can initiate from dsDNA
RecA protein can polymerize on dsDNA to produce a nucleoprotein filament. RecA protein in this filament is in the high-affinity DNA-binding state, implying that it could be active for DNA pairing (Zaitsev and Kowalczykowski 1998). However, within the linear dsDNA–RecA protein filament, each strand of dsDNA is not bound equivalently (Chow et al. 1988; Lindsley and Cox 1989). The two DNA strands in this filament show asymmetry with regard to nuclease sensitivity. The loading strand, defined as the strand containing a single-stranded region that permits the 5′ → 3′ polymerization of RecA protein into the dsDNA region, is about two times more resistant than the nonloading strand; furthermore, there is a slight difference in the susceptibility of each strand to attack by KMnO4 (Adzuma 1992). This nonequivalent disposition of DNA strands in the RecA protein–dsDNA filament suggested to us that DNA strand exchange carried out by this RecA nucleoprotein filament might also display asymmetry in a pairing reaction with ssDNA. DNA strand exchange could depend on whether the pairing ssDNA was complementary to the loading strand of the duplex DNA or to the nonloading oligonucleotide strand (Fig. 1).
Figure 1.
The inverse DNA strand exchange reactions. Thick circles designate M13 ssDNA, which is used as the loading DNA strand for RecA protein polymerization; thick lines represent the ssDNA oligonucleotide (63-mer 1), which is identical to M13 ssDNA sequence at the duplex DNA locus. Thin lines designate oligonucleotide 63-mer 2, which is complementary to the M13 ssDNA at the same duplex locus. (A) Inverse DNA strand exchange with oligonucleotide 1 (complementary to the oligonucleotide strand of the duplex DNA substrate), resulting in a dsDNA oligonucleotide product. (B) Inverse DNA strand exchange with oligonucleotide 2 (complementary to the loading DNA strand), resulting in a displaced ssDNA product.
Loading of RecA protein onto the dsDNA with the predicted asymmetry was achieved by using circular M13 ssDNA as the loading strand for the 63-bp duplex region that was formed by annealing an oligonucleotide to the M13 ssDNA. The circularity of M13 ssDNA is an additional advantage, because it stabilizes the RecA nucleoprotein complex (Lindsley and Cox 1989; Shan and Cox 1997). Figure 1 illustrates the types of products expected for the inverse reactions, depending on which complementary ssDNA is used. To facilitate description, the M13 ssDNA with the annealed 63-mer oligonucleotide 2 is simply referred to as the duplex DNA substrate. When RecA protein is assembled on the duplex DNA substrate, the loading strand is the M13 ssDNA. Oligonucleotide 1 is complementary to the 63-mer oligonucleotide 2, and oligonucleotide 2 is complementary to the loading M13 ssDNA strand; both oligonucleotides 1 and 2 are exactly the same length as the dsDNA region in the duplex DNA substrate. Because the 63-mer oligonucleotide of the duplex DNA substrate is radioactively labeled, the expected product for the reaction involving oligonucleotide 1 is a labeled double-stranded 63-mer (composed of oligonucleotides 1 and 2; Fig. 1A). For oligonucleotide 2 (which is complementary to the loading strand) the expected product is the displaced, labeled single-stranded 63-mer (2, Fig. 1B).
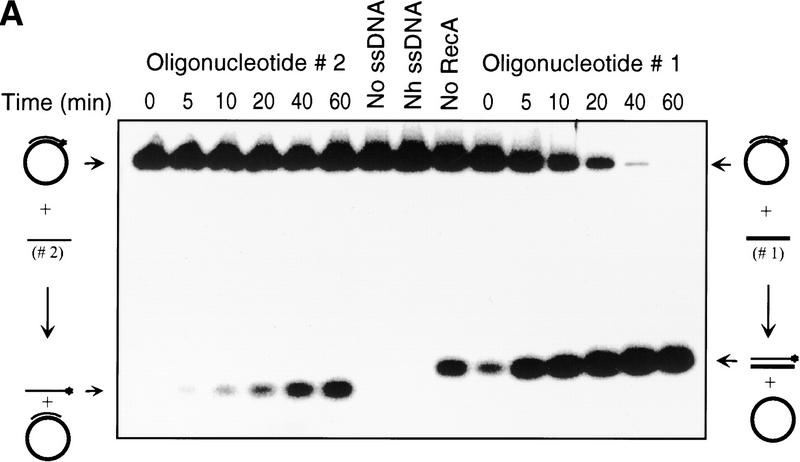
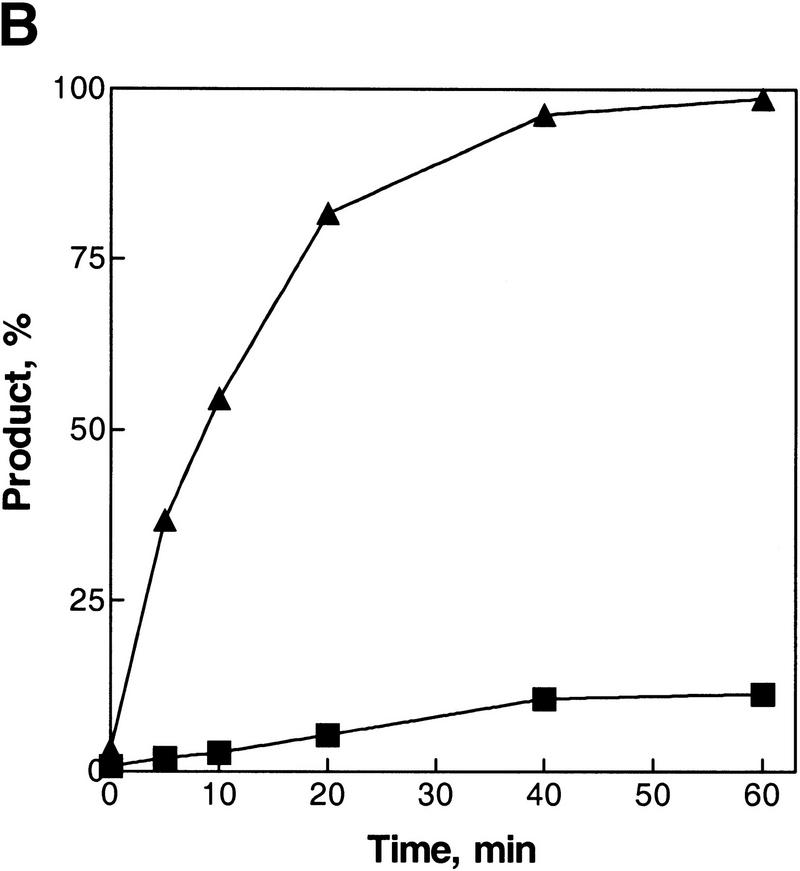
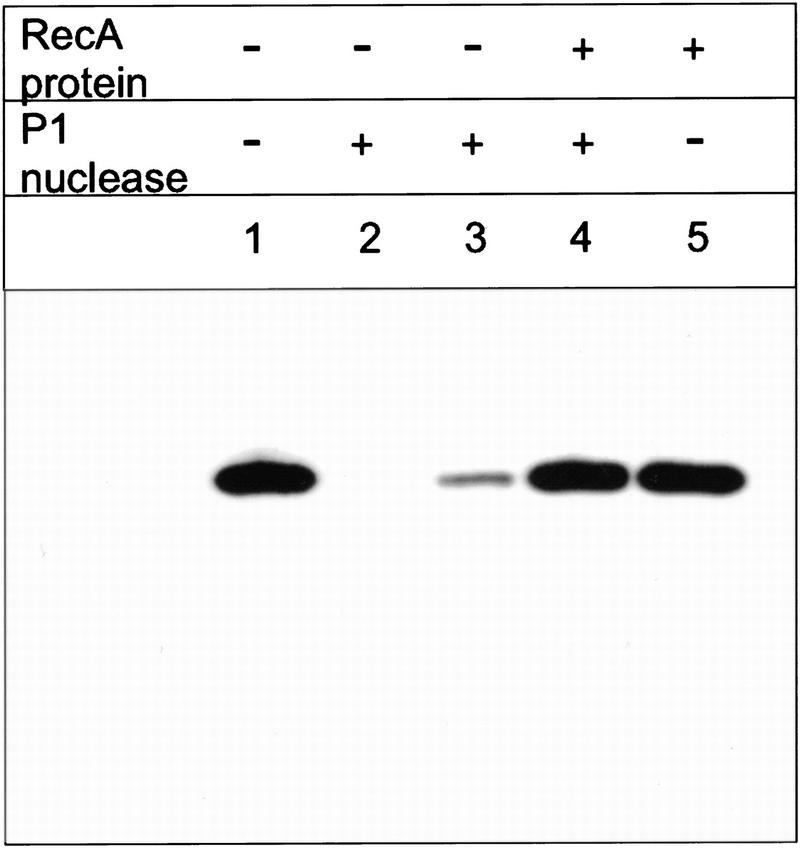
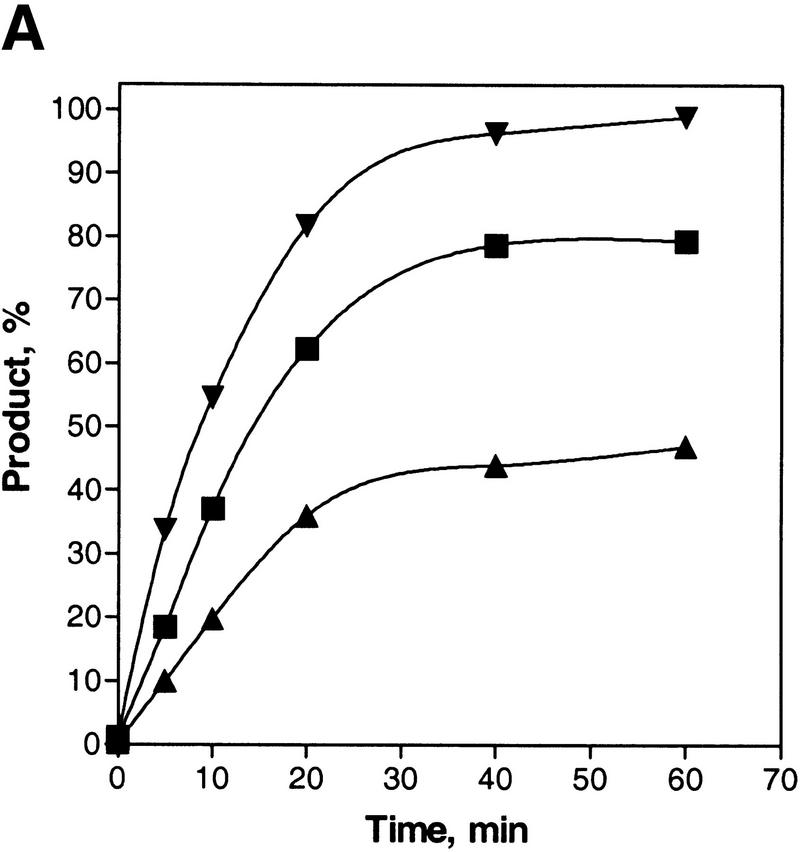
Figure 2A shows a time course for the inverse DNA strand exchange reactions that were depicted in Figure 1, in which RecA protein is loaded onto the dsDNA before initiation of DNA strand exchange. The results of this experiment show that DNA strand exchange does occur. For oligonucleotide 1 there is a time-dependent increase in dsDNA product formed: ∼98% of the duplex DNA substrate is converted into product (Fig. 2B). In contrast, when oligonucleotide 2 is used as a pairing partner, much less exchange of DNA strands occurs; only ∼11% of oligonucleotide 2 from the original duplex DNA substrate is exchanged for the unlabeled oligonucleotide 2 to produce the labeled ssDNA product. This asymmetry in product formation parallels the predicted asymmetry based on the loading of RecA protein onto duplex DNA. Figure 2A also shows that the reaction requires both DNA sequence homology and RecA protein. The dsDNA produced in the lane designated No RecA represents the level of spontaneous exchange between these DNA substrates, which is 10-fold less that the amount produced in the RecA protein-mediated reaction with oligonucleotide 1. Figure 2A also shows that, in the presence of RecA protein, there is no labeled oligonucleotide 2 released from duplex DNA substrate (see lanes designated No ssDNA or Nh ssDNA), although an excess of cold oligonucleotide was added just before deproteinization to prevent hybridization back to the M13 DNA of any ssDNA that may have formed. Furthermore, in agreement, none of oligonucleotide 2 within the duplex substrates is susceptible to degradation by P1 nuclease when protein is present (Fig. 3, lane 4), demonstrating that free ssDNA is not being generated by the action of RecA protein. In contrast, free ssDNA is completely degraded (lane 2) and free dsDNA is almost fully degraded (lane 3) under the same conditions. These results demonstrate that, within the RecA nucleoprotein filament, oligonucleotide 2 is not even partially displaced (Fig. 3); had any free ssDNA been produced, this would have been detected as sensitivity to the P1 nuclease because, in the presence of ATP, the limiting concentration of RecA protein would have quickly redistributed to bind preferentially to the longer M13 ssDNA substrate. This failure to detect strand separation in the presence of RecA protein is also consistent with our inability to detect strand separation of dsDNA that was bound to the secondary DNA-binding site of RecA protein, using both chemical and enzymatic probes (Mazin and Kowalczykowski 1999). Thus, the 63-mer dsDNA produced is not the trivial product of annealing between free oligonucleotide 1 and RecA protein-displaced oligonucleotide 2. All subsequent experiments will focus on the more efficient inverse DNA strand exchange reaction that is represented in Figure 1A.
Figure 2.
Asymmetry of inverse DNA strand exchange. (A) Time courses for the inverse reactions depicted in Fig. 1. The reactions were carried out as described under Materials and Methods. Concentrations of oligonucleotides 1 and 2 were 10-fold higher than the molar (molecule) concentration of the M13 ssDNA–oligonucleotide 2 duplex substrate. The lanes marked No RecA and No ssDNA designate control reactions performed without either RecA protein or oligonucleotide 1, respectively. The lane marked Nh ssDNA designates the control reaction using nonhomologous ssDNA (63-mer oligonucleotide 3) instead of the homologous ssDNA. Control reactions were conducted for 60 min at 37°C. To prevent annealing of the potentially displaced oligonucleotide 2 back to the M13 ssDNA, a 10-fold molar (molecular) excess of cold oligonucleotide 2 was added to the control reactions just before deproteinization. (B) Data from A, quantified and plotted (▴) Oligonucleotide 1; (█) oligonucleotide 2.
Figure 3.
Free ssDNA is not produced as an intermediate of inverse DNA strand exchange. The dsDNA substrate (30 ng in oligonucleotide 2) was treated with 1 munit of P1 nuclease either in the presence or absence of RecA protein (as indicated) for 60 min at 37°C (lanes 2,3,4). (Lanes 1,5) Negative controls in which the P1 nuclease was omitted. (Lane 2) The positive control demonstrating degradation of the ssDNA; in this lane, oligonucleotide 2 was mixed with noncomplementary φX174 ssDNA, instead of the complementary M13 ssDNA.
The yield of inverse DNA strand exchange products depends on the concentrations of homologous ssDNA and RecA protein
To determine whether a change in concentration of a pairing partner affected the DNA strand exchange process, reactions were examined at various ssDNA concentrations. As shown in Figure 4A, changing the concentration of oligonucleotide 1 over a 10-fold range increases both the extent of product formation and the initial reaction rate by slightly more than 2-fold. Even at the lowest concentration tested, DNA strand exchange yield is fivefold greater than for the spontaneous process. The yield of product is proportional to the amount of ssDNA relative to the total DNA (molecule) concentration, suggesting that inverse DNA strand exchange achieves an apparent equilibrium (or more accurately, steady-state) between substrate and product formation: for example, with a fourfold excess of ssDNA (more than duplex DNA substrate), the yield of reaction product is 80%, which agrees with the predicted yield of 4/5. This finding suggests that the inverse DNA strand exchange reaction is reversible; in agreement, we find that the dsDNA product (i.e., the 63-mer duplex) of DNA strand exchange remains complexed with RecA protein (E.N. Zaitsev and S.C. Kowalczykowski, unpubl.), implying that this product could participate in DNA strand exchange with the ssDNA (product) produced by inverse exchange, although this inference was not directly tested.
Figure 4.
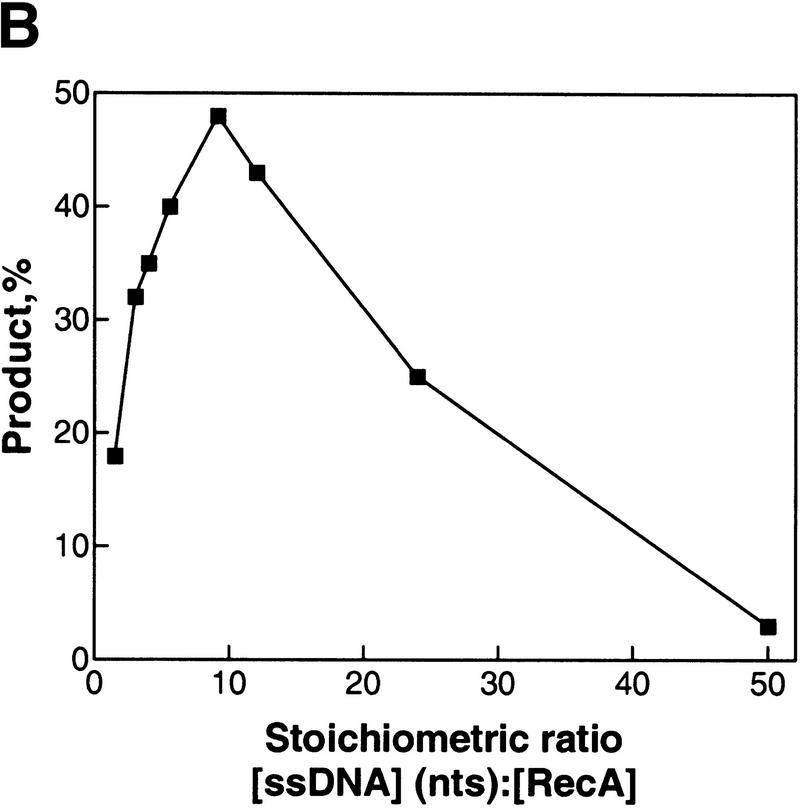
The yield of heteroduplex DNA formed by inverse DNA strand exchange increases with ssDNA concentration and has an optimal RecA protein concentration. (A) The inverse reaction was performed the standard way except that three different concentrations of ssDNA (oligonucleotide 1) were used: Equimolar (in molecules) to the duplex DNA substrate (1×, ▴), a fourfold excess concentration (4×, █), or a 10-fold excess concentration (10×, ▾); the latter concentration corresponds to the experimental conditions used in Fig. 2. (B) The inverse reaction performed the standard way, except using an equimolar (i.e., 1×) concentration of dsDNA and ssDNA (6.3 nm molecules), as a function of RecA protein concentration. The x-axis is plotted as the ratio of ssDNA (in nucleotides) to RecA protein.
Conventional DNA strand exchange requires the formation of a contiguous presynaptic filament that is saturated with respect to RecA protein. Therefore, the RecA protein concentration dependence of inverse strand exchange was examined (Fig. 4B). Optimal exchange occurs at an apparent ratio of one RecA protein monomer per 10 nucleotides of M13 ssDNA [note that the lowest (i.e., equimolar; 1X in Fig. 4A) concentration of homologous 63-mer was used, accounting for the lower yield relative to Fig. 1]. Given that the M13 ssDNA possesses extensive secondary structure under these conditions, this amount of RecA protein represents the amount needed to saturate the available ssDNA (Kowalczykowski and Krupp 1987). However, in contrast to the conventional reaction, excess RecA protein is inhibitory, presumably due to binding of RecA protein to the homologous target ssDNA. Although seemingly unusual, this behavior has an interesting counterpart in the behavior of the eukaryotic homolog, Rad51 protein; Rad51 protein shows an optimum in conventional DNA strand exchange because excess Rad51 protein binds efficiently the homologous DNA partner, in this case duplex DNA, thereby blocking DNA pairing (Sung and Robberson 1995).
An additional hallmark of RecA protein-promoted DNA strand exchange is its characteristic dependence on the magnesium ion concentration (Cox and Lehman 1982). The inverse reaction displays a very similar, although not identical, dependence; there is little inverse DNA strand exchange until a minimum concentration of 8 mm is exceeded; the optimum occurs at 10 mm; and then it declines (by ∼50%) at 15 mm magnesium acetate (R. Ando and S.C. Kowalczykowski, unpubl.); this dependence is very similar to that reported for an inverse R-loop formation reaction mediated by RecA protein that was discovered independently (Kasahara et al. 2000). Thus, inverse DNA strand exchange shares many of the features seen for conventional DNA strand exchange but, not unexpectedly, some of the characteristics are unique.
Inverse DNA strand exchange occurs in the presence of an excess of nonhomologous ssDNA
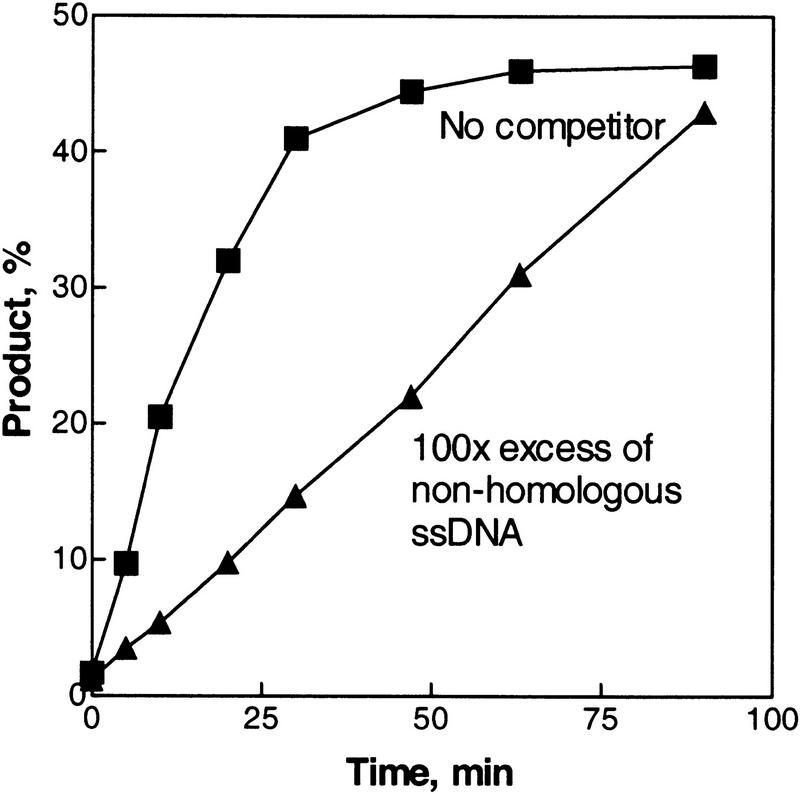
In conventional DNA strand exchange, the presynaptic RecA protein–ssDNA filament must find its homologous dsDNA target among a vast excess of nonhomologous dsDNA “competitor sequences.” In the case of inverse DNA strand exchange, the RecA protein–dsDNA filament faces a similar challenge; it requires a similar search capacity to distinguish its target among an excess of nonhomologous ssDNA. To assess the sensitivity of the inverse DNA strand exchange reaction to high concentrations of nonhomologous ssDNA, the reaction was performed in the presence of a 100-fold excess of nonhomologous ssDNA (oligonucleotide 3). As shown in Figure 5, the RecA protein–dsDNA filament can locate a homologous ssDNA partner, even in the presence of a 100–fold excess of nonhomologous ssDNA. The presence of the nonhomologous competitor affects the rate of the reaction by only about twofold, but not the yield. This result also argues that inverse DNA strand exchange does not proceed by dissociation of RecA protein from the dsDNA, followed by assembly on the ssDNA and subsequent conventional DNA strand exchange, because the excess ssDNA would block that process. This issue is addressed more directly in the next section.
Figure 5.
Inverse DNA strand exchange proceeds in the presence of an excess of nonhomologous competitor ssDNA. The reactions were carried out in the standard way, except that an equimolar (in molecules) concentration of homologous ssDNA (63-mer 1) was used. The reaction performed in the presence of competitor contained a 100-fold molar excess of nonhomologous ssDNA (63-mer 3) in addition to the homologous ssDNA.
Inverse DNA strand exchange is not due to conventional DNA strand exchange after redistribution of the RecA protein
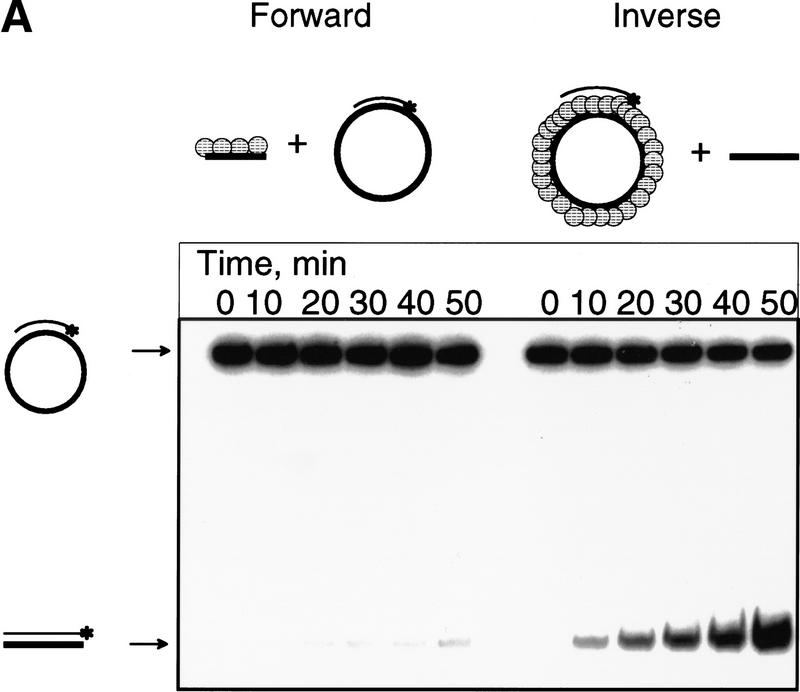
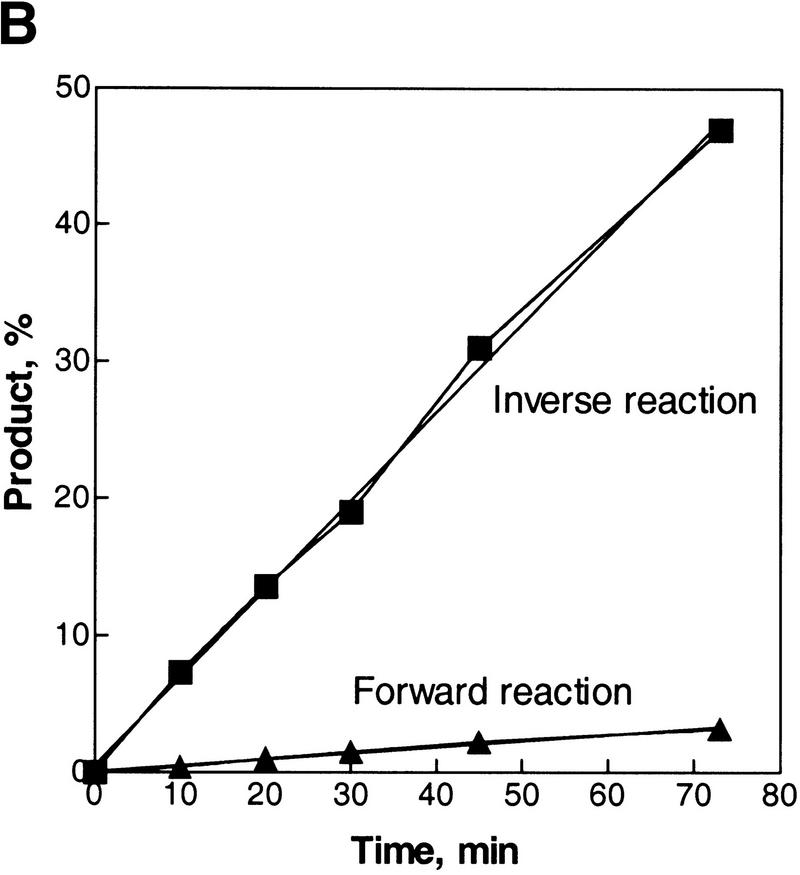
The experiments described thus far leave room for an alternative explanation of product formation. RecA protein could dissociate from the duplex DNA substrate and bind the ssDNA oligonucleotide directly; afterward, the RecA protein–ssDNA complex could participate in the standard forward reaction. To eliminate this possibility, we compared this potential “artifactual” forward DNA strand exchange to the inverse reaction under the same conditions. Reactions were performed in the presence of both a 100-fold excess (molecules) of nonhomologous oligonucleotide (3) and a 100-fold excess (nucleotides) of φX174 ssDNA to sequester any RecA protein that might dissociate from the initial nucleoprotein complex. Figure 6 shows the results of such a comparison. The forward reaction in which RecA protein is assembled in the presence of the competing ssDNA is severely inhibited (∼1.5% product formation), whereas the inverse reaction is still effective (∼46% product formation; Fig. 6B); as demonstrated in Figures 4 and 5, the lower concentration (i.e., equimolar) of ssDNA used accounts for the reduced yield relative to Figure 1. These results eliminate the trivial explanation that the inverse DNA strand exchange reaction is simply a consequence of RecA protein dissociation from the initial duplex DNA substrate, binding to the oligonucleotide, and then promoting the forward reaction; if it were, then the inverse reaction would have been inhibited by the excess ssDNA to the same degree as the forward reaction that could have occurred after redistribution of the RecA protein.
Figure 6.
Comparison of the forward and the inverse DNA strand exchange reactions. The reactions were carried out as described In the Materials and Methods, except that a 100fold excess (nucleotides) each of oligonucleotide 3 and φX174 ssDNA was present as a nonhomologous competitor and an equimolar (molecules) concentration of ssDNA (oligonucleotide 1) was used. For the inverse reaction, RecA protein was preincubated first with the duplex DNA substrate for 5 min at 37°C; afterward oligonucleotide 1, in the presence of a 100-fold excess of each nonhomologous oligonucleotide 3 and φX174 ssDNA, was added. For the forward reaction, RecA protein was preincubated first with the ssDNA (oligonucleotide 1), in the presence of a 100-fold excess of each nonhomologous oligonucleotide 3 and φX174 ssDNA, and then the forward reaction was initiated by adding the duplex DNA (M13 ssDNA/63-mer 2). (A) Time courses for both the forward and the inverse reactions. (B) Data from A, quantified and plotted.
The reactions described above were performed at pH 6.2 because of the initial concern that the RecA protein–dsDNA complex might be too unstable at pH 7.5. However, inverse DNA strand exchange reactions performed at pH 7.5 in Tris acetate buffer were only twofold slower, and produced the same yield of product as the reactions conducted at the lower pH (data not shown). This small difference likely stems from a decreased stability of the dsDNA–RecA protein filament at the higher pH conditions (Lindsley and Cox 1989, 1990).
Inverse DNA strand exchange occurs in the absence of protein dissociation associated with ATP hydrolysis
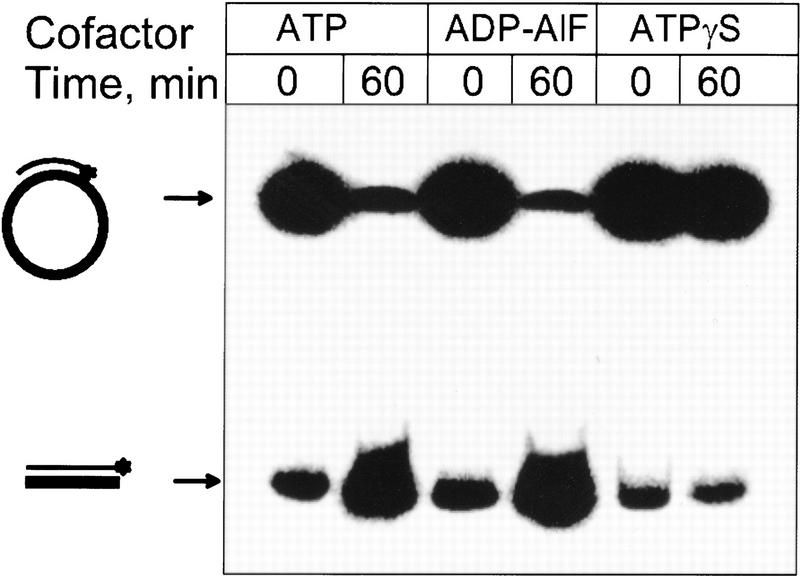
RecA protein can promote conventional DNA strand exchange in the complete absence of ATP hydrolysis, occurring in the presence of either ATPγS (Menetski et al. 1990) or ADP · AlF4 (Kowalczykowski and Krupp 1995). More important, the RecA protein–DNA complexes formed in the presence of either ATPγS or ADP · AlF4 are highly stable (Menetski and Kowalczykowski 1985; Kowalczykowski and Krupp 1995), and effectively do not transfer to a second DNA molecule (Menetski and Kowalczykowski 1987). To examine whether the inverse reaction could occur in the absence of RecA protein transfer and ATP hydrolysis, inverse DNA strand exchange was performed with substitution of either for ATP (Fig. 7). To ensure contiguous formation of the RecA nucleoprotein filaments, these reactions were performed by initially forming the filaments in the presence of 1 mm Mg acetate, and then initiating DNA strand exchange by raising the magnesium acetate concentration to 10 mm (Muniyappa et al. 1984). The results show that ADP · AlF4 supports inverse DNA strand exchange as efficiently as ATP; because the RecA protein–DNA complexes formed in the presence of ADP · AlF4 are highly stable (Kowalczykowski and Krupp 1995), this finding also substantiates our claim that inverse DNA strand exchange occurs without net transfer of RecA protein to the ssDNA. However, ATPγS did not support inverse DNA strand exchange (Fig. 7), at any Mg2+ ion concentration tested (1–15 mm; data not shown). This difference in the nucleotide cofactor utilization additionally differentiates the forward and inverse DNA strand exchange reactions, and its possible basis is considered in the Discussion.
Figure 7.
Inverse DNA strand exchange can occur in the absence of ATP hydrolysis. Reactions were carried out as described in the Materials and Methods except that, instead of ATP and the ATP regeneration system used for the reactions with ATP, either 1 mm ATPγS or 5 mm ADP together with 0.4 mm Al(NO3)3 and 10 mm NaF (Kowalczykowski and Krupp 1995) were used in the reactions containing ATPγS and the ADP · AlF4, respectively. Also, for all reactions, RecA protein was loaded onto the dsDNA at 1 mm magnesium acetate; after a 10-min incubation, magnesium acetate was added to a final concentration of 10 mm, and DNA strand exchange was then initiated by addition of the homologous ssDNA.
The RecA protein–dsDNA filament can participate in exchange with RNA
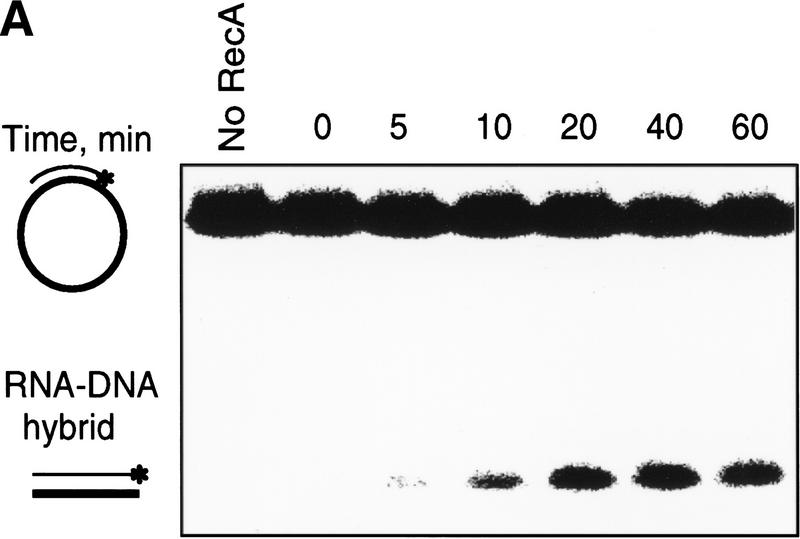
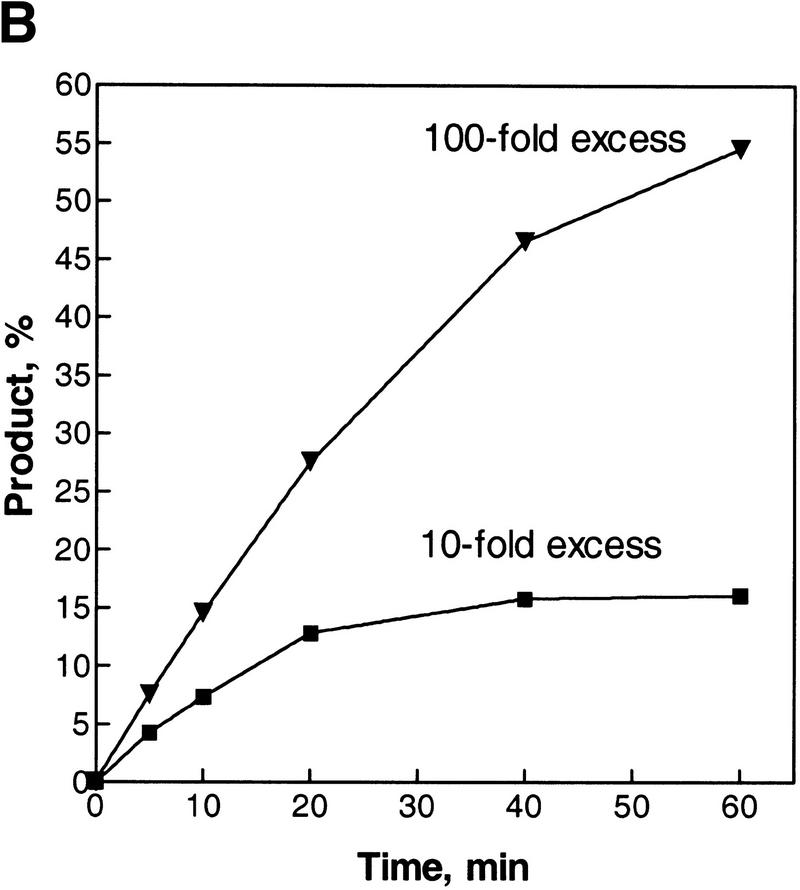
Constitutive stable DNA replication (cSDR) was proposed by Kogoma and colleagues to initiate from an R-loop that was hypothesized to form by invasion of dsDNA by an RNA transcript (Hong et al. 1995; Kogoma 1997). However, invasion of dsDNA by a RecA protein–ssRNA filament has never been achieved (see Discussion). Recently, the Kogoma laboratory showed that RecA protein could produce R-loop structures in a reaction whose characteristics were strikingly similar to the inverse DNA strand exchange reaction that we described here (Kasahara et al. 2000). To determine whether RecA protein could pair RNA with dsDNA by means of the inverse strand exchange process, experiments similar to those presented in Figure 2B were conducted, except that a 63-mer ribo-oligonucleotide (1R) was used instead of the ssDNA 63-mer oligonucleotide 1. As shown in Figure 8A, a RecA–dsDNA nucleoprotein filament is able to recombine with complementary RNA. The reaction is dependent on RecA protein, and proceeds with a similar time course as the inverse strand exchange reaction with ssDNA. Using standard reaction conditions (a 10-fold molecular excess of ssRNA) the yield of RNA strand exchange product is ∼15% (Fig. 8B). Thus, the RNA strand exchange is less efficient than DNA strand exchange (compare to Fig. 4). However, a 10-fold increase in ssRNA concentration (to a 100-fold molecular excess) raises the yield of the RNA–DNA hybrid product of this nucleic acid strand exchange reaction to ∼55%. Because RecA protein does not promote a conventional forward strand exchange reaction with RNA as a result of poor binding to the RNA (Kirkpatrick and Radding 1992; Kirkpatrick et al. 1992), our observations cannot be explained by redistribution of RecA to the RNA, followed by conventional strand exchange. Consistent with this view, a 100-fold molar excess of nonhomologous 63-mer ssDNA did not inhibit the inverse RNA strand exchange reaction (although the reaction rate was slowed by an amount comparable to the reduction seen in Fig. 5 for the inverse DNA strand exchange process; data not shown). Furthermore, the lower product yield seen with ssRNA as a partner argues against the notion that some form of free ssDNA is produce by the interaction of RecA protein with the duplex DNA substrate; if ssDNA were being produced, then its annealing with either ssDNA or ssRNA would be comparable. Thus, RecA protein is clearly capable of promoting inverse strand exchange with RNA, and we suggest that the R-loop formation observed by the Kogoma laboratory occurs by the same mechanism.
Figure 8.
Inverse exchange of strands between the RecA protein–dsDNA filament and ssRNA. The reaction was conducted as described under Materials and Methods, but using ssRNA (oligoribonucleotide 1R) instead of ssDNA (oligodeoxyribonucleotide 1). (A) Time course of the reaction. (B) The data from A were quantified and plotted (▾). Also plotted are the data for a reaction using a 100-fold molar excess of ssRNA (█).
Discussion
In this paper we describe a new nucleic acid strand-exchange activity of the RecA protein. We show that the dsDNA–RecA nucleoprotein filament can search for homology, pair with either homologous ssDNA or ssRNA, and promote the exchange of strands between the initial duplex DNA and the single-stranded nucleic acid. This is the “inverse” of the conventional DNA strand exchange reaction. Rather than the typical interaction of a ssDNA–RecA nucleoprotein filament with dsDNA, a dsDNA–RecA nucleoprotein filament interacts with a single-stranded nucleic acid. This reaction is both RecA protein and homology dependent (see Fig. 2). Also, this reaction shows asymmetry with respect to the strand of the dsDNA that is the complementary participant in the reaction (see Figs. 1 and 2).
Superficially, the inverse reaction appears to be simply conventional DNA strand exchange progressing in the “opposite” direction. It is initiated by a dsDNA–RecA filament that could be identical to the final product of the forward reaction (Register et al. 1987; Stasiak and Egelman 1988, 1994; Ullsperger and Cox 1995). However, inverse DNA strand exchange is not just a reversal of the forward process, nor is it occurring by redistribution of the RecA protein to the ssDNA or ssRNA. These conclusions are based on a number of clear experimental distinctions. First, inverse DNA strand exchange is relatively unaffected by an excess of nonhomologous ssDNA (∼46% exchange), which is in marked contrast to conventional DNA strand exchange (only ∼1.5% exchange). Second, inverse DNA strand exchanges does not occur with ATPγS, but does with either ATP or ADP · AlF4, whereas conventional DNA strand exchange occurs with all three nucleotide cofactors. Our finding with ATPγS confirms a previous report that the ATPγS–RecA protein–dsDNA filament interacts with ssDNA only in a homology-independent manner and is incapable of homologous pairing (Muller et al. 1990a). Our explanation for this failing is that ATPγS induces a very high affinity for DNA in both the primary and secondary DNA-binding sites of RecA protein (Kowalczykowski 1986; Mazin and Kowalczykowski 1996, 1998; Zaitsev and Kowalczykowski 1998, 1999), which kinetically restricts an efficient homology search. Because the affinity for ssDNA is greater than for dsDNA, the search for homology within dsDNA by a ssDNA presynaptic filament (this is the conventional reaction) is possible in the presence of ATPγS, whereas the search for homology within ssDNA by a dsDNA presynaptic filament (inverse DNA strand exchange) is kinetically constrained. Because the affinity of RecA protein for RNA is lower than for DNA, we also suggest that the ability of ATPγS to support R-loop formation in the reaction described by Kasahara et al. (2000) is due to this lower affinity that permits an iterative sampling of the ssRNA for homology (while bound to the secondary binding site) that is not possible with the more stable complexes formed with ssDNA; this pseudoequilibrium view also explains the lower yield of inverse strand exchange product formed using ssRNA as compared to the identical concentration of ssDNA. Consistent with this view, the lower affinities for DNA induced by ADP · AlF4 permit the search to occur in both the forward (Kowalczykowski and Krupp 1995) and inverse reactions. The third distinction between the inverse and forward reaction is that the inverse reaction is quite sensitive to an excess of RecA protein over the dsDNA concentration, whereas the forward reaction easily tolerates this excess. This difference likely stems from the different affinities of RecA protein for ssDNA versus dsDNA. Because the RecA protein binds dsDNA slowly at normal conditions, an excess of RecA protein is not inhibitory in the forward reaction. In the case of the inverse reaction, however, excess RecA protein easily binds ssDNA, thus inactivating the second partner of DNA strand exchange. Fourth, the apparent equilibrium established between substrate and product yield suggests that the inverse reaction may be reversible; this is distinct from the forward reaction where some combination of the binding of SSB protein to the displaced ssDNA, and the nonsymmetric binding of RecA protein to the dsDNA product (rather than to the ssDNA product) makes this reaction appear irreversible. We imagine that in the absence of SSB protein (and DNA secondary structure), the conventional reaction could be pseudoreversible. Finally, and perhaps most important, the inverse reaction accepts RNA as a homologous partner. Because strand exchange between ssRNA and dsDNA has never been achieved in vitro by the conventional reaction, the reaction that we are observing represents a newly described reaction for RecA protein.
What is the biological function of inverse DNA strand exchange? Although we could imagine a minor pathway of DNA repair that might involve the pairing of gapped or tailed dsDNA with ssDNA as the first step in the pairing process, we do not foresee this as a major function. Instead, we would propose that the inverse DNA strand exchange process could play a role in the maturation of single-end joint molecules into double-ended joint molecules in an asymmetric variation of the classic double-strand break repair (DSBR) model (Szostak et al. 1983). This model requires invasion by both processed ssDNA ends of a dsDNA break (Fig. 9). Although illustrations of this model show the invasion events as symmetric (i.e., both ends invading at the same time by presumably the same process), this is not an obligatory component of the model. If instead, the invasions occurred in two separate steps (i.e., asymmetrically), then the first pairing event could occur by a conventional forward reaction promoted by RecA protein assembling on one of the ssDNA tails to mediate formation of a joint molecule (D-loop). Because formation of a D-loop is known to stimulate polymerization of RecA protein onto the duplex DNA recipient (Shibata et al. 1982), the resultant formation of a RecA protein–dsDNA filament creates the nucleoprotein filament that is the active species in the inverse DNA strand exchange reaction that we described here. This filament can now pair with the processed ssDNA from the other end of the dsDNA break. Thus, sequential steps involving conventional DNA strand exchange followed by inverse DNA strand exchange can effect formation of the double-ended joint molecules that are intermediates of DSBR.
Figure 9.
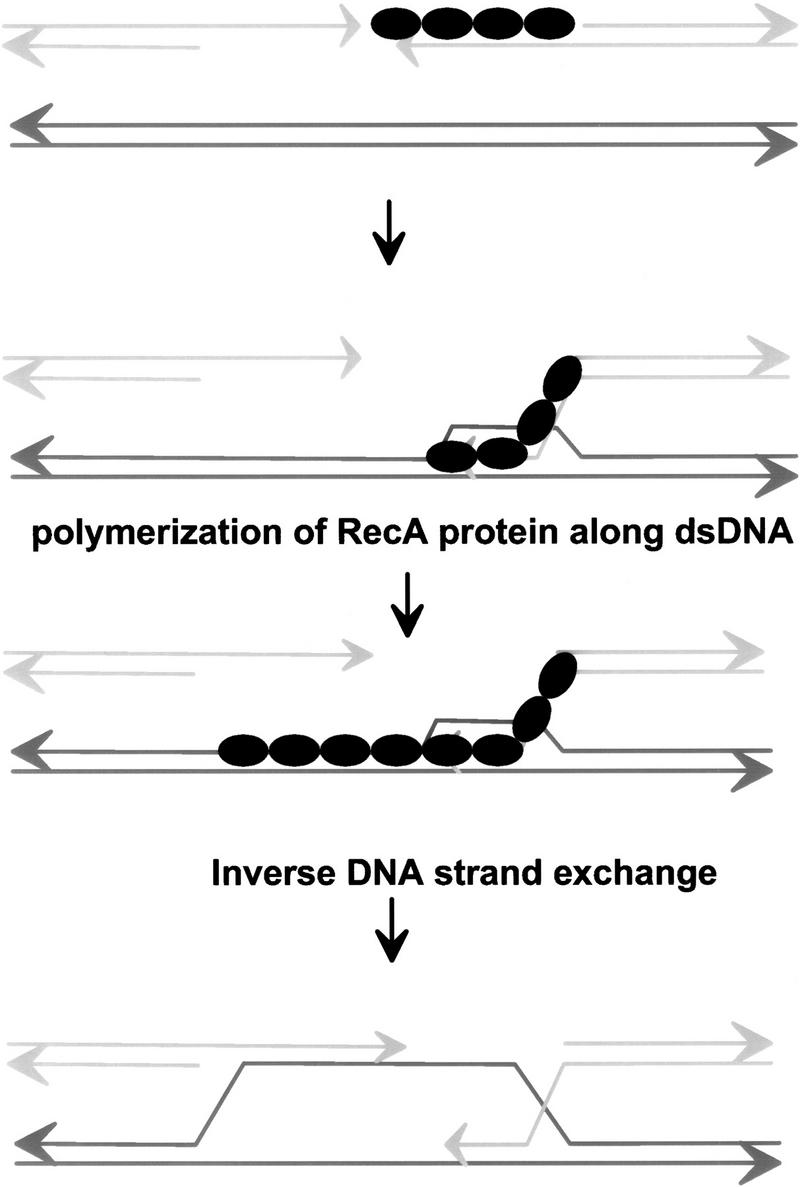
A proposed function for inverse DNA strand exchange in DSBR. Binding of RecA protein to ssDNA is produced by nucleolytic resection at one end of the dsDNA break. Conventional DNA strand invasion ensues, producing a single-ended joint molecule. Joint molecule formation allows the polymerization of RecA protein onto the homologous dsDNA target, to form a RecA–dsDNA nucleoprotein filament. This filament then promotes an inverse DNA strand exchange reaction to form a double-ended joint molecule.
Finally, the unprecedented capability of the dsDNA–RecA nucleoprotein filament to promote pairing and exchange with homologous RNA has a clear and obvious role in recombination-dependent replication processes (Kogoma 1996, 1997). In the mechanism of constitutive stable DNA replication, as proposed by Kogoma (Kogoma et al. 1979; Kogoma 1996, 1997), the formation of a RecA-dependent R-loop is an important step. Yet, despite the compelling in vivo evidence, published in vitro experiments have failed to produce RecA protein-mediated R-loops. Although the RecA nucleoprotein filament containing ssDNA can bind to and anneal with complementary ssRNA, thus promoting RNA–DNA hybridization, the equivalent nucleoprotein complex assembled on ssRNA is unable to promote strand exchange (Kirkpatrick and Radding 1992; Kirkpatrick et al. 1992). Therefore, the only demonstrated way to produce RecA protein-mediated R-loops is to use the inverse nucleic acid strand exchange process, a conclusion that is corroborated by recent work from the Kogoma laboratory (Kasahara et al. 2000). We propose that this is the biological function of this unique inverse RNA strand exchange reaction.
Materials and methods
DNA and RNA
The two 63-mer oligodeoxyribonucleotides, which are either identical (oligonucleotide 1: 5′-ACAGCACCAGATTCAGCAATTAAGCTCTAAGC CATCCGCAAAAATGACCTCTTATCAAAAGGA-3′) or complementary (oligonucleotide 2: 5′-TCCTTTTGATAAGAGGTCATTTTTGCGGATGG CTTAGAGCTTAATTGCTGAATCTGGTGCTGT-3′) to ssDNA of the bacteriophage M13, have been described (Mazin and Kowalczykowski 1996). Oligodeoxyribonucleotide 3 is the 63-mer (5′-TACACGACGGGGAGTCAGGCAACTATGGAGAACGAATAGACAGATCGCTGAGATAGGTGCC-3′). Oligoribonucleotide 1R has the same nucleotide sequence as 1, and was purchased from Oligos Etc., Inc. & Oligo Therapeutics. The 54-mer dsDNA was produced by annealing two complementary oligonucleotides: SKBT45 (5′-CGGCCAGTGAATTCTTTTATTGGCTGGTGGTGGGATCCCTGCA GGCATGCAAGC-3′) and SKBT46 (5′-GCTTGCATGCCTGCAGGGATCCCACCACCAGCCAATAAAAGAATTCACTGGCCG-3′).
Tailed dsDNA was prepared by annealing oligonucleotide 2 and M13 mp7 ssDNA. Oligonucleotide 2 (0.3 μg) was labeled with 32P using T4 polynucleotide kinase in polynucleotide kinase buffer (NEB) and mixed with an equimolar (molecule) amount (34 μg) of M13 mp7 ssDNA. The mixture (40 μl) was annealed by heating at 95°C for 5 min, and then cooled slowly (∼40 min) to room temperature. The annealed DNA was purified from unincorporated [32P]ATP and from free 63-mer oligonucleotide 2 using S-400 Spin columns (Pharmacia). The annealed products were analyzed by electrophoresis using native 10% (19:1) polyacrylamide gels that were dried on DEAE paper and quantified using a Betascope 603 radioisotope analyzer (Betagen). The concentration of the annealed M13 ssDNA/63-mer oligonucleotide 2 duplex DNA substrate was calculated by comparing the radioactivity of the purified labeled duplex DNA substrate to a known amount of 5′-end-labeled oligonucleotide 2 after electrophoresis on the same gel. The purified M13 ssDNA/63-mer 2 duplex DNA substrate was contaminated by <1% of free 63-mer 2. Oligonucleotide 3 and phage φX174 ssDNA were used as nonhomologous competitors in the DNA strand exchange reactions.
RecA protein
RecA protein was purified using a procedure based on spermidine precipitation (Griffith and Shores 1985) (S.C. Kowalczykowski, unpubl.). Protein concentration was measured using an extinction coefficient of 2.7 × 104 m/cm at 280 nm.
Inverse DNA strand exchange reaction
The inverse DNA strand exchange reaction was performed in buffer containing 25 mm MES-NaOH (pH 6.2), 10 mm Mg acetate, 0.1 mm DTT, 4.5 mm PEP, 10 U/ml pyruvate kinase, and 3 mm ATP. The concentrations of M13 ssDNA/63-mer 2 duplex DNA and 63-mer ssDNA (oligonucleotide 1 or 2) were 6.3 nm (molecules) and 63 nm (molecules), which is equivalent to 46.5 μm and 4 μm (nucleotides), respectively. The concentration of RecA protein was 4.65 μm, which corresponds to a stoichiometry of 1 RecA monomer per 10 nucleotides of M13 ssDNA. Changes to these conditions are described in figure legends 4–8. The inverse strand exchange reactions with RNA were conducted using the same conditions as for the reaction with DNA, but oligoribonucleotide 1R was used instead of oligodeoxyribonucleotide 1. Components were added as follows: RecA protein was added to the duplex DNA substrate in complete reaction buffer and was incubated for 5 min at 37°C. Afterward, DNA strand exchange was initiated by adding ssDNA (oligonucleotides 1, 2, or 3). Portions (10 μl) were removed at various times and the reaction was stopped by addition of 2.5 μl of loading solution (2.5% SDS, 0.125 m EDTA, 15% Ficoll). The samples were then run on a 6% polyacrylamide gel (19:1) in Tris-borate buffer for 2 hr at 20 V/cm. Gels were dried on DEAE paper and the bands were analyzed using either a Betascope 603 or a Storm PhosphorImager (Molecular Dynamics, Inc.). The amount of radioactivity in the reaction product and in the remaining substrate was measured and expressed as a percentage.
Conventional (forward) DNA strand exchange was performed in the same way, except that RecA protein was preincubated with the 63-mer ssDNA (oligonucleotide 1) first and the reaction was initiated by adding the duplex DNA substrate (M13 ssDNA/63-mer 2).
Acknowledgments
We are grateful to Tokio Kogoma and David Bates for providing data before publication and to the many members of this laboratory for their critical reading of this manuscript: Frederic Chedin, Dan Anderson, Deana Arnold, Rick Ando, Piero Bianco, Carole Bornarth, Frank Harmon, Noriko Kantake, Julie Kleiman, Alex Mazin, Jim New, Erica Seitz, Susan Shetterly, and Tomohiko Sugiyama. This work was supported by NIH grant AI-18987.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sckowalczykowski@ucdavis.edu; FAX (530) 752-5939.
References
- Adzuma K. Stable synapsis of homologous DNA molecules mediated by the Escherichia coli RecA protein involves local exchange of DNA strands. Genes & Dev. 1992;6:1679–1694. doi: 10.1101/gad.6.9.1679. [DOI] [PubMed] [Google Scholar]
- Bedale WA, Inman RB, Cox MM. RecA protein-facilitated DNA strand breaks. A mechanism for bypassing DNA structural barriers during strand exchange. J Biol Chem. 1991;266:6499–6510. [PubMed] [Google Scholar]
- ————— A reverse DNA strand exchange mediated by recA protein and exonuclease I. The generation of apparent DNA strand breaks by recA protein is explained. J Biol Chem. 1993;268:15004–15016. [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Chiu SK, Wong BC, Chow SA. Homologous pairing in duplex DNA regions and the formation of four-stranded paranemic joints promoted by RecA protein. Effects of gap length and negative superhelicity. J Biol Chem. 1990;265:21262–21268. [PubMed] [Google Scholar]
- Chow SA, Honigberg SM, Radding CM. DNase protection by recA protein during strand exchange. Asymmetric protection of the Holliday structure. J Biol Chem. 1988;263:3335–3347. [PubMed] [Google Scholar]
- Clark AJ, Sandler SJ. Homologous genetic recombination: The pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- Conley EC, West SC. Homologous pairing and the formation of nascent synaptic intermediates between regions of duplex DNA by RecA protein. Cell. 1989;56:987–995. doi: 10.1016/0092-8674(89)90632-6. [DOI] [PubMed] [Google Scholar]
- Conley EC, West SC. Underwinding of DNA associated with duplex-duplex pairing by RecA protein. J Biol Chem. 1990;265:10156–10163. [PubMed] [Google Scholar]
- Cox MM. Alignment of 3 (but not 4) DNA strands within a RecA protein filament. J Biol Chem. 1995;270:26021–26024. doi: 10.1074/jbc.270.44.26021. [DOI] [PubMed] [Google Scholar]
- Cox MM, Lehman IR. RecA protein-promoted DNA strand exchange: Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982;257:8523–8532. [PubMed] [Google Scholar]
- Griffith J, Shores CG. RecA protein rapidly crystallizes in the presence of spermidine: A valuable step in its purification and physical characterization. Biochemistry. 1985;24:158–162. doi: 10.1021/bi00322a022. [DOI] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, Kogoma T. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 1995;14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Clikeman JA, Bates DB, Kogoma T. RecA protein-dependent R-loop formation in vitro. Genes & Dev. 2000;14:360–365. [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DP, Radding CM. RecA protein promotes rapid RNA-DNA hybridization in heterogeneous RNA–mixtures. Nucleic Acids Res. 1992;20:4347–4353. doi: 10.1093/nar/20.16.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DP, Rao BJ, Radding CM. RNA–DNA hybridization promoted by E. coli RecA protein. Nucleic Acids Res. 1992;20:4339–4346. doi: 10.1093/nar/20.16.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- ————— Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Torrey TA, Connaughton MJ. Induction of UV-resistant DNA replication in Escherichia coli: Induced stable DNA replication as an SOS function. Mol Gen Genet. 1979;176:1–9. doi: 10.1007/BF00334288. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Interaction of recA protein with a photoaffinity analogue of ATP, 8-azido- ATP: Determination of nucleotide cofactor binding parameters and of the relationship between ATP binding and ATP hydrolysis. Biochemistry. 1986;25:5872–5881. doi: 10.1021/bi00368a006. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Krupp RA. Effects of the Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein: Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J Mol Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- ————— DNA-strand exchange promoted by RecA protein in the absence of ATP: Implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc Natl Acad Sci. 1995;92:3478–3482. doi: 10.1073/pnas.92.8.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley JE, Cox MM. Dissociation pathway for recA nucleoprotein filaments formed on linear duplex DNA. J Mol Biol. 1989;205:695–711. doi: 10.1016/0022-2836(89)90315-x. [DOI] [PubMed] [Google Scholar]
- ————— On RecA protein-mediated homologous alignment of two DNA molecules. Three strands versus four strands. J Biol Chem. 1990;265:10164–10171. [PubMed] [Google Scholar]
- Mazin AV, Kowalczykowski SC. The specificity of the secondary DNA binding site of RecA protein defines its role in DNA strand exchange. Proc Natl Acad Sci. 1996;93:10673–10678. doi: 10.1073/pnas.93.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 1998;17:1161–1168. doi: 10.1093/emboj/17.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A novel property of the RecA nucleoprotein filament: Activation of double-stranded DNA for strand exchange in trans. Genes & Dev. 1999;13:2005–2016. doi: 10.1101/gad.13.15.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- ————— Transfer of recA protein from one polynucleotide to another: Effect of ATP and determination of the processivity of ATP hydrolysis during transfer. J Biol Chem. 1987;262:2093–2100. [PubMed] [Google Scholar]
- Menetski JP, Bear DG, Kowalczykowski SC. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc Natl Acad Sci. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Koller T, Stasiak A. Characterization of the DNA binding activity of stable RecA-DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J Mol Biol. 1990a;212:97–112. doi: 10.1016/0022-2836(90)90307-8. [DOI] [PubMed] [Google Scholar]
- ————— Characterization of the DNA binding activity of stable RecA–DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J Mol Biol. 1990b;212:97–112. doi: 10.1016/0022-2836(90)90307-8. [DOI] [PubMed] [Google Scholar]
- Muniyappa K, Shaner SL, Tsang SS, Radding CM. Mechanism of the concerted action of recA protein and helix destabilizing proteins in homologous recombination. Proc Natl Acad Sci. 1984;81:2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Register JC, III, Christiansen G, Griffith J. Electron microscopic visualization of the RecA protein-mediated pairing and branch migration phases of DNA strand exchange. J Biol Chem. 1987;262:12812–12820. [PubMed] [Google Scholar]
- Roca AI, Cox MM. RecA protein: Structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Shan Q, Cox MM. RecA filament dynamics during DNA strand exchange reactions. J Biol Chem. 1997;272:11063–11073. doi: 10.1074/jbc.272.17.11063. [DOI] [PubMed] [Google Scholar]
- ————— On the mechanism of RecA-mediated repair of double-strand breaks: No role for four-strand DNA pairing intermediates. Mol Cell. 1998;1:309–317. doi: 10.1016/s1097-2765(00)80031-3. [DOI] [PubMed] [Google Scholar]
- Shaner SL, Flory J, Radding CM. The distribution of Escherichia coli recA protein bound to duplex DNA with single-stranded ends. J Biol Chem. 1987;262:9220–9230. [PubMed] [Google Scholar]
- Shibata T, Ohtani T, Iwabuchi M, Ando T. D-loop cycle: A circular reaction sequence which comprises formation and dissociation of D-loops and inactivation of superhelical closed DNA promoted by recA protein of Escherichia coli. J Biol Chem. 1982;257:13981–13986. [PubMed] [Google Scholar]
- Stasiak A, Egelman EH. Visualization of recombination reactions. In: Kucherlapati R, Smith GR, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 265–308. [Google Scholar]
- ————— Structure and function of RecA–DNA complexes. Experientia. 1994;50:192–203. doi: 10.1007/BF01924002. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51–ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Ullsperger CJ, Cox MM. Quantitative RecA protein binding to the hybrid duplex product of DNA strand exchange. Biochemistry. 1995;34:10859–10866. doi: 10.1021/bi00034a019. [DOI] [PubMed] [Google Scholar]
- West SC, Howard-Flanders P. Duplex–duplex interactions catalyzed by recA protein allow strand exchanges to pass double-strand breaks in DNA. Cell. 1984;37:683–691. doi: 10.1016/0092-8674(84)90401-x. [DOI] [PubMed] [Google Scholar]
- Zaitsev EN, Kowalczykowski SC. Binding of double-stranded DNA by Escherichia coli RecA protein monitored by a fluorescent dye displacement assay. Nucleic Acids Res. 1998;26:650–654. doi: 10.1093/nar/26.2.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The simultaneous binding of two double-stranded DNA molecules by Escherichia coli RecA protein. J Mol Biol. 1999;287:21–31. doi: 10.1006/jmbi.1998.2580. [DOI] [PubMed] [Google Scholar]