Abstract
NAC1, a BTB/POZ family member, has been suggested to participate in maintaining stemness of embryonic stem cells and has been implicated in the pathogenesis of human cancer. In ovarian cancer, NAC1 upregulation is associated with disease aggressiveness and with the development of chemoresistance. Like other BTB/POZ proteins, NAC1 forms discrete nuclear bodies in non-dividing cells. To investigate the biologic role of NAC1 nuclear bodies, we characterized the expression dynamics of NAC1 nuclear bodies during different phases of the cell cycle. Fluorescence recovery after photobleaching assays revealed that NAC1 was rapidly exchanged between the nucleoplasm and NAC1 nuclear bodies in interphase cells. The number of NAC1 bodies significantly increased and their size decreased in S-phase as compared to G0/G1 and G2 phases. NAC1 nuclear bodies disappeared and NAC1 became diffuse during mitosis. NAC1 nuclear bodies reappeared immediately after completion of mitosis. These results indicate that a cell cycle-dependent regulatory mechanism controls NAC1 body formation in the nucleus and suggest that NAC1 body dynamics are associated with mitosis or cytokinesis.
Keywords: NAC1, Kinetics, cell cycle, carcinogenesis
INTRODUCTION
Currently, ovarian cancer is one of the most lethal gynecological malignant diseases in the United States (1). The gene encoding the nucleus accumbens associated protein-1 (NAC1) has been recently discovered to be one of the genes that is up-regulated during tumorigenesis of several types of gynecological cancers including ovarian carcinoma, endometrioid carcinoma of the uterus, and cervical adenocarcinoma (2). NAC1 is located in the nucleus where it forms homodimers via the BTB/POZ domain, which is reported to be an essential protein motif mediating survival and growth in tumor cells (2). Over-expression of NAC1 has been reported to contribute to the development of chemoresistance in ovarian cancer cells (3, 2, 4). Interestingly, a high expression level of NAC1 causes the formation of distinct, dense body-like structures in the nucleus (2). Nuclear organelles such as the nucleoli, nuclear speckles, Cajal bodies (5) and PML bodies (6, 7), are all believed to be associated with gene regulation governing important cell functions (8, 5, 9, 10). Given their nuclear location, the newly discovered NAC1 nuclear bodies (NBs) may be also involved with gene or genome regulation.
Recently, two Gadd45 pathway proteins, Gadd45-γ and Gadd45-γ-interacting protein 1 (Gadd45gip1), have been identified as regulatory targets of NAC1 (3, 4). Expression of these two proteins can decrease cell proliferation and increase apoptosis. When a dominant–negative form of the NAC1 protein containing only the BTB/POZ domain is introduced into cells, NAC1 NBs are disrupted, and the expression of both Gadd45gip1 and Gadd45-γ increases (4). This suggests that the NAC1 protein and its intact structure may be involved in the regulation of transcription of these two proteins. However, the details of the mechanism by which NAC1 NB regulate gene transcription is unknown.
The nucleus comprises a heterogeneous environment, in part due to the spatial distribution of chromatin structures. Individual genes are separated to distinct locations, where transcription can be specifically regulated (11). Transcription of a gene is governed not only by interactions between the gene and its transcription factors, but also by the temporal and spatial distribution of components of the transcription system (12). In the nucleus, the NAC1 NBs may use their specific affinity for NAC1 protein to create a spatial discrepancy that may regulate transcription. Based on this reasoning, we have deliberately characterized the morphological changes in NAC1 NB during different phases of the cell cycle. In this study, we explored NAC1 NB morphology throughout the cell cycle in ovarian cancer cells in which NAC1 proteins were highly expressed because this might indicate the extent to which NAC1 NB could be a regulatory factor. Our results showed that the morphology of NAC1 NBs varied dramatically during different phases of the cell cycle. Based on these observations we have proposed a molecular mechanism to explain the formation of NAC1 NBs.
MATERIALS AND METHODS
Cell culture and cell cycle synchronization
RK3E/GFP-NAC1 cells were derived from the RK3E cell line (American Type Culture Collection, Manassas, VA) in which the GFP-NAC1 protein was stably expressed. Cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT), 100 IU/ml penicillin, and 100 μg/ml streptomycin, in 10-cm cell culture dishes in a water-jacketed, 5%-CO2 incubator, and passed at a concentration of approximately 1×104 cells/ml every 3–4 days. For experiments, cells were re-plated in glass bottom dishes (In Vitro Scientific, Sunnyvale, CA) pre-treated with 0.01% poly-l-lysine (Sigma, St. Louis, MO). To obtain synchronized cultures, cells were arrested in early S phase by addition of thymidine (2 mM) to the culture medium. After 18 hours the culture media was changed back to normal conditions and cells were allowed to propagate for 9 hours through S phase and to enter G2 phase. At this point, thymidine concentration was returned to 2 mM for another 16 hours to arrest cells in early S phase of next cell cycle.
Microscopy system and image acquisition
A Cascade:1K EMCCD camera (Roper scientific, Tucson, AZ), mounted on a Nikon TE 2000-E inverted microscope (Nikon, Melville, NY), was used to acquire wide-field fluorescence images. To determine the intensity profile of Hoechst 33342-stained nuclei, nuclear images were acquired with a 10× objective lens (N.A. = 0.45) (Nikon). To assess the z-direction projection of NAC1 NB with high spatial resolution, NAC1 NB images were acquired using a 0.2 μm step-size in the z-direction to create a Z-stacked image with a 60× oil-immersion objective lens (N.A. = 1.4) (Nikon). The intensity profile of the NAC1 NB at a pixel was reconstructed by the standard deviation of the intensity profile of the Z-stacked images at that particular pixel to describe the relative intensity of the object. Time-resolved NAC1 NB images were obtained using incident light at 12% of full illumination power to avoid phototoxicity and photobleaching effects. To compensate for the decrease in light intensity, the EM-gain was activated to half of the maximum setting (2000 au) (13).
Immunostaining
SKOV3 ovarian cancer cells were grown in 8-well chamber slides. Cells were fixed in 3% paraformaldehyde and washed in phosphate buffer solution with Triton X-100 (0.5%). The cells were then incubated with anti-NAC1 antibody (Novus Biologicals) at 1:300 dilution followed by a Rhodamine-conjugated anti-mouse antibody (Jackson ImmunoResearch Lab., West Grove, PA). Cells were then co-stained with FITC-conjugated anti-alpha-tubulin antibody at a dilution of 1:150. The incubation time for all antibodies was 2 hours at room temperature. Cells were counter-stained with Hoechst 33342.
Nuclear DNA content and NB size analysis
The intensity of a Hoechst 33342-stained nucleus was analyzed using a custom-made program in MATLAB (Mathworks, Natick, MA). A 19 pixel-by-19 pixel Gaussian kernel was applied to the background-subtracted image to filter the imaging noise (14, 15) and to identify the region of the nucleus. Thereafter, the total intensity (or the sum of the intensity) of the pixels where the nucleus was located was collected to represent relative DNA content in the nucleus. To estimate the size of a NB in a z-stack image, the image backgrounds of individual images were separately subtracted, then the particle intensity of the images was fitting utilizing a least-square algorithm to locate the position of the particle from its local maximum intensity pixel and its adjacent 4 pixels (16). For the purpose of increasing the computing performance, the fitting was achieved by a logarithmic 2-dimensional Gaussian distribution formula,
| (1) |
where IP represents the pixel intensity of an image and the fitted parameters, I′, and , represent the particle peak intensity, particle image radius, and center position of the particle in x- and y-direction, respectively. For an object, the parameters, I′ and , negatively correlated with the distance of a particle to its focal plane. Therefore, the minimum of a particle at the sequential z-stack was used to represent its in-focus size.
Fluorescence recovery after photobleaching
A Nikon EZ-C1 confocal system (Nikon) on the TE-2000 microscope was used to perform the fluorescence recovery after photobleaching (FRAP) experiment through a 60× oil-immersion objective lens. In addition, an incubator (In Vivo Scientific, St. Louis, Missouri) mounted on the microscope stage maintained culture conditions at 5% CO2 and 37°C to allow an extended time period of observation. In each FRAP experiment, a selected intact NB was bleached, and the intensity recovery was monitored every 5 sec for 10 min with a 150 μm pinhole size, which limited the intensity fluctuation that resulted from NB movement in the z-direction. Each FRAP experiment was then analyzed by the following process: The original (pre-photobleaching) intensity of the monitored NB was estimated from five sequential images and denoted Ipre The NB intensity immediately after bleaching was denoted Ibl The recovering intensity of the bleached NB, denoted IP, was normalized to In from the following calculation.
| (2) |
The half-life of fluorescence recovery (τ1/2) was obtained as the time required to recover half of the plateau intensity.
RESULTS
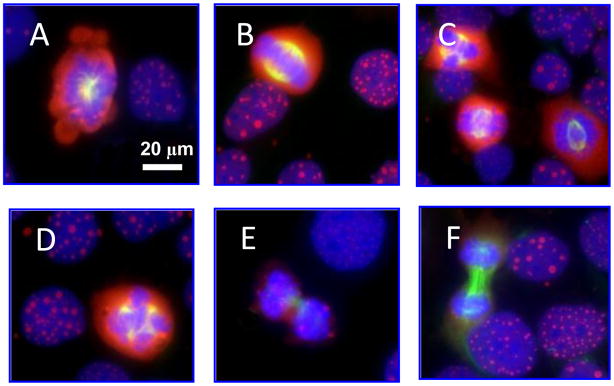
It has been demonstrated that over-expression of NAC1 proteins gives rise to the formation of dense body-like structures in the nucleus termed NAC1 NBs (2). More importantly, we observed a heterogeneous distribution in size and number of NAC1 NBs among ovarian cancer cells, suggesting an underlying dynamics in the genesis of NAC1 NBs. One of the most striking findings was that NAC1 NBs were not found in mitotic cells (Figure 1), raising the possibility that the formation of NAC1 NB might be dependent on the cell cycle.
Figure 1. Patterns of NAC1 NB in different phases of cell cycle.
Immunofluorescent micrographs show diffuse NAC1 (red fluorescence) distribution in cells undergoing mitosis, whereas NAC1 bodies are evident for cells in interphase. Cells were co-stained with anti-α-tubulin antibody (green fluorescence) and were counterstained with DAPI (blue fluorescence). (A) prophase; (B–D) metaphase; (E) anaphase; (F) telophase. Interphase cells are present in all panels.
To explore the development of NAC1 NBs in nuclei, we correlated the size distribution of NAC1 NBs with the cell cycle. Phases of the cell cycle were determined by quantifying the DNA content of the nucleus using a fluorescent DNA probe. This method has been widely used in flow cytometry to estimate cell populations in particular phases of the cell cycle. Following this concept, we developed a single-cell assay using wide-field fluorescence microscopy to determine cell cycle phase and simultaneously quantify the size distribution of the NAC1 NBs in the nucleus.
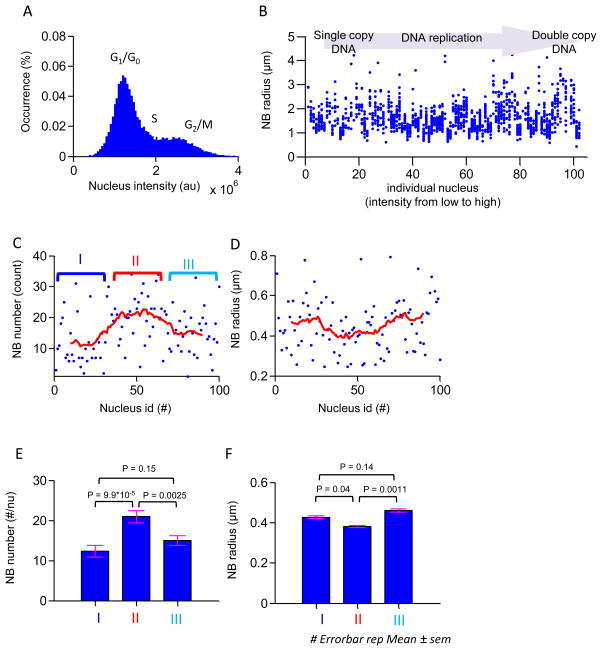
Nuclear DNA content was determined by analyzing a series of microscopic images containing more than 10,000 Hoechst 33342-stained nuclei (see methods) (Figure 2 A). The distribution of nuclear DNA content shown in the histogram was typical of data from flow cytometry experiments, and showed two subgroups of nuclear DNA content in which one group had a peak fluorescence intensity two times greater than the other. Cells in the lower intensity subgroup were in G0/G1, and cells in the higher intensity subgroup were in the G2/M phase (Figure 2 A). Because our single cell assay permitted determining both fluorescence intensity and size of NAC1 NB, the sizes of the NAC1 NBs could be correlated with cell cycle in these cells.
Figure 2. NAC1 NB morphology analysis during interphase.
(A) A histogram of nuclear intensities (N = 18,485) estimated by wide-field fluorescence microscopy in cultured cells. Two peaks in this distribution indicate the mean cell nucleus intensity at G1/G0 phase and G2 phase from left to right. (B) Scatter plot of NB size versus nuclear fluorescence intensity. Cells are arranged in order of increasing nuclear fluorescence intensity. Each column of dots represents size distribution of NB of an individual nucleus. This plot represents estimated NAC1 NB size propagation throughout interphase. Intensities of nuclei are sub-grouped into three equal partitions representing the cell cycle in G1, S, and G2 phases. Differences in mean size of NB in the three groups were assessed by Student t-test. (C) NB number in each nucleus is plotted against individual nuclei arranged in order of increasing fluorescence intensity. Each dot represents average number of NB in an individual nucleus. Solid line indicates moving average number of NB within a window of 20 nuclei. (D) Average NB size in each nucleus is plotted against individual nuclei arranged in order of increasing fluorescence intensity. Each dot represents the average size of NB in an individual nucleus. Solid line indicates moving average size of NB within a window of 20 nuclei. (E–F) The mean value of NB number and average size in three groups is shown by bar plots. Significance of differences between NB number or average size during different phases of cell cycle were determined by Student’s t-test. Errorbars represent the standard error of the mean.
The size of each NAC1 NB was estimated within its focal plane, determined by the narrowest apparent radius in a z-stack scan using a high-magnification objective lens (see methods). The NAC1 NB size distribution was collected from NBs in 100 randomly selected cells; cell cycle phases as judged by nuclear intensity were continuously distributed from single copies of DNA (G0/G1) to double copies of DNA (G2/M) (data not shown). The sizes of NBs in each nucleus were then sequentially plotted against the corresponding nuclear intensity, sorted from low to high, representing the propagation of the cell cycle from G1-S-G2 under normal growth conditions. The size distribution of the NAC1 NBs in these nuclei revealed the size propagation of NAC1 NBs throughout the cell cycle (Figure 2 B). The morphology of NAC1 NBs in individual nuclei was further characterized by estimating the corresponding number of NBs and the average size of NBs in these 100 nuclei (Figure 2 C and D). To further evaluate morphology dynamics of NAC-1 in interphase we adopted the moving average, calculated using total NBs in a window size of 20 nuclei to overcome the estimation noise in DNA content and cellular heterogeneity. Interestingly, results showed that the NB number and size had three distinct states corresponding to the early interphase (phase I), mid interphase (phase II) and late interphase (phase III), which are possibly corresponding to G1/G0, S, and G2/M phases of the cell cycle. As the cell cycle propagates through the cycle, NB number dramatically increases at stage II and then decreases afterward, while NB size has the opposite trend. Cells in phase I had an average of ~13 NBs and the average size of each NB was ~0.43 μm (Figure 2 E and F). The number of NBs increased ~2 times to ~21 per nucleus in S phase, but the average size decreased to ~0.38 μm. In the phase III, the average size of a NAC1 NB increased and returned to a level similar to the phase I; meanwhile, the number of NBs dropped to ~15 per nucleus. Analysis by Student’s t-test showed that differences in average NB radius and NB number in between phase I- phase II and phase II-phase III were significant; differences between phase I-phase III were not significant. These results suggested that the morphology of NAC1 NB was tightly associated with the cell cycle. Together, the results suggested NB morphology undergoes a major change associated with S phase.
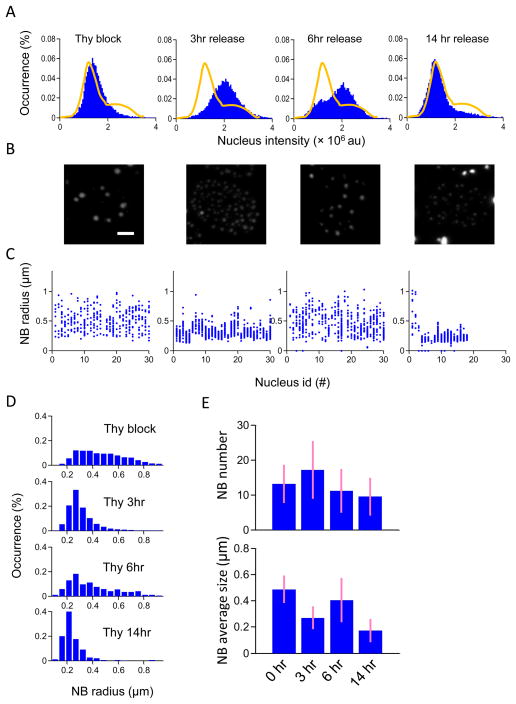
To further study changes in NB size throughout the cell cycle, thymidine was used to arrest cells at the early S phase. Release of the cell culture from thymidine enables the cells to re-enter the cell cycle. Propagation of synchronized cells through the cell cycle to the late S, G2/M and G0/G1 phase is shown in histograms of the intensities of Hoechst 33342-stained nuclei of cells grown for 3 hours, 6 hours and 14 hours after thymidine was released from the culture medium (Figure 3 A). The appearance of NAC1 NBs in the nucleus was clearly different at each time after release from thymidine (Figure 3 B). The corresponding sizes and amounts of the NAC1 NBs were quantified at each time point (Figure 3 C). The results showed that in arrested cells the sizes of NAC1 NBs were distributed from ~0.27 μm to 1.08 μm with a mean of ~0.49 μm, and that there were ~12 NBs per nucleus. After 3 hours release from thymidine arrest, the average size of NBs was ~0.27 μm and was more or less homogeneously distributed between 0.22 and 0.54 μm. The average number of NBs increased to ~18 per nucleus. After 6 hours release, the number of NBs had dropped to ~11 per nucleus and the average size had increased to ~0.47 μm. By 14 hours after release cells had passed from M phase to the next stage, and the NBs had a smaller average size ~ 0.21 μm and the number of NBs was ~10 per nucleus. At this stage, NB size is smaller than the NBs size in S phase, the NB number is ~ 2 times less than NB number in S phase but more close to the thymidine arrested state. Therefore, together with the nucleus intensity distribution these results suggest after 14hr release from thymidine, the cell cycle is in G1 phases and the NBs are propagating in size. It is worth noting that the size and number of NAC1 NBs were similar in arrested cells (early S-phase) and 6 hours after release (Figure 3 D – E). Together, these results demonstrated that morphologic variation in NAC1 NBs was associated with the cell cycle.
Figure 3. NAC1 NB morphological changes from early S phase in cell cycle-synchronized cells.
(A) Histograms of cell nuclear fluorescence intensity. (Left to Right) before thymidine release, 3 hours, 6 hours, and 14 hours after thymidine release. Solid lines indicate the distribution of cell nuclear fluorescence intensities under normal growth conditions showing G1/G0 and G2 phases. (B) Projected images in the z-direction from a set of z-stack confocal images, showing representative NAC1 NB morphology at different cell cycle phases. The scale bar represents the length of 5 μm. (C) NB size distribution in nuclei shown by plotting the size of each NB against individual nuclei arranged in order of increasing fluorescence intensity. The four different cell cycle conditions are shown respectively from left to right. (D) Overall NB distributions in the four conditions over different nuclei are shown in histogram plots. (E) Bar plots showing mean NB numbers and average NB size in a nucleus at different times after thymidine release.
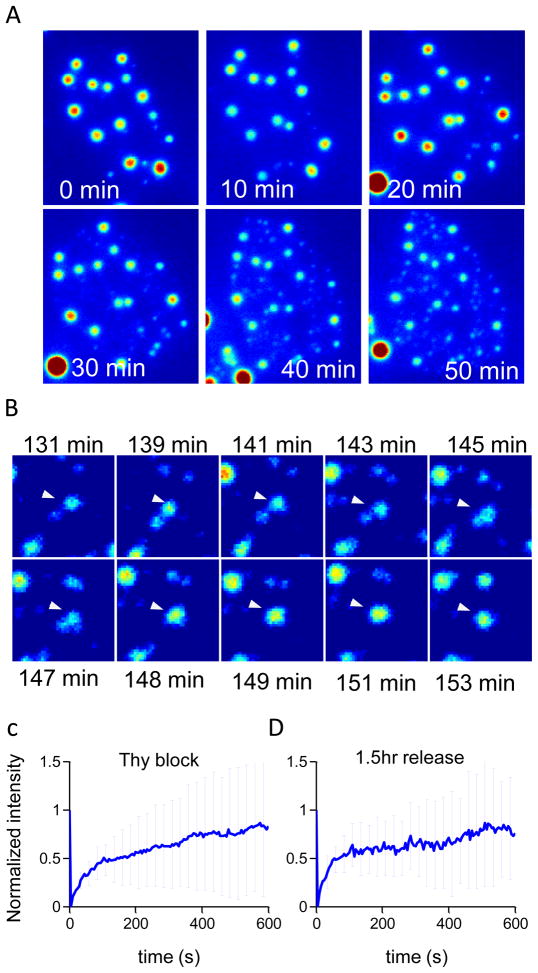
We next investigated real-time morphological changes in NAC1 NB during progression of the S phase. NAC1 NBs were monitored for 3 hours after the cell culture was released from thymidine. In the first 50 minutes, multiple new NBs formed in locations distinct from sites of pre-existing NBs, and the size of pre-existing NBs decreased. Thus, the sizes of NAC1 NB in a nucleus became more homogeneous (Figure 4 A). Approximately 2 hours after thymidine release, as the cell cycle progressed toward the G2 phase, adjacent NBs began to aggregate, generating larger NBs (Figure 4 B). This result suggested that the apparent NB size change across the S phase involved a two-step process of 1) formation of new NBs in distinct locations and 2) later fusion to form larger NBs. To further investigate whether underlying NAC1 molecular dynamics changed across S phase, a FRAP experiment was performed on similarly sized NAC1 NBs before thymidine block and approximately 1.5 hours after thymidine release. There were no significant differences between these two recovery curves, but the recovery half time (τ1/2) for a NAC1 NB was ~ 80 seconds during thymidine arrest and ~ 40 seconds after 1.5 hours release from thymidine arrest (Figure 4 C, D). Thus, the exchange of NAC1 between NBs and the nucleus was more rapid in S-phase than under arrest in G0/G1. This further suggested that the formation of NBs in S phase was through a dynamic exchange in which newly synthesized NBs competed with pre-existing NBs for available nuclear NAC1 proteins until both reached a balanced state in which the association and dissociation rates of all NBs were equilibrated.
Figure 4. New NAC1 NB formation in S phase occurs through a nucleation-propagation process and NBs form larger clusters through a fusion process.
(A) Time lapse Z-projected reconstructed fluorescent image showing new NB synthesis during S phase. (B) Time-sequence of a z-projected image showing the fusion process of NBs approximately 2 hours after release. Arrows indicate two individual NBs undergoing fusion. (C–D) Intensity recovery plot from FRAP experiment for thymidine arrested cells and 1.5 hours after release. Graphs show mean normalized fluorescence intensity (error bars indicate standard deviation of at least 10 FRAP experiments).
DISCUSSION
It has been shown BTB/POZ domain mediates the homodimerizaion (17–19). Furthermore, the BTB/POZ domain was also involved in self-multerimerization in a number of other BTB/POZ domain containing proteins (20–22) possibly through dimer-dimer interactions by the BTB/POZ domain (19). Therefore, it is suggested that the ability to form discrete nuclear bodies (NBs) when over-expressed in NAC1 is due to the BTB/POZ domain (2, 23). In this report, we used an engineered cell line that can constitutively express EGFP-tagged NAC1, a member of the BTB/POZ domain-containing protein family, to characterize the biophysical properties of NAC1 NBs. This cell line was derived from the non-transformed epithelial cell line, RK3E. First, we quantified the number of NAC1 NBs in individual cells within un-synchronized cells. The results suggested that the quantity of NAC1 NBs varied substantially among cells. To investigate this variation, we developed a procedure using quantitative live-cell fluorescence microscopy that permitted simultaneous identification of cell cycle phase and analysis of NAC1 NBs in individual cells. Using this method, we observed that the amount of NAC1 NBs in a nucleus correlated with the cell cycle. Interestingly, the average number of NAC1 NBs in a nucleus increased from 13 to 21 through the transition from G0/G1 to S phase, and then decreased to 15 during transition from S phase to G2 phase. In addition, size variation in NAC1 NBs in cells in G0/G1 phase or G2/M phase was more significant than in S phase. Finally, FRAP experiments on NAC1 NBs showed that NAC1 proteins were rapidly exchanged between the nucleoplasm and NAC1 NBs in interphase cells.
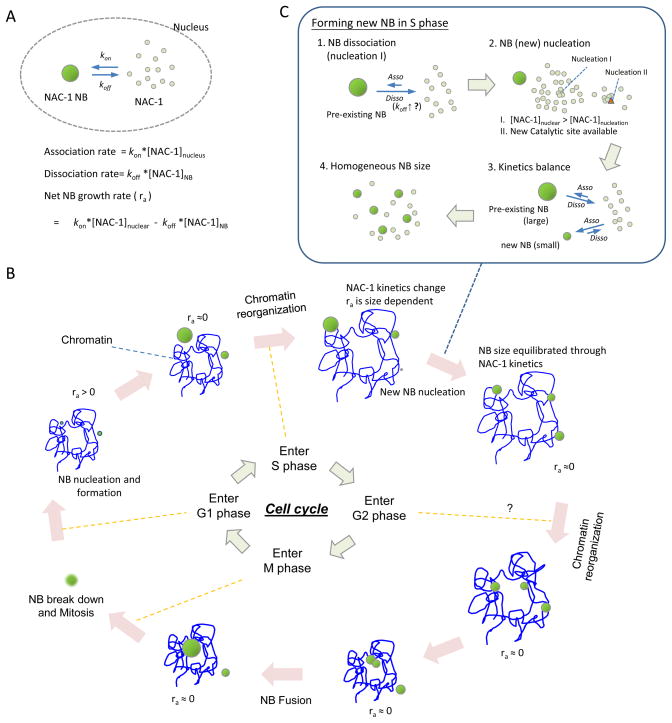
Based on our observations, we propose a simple kinetic model (Figure 5) that helps explain the morphologic change of NAC1 NBs across different phases of the cell cycle. Because mitosis dissolves NBs, the concentration of free NAC1 proteins is above the critical nucleation concentration (i.e., the minimal concentration to form a new NB) in a normal nuclear environment. Hence, NAC1 nucleation in a nucleus can occur immediately after cell division is completed. Thus, at the G1 phase, the NAC1 protein association rate (= kon[NAC1]nuclear, where kon is the association constant of NAC1 proteins in a nucleus) is larger than its dissociation rate (= koff [NAC1]NB, where koff is the dissociation constant of NAC1 from a NAC1 NB). The presence of the [NAC1]NB, is the consequence of assuming [NAC1] at the NAC1 NB surface is a variable to provide a general empirical kinetic model since there is no experimental evidence to suggest the reaction order of a kinetics model. The net association rate, Ra (mole/cm3·s), for NAC1 attachment to a nuclear body can be represented as
Figure 5.
Schematic diagram of NAC1 NB kinetic assembly cycle in cell cycle interphase.
| (3) |
NAC1 NBs can grow continuously until the association/dissociation rate is balanced (Ra = 0). T he growth rate of NB is determined by the molar shell balance on the NB surface, which can be represented as
| (4) |
where r is the radius (cm) of NB, ρ is molar density (mole/cm3) and φ is NAC1 volume ratio in the NB. This expression can be further simplified to
| (5) |
The NB radius at any timerτ, after its formation, is the result of integrating the net association rate, i.e.,
| (6) |
Based on this equation, the size of NAC1 NB is least dependent on the NAC1 volume ratio, molar density, and net association rate of each NB. Therefore, under the assumption that ρ and φ are constant, and each Ra of the NAC1 NBs in the same nucleus is uniform, early NB formation leads to a relatively larger size. This conclusion is in agreement with the observation that the continuation of NB formation throughout the G1 phase until the G1/S transition results in various sizes of NBs.
In S phase, the nucleus undergoes drastic chemical and physical changes associated with chromosome duplication, condensation, and separation (24). These events may modulate the kinetic state of NAC1 proteins, and ultimately may result in morphological transformation of a NAC1 NB into multiple smaller NBs. Two possible molecular mechanisms can be responsible for this phenomenon. One mechanism is that, from G1 to S phase, the increase in the dissociation constant (koff) is a characteristic of a larger NB. This accelerates protein dissociation from NAC1 NBs when they are larger than a critical size. As a result, NAC1 concentration is elevated in the nucleoplasm facilitating new NAC1 NB formation. The second possible mechanism is that the phase change from G1 to S alters the nuclear environment, which directly facilitates new NB formation. With either mechanism, the appearance of a more homogeneous size distribution of NAC1 NBs during S-phase suggests that the newly formed NBs have a positive net association rate, but that the net association rate for pre-existing NBs is negative under the same level of [NAC1]nucleus.
As the cell cycle progresses to G2, at which point chromosome replication ceases, the nuclear environment returns to a state similar to that of G0/G1 phase. This change, along with nuclear re-organization, could encourage the clustering of adjacent NBs and their subsequent fusion (as documented in Figure 3 B). Finally, NAC1 NBs break down in a manner that is plausibly associated with mitosis. This model postulates that the functional role of NAC1 is repeated every cell cycle, and that the regulatory role of NAC1 is distinct during different phases of the cell cycle.
The correlation between the number of NBs and phase of the cell cycle that we have observed for NAC1 NBs is similar to that reported in a previous study on PML NBs by Bazett-Jones and colleagues (25). The number of PML NBs also increases approximately 2 fold during S phase compared to the rest of the cell cycle, i.e., G0/G1 and G2. In contrast to NAC1 NBs, the quantity of PML NBs increased during S phase through nuclear body fission. FRAP experiments on PML NBs have shown that PML-V, one of the components in PML NBs, possesses a very slow exchange rate (26). This suggests that PML-V could play a structural role in building the basic frame of a stable PML NB. In addition, an electron microscopic study showed that PML NBs are physically associated with chromatin (2). Therefore, even though both PML and NAC1 NBs increase in number during S phase, fundamental mechanisms and physical properties could be different. For example, nuclear fission of PML NBs could occur as a result of physical reorganization of the nucleus in the S phase rather than through chemical control as seen in NAC1 NBs.
The high expression level of NAC1 in ovarian cancer cells is potentially linked to cancer cell phenotypes, including the high resistance to paclitaxel (a cancer drug), the elevation of cell proliferation, and the prevention of apoptosis. These phenotypes can, in part, be explained from biochemical studies of the NAC1 protein, which showed that NAC1 protein can suppress the Gadd45 pathway when exogenously expressed in NIH3T3 and RK3E cells (3, 4). When the Gadd45 pathway is suppressed, these cells effectively reduce apoptosis, increase proliferation, and become resistant to paclitaxel. Even though NAC1 NBs are only present when the NAC1 protein is over-expressed, the morphological change of NAC1 NBs throughout different phases of the cell cycle offers us a strong indication that NAC1 NBs could be involved in nuclear activities throughout the cell cycle by controlling the interaction among NAC1 proteins in the nucleoplasm.
The transformation of NAC1 NBs to a diffuse pattern during mitosis is highly interesting. It is possible that the increased concentration of soluble NAC1 is responsible for the completion of mitosis or cytokinesis, as soluble NAC1 proteins may interact with other proteins involved in these processes. To this end, we have recently demonstrated that NAC1 knockdown or prevention of NAC1 homodimerization resulted in a defect in late cytokinesis involving an actin-dependent mechanism (Yap et al., unpublished data). Elucidation of the biological roles of NAC1 during mitosis is critical to understanding the importance of the dynamics of NAC1 NBs, which may serve as a reservoir to prevent untimely increases in soluble NAC1 proteins in non-dividing cells.
In this report, we identify the basic biophysical properties of NAC1 NBs and correlate these properties with the cell cycle. We also propose a kinetic mechanism to explain the correlation. Even though the details of the NAC1 kinetic mechanism are not fully understood, our results firmly support the interpretation that the activity of NAC1 proteins is highly coupled with the progression of the cell cycle. This conclusion also leads to reasonable speculation that NAC1 function in cells is tightly bound to its kinetic mechanism. Therefore, the cell cycle should be considered an important factor in studies evaluating the role of NAC1 in the nucleus. The interaction between NAC1 NBs and cellular proteins involved in mitosis and cytokinesis is likely to be relevant and should be further investigated.
Acknowledgments
This study was supported in part by grants from NIH/NCI CA103937, NIH/NCI U54CA143868, and NIH/NIBIB R01EB004416.
Abbreviations
- FRAP
fluorescence recovery after photobleaching
- NAC1
nucleus accumbens associated protein-1
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama K, Nakayama N, Davidson B, Sheu JJ, Jinawath N, Santillan A, Salani R, Bristow RE, Morin PJ, Kurman RJ, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc Natl Acad Sci U S A. 2006;103:18739–18744. doi: 10.1073/pnas.0604083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinawath N, Vasoontara C, Yap KL, Thiaville MM, Nakayama K, Wang TL, Shih IM. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene. 2009;28:1941–1948. doi: 10.1038/onc.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama K, Nakayama N, Wang TL, Shih Ie M. NAC-1 controls cell growth and survival by repressing transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer Res. 2007;67:8058–8064. doi: 10.1158/0008-5472.CAN-07-1357. [DOI] [PubMed] [Google Scholar]
- 5.Branco MR, Pombo A. Chromosome organization: new facts, new models. Trends Cell Biol. 2007;17:127–134. doi: 10.1016/j.tcb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 7.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 9.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 10.Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 11.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu PH, Nelson N, Tseng Y. A general method for improving spatial resolution by optimization of electron multiplication in CCD imaging. Optics Express. 2010;18:5199–5212. doi: 10.1364/OE.18.005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez RC, Woods RE. Digital image processing. Prentice Hall; Upper Saddle River, N.J: 2002. [Google Scholar]
- 15.Wu PH, Arce SH, Burney PR, Tseng Y. A novel approach to high accuracy of video-based microrheology. Biophys J. 2009;96:5103–5111. doi: 10.1016/j.bpj.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Zollman S, Couderc JL, Laski FA. Retraction. The BTB domain of bric a brac mediates dimerization in vitro. Mol Cell Biol. 1997;17:6772. doi: 10.1128/mcb.17.11.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Lopez-Guisa JM, Ninan N, Weiner EJ, Rauscher FJ, 3rd, Marmorstein R. Overexpression, purification, characterization, and crystallization of the BTB/POZ domain from the PLZF oncoprotein. J Biol Chem. 1997;272:27324–27329. doi: 10.1074/jbc.272.43.27324. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, 3rd, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–5282. [PubMed] [Google Scholar]
- 20.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Zollman S, Couderc JL, Laski FA. The BTB domain of bric a brac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Dhordain P, Albagli O, Ansieau S, Koken MH, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert JP, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 23.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 25.Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci. 2006;119:1026–1033. doi: 10.1242/jcs.02816. [DOI] [PubMed] [Google Scholar]
- 26.Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]