Abstract
Retinoic acid (atRA) signaling is essential for regulating embryonic development, and atRA levels must be tightly controlled in order to prevent congenital abnormalities and fetal death which can result from both excessive and insufficient atRA signaling. Cellular enzymes synthesize atRA from Vitamin A, which is obtained from dietary sources. Embryos express multiple enzymes that are biochemically capable of catalyzing the initial step of Vitamin A oxidation, but the precise contribution of these enzymes to embryonic atRA synthesis remains unknown. Using Rdh10trex-mutant embryos, dietary supplementation of retinaldehyde, and retinol dehydrogenase (RDH) activity assays, we demonstrate that RDH10 is the primary RDH responsible for the first step of embryonic Vitamin A oxidation. Moreover, we show that this initial step of atRA synthesis occurs predominantly in a membrane-bound cellular compartment, which prevents inhibition by the cytosolic cellular retinol binding protein (RBP1). These studies reveal that widely-expressed cytosolic enzymes with RDH activity play a very limited role in embryonic atRA synthesis under normal dietary conditions. This provides a breakthrough in understanding the precise cellular mechanisms that regulate Vitamin A metabolism and the synthesis of the essential embryonic regulatory molecule atRA.
Keywords: Retinol, retinoic acid, retinol dehydrogenase, alcohol dehydrogenase, retinoid metabolism, Vitamin A
Introduction
The Vitamin A derivative, all-trans retinoic acid (atRA), is a potent signaling molecule necessary for patterning, morphogenesis, and organogenesis during embryonic development (Clagett-Dame and DeLuca, 2002; Duester, 2008; Ross et al., 2000; Zile, 1998). The level and tissue distribution of atRA must be tightly regulated during embryogenesis as excessive or insufficient atRA signaling can cause a variety of congenital abnormalities and fetal death (Clagett-Dame and DeLuca, 2002; Duester, 2008; Ross et al., 2000; Zile, 1998). During embryogenesis the precise regulation of atRA levels and tissue distribution are accomplished through a balance of synthesis and degradation. Because atRA signaling controls transcription of numerous essential developmental genes, it is important to gain a clear understanding of the enzymes, binding proteins and cellular conditions that impact the metabolism of this essential compound.
Synthesis of atRA from its precursor Vitamin A (all-trans-retinol; atROL) occurs in a two-step process. First, atROL is oxidized to generate all-trans-retinal (atRAL), a reaction facilitated by an enzyme with retinol dehydrogenase (RDH) activity. Once atRAL is generated, it is subsequently oxidized to form retinoic acid, a reaction facilitated by an enzyme with retinaldehyde dehydrogenase (RALDH) activity. RALDH activity is known to be mediated by the three members of the RALDH family, RALDH1, RALDH2, and RALDH3 (Niederreither et al., 2002a; Niederreither et al., 1997). Each of the RALDH enzymes is expressed in a distinct spatiotemporal pattern during embryogenesis and is required for synthesis of atRA in distinct tissues. RALDH2 is the most widely expressed of the RALDHs, and is most crucial for embryonic atRA synthesis, as Raldh2−/− embryos die around E10.5 and lack axial rotation, neural tube closure, heart looping and cardiac atrial chamber formation (Niederreither et al., 1999). Raldh1−/− mice exhibit normal survival to adulthood, are fertile, and exhibit no obvious defects (Fan et al., 2003). Raldh3−/− mice exhibit mild ocular defects and lethality at birth due to a blockage of the nasal passages, which prevents efficient respiration (Dupe et al., 2003).
Whereas the enzymes responsible for the RALDH oxidation of atRAL to atRA have been definitively known for some time, the identity of the enzymes essential for the oxidation of Vitamin A (atROL) to atRAL during embryogenesis have remained elusive. Biochemically, members of two large enzyme families are capable of catalyzing RDH activity in vitro: the microsomal short-chain dehydrogenase/reductases (SDR) and the cytosolic medium-chain alcohol dehydrogenases (ADH) (Duester, 2008; Pares et al., 2008). Because several ADHs are widely or ubiquitously expressed during embryogenesis it was originally proposed that the initial oxidation of Vitamin A during retinoic acid synthesis was carried out ubiquitously by ADH family members (Ang et al., 1996a; Ang et al., 1996b; Duester et al., 2003; Molotkov et al., 2002). However, our recent finding that a point mutation in Rdh10 (Rdh10trex) impairs RA synthesis and causes numerous embryonic abnormalities resulting in embryonic lethality between E10.5–E13.0 demonstrates clearly that RDH10 plays a predominant role in the first reaction of the two-step process of atRA synthesis (Sandell et al., 2007). The phenotype of the Rdh10trex mutant is consistent with the known phenotypes of retinoid deficiency, although not quite as severe as the phenotype of Raldh2−/− embryos. This is likely due to the reduced enduring level of atRA produced in the Rdh10trex mutant embryos, which may result from RDH activity of other enzymes or from residual activity of the destabilized RDH10 protein produced by the point mutation.
RDH10 and ADH family members could each conceivably contribute to embryonic Vitamin A metabolism. RDH10 is expressed in a spatiotemporally-restricted pattern within the embryo, coinciding closely with sites of RALDH gene expression (Cammas et al., 2007; Romand et al., 2008; Sandell et al., 2007). ADH1 (a class I enzyme) and ADH7 (a class IV enzyme with robust RDH activity in vitro) are expressed in distinct tissue-specific patterns in mouse embryos while ADH5 (a class III ADH enzyme with low RDH activity) is found broadly expressed throughout the embryo (Duester et al., 1995; Molotkov et al., 2002). Given that RDH10 and ADH family members are each present within embryonic tissues and are each capable of RDH activity in vitro, we sought to understand why ADHs have little or no physiologically-relevant RDH activity in embryos and what distinguishes RDH10, making it the dominant RDH in atRA metabolism during embryogenesis.
The precise enzymatic role of RDH10 in embryonic atRA synthesis has not been fully demonstrated, although our previous work showed that RDH10 catalyzes the oxidation of atROL to atRAL in vitro (Wu et al., 2002). Since that reaction is the first step of atRA synthesis, and Rdh10trex mutants have a deficiency in atRA synthesis, we proposed that RDH10 is necessary to produce sufficient levels of atRAL to serve as substrate for RALDH enzymes that catalyze the second step of atRA synthesis. However, it was surprising that the widely expressed ADHs, which have RDH activity, did not compensate for the loss of RDH10. The inability of ADHs to compensate for the loss of RDH10 could indicate that ADHs do not normally function in the capacity of an RDH in vivo despite in vitro evidence to the contrary. An alternative explanation could be that RDH10 is essential for both the first and second oxidative steps of atRA synthesis. Although SDRs and ALDHs utilize completely distinct modes of catalysis and the alcohol oxidation is chemically reversible, whereas aldehyde oxidation is not, we decided to systematically explore all possible developmental reasons behind the embryonic requirement for RDH10. Thus, in order to fully explore the precise enzymatic activity of RDH10 it was necessary to determine the capacity of RDH10 to catalyze the oxidation of atRAL.
The importance of resolving the precise role of RDH10 in embryonic atRA synthesis is underscored by studies in RARE-lacZ reporter mice that have indicated that there are sites of RALDH-independent atRA synthesis in the developing embryo, which include areas in the forebrain, hindbrain, spinal cord, neural tube and heart (Mic et al., 2002; Niederreither et al., 2002b). Such areas of “RALDH-independent” activity could be accounted for by unidentified RALDH(s). Alternatively, several studies have identified non-RALDH enzymes with the capability of synthesizing atRA in vitro, including members of the Cytochrome P450 (CYP) family (CYP1A1, CYP1A2, CYP3A6, and CYP1B1) (Chambers et al., 2007; Ding et al., 2001; Roberts et al., 1992; Tomita et al., 1996; Tomita et al., 1993), and these enzymes may account for RALDH-independent atRA synthesis. CYP1B1 is particularly interesting, because it possesses the unique enzymatic capacity to catalyze the stepwise oxidation of atROL to atRA in vitro, and thus combines the function of a RDH and a RALDH into one enzyme (Chambers et al., 2007). However, it remains to be determined if CYP1B1 can directly synthesize atRA from atROL in vivo. The CYP enzymes generally have much lower activity than the RALDHs, but they may contribute to sites of RALDH-independent atRA synthesis during embryogenesis. Most interestingly, RDH10 is expressed in at least two areas which have RALDH-independent atRA synthesis, including the floor plate of the neural tube and the hindbrain (Romand et al., 2008; Sandell et al., 2007), raising the possibility that RDH10 may contribute to RALDH-independent atRA synthesis, either on its own or in concert with CYPs.
Another aspect of atRA metabolism that is not well understood is the sub-cellular location of various reactions and intermediates. The precursor molecule atROL is derived initially from the maternal diet, distributed to the embryo through the circulation and transported into embryonic cells via the cell surface receptor STRA6 (Bouillet et al., 1997; Kawaguchi et al., 2007). The second reaction in atRA synthesis, the conversion of atRAL to atRA by the RALDH family of enzymes takes place in the cytosol. Thus, it is reasonable to expect that the initial RDH reaction, the conversion of atROL to atRAL, would also occur within the cytosol. However, the finding that RDH10, which is membrane-bound, is the dominant embryonic RDH, and the fact that ADHs, which are cytosolic, do not apparently contribute substantially to the process, led us to hypothesize that the first enzymatic step of atRA synthesis occurs not in the cytosol but, instead, takes place primarily in a membrane compartment. We therefore sought to define the sub-cellular location of the first step of atRA synthesis and determine the relative contribution of ADHs and RDH10 to this important process.
In the present study we investigated the role of RDH10 in the stepwise synthesis of atRA during embryogenesis. We demonstrated that RDH10 does not catalyze conversion of atRAL to atRA in vitro and rescued Rdh10trex mutants by supplementation of the maternal diet with atRAL. Taken together these data demonstrate that RDH10 is essential only for the first step of embryonic atRA synthesis. We measured total embryonic RDH activity and showed that membrane-bound RDH activity is abolished in Rdh10trex mutants, demonstrating that RDH10 is the primary enzyme carrying out RDH activity in the membrane compartment within embryos.
We show that RDH10 is unable to oxidize atROL bound to the cytosolic cellular retinol-binding protein (RBP1) in vitro and demonstrate this inhibitory protein is expressed during the stages of development when RDH10 is active. We also demonstrate that ADH7, which exhibits high RDH activity on atROL in aqueous solution, is unable to oxidize atROL that has been incorporated into liposomes. Taken together, these data suggest that RDH10 oxidizes a RBP1-free pool of atROL present in cellular membranes in vivo, and that the ADH enzymes are unable to easily access membrane pools of atROL. Thus, we speculate that ADHs do not contribute to embryonic synthesis of atRA because they may function only in the aqueous cytosolic compartment where inhibitory RBP1 is present. Importantly, the data indicate that that the first step in the two-step embryonic atRA synthesis is carried out predominantly by RDH10 in a membrane-bound environment protected from inhibitory RBP1.
Materials and Methods
Construction of Expression Vectors
A human RALDH2 mammalian expression vector was constructed by PCR amplification from a commercially-available RALDH2 cDNA clone (CLONE ID 30471325, Open Biosystems, Huntsville, AL) using the following primers: forward, 5’- ATGGATCCGCCACCATGGCCTCGCTGCAGCTC-3’ containing a BamHI site (underline) and reverse, 5’- ACGAATTCGGAGTTCTTCTGGGGGATCTTCA-3’ containing an EcoRI site (underline). The PCR product was digested with BamHI and EcoRI and inserted into the pcDNA6/V5/His mammalian expression vector (Invitrogen, Carlsbad, CA, U.S.A.) to express RALDH2-V5-His protein. The bacterial expression vector pQE30-CRBP-His was kindly provided by Dr. Kris Palczewski at Case Western Reserve University, and the bacterial expression vector pet19b-CRALBP-His was a kind gift from Dr. John Crabb at the Cleveland Clinic Foundation (Crabb et al., 1998). The pcDNA6-RDH10 mammalian expression vector was described previously (Takahashi et al., 2008; Wu et al., 2002).
A human ADH7 full-length mammalian expression vector was constructed by PCR amplification from a commercially-available ADH7 cDNA clone corresponding to NCBI reference sequence NM_000673 (CLONE ID LIFESEQ3287393, Open Biosystems, Huntsville, AL) using the following primers: forward, 5’-TACAAAGACGATGACGACAAGATGGGCACTGCTGGAAAAG-3’ containing a partial Flag tag sequence (italic) and reverse, 5’-ACGAATTCTCAAAACGTCAGGACCGTTCGAATG-3’ containing and EcoRI site (underline). After the first PCR amplification, the ADH7 PCR product was inserted into pGEM-TEasy vector (Promega) by TA-cloning. Then a second PCR was performed using the original reverse primer and a new forward primer in order to add residues of the Flag tag and a BamHI site: forward, 5’-AAGGATCCGCCACCATGGACTACAAAGACGATGACGACAAG-3’ containing a BamHI site (underline) and a full Flag tag sequence (italic). Then the PCR product was digested with BamHI and EcoRI and inserted into the pcDNA3.1(+) mammalian expression vector (Invitrogen, Carlsbad, CA, U.S.A.) to express ADH7 with an N-terminal Flag tag.
In vitro RALDH Activity Assays
Whole cell lysates were prepared after COS1 cells were either transiently transfected with either pcDNA6-RDH10 or pcDNA6-RALDH2-His expression vectors using FuGENE 6 reagent (Roche, Indianapolis, IN, U.S.A.). At 48 hrs after transfection, cells were lysed by sonication in either RDH activity buffer (0.1 M sodium phosphate pH 7.4) or RALDH Activity Buffer (20 mM HEPES pH 8.5 with 150 mM KCl, 1 mM EDTA and 2mM DTT) and stored at −80°C.
All the following procedures were performed under dim red light. To measure RALDH activity in vitro, the whole cell lysates of COS1 cells expressing either recombinant RDH10 or RALDH2 were used as a source of the enzymes. A total of 200 µg of cell lysate proteins were suspended in a final volume of 200 µl of either RDH activity buffer or RALDH activity buffer containing 0.5% BSA, and 2 mM nicotinamide adenine dinucleotide (NAD+) or nicotinamide adenine dinucleotide phosphate (NADP+). The reaction was initiated by adding 2 µl of atRAL dissolved in DMF (dimethyl formamide) at a final concentration of 5 µM. The reaction mixtures were incubated at 37°C for 30 min with gentle agitation. The reaction was terminated by the addition of 200 µl of methanol. To extract non-polar retinoids, 4 µl of 0.1M sodium hydroxide was added followed by 600 µl of hexane. After removal of non-polar retinoids, retinoic acid was extracted by the addition of 16 µl of 5M HCl and 600 µl of hexane. Samples were then dried under argon and dissolved in methanol for high performance liquid chromatography (HPLC) analysis (Waters 515 HPLC pump and 2996 Photodiode Array Detector; Waters Corporation, Milford, MA) using a reverse phase 3.5 µm column (Waters C18; Waters Corporation, Milford, MA). Retinoids were separated by a gradient of 50% to 100% methanol in water plus 0.1% trifluoroacetic acid. The peak of each retinoid isomer was identified based on its absorption spectrum and retention time on the column compared to pure retinoid standards. The enzymatic activity was calculated from the area of the atRA peak using synthetic purified atRA as a standard for calibration with Empower software (Waters Corporation, Milford, MA). Kinetic parameters were calculated using EnzFitter program (Biosoft, Cambridge, UK).
Rescue of Rdh10trex-mutant Embryos with atRAL Supplementation
Timed matings were performed to define E0 as 12:00 a.m. of the morning when a mating semen plug was observed. For rescues experiments, atRAL was dissolved in corn oil and administered approximately every 12 hours by oral gavage to pregnant females from E7.25 up to E12.25 at a final concentration of 12.5 mg atRAL/kg maternal body weight. Mock-gavaged animals received corn oil alone, and all gavage doses were given at a volume of 100 µl. Embryos were collected at various stages and genotyped to identify the Rdh10trex allele point mutation as described previously (Sandell et al., 2007). For morphological analyses, embryos were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin. Alcian blue cartilage staining and β-galactosidase staining of embryos were performed as previously described (Sandell et al., 2007).
Embryonic RDH Activity Assays
Whole embryos were resuspended in 4 volume equivalents of 0.1M sodium phosphate pH 7.4 with 0.25 M sucrose and homogenized by sonication. Lysates were centrifuged at 10,000×g for 10 minutes to remove cell debris. The supernatant was centrifuged at 100,000×g for 1 hour to separate cytosolic and membrane fractions. The pellet (membranes) was resuspended in RDH activity buffer, analyzed for protein concentration by Bradford analysis, and frozen at −80°C until time of analysis.
Under dim red light, a total of 25 µg or 50 µg of embryo membrane proteins were suspended in a final volume of 200 µl of RDH activity buffer containing 1% BSA, and 1 mM NAD+ or NADP+. The reaction was initiated by adding 2 µl of 11,12-3H-atROL (1 mCi/ml, 52 Ci/mmol, PerkinElmer) dissolved in DMF at a final concentration of 0.2 µM. After incubation at 37°C for 1 hour with gentle agitation, the reaction was terminated by the addition of 300 µl of methanol, and retinoids were extracted with 300 µl of hexane for normal phase HPLC analysis coupled to Radiomatic 610TR Flow Scintillation Analyzer (PerkinElmer). The peak of each retinoid isomer was identified based on the retention time on the column compared to pure retinoid standards, and enzymatic activity was calculated from the area of the 3H-atRAL peak using synthetic purified 3H-atRAL as a standard for calibration with Radiomatic 610TR software.
Nickel-affinity chromatography
Bacterial cells expressing Rbp1-His were suspended in cell lysis buffer (50 mM NaH2PO4, 300 mM NaCl pH 8.0) supplemented with 1 mM PMSF, 25 µg/ml of aprotinin, leupeptin, and pepstatin, and 10 µg/ml DNase and sonicated on ice. The cell lysates were incubated with Ni resin for 2 h at 4°C. After adsorption, unbound proteins were removed, and the beads were washed with cell lysis buffer containing 20 mM imidazole. Bound proteins were eluted with cell lysis buffer containing 250 mM imidazole, and then imidazole was removed by dialysis into PBS.
RDH10 Activity Assays with RBP1
The pcDNA6-RDH10 expression vector was transfected into COS1 cells using FuGENE 6 reagent. At 48 hours after transfection, cells were lysed by sonication in RDH activity buffer. The cell lysate was centrifuged at 100,000×g for 1 hour to separate the cytosolic and membrane fractions. The supernatant was removed, and the pellet was resuspended in RDH activity buffer and briefly centrifuged at 18,000×g to wash away remaining cytosolic proteins. The final membrane fraction was resuspended in RDH activity buffer, analyzed for protein concentration by Bradford analysis, and stored at −80° C.
Under dim red light, a total of 32 µg of membrane proteins were suspended in a final volume of 200 µl of RDH activity buffer containing 1% BSA and 1 mM NADP+ in the absence or presence of increasing concentrations of purified recombinant RBP1-His. The reaction was initiated by adding 2 µl of atROL dissolved in DMF at a final concentration of 5.5 µM. The reaction mixtures were incubated at 37°C for 30 minutes with gentle agitation. The reaction was terminated by the addition of 300 µl of methanol, and retinoids were extracted with 300 µl of hexane for HPLC analysis using a normal phase 5 µm column (Lichrosphere SI-60; Alltech, Deerfield, IL) (Wu et al., 2004). The enzymatic activity was calculated from the area of the atRAL peak using synthetic purified atRAL as standard for calibration with Empower software.
ADH7 Activity Assays
COS1 cells were transfected with pcDNA3.1-Flag-ADH7 expression vector using FuGENE 6 reagent. At 48 hours post-transfection, cells were lysed in RDH activity buffer by 5 cycles of freeze-thawing, and then cytosolic fractions were prepared by centrifuging at 18,000×g for 10 minutes. The pellets were discarded, and the supernatant (cytosolic fraction) was analyzed for protein concentration by Bradford analysis, and stored at −80°C.
Under dim red light, a total of 0.7 µg of cytosolic proteins were suspended in a final volume of 200 µl of RDH activity buffer containing 1% BSA, and 1 mM NAD+. The reaction was initiated by adding either 2 µl of either atROL dissolved in DMF or atROL incorporated into liposomes, both at a final concentration of 5.5 µM atROL in the reaction. The reaction mixtures were incubated at 37°C with gentle agitation for the specified incubation periods. The reactions were terminated by the addition of 300 µl of methanol, and retinoids were extracted and analyzed by HPLC as described above for RDH10 activity assays.
Results
RDH10 is Only Necessary for the First Step of Embryonic atRA Synthesis
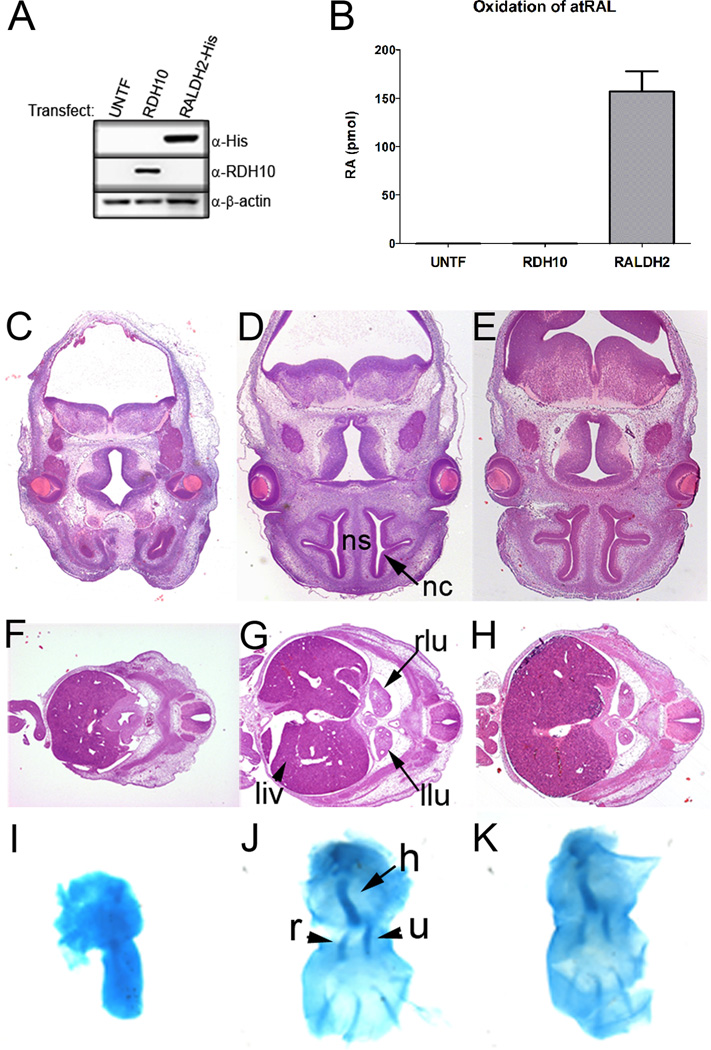
The developmental importance of resolving the precise role of RDH10 in embryonic atRA synthesis is highlighted by the observations that widely-expressed ADHs do not compensate for the loss of RDH10 in Rdh10trex mutants (Sandell et al., 2007), and that the developing embryos has RALDH-independent sites of atRA synthesis (Mic et al., 2002; Niederreither et al., 2002b). We therefore tested the capacity of RDH10 to catalyze the second oxidation of atRAL to atRA in vitro. Recombinant RDH10 was expressed in COS1 cells, and cell lysates were prepared to assay RDH10 activity with atRAL substrate and either NAD+ or NADP+ cofactor. Neither untransfected nor RDH10-expressing cell lysates produced any detectable atRA (Fig. 1 A and B). In contrast, cell lysates expressing recombinant RALDH2 produced a significant amount of atRA under the same conditions. These data indicate that as anticipated RDH10 lacks the ability to oxidize atRAL in vitro and is therefore not likely involved in the second oxidative step of atRA synthesis.
Figure 1. RDH10 is strictly required for the first step of embryonic atRA synthesis.
(A) Western blot of COS1 whole cell lysates shows expression of either RDH10 or RALDH2-His following transfection (UNTF, untransfected cells). (B) COS1 whole cell lysates shown in (A) were used for in vitro atRAL oxidation assays. The graph shows the amount of atRA generated from each cell lysate and represents the mean +/− s.d. from three independent experiments. (C–K) Embryos were collected at E12.5. The left (C, F, I) and middle (D, G, J) columns show embryos processed after mock supplementation which are either Rdh10trex or wild type, respectively. The right column (E, H, K) shows Rdh10trex embryos processed after maternal supplementation with all-trans-retinal. H&E-stained transverse sections are shown at the level of: (C–E) the frontonasal process, dorsal top, ventral bottom. (ns, nasal septum; nc, nasal cavity); (F–H) the liver and lung bud, dorsal right, ventral left. (liv, liver; rlu, right lung bud; llu, left lung bud). (I–K) Alcian blue cartilage staining of forelimbs (h, humerus; r, radius; u, ulna). These histological analyses show that atRAL supplementation is sufficient to rescue craniofacial, lung, liver and forelimb development in Rdh10trex embryos.
To address the question of whether the essential contribution of RDH10 to embryonic atRA synthesis is limited to the oxidization of atROL to atRAL, thus providing a substrate for RALDH enzymes, we tested if supplementation of the maternal diet with atRAL could prevent developmental malformations in Rdh10trex embryos. Following atRAL or mock diet supplementation, embryos were collected at E12.5 and histological analysis was performed to evaluate the morphological development of the lungs, liver, forelimbs and craniofacial anatomy. Rdh10trex embryos typically exhibit a midline facial cleft with agenesis of the nasal septum and nasal cavity (Fig. 1 C). The eyes are malformed and submerged within head tissues (Fig. 1C). More caudally, there is little to no evidence for lung bud formation and growth, and the liver is hypoplastic (Fig. 1 F). The forelimbs are always truncated with ulna agenesis (Fig. 1I). Comparisons with wild-type embryos (Fig. 1 D, G, J) demonstrate that atRAL supplementation to a large extent rescues the lung, liver, forelimb, and craniofacial abnormalities that are otherwise present in mock-rescued Rdh10trex embryos supplemented with corn oil alone (Fig. 1 C–K). Rescued embryos (Fig. 1 E, H, K) exhibit a clear nasal septum in the midline of the face, flanked bilaterally by well developed nasal cavities and the eyes are no longer occluded within head mesenchyme (Fig. 1 E). The atRAL supplementation rescued lung bud induction and growth of both the lungs and liver (Fig. 1 H). Forelimb development and patterning were also restored. Not only were the forelimbs in rescued embryos equivalent in size to controls, but agenesis of the ulna was prevented (Fig. 1 K). The finding that atRAL supplementation is sufficient to restore development in Rdh10trex embryos establishes that the essential role of RDH10 during embryogenesis is in catalysis of the first step of Vitamin A metabolism and atRA synthesis. This disproves the previously-accepted theory that the tissue specificity of atRA synthesis from atROL is dictated solely by RALDH activity (Duester et al., 2003; Molotkov et al., 2002). Both RDH and RALDH activities are critical for the spatiotemporal metabolism of Vitamin A and generation of atRA. Furthermore our results demonstrate that there is an absence of widespread, redundant RDH activity in the developing embryo, despite the widespread expression of ADHs.
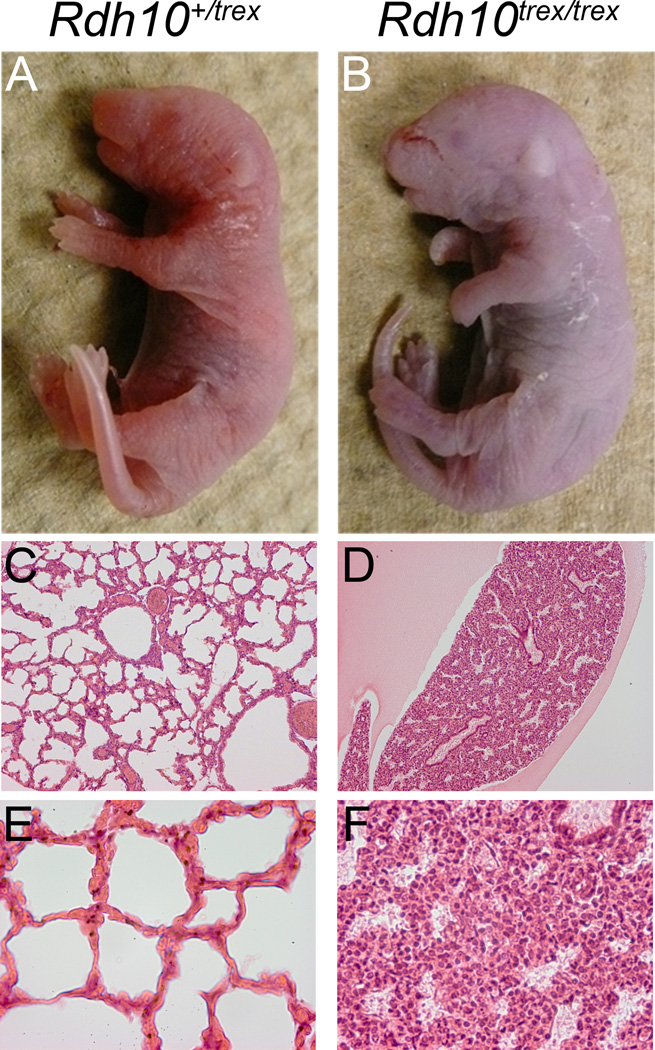
We extended atRAL supplementation for the remainder of gestation, which rescued Rdh10trex mutants to birth (Fig. 2 A and B). The size of Rdh10trex pups appeared normal, however they died shortly thereafter. Despite evidence for normal eye, limb and lung development and growth, as well as proper nasal cavity formation and the absence of facial clefting during embryogenesis, it is apparent that postnatally the lungs never inflate (Fig. 2 C–F). It is unclear why Rdh10trex pups do not breathe and fail to survive, but it is likely a consequence of incomplete nasal slit formation and connection to the nasal cavity or alternatively a problem in diaphragm development. These residual phenotypes reflect the challenges of dietary supplementation regimes in rescuing developmental anomalies and result from variations in dosage or tissue distribution of supplemental atRAL relative to endogenously produced atRAL.
Figure 2. atRAL supplementation rescues Rdh10trex mutants to birth.
Pregnant females were supplemented with atRAL by oral gavage every 12 hours as described in material and methods, beginning at E7.25 and extending up until birth. (A and B) Pups were collected and photographed directly after birth. (C–F) H&E stained transverse sections through the lung tissue are shown at 10X (C and D) and 40X (E and F) magnification. Note the expanded lung tissue morphology, indicative of lung inflation, in the Rdh10+/trex pup. In contrast, the Rdh10trex/trex lung tissue remains condensed.
RDH10 Accounts for the Majority of Embryonic RDH Activity
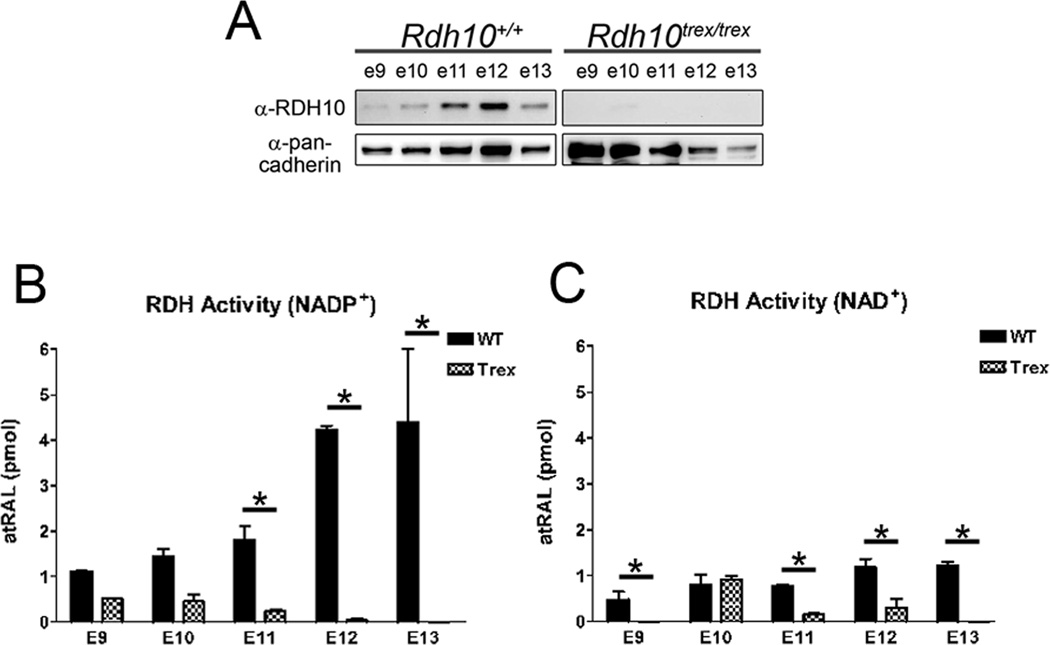
To understand why enzymes with RDH activity cannot fully compensate for the loss of RDH10 during embryogenesis, we evaluated the contribution of RDH10 to total embryonic RDH activity. In Rdh10trex embryos, RDH10 protein was not detected at any of the developmental stages examined (Fig. 3 A), indicating that the point mutation in the Rdh10trex allele severely reduces RDH10 protein stability to undetectable levels. Thus, we compared the RDH activity of Rdh10trex embryos to wild-type embryos to determine the contribution of RDH10 to total embryonic RDH activity. Embryonic whole cell lysates had very little RDH activity, due to the neutralizing effects of reducing factors in the cytosol (data not shown). Therefore, to measure membrane-bound RDH activity in the absence of cytosolic factors, membrane fractions were prepared from whole embryos. Membrane fractions displayed RDH activity with both NAD+ and NADP+ cofactors, although NADP+-dependent RDH activity was 2 to 8-fold higher than NAD+-dependent activity, which indicates that the majority of membrane-bound RDH activity is NADP+-dependent (Fig. 3 B and C). Rdh10trex mutants have a significant reduction in RDH activity from E9 to E13 compared to wild-type. The most sizeable difference occurs in NADP+-dependent RDH activity at E12 and E13, the stage at which wild-type embryos have the highest level of activity, while in contrast Rdh10trex embryos have nearly undetectable activity (Fig. 3 B). We have previously shown that RDH10 prefers NADP+ cofactor in vitro (Wu et al., 2002; Wu et al., 2004), which explains why NADP+-dependent activity is severely reduced in Rdh10trex mice. These data demonstrate that RDH10 accounts for the majority of membrane-bound RDH activity in the developing embryo.
Figure 3. Rdh10trex embryos have a significant reduction in total RDH activity.
(A) Whole embryo membrane fractions were prepared for RDH activity assays and analyzed by Western blotting (20 µg of protein per lane) using an anti-RDH10 antibody and an anti-pan-cadherin antibody as a loading control. (B and C) RDH activity was measured with either (B) NADP+ or (C) NAD+ cofactor. The bar graphs represent the mean +/− s.d. from at least three independent experiments (*p<0.05 by two-way ANOVA).
Consistent with our previous study, we show that a small amount of microsomal RDH activity remains in Rdh10trex mutant embryos from E9 to E11 (Fig. 3 B and C). This residual RDH activity could be due to a small amount of persistent RDH10 activity stemming from the presence of a low level of RDH10 protein that is beyond the limits of detection by immunoblotting. Alternatively, the residual RDH activity may be due to the function of one or more other membrane-bound RDHs, the identity of which is unknown. Interestingly, RDH10-independent activity is completely absent at E12 and E13, indicating that RDH10 is the only membrane-bound RDH expressed during this stage of development, which may account for the timely death of Rdh10trex embryos at E12.5/E13. These results strongly suggest that RDH10 is essential for embryonic RDH activity, not simply because it generates pools of atRAL in specific tissues, but rather because it accounts for the majority of the RDH activity throughout the developing embryo. In contrast to previous dogma, these data show that an NADP+-dependent membrane-bound RDH10 is the primary embryonic RDH, whereas NAD+-dependent cytosolic ADHs do not contribute significantly to embryonic RDH activity.
Holo-RBP1 is Not a Substrate for RDH10
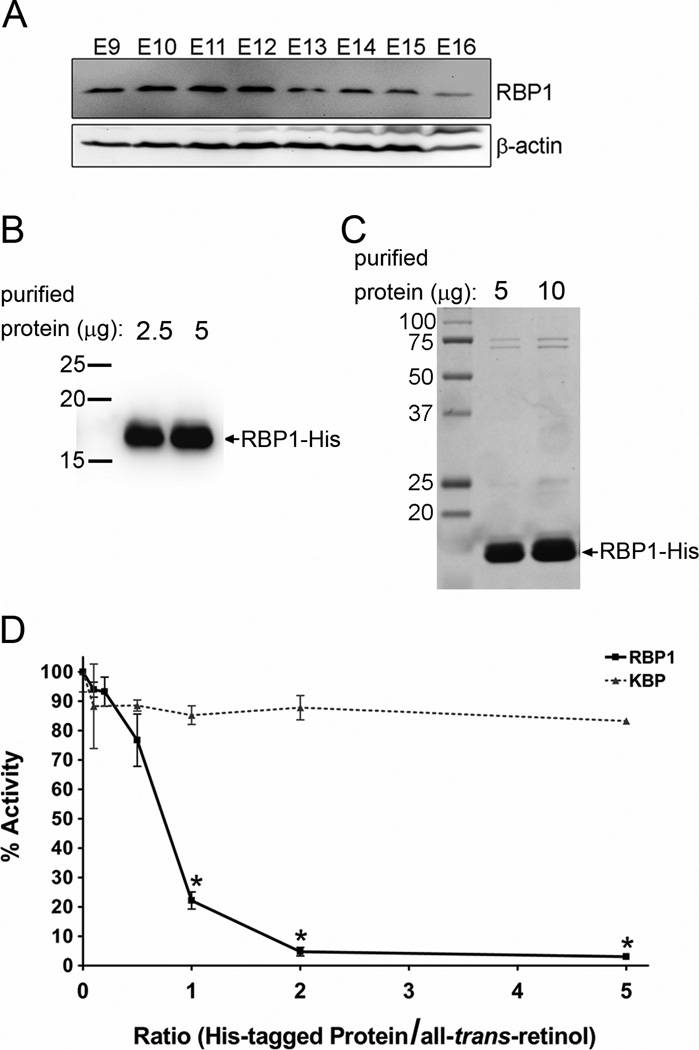
Cellular retinol binding protein (RBP1) is a cytosolic protein which binds to atROL with high affinity and specificity (MacDonald and Ong, 1987; Noy, 2000). Consequently, it has been proposed that physiologically-relevant RDHs must be able to utilize holo-RBP1 as substrate, yet all RDHs studied to date lack the ability to efficiently oxidize holo-RBP1 (Belyaeva et al., 2008; Gallego et al., 2006; Kedishvili et al., 1998; Napoli, 1999). We performed immunoblotting on whole embryo homogenates and found that RBP1 is expressed as early as E9 and up to E16 (Fig. 4 A). This demonstrates that RBP1 expression coincides temporally with RDH10 expression (Fig. 3 A) and ongoing embryonic atRA synthesis. This also suggests that RDH10 must be able to function in the presence of RBP1.
Figure 4. RDH10 does not utilize holo-RBP1 as substrate.
(A) Western blotting with an anti-RBP1 antibody was performed on whole embryo homogenates (50 µg protein per lane) from stage E9 to E16. RBP1 expression was detected at all developmental stages form E9 to E16. (B and C) Recombinant His-RBP1 was expressed in E. Coli and then purified by nickel affinity chromatography for use in in vitro RDH activity assays. (B) Purified His-RBP1 (2.5 µg in lane 1 and 5 µg in lane 2) was analyzed by western blotting using an anti-RBP1 antibody. (C) The purity of the His-RBP1 was evaluated by Coomassie staining. Lanes 1 and 2 were loaded with 5 µg and 10 µg of purified His-RBP1, respectively. (D) RDH activity assays were performed with atROL substrate in the absence or presence of increasing concentrations of either purified recombinant His-RBP1 or His-KBP (non-retinoid-binding protein, negative control). The graph represents the mean +/− s.d. from at least three independent experiments (*p<0.05 by student’s t-test analysis).
To determine if RDH10 can oxidize atROL from holo-RBP1, RDH10 activity was measured in vitro with increasing concentrations of purified recombinant His-tagged RBP1 (Fig. 4 B and C). As a negative control, the assay was performed with a purified recombinant His-tagged protein that does not bind to retinoids, kallikrein-binding protein (KBP). At a 1:1 molar ratio of RBP1 to atROL, atRAL production was reduced to 22% of the amount of atRAL produced in the absence of RBP1, and at a 2:1 molar ratio of RBP1:atROL only 5% activity remained (Figure 4 D). This remaining RDH10 activity is due to dissociated free atROL which is in equilibrium with RBP1, and is consistent with a recently published report (Belyaeva et al., 2008). In contrast, non-specific inhibition by KBP only reduced RDH10 activity to 90% of its normal level, even at a 5:1 molar ratio of KBP:atROL. These data demonstrate that RBP1 inhibits RDH10 activity, and thus RDH10 does not utilize holo-RBP1 as substrate.
ADH7 Cannot Efficiently Oxidize atROL from Phospholipid Membranes
RDH10 is a membrane-bound protein, and we have previously shown that binding to membrane is essential for RDH10 activity (Takahashi et al., 2009). The substrate atROL is a relatively hydrophobic molecule, which is localized in cellular membranes in vivo (Noy and Xu, 1990). Binding to RBP1 provides atROL with a hydrophobic binding pocket within the aqueous cytosolic compartment, but it does not preclude accumulation of RBP1-free atROL in cellular membranes. We speculate that cellular membranes contain a pool of RBP1-free atROL which allows membrane-bound RDH10 to oxidize atROL in vivo, despite the cytosolic expression of RBP1. We hypothesize that cytosolic ADHs do not contribute significantly to embryonic RDH activity because cytosolic atROL is in the form of holo-RBP1, which inhibits ADH activity, and ADHs have limited access to the RBP1-free pools of atROL in cellular membranes.
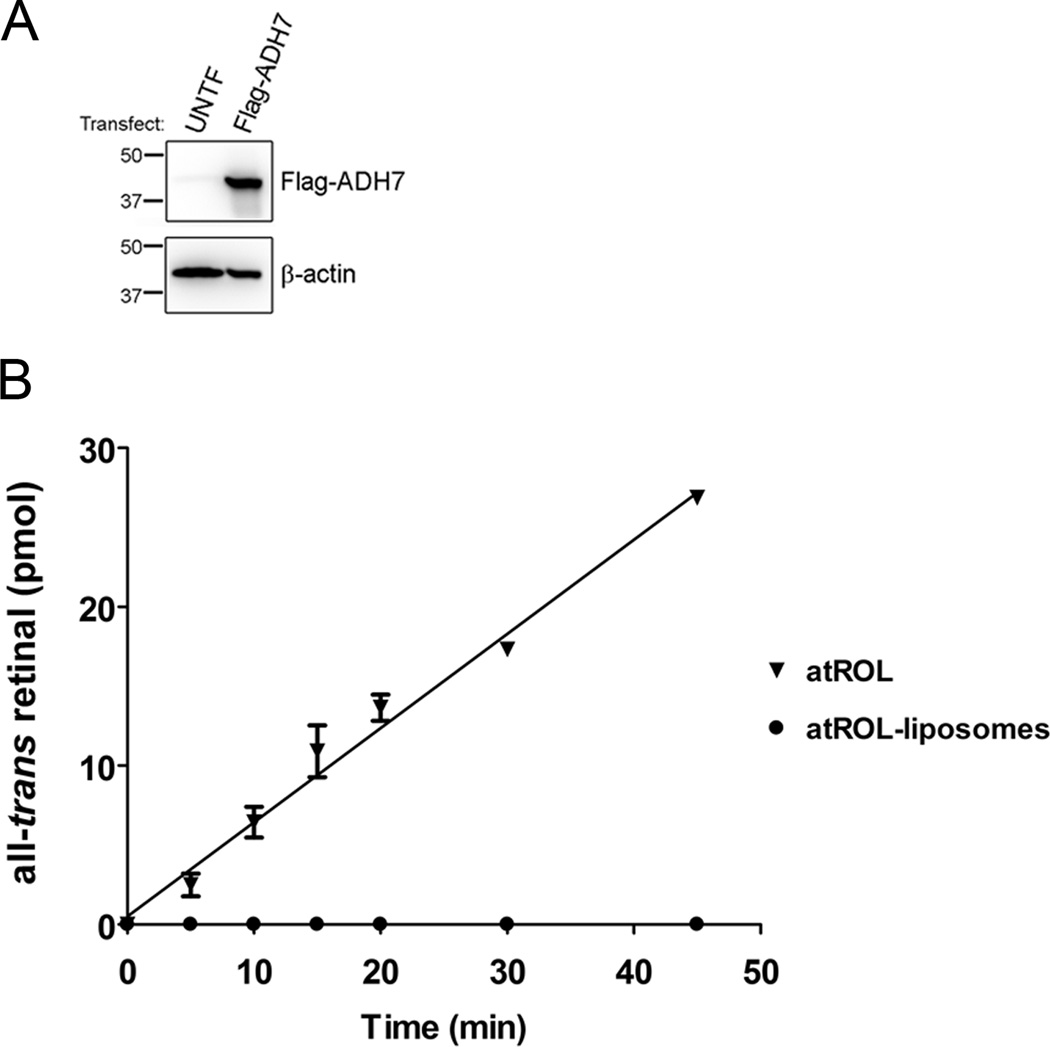
To test this theory, we measured recombinant ADH7 activity in vitro using equimolar concentrations of either atROL in aqueous solution or atROL incorporated into phospholipid membranes (liposomes) as substrate. With increasing reaction time, ADH7 generated increasing amounts of atRAL when provided with atROL in aqueous solution (Fig. 5 B). In contrast, atRAL generation was undetectable over the entire incubation period when ADH7 was provided with an equimolar amount of atROL incorporated in liposomes. This demonstrates that ADH7 has limited access to atROL localized in membranes, and supports the idea that ADHs are unable to compensate for the loss of RDH10 because ADHs have limited access to RBP1-free atROL in cellular membranes.
Figure 5. ADH7 cannot efficiently oxidize atROL from liposomal membranes.
(A) Western blot of COS1 cell lysates shows expression of Flag-ADH7 following transfection. (B) ADH7 activity was quantified using either atROL in aqueous solution or atROL incorporated into liposomes as substrate. The graph represents the mean +/− s.d. from at least three independent experiments.
Discussion
Retinoic acid is a potent signaling molecule necessary for patterning, morphogenesis, and organogenesis during embryonic development. Retinoic acid is synthesized from Vitamin A (all-trans-retinol; atROL) via a two-step oxidation process, and our results clearly demonstrate that the first and second enzymatic steps of embryonic Vitamin A metabolism and atRA synthesis are largely confined to separate cellular compartments. This may play a critical role in regulating the precise spatiotemporal control of retinoic acid synthesis during embryogenesis, which is important because both excessive and insufficient atRA signaling can result in congenital abnormalities and fetal death.
Although the RALDH enzymes responsible for the second oxidation reaction of atRAL to atRA have been definitively known for some time, the identity of the enzymes essential for the first oxidation reaction of Vitamin A (atROL) to atRAL during embryogenesis have remained elusive. Our study reveals that membrane-bound RDH10 is the primary embryonic RDH essential for the oxidation of atROL during the generation of atRA in the developing embryo. Furthermore we demonstrated that the membrane localization of RDH10 is functionally important to allow RDH10 to oxidize RBP1-free pools of atROL stored in the cellular membrane. This explains why widely-expressed cytosolic ADHs cannot compensate for the loss of RDH10, because RBP1 inhibits oxidation of cytosolic pools of atROL and ADHs cannot efficiently extract and oxidize RBP1-free atROL from membranes.
Our present study, combined with previous studies showing ADH-deficient mice develop normally unless reared under conditions of either Vitamin A deficiency or excess, strongly suggests that cytosolic ADHs do not contribute significantly to the oxidation of atROL during normal embryonic development. However, ADHs may be important in the event of gestational exposure to excessive levels of either atROL or ethanol, both of which can be toxic in high doses. This theory is supported by previous studies which have shown that Adh1−/− and Adh4−/− mice have delayed metabolic clearance of excess doses of atROL and ethanol (Deltour et al., 1999a, b). Since cytosolic RBP1 binds atROL at a 1:1 molar ratio, excessive levels of atROL that are in molar excess of RBP1 may limit RBP1-mediated inhibition of ADH activity and thereby increase ADH-mediated oxidation of atROL under such circumstances. It will be interesting in future studies to test this genetically in Rdh10/Adh compound mutants.
Conclusions
In summary, this study reveals that membrane-bound RDH10 is the primary embryonic RDH essential for atRA generation in the developing embryo, and that the first and second enzymatic steps of embryonic atRA synthesis are largely confined to separate cellular compartments. Membrane localization of RDH10 is important to allow RDH10 to oxidize RBP1-free pools of atROL stored in the cellular membrane. Widely-expressed cytosolic ADHs cannot compensate for the loss of RDH10, because RBP1 inhibits oxidation of cytosolic pools of atROL and ADHs cannot efficiently extract and oxidize RBP1-free atROL from membranes. These breakthrough studies provide a foundation for expanding our understanding of the cellular mechanisms which modulate atRA metabolism and signaling during embryogenesis.
Highlights.
Retinal supplementation rescues embryonic atRA synthesis in Rdh10Trex mutants
Oxidation of retinol to retinal occurs primarily in a membrane-bound compartment
RDH10 accounts for the majority of retinol oxidation during embryogenesis
Membrane localization of RDH10 prevents inhibition by cytosolic RBP1
We resolve why cytosolic ADHs have a limited contribution to embryonic atRA synthesis
Acknowledgements
We thank the Imaging Core Facility at the Oklahoma Medical Research Foundation, in particular Ben Fowler, Aaron Dockins, and Didier Nuno for meticulously processing histological specimens. This study was supported by NIH grants EY018659, EY012231, EY019309 awarded to JM, a grant (P20RR024215) from the National Center for Research Resources awarded to JM, and a grant from the Oklahoma Center for the Advancement of Science and Technology awarded to KF. Research in the Trainor laboratory is supported by the Stowers Institute for Medical Research and the National Institute of Dental and Craniofacial Research (RO1 DE 016082).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Krysten M. Farjo, Email: Krysten-farjo@ouhsc.edu.
Gennadiy Moiseyev, Email: Gennadiy-moiseyev@ouhsc.edu.
Olga Nikolaeva, Email: Olga-nikolaeva@ouhsc.edu.
Lisa L. Sandell, Email: LLS@Stowers.org.
Paul A. Trainor, Email: PAT@Stowers.org.
References
- Ang HL, Deltour L, Hayamizu TF, Zgombic-Knight M, Duester G. Retinoic acid synthesis in mouse embryos during gastrulation and craniofacial development linked to class IV alcohol dehydrogenase gene expression. J Biol Chem. 1996a;271:9526–9534. doi: 10.1074/jbc.271.16.9526. [DOI] [PubMed] [Google Scholar]
- Ang HL, Deltour L, Zgombic-Knight M, Wagner MA, Duester G. Expression patterns of class I and class IV alcohol dehydrogenase genes in developing epithelia suggest a role for alcohol dehydrogenase in local retinoic acid synthesis. Alcohol Clin Exp Res. 1996b;20:1050–1064. doi: 10.1111/j.1530-0277.1996.tb01946.x. [DOI] [PubMed] [Google Scholar]
- Belyaeva OV, Johnson MP, Kedishvili NY. Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase. J Biol Chem. 2008;283:20299–20308. doi: 10.1074/jbc.M800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva OV, Korkina OV, Stetsenko AV, Kim T, Nelson PS, Kedishvili NY. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry. 2005;44:7035–7047. doi: 10.1021/bi050226k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev. 1997;63:173–186. doi: 10.1016/s0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- Cammas L, Romand R, Fraulob V, Mura C, Dolle P. Expression of the murine retinol dehydrogenase 10 (Rdh10) gene correlates with many sites of retinoid signalling during embryogenesis and organ differentiation. Dev Dyn. 2007;236:2899–2908. doi: 10.1002/dvdy.21312. [DOI] [PubMed] [Google Scholar]
- Chambers D, Wilson L, Maden M, Lumsden A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development. 2007;134:1369–1383. doi: 10.1242/dev.02815. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, DeLuca HF. The role of vitamin A in mammalian reproduction and embryonic development. Annu Rev Nutr. 2002;22:347–381. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- Crabb JW, Chen Y, Goldflam S, West K, Kapron J. Methods for producing recombinant human cellular retinaldehyde-binding protein. Methods Mol Biol. 1998;89:91–104. doi: 10.1385/0-89603-438-0:91. [DOI] [PubMed] [Google Scholar]
- Deltour L, Foglio MH, Duester G. Impaired retinol utilization in Adh4 alcohol dehydrogenase mutant mice. Dev Genet. 1999a;25:1–10. doi: 10.1002/(SICI)1520-6408(1999)25:1<1::AID-DVG1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Deltour L, Foglio MH, Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice. Overlapping roles of Adh1 and Adh4 in ethanol clearance and metabolism of retinol to retinoic acid. J Biol Chem. 1999b;274:16796–16801. doi: 10.1074/jbc.274.24.16796. [DOI] [PubMed] [Google Scholar]
- Ding J, Ichikawa M, Furukawa A, Tomita S, Tanaka K, Ichikawa Y. Low synthesis of retinoic acid due to impaired cytochrome P450 1a1 expression in mouse xeroderma pigmentosum fibroblasts. Int J Biochem Cell Biol. 2001;33:603–612. doi: 10.1016/s1357-2725(01)00037-1. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G, Ang HL, Deltour L, Foglio MH, Hayamizu TF, Zgombic-Knight M. Class I and class IV alcohol dehydrogenase (retinol dehydrogenase) gene expression in mouse embryos. Adv Exp Med Biol. 1995;372:301–313. doi: 10.1007/978-1-4615-1965-2_36. [DOI] [PubMed] [Google Scholar]
- Duester G, Mic FA, Molotkov A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem Biol Interact. 2003;143–144:201–210. doi: 10.1016/s0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci U S A. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo KM, Moiseyev G, Takahashi Y, Crouch RK, Ma JX. The 11-cis-retinol dehydrogenase activity of RDH10 and its interaction with visual cycle proteins. Invest Ophthalmol Vis Sci. 2009;50:5089–5097. doi: 10.1167/iovs.09-3797. [DOI] [PubMed] [Google Scholar]
- Gallego O, Belyaeva OV, Porte S, Ruiz FX, Stetsenko AV, Shabrova EV, Kostereva NV, Farres J, Pares X, Kedishvili NY. Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids. Biochem J. 2006;399:101–109. doi: 10.1042/BJ20051988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F, Jang GF, Imanishi Y, Driessen CA, Matsumura M, Nelson PS, Palczewski K. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J Biol Chem. 2002;277:45537–45546. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasus-Jacobi A, Ou J, Bashmakov YK, Shelton JM, Richardson JA, Goldstein JL, Brown MS. Characterization of mouse short-chain aldehyde reductase (SCALD), an enzyme regulated by sterol regulatory element-binding proteins. J Biol Chem. 2003;278:32380–32389. doi: 10.1074/jbc.M304969200. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- Kedishvili NY, Gough WH, Davis WI, Parsons S, Li TK, Bosron WF. Effect of cellular retinol-binding protein on retinol oxidation by human class IV retinol/alcohol dehydrogenase and inhibition by ethanol. Biochem Biophys Res Commun. 1998;249:191–196. doi: 10.1006/bbrc.1998.9105. [DOI] [PubMed] [Google Scholar]
- MacDonald PN, Ong DE. Binding specificities of cellular retinol-binding protein and cellular retinol-binding protein, type II. J Biol Chem. 1987;262:10550–10556. [PubMed] [Google Scholar]
- Marchette LD, Thompson DA, Kravtsova M, Ngansop TN, Mandal MN, Kasus-Jacobi A. Retinol dehydrogenase 12 detoxifies 4-hydroxynonenal in photoreceptor cells. Free Radic Biol Med. 2009;48:16–25. doi: 10.1016/j.freeradbiomed.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, Fan X, Deltour L, Foglio MH, Martras S, Farres J, Pares X, Duester G. Stimulation of retinoic acid production and growth by ubiquitously expressed alcohol dehydrogenase Adh3. Proc Natl Acad Sci U S A. 2002;99:5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim Biophys Acta. 1999;1440:139–162. doi: 10.1016/s1388-1981(99)00117-1. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Fraulob V, Garnier JM, Chambon P, Dolle P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002a;110:165–171. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Fraulob V, Chambon P, Dolle P. Retinaldehyde dehydrogenase 2 (RALDH2)- independent patterns of retinoic acid synthesis in the mouse embryo. Proc Natl Acad Sci U S A. 2002b;99:16111–16116. doi: 10.1073/pnas.252626599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348(Pt 3):481–495. [PMC free article] [PubMed] [Google Scholar]
- Noy N, Xu ZJ. Thermodynamic parameters of the binding of retinol to binding proteins and to membranes. Biochemistry. 1990;29:3888–3892. doi: 10.1021/bi00468a014. [DOI] [PubMed] [Google Scholar]
- Pares X, Farres J, Kedishvili N, Duester G. Medium- and short-chain dehydrogenase/reductase gene and protein families : Medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell Mol Life Sci. 2008;65:3936–3949. doi: 10.1007/s00018-008-8591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ES, Vaz AD, Coon MJ. Role of isozymes of rabbit microsomal cytochrome P-450 in the metabolism of retinoic acid, retinol, and retinal. Mol Pharmacol. 1992;41:427–433. [PubMed] [Google Scholar]
- Romand R, Kondo T, Cammas L, Hashino E, Dolle P. Dynamic expression of the retinoic acid-synthesizing enzyme retinol dehydrogenase 10 (rdh10) in the developing mouse brain and sensory organs. J Comp Neurol. 2008;508:879–892. doi: 10.1002/cne.21707. [DOI] [PubMed] [Google Scholar]
- Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol Rev. 2000;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007;21:1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Moiseyev G, Farjo K, Ma JX. Characterization of key residues and membrane association domains in RDH10. Biochem J. 2008 doi: 10.1042/BJ20080812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Moiseyev G, Farjo K, Ma JX. Characterization of key residues and membrane association domains in retinol dehydrogenase 10. Biochem J. 2009;419:113–122. doi: 10.1042/BJ20080812. 111 p following 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Okuyama E, Ohnishi T, Ichikawa Y. Characteristic properties of a retinoic acid synthetic cytochrome P-450 purified from liver microsomes of 3-methylcholanthrene-induced rats. Biochim Biophys Acta. 1996;1290:273–281. doi: 10.1016/0304-4165(96)00030-x. [DOI] [PubMed] [Google Scholar]
- Tomita S, Tsujita M, Matsuo Y, Yubisui T, Ichikawa Y. Identification of a microsomal retinoic acid synthase as a microsomal cytochrome P-450-linked monooxygenase system. Int J Biochem. 1993;25:1775–1784. [PubMed] [Google Scholar]
- Wu BX, Chen Y, Chen Y, Fan J, Rohrer B, Crouch RK, Ma JX. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest Ophthalmol Vis Sci. 2002;43:3365–3372. [PubMed] [Google Scholar]
- Wu BX, Moiseyev G, Chen Y, Rohrer B, Crouch RK, Ma JX. Identification of RDH10, an All-trans Retinol Dehydrogenase, in Retinal Muller Cells. Invest Ophthalmol Vis Sci. 2004;45:3857–3862. doi: 10.1167/iovs.03-1302. [DOI] [PubMed] [Google Scholar]
- Zile MH. Vitamin A and embryonic development: an overview. J Nutr. 1998;128:455S–458S. doi: 10.1093/jn/128.2.455S. [DOI] [PubMed] [Google Scholar]