Abstract
DBP, the founding member of the PAR leucine zipper transcription factor family, is expressed according to a robust daily rhythm in the suprachiasmatic nucleus and several peripheral tissues. Previous studies with mice deleted for the Dbp gene have established that DBP participates in the regulation of several clock outputs, including locomotor activity, sleep distribution, and liver gene expression. Here we present evidence that circadian Dbp transcription requires the basic helix–loop–helix–PAS protein CLOCK, an essential component of the negative-feedback circuitry generating circadian oscillations in mammals and fruit flies. Genetic and biochemical experiments suggest that CLOCK regulates Dbp expression by binding to E-box motifs within putative enhancer regions located in the first and second introns. Similar E-box motifs have been found previously in the promoter sequence of the murine clock gene mPeriod1. Hence, the same molecular mechanisms generating circadian oscillations in the expression of clock genes may directly control the rhythmic transcription of clock output regulators such as Dbp.
Keywords: PAR factors, circadian transcription, clock, molecular oscillator, output
In mammals, many physiological processes are subject to circadian regulation. These include sleep–wake cycles, body temperature, heartbeat, blood pressure, endocrine functions, renal activity, and liver metabolism (for review, see Schibler and Lavery 1999). During the past few years impressive progress has been made in the elucidation of molecular mechanisms generating circadian oscillations in animal systems. In both Drosophila and mammals, a feedback loop in gene expression is thought to drive circadian oscillations (Rosbash et al. 1996; Brown and Schibler 1999; Dunlap 1999; Young 1999). In the mouse, the negative limb of this feedback loop is believed to be comprised of mPer1, mPer2, and mPer3, three murine homologs of the Drosophila period gene; Cry1 and Cry2, two homologs of the Drosophila cryptochrome gene; and perhaps mTim, a homolog of the Drosophila timeless gene. The positive limb of this feedback loop contains the two basic helix-loop-helix–PAS (bHLH) proteins CLOCK and BMAL1 in both Drosophila and mammals. The currently held model for the mammalian feedback loop poses that one or multiple mPer and Cry genes are activated by a CLOCK/BMAL1 heterodimer via an E-box motif (Kume et al. 1999). The three PER proteins encoded by these genes undergo interactions with themselves and with the two CRY proteins. Once these heteromultimeric complexes reach a critical concentration, they repress transcription of mPer genes, probably by attenuating the activation potential of the CLOCK/BMAL1 heterodimer. Upon decay of PER and/or CRY proteins, repression is relieved, and a new wave of mPer and Cry expression can be initiated. The oscillation of gene expression generated by this feedback loop has a period length of about 24 hr and can thus account for the circadian timing observed in overt rhythms in physiology and behavior. Initially, it was believed that oscillations of clock proteins were present only in specialized pacemaker neurons within the suprachiasmatic nucleus (SCN). Recently, however, molecular clocks similar to those operating in SCN neurons have been uncovered in peripheral cell types (Zylka et al. 1998) and even in immortalized fibroblast cell lines kept in tissue culture (Balsalobre et al. 1998).
An important question to be answered is how the central negative feedback loop drives circadian expression of clock-controlled genes. In this investigation we approached this issue by studying Dbp, a clock-controlled gene whose expression oscillates with a very high circadian amplitude (Wuarin and Schibler 1990). DBP (albumin D-element binding protein; Mueller et al. 1990) is the founding member of the PAR family of basic leucine zipper (bZip) transcription factors. Other members of this family include TEF (thyroid embryonic factor) (Drolet et al. 1991), its avian ortholog VBP (vitellogenin promoter-binding protein) (Iyer et al. 1991), and HLF (hepatocyte leukemia factor) (Hunger et al. 1992; Inaba et al. 1992). All of these proteins share high amino acid sequence similarities within a amino-terminal activation domain, a PAR domain rich in proline and acidic amino acid residues, and a carboxy-terminal moiety encompassing the bZip region necessary for DNA binding and dimerization. In vitro all PAR bZip proteins avidly bind the consensus DNA recognition sequence 5′-RTTAYGTAAY-3′ as homo- or heterodimers (Falvey et al. 1996).
In rat and mouse liver the expression of all three PAR bZip proteins is subject to strong circadian regulation, peak and trough levels being reached in the early evening and morning, respectively (Wuarin and Schibler 1990; Falvey et al. 1995; Fonjallaz et al. 1996; Lopez-Molina et al. 1997). In the case of Dbp the amplitude of circadian mRNA oscillation can largely account for the daily amplitude in protein oscillation (Fonjallaz et al. 1996). The mRNA accumulation oscillates not only in peripheral tissues such as liver, but also in neurons of the SCN, believed to harbor the central circadian pacemaker (Ralph et al. 1990). Moreover, run-on experiments in isolated nuclei (Wuarin and Schibler 1990; Lavery and Schibler 1993) and physical mapping of nascent RNA chains (Wuarin and Schibler 1994) suggest that circadian transcription plays a pivotal role in rhythmic DBP expression.
Genetic loss-of-function experiments have begun to illuminate some physiological roles of DBP (Lopez-Molina et al. 1997). Although this transcription factor is not essential for embryonic development and survival during adulthood, it is involved in the control of several circadian outputs. Mice homozygous for a Dbp null allele differ from wild-type mice in the period length and the amplitude of circadian locomotor activity (Lopez-Molina et al. 1997), in several electroencephalogram (EEG) parameters of sleeping behavior (Franken et al. 2000), and in the circadian expression of some liver genes, such as the ones specifying steroid 15α hydroxylase, coumarin 7 hydroxlase, and cholesterol 7α hydroxylase (Lavery et al. 1999; L. Lopez-Molina, D.J. Lavery, and U. Schibler, unpubl.). However, in contrast to mice with mutations in Clock (Antoch et al. 1997; King et al. 1997b) or mPer2 (Zheng et al. 1999) and to Cry1 and Cry2 double knock-out mice (van der Horst et al. 1999), mice with a deletion of Dbp still display rhythmic wheel-running behavior when examined under constant dark conditions. It thus has been concluded that DBP is a participant in output pathways rather a central clock component (Lopez-Molina et al. 1997).
Because the high variation of daily Dbp mRNA accumulation was found to be controlled mainly at the transcriptional level (Wuarin and Schibler 1990, 1994; Lavery and Schibler 1993), we wished to use this system to examine how the central oscillator mechanism drives circadian transcription of output genes. To identify upstream regulators of Dbp we localized putative circadian regulatory elements within the Dbp gene by mapping DNase I hypersensitive sites in mouse liver chromatin. Here we report the identification of E-box motifs within DNase I hypersensitive regions located in the first and second introns and show that these elements can bind CLOCK, an essential component of the molecular oscillator. Moreover, in Clock mutant mice circadian Dbp expression is abolished in the liver and severely dampened in the SCN. These experiments suggest a direct coupling of Dbp expression to the molecular oscillator in both SCN neurons and peripheral cell types.
Results
Intragenic regions are required for circadian Dbp expression
To identify possible functions of Dbp in the circadian time-keeping system, two different strains of Dbp−/− mice, Dbpdim and Dbpnull, have been generated (Lopez-Molina et al. 1997). Strain Dbpdim contains a neomycin-resistance cassette insertion into exon 4 and encodes a truncated and unstable protein devoid of a leucine zipper dimerization domain (Lopez-Molina et al. 1997). Although the protein issued by this mutant allele did not accumulate to detectable levels in liver nuclei, the Dbpdim mRNA displayed a similarly robust circadian accumulation in both SCN neurons and hepatocytes (Lopez-Molina et al. 1997; L. Lopez-Molina and U. Schibler, unpubl.). It thus was concluded that DBP was not critical for the regulation of its own circadian expression. In the second Dbp mutant strain, Dbpnull, most intragenic sequences that encompass the entire open reading frame (ORF) of Dbp were replaced by a bacterial lacZ cassette (Fig. 1A) to monitor tissue specific gene expression. However, in situ hybridization revealed that the Dbp–lacZ fusion gene, contrary to the wild-type Dbp allele, was expressed in only a small subset of brain structures (e.g., hippocampus and dentate gyrus, see Lopez-Molina et al. 1997). Importantly, little if any Dbp–lacZ mRNA could be detected in the SCN even at times during which Dbp mRNA accumulates to massive amounts in wild-type animals. Taken together, the observations described above strongly suggested that intragenic Dbp sequences are essential for high-amplitude circadian expression of Dbp mRNA in the SCN.
Figure 1.
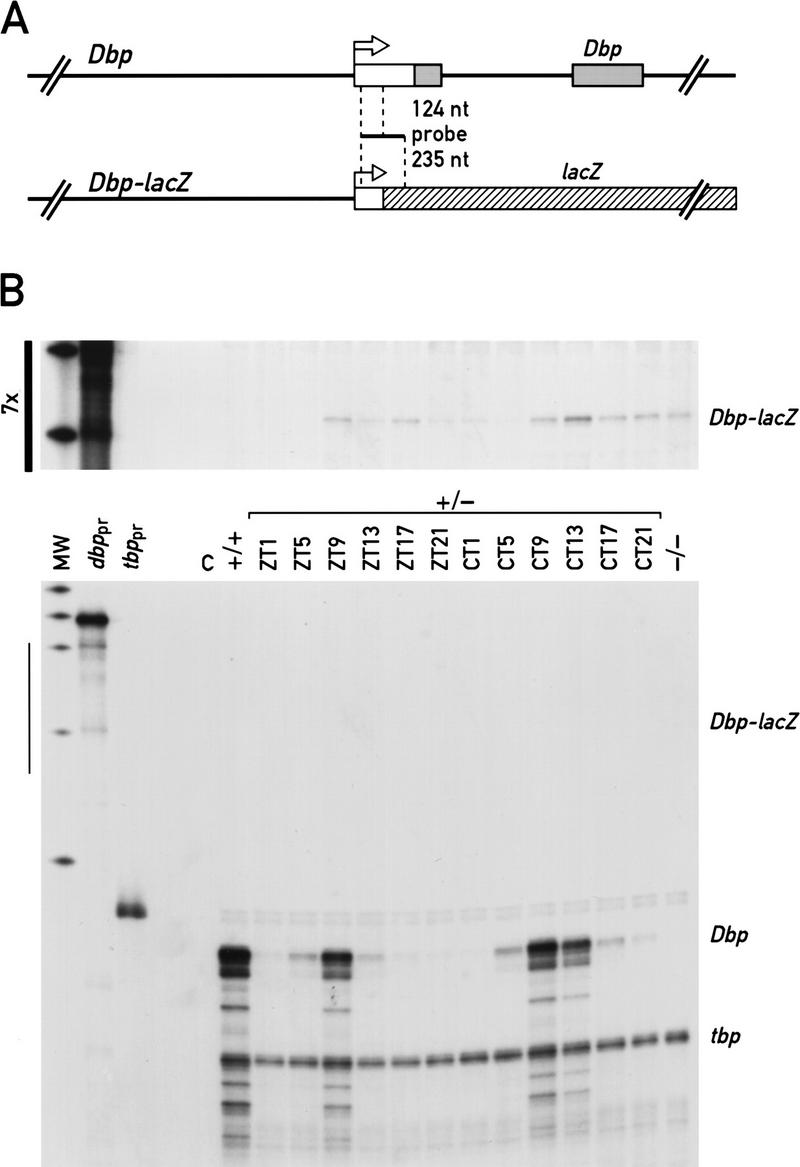
Intragenic regulatory sites are important for high-level expression of Dbp in mouse liver. (A) Schematic representations of the Dbp wild-type and Dbp–lacZ fusion alleles. The position of the antisense RNA probe used in the ribonuclease protection experiments shown in B is indicated. Upon hybridization with Dbp mRNA or Dbp–lacZ mRNA 124 nucleotides or 235 nucleotides, respectively, are protected from ribonuclease digestion. (B) Ribonuclease protection experiments were performed with whole-cell liver RNA from Dbp+/− mice extracted at the indicated time points from animals kept under 12:12 hr light/dark conditions (ZT) or animals kept for 10 days under constant conditions (CT). The RNA samples were hybridized to specific probes for Dbp–lacZ mRNA and tbp mRNA. tbp mRNA served as an internal control for a transcript that is constitutively expressed throughout the day. Autoradiographs were exposed for 15 hr (bottom) or 105 hr (top, see bar at left), and the resulting bands were quantified with a scanner. (MW) Molecular weight marker; (Dbppr, tbppr) undigested probes for Dbp–lacZ and tbp, respectively; (C) negative control with an equivalent amount of yeast tRNA; (+/+, −/−) whole-cell liver RNAs from wild-type or Dbp−/− mice, respectively, at ZT9.
To examine the importance of intragenic sequences for rhythmic transcription in peripheral organs, the levels of mRNAs issued by either Dbp or the Dbp–lacZ were compared by RNase protection experiments with whole-cell liver RNA from heterozygous mice containing one each of these alleles. As shown in Figure 1B, the wild-type Dbp allele produces at least 100-fold more transcripts than the Dbp–lacZ gene. This is unlikely to be due to an intrinsically low stability of Dbp–lacZ mRNA, as this transcript accumulates to relatively high levels in certain brain structures (see above) and other fusion mRNAs carrying an identical lacZ moiety are readily detected in liver and other peripheral organs (F. Damiola, N. Preitner, and U. Schibler, unpubl.). Hence, intragenic enhancer sequences that have been deleted in constructing the Dbp–lacZ allele appear to play an important role for high-amplitude circadian Dbp transcription in both the SCN and peripheral organs.
The mapping of DNase I hypersensitive chromatin regions within the Dbp locus
The binding of transcription factors to specific cis-acting DNA sequences frequently causes distortions of the surrounding chromatin, which in turn leads to DNA sites that are particularly sensitive to the attack of nucleases such as DNase I. Hence, nuclease hypersensitive sites are often diagnostic for cis-acting regulatory sequences, such as promoter, enhancer, and silencer elements (see Boyes and Felsenfeld 1996, and references therein).DNase I hypersensitive sites within the Dbp locus were determined in liver nuclei harvested at intervals of four hr around the clock. As shown in Figure 2A, seven hypersensitive regions can be identified in the Dbp gene. Region 1, located at the end of intron 2, and region 3, located at the end of exon 2, seem to be present at all times of day (Fig. 2B). In contrast, regions 4 and 5 around positions +900 and +1, respectively, are much more susceptible to DNase I digestions at times of day when Dbp is maximally transcribed [Zeitgeber time (ZT) 7 to ZT 11; Fig. 2C]. The DNase I sensitivity of region 7, around position −900 (Fig. 2C), and the upper band of region 2 (Fig. 2B), located within the intron 2, oscillate in a similar manner, albeit with a lower amplitude. Region 6, around position −400, behaves in a more complex fashion, with some bands being present in phase and others in antiphase to the regulation of Dbp transcription (Fig. 2C). Very similar temporal patterns of DNase I hypersensitive sites were observed under light-entrained and constant dark conditions, and the sites were also detected in DNA isolated from nuclei obtained from the kidney (data not shown). No additional hypersensitive sites are detected in the 3′ moiety of Dbp or downstream of the polyadenylation site (Fig. 2D). Circadian hypersensitive sites were found to correlate tightly with the daily phase of Dbp transcription, and when the same DNA was probed for the constitutively active C/ebpα gene (see Wuarin and Schibler 1990), no daytime-related changes in DNase I hypersensitive sites could be detected (Fig. 2E).
Figure 2.
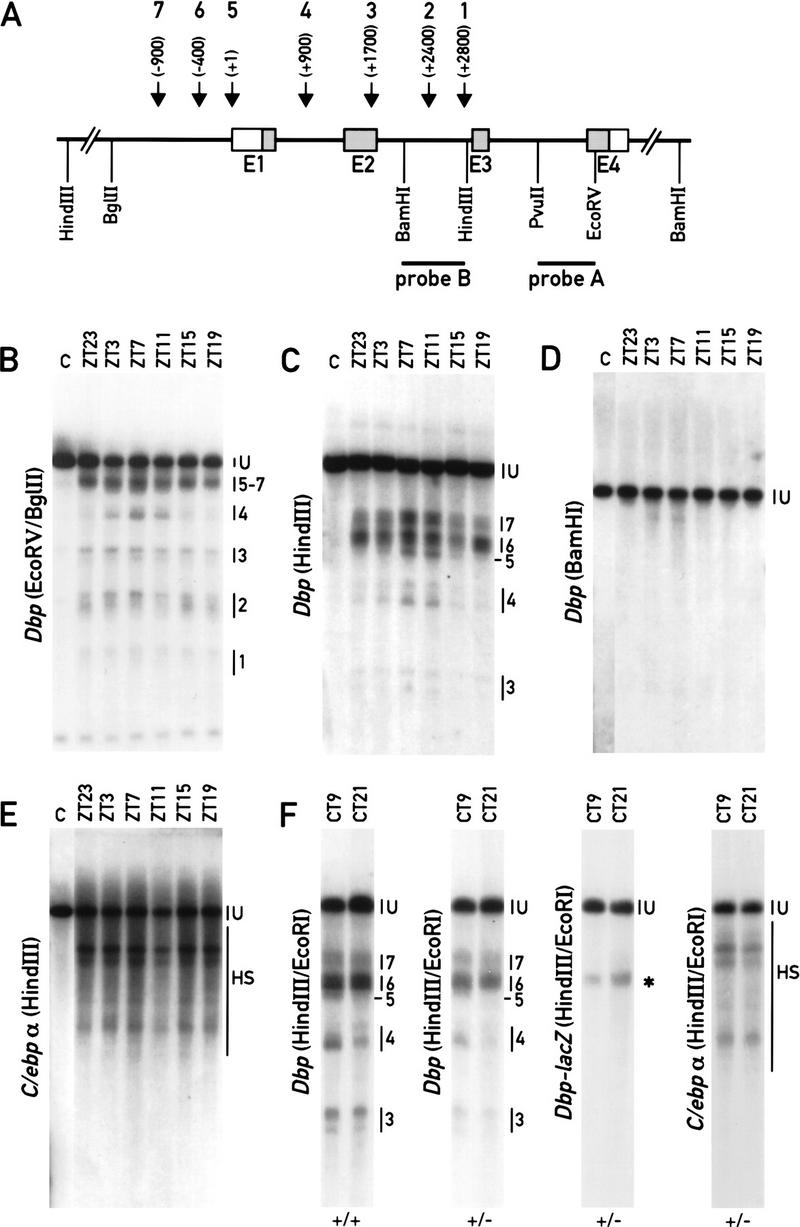
DNase I hypersensitive sites within the Dbp locus. (A) Schematic representation of the Dbp gene with its four exons (E1–E4) and three introns. The positions of the DNA hybridization probes and the restriction fragments used in the indirect end-labeling experiments are indicated. The approximate positions of the seven DNase I hypersensitive regions detected in B and C are depicted on top of the cartoon. (B) Mapping of DNase I hypersensitive sites starting from exon 4. Equal aliquots of liver nuclei harvested at the indicated times were treated with DNase I (ZT23 to ZT19) or without DNase I (lane C, derived from ZT7). After exhaustive digestion with EcoRV and BglII, the fragments were visualized using probe A. (C) Fine mapping of DNase I hypersensitive sites in the 5′ moiety of the Dbp locus. The DNA was digested with HindIII and probed with probe B. (D) Mapping of DNase I hypersensitive sites in the 3′ moiety of the Dbp locus. The same DNA was digested to completion with BamHI and processed as described in C. A hypersensitive site, mapping to about +500 from the polyadenylation site of Dbp, was not reproducibly observed in other experiments. (E) DNase I hypersensitive sites within the constitutive C/ebpα locus. DNase I/HindIII-digested DNAs were probed with a NcoI/HindIII fragment encompassing the ORF of the intron-less C/ebpα gene. (U) Full-length genomic fragment. (F) Comparison of DNase I hypersensitive sites within the Dbp wild-type allele (panels 1 and 2) and the Dbp–lacZ knockout allele (panel 3). Panel 4 shows DNase I hypersensitive sites within the C/ebp locus as a control. (+/+) Dbp wild-type mice; (+/−) Dbp heterozygous mice. The asterisk marks the position of the hypersensitive region present in the Dbp–lacZ allele.
As shown in Figure 1, Dbp–lacZ is transcriptionally nearly inactive in spite of the fact that it contains all of the Dbp 5′-flanking sequences. We thus wanted to determine whether the DNase I hypersensitive sites (5–7) can also been observed within the Dbp–lacZ promoter region. Figure 2F shows that a single weak hypersensitive region at position −400 could be observed in this region, whereas the hypersensitive sites 5 (transcriptional start site) and 7 (−900) are lacking. Hence, the formation of these two hypersensitive regions may require interactions with intragenic regulatory sequences.
Intronic DNase I hypersensitive regions encompass E-box motifs that bind CLOCK
Because intragenic enhancer sequences appear to be essential for robust circadian Dbp expression (see above), we decided to investigate the hypersensitive regions located downstream of the cap site in greater detail. A precise mapping using internal size markers (data not shown) and subsequent sequence inspection of these regions revealed potential protein–DNA-binding sites located within these sites. We paid particular attention to the presence of E-box motifs, as this type of cis-acting element has already been shown to drive circadian period expression in Drosophila (Hao et al. 1997, 1999) and has been proposed to mediate circadian expression of the mouse period1 (Gekakis et al. 1998) and vasopressin genes (Jin et al. 1999). Two E-box motifs resembling the E-box consensus motif CACGTG (Gekakis et al. 1998; Hogenesch et al. 1998) were found to be present at positions +2398 and +2510, mapping very close to the two bands observed within region 2. Another E-box motif with the related core sequence CACATG (located at +857 in the center of the circadian region 4) was identified by an in vitro DNase I footprinting approach (data not shown). A related E-box motif was found in the hypersensitive region 7 at −888. These E-box motifs may be direct targets for CLOCK, an essential pacemaker component (for review, see Schibler 1998). As a large number of bHLH proteins can bind E-box motifs in vitro, simple electromobility shift assays (EMSA) were inadequate to resolve all complexes obtained with these DNA sequences (data not shown). We thus resorted to a two-dimensional high-resolution EMSA technique (Ossipow et al. 1993) and adapted it to the analysis of large DNA-binding proteins (see Materials and Methods). Briefly, after running the protein–DNA complexes in the first dimension, they were cross-linked in the gel and placed on top of a second, denaturing gel. After their final separation these complexes were visualized by autoradiography. To identify potential CLOCK-containing complexes, these two-dimensional EMSA experiments were performed in the presence and absence of CLOCK antiserum.
As shown in Figure 3A, the obtained pattern of protein–DNA complexes was found to be complex. However, two protein–DNA complexes disappear when CLOCK antiserum is included in the first dimension of the EMSA reaction using the E-box motif from DNase I hypersensitive region 4 (+857) as a binding site. As these two complexes migrate at the same position in their first dimension, they most probably formed a heterodimeric complex before denaturation with SDS. The nature of the interaction partner for CLOCK is currently unknown, but it might be related to BMAL1, known to form a complex with CLOCK in vitro (Gekakis et al. 1998; Hogenesch et al. 1998). No attempts have been made to identify the major cross-linked complexes. According to their approximate molecular mass and their high abundance, they may represent protein–DNA complexes containing upstream stimulating factor (USF) isoforms (see Potter et al. 1991; Viollet et al. 1996).
Figure 3.
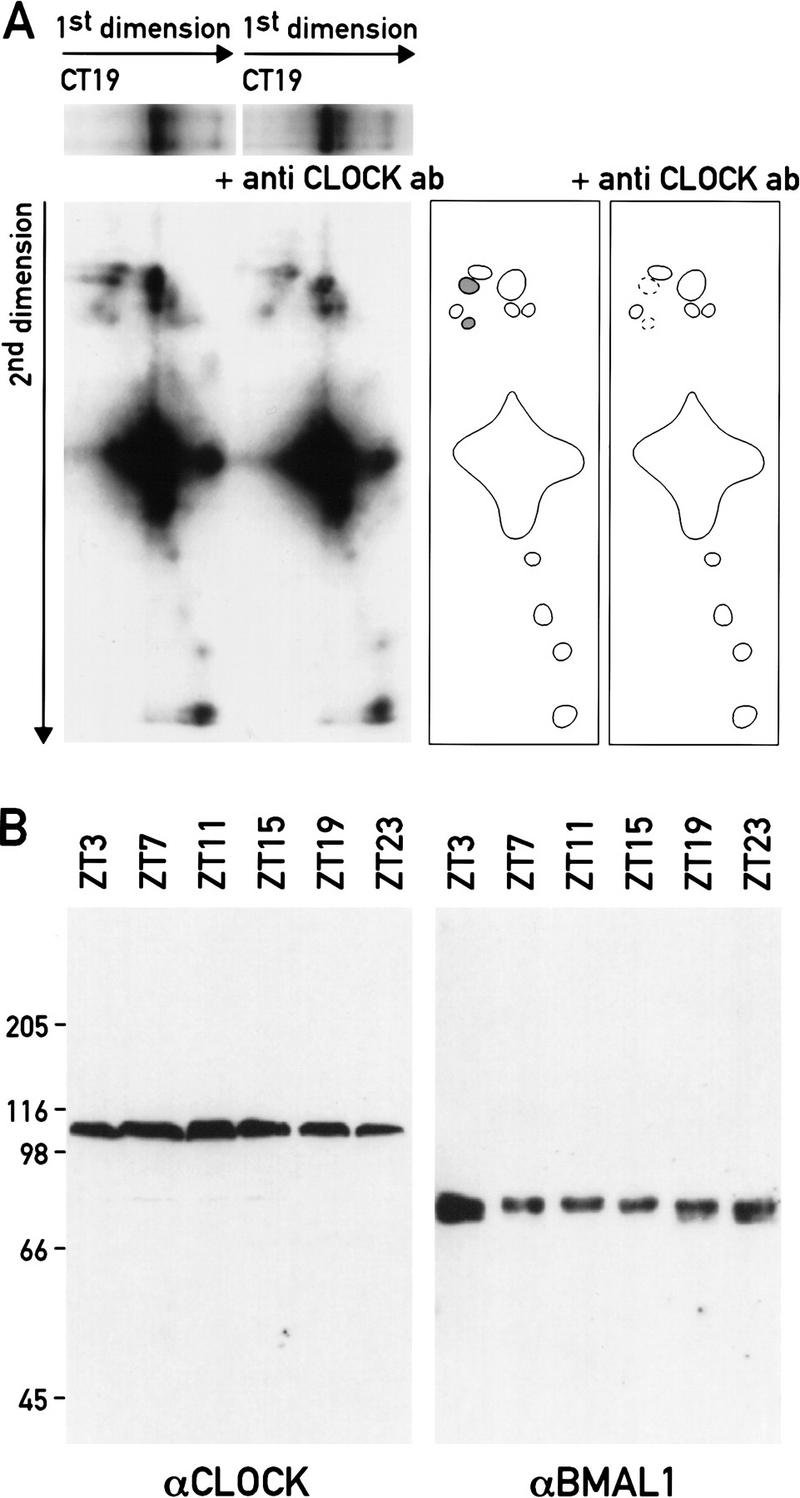
An intronic E-box motif that binds CLOCK in vitro. (A) Two-dimensional gel EMSA. Nuclear extracts from mouse livers harvested at CT19 were incubated with an oligonucleotide containing the intronic E-box motif (+857) of Dbp. After separation on a 4% polyacrylamide/0.5% agarose composite gel (1st dimension) the protein–DNA complexes were UV cross-linked, size-fractionated by electrophoresis through a 7% SDS–polyacrylamide gel (2nd dimension), blotted onto a membrane, and visualized by autoradiography. In one reaction (+ antiCLOCK ab), the liver nuclear extract was incubated with a CLOCK-specific antiserum before the two-dimensional EMSA analysis was performed. The contours of the spots corresponding to radiolabeled cross-linked protein–DNA complexes are depicted in the drawings at the right-hand site of the autoradiographs. Two spots are lacking in the experiments with CLOCK-specific antiserum. Their vertical alignment in the second dimension indicates that the two complexes were part of a heterodimeric complex in the first dimension. (B) Daily accumulation of CLOCK and BMAL1 in liver nuclei. Nuclear proteins harvested at the indicated time points were separated on a 7% SDS-polyacrylamide gel and probed with CLOCK- or BMAL1-specific antibodies. The migrations of standard proteins according to their molecular mass are indicated on the left (in kD).
In contrast to the highly circadian hypersensitivity of the +857 binding site in liver chromatin (see above), qualitatively and quantitatively very similar CLOCK/DNA complexes could be formed in vitro with liver nuclear proteins harvested throughout the day (data not shown). Furthermore, a nearly identical pattern of protein–DNA complexes and similar CLOCK-containing complexes could also be observed with either of the two E-box motifs resident in the DNase I hypersensitive region 2 from positions +2398 and +2510 (data not shown). As judged by Western blot analysis, both CLOCK and BMAL1 were present at nearly invariable levels in liver nuclear extracts at all times of day (Fig. 3B). The molecular basis for the cycling nature of hypersensitive region 4 thus cannot readily be explained solely by a cyclic accumulation of CLOCK protein. Conceivably, however, components of the negative limb of the feedback loop, such as PER and CRY proteins, sequester CLOCK in a complex unable to bind to its cognate E box during times at which Dbp is not transcribed. The phase of circadian CRY protein accumulation would be in keeping with such a scenario (J.A. Ripperger and U. Schibler, unpubl.).
In conclusion, the DNase I mapping experiments in conjunction with the two-dimensional EMSA assays presented in this section unveiled six putative Dbp regulatory sequences, of which two are located upstream (6 and 7) and four downstream (1–4) of the transcription initiation site. The sensitivity towards DNase I digestion of four of these regions (2, 4, 6, and 7) and that of the promoter region encompassing the transcription initiation site oscillate with the same phase as Dbp transcription. Three of the putative regulatory sequences (2, 4, and 7) contain E boxes that are potential binding sites for CLOCK.
The intronic Dbp E box in DNase I hypersensitive site 4 acts as a CLOCK/BMAL1 target sequence in cotransfection experiments
All four identified E-box motifs were found to function as cis-acting elements when they were linked to a luciferase reporter gene and transfected into murine LTK− fibroblasts (Fig. 4A). To examine whether CLOCK/BMAL1 can activate transcription of a cis-linked luciferase reporter gene via the Dbp E-box motif from intron 1, a series of cotransfection experiments were performed. As shown in Figure 4B, CLOCK and BMAL1 increase E box-driven reporter gene activity in a dose-dependent manner. Transfection of either expression vector alone did not significantly influence the basal reporter gene activity (data not shown). In contrast, cotransfection of a mutated form of CLOCK, CLOCKΔ19 (obtained from Clock/Clock mutant mice and supposed to act as a dominant-negative inhibitor; Antoch et al. 1997; King et al. 1997a), repressed basal reporter gene activity. Conceivably, this down-regulation is the consequence of a competition of CLOCKΔ19 with endogenous wild-type CLOCK and/or other related bHLH proteins. No effects of CLOCK on luciferase expression have been observed in experiments with reporter genes carrying a mutated version of the E-box motif (Fig. 4B). In conclusion, the cotransfection experiments presented in this section are compatible with the speculation that the intronic Dbp E box may serve as a functional CLOCK/BMAL1 recognition sequence. Qualitatively similar experiments were also obtained with either of the two E boxes resident in intron 2 (data not shown) and with E boxes for different clock-related genes (Gekakis et al. 1998; Kume et al. 1999).
Figure 4.
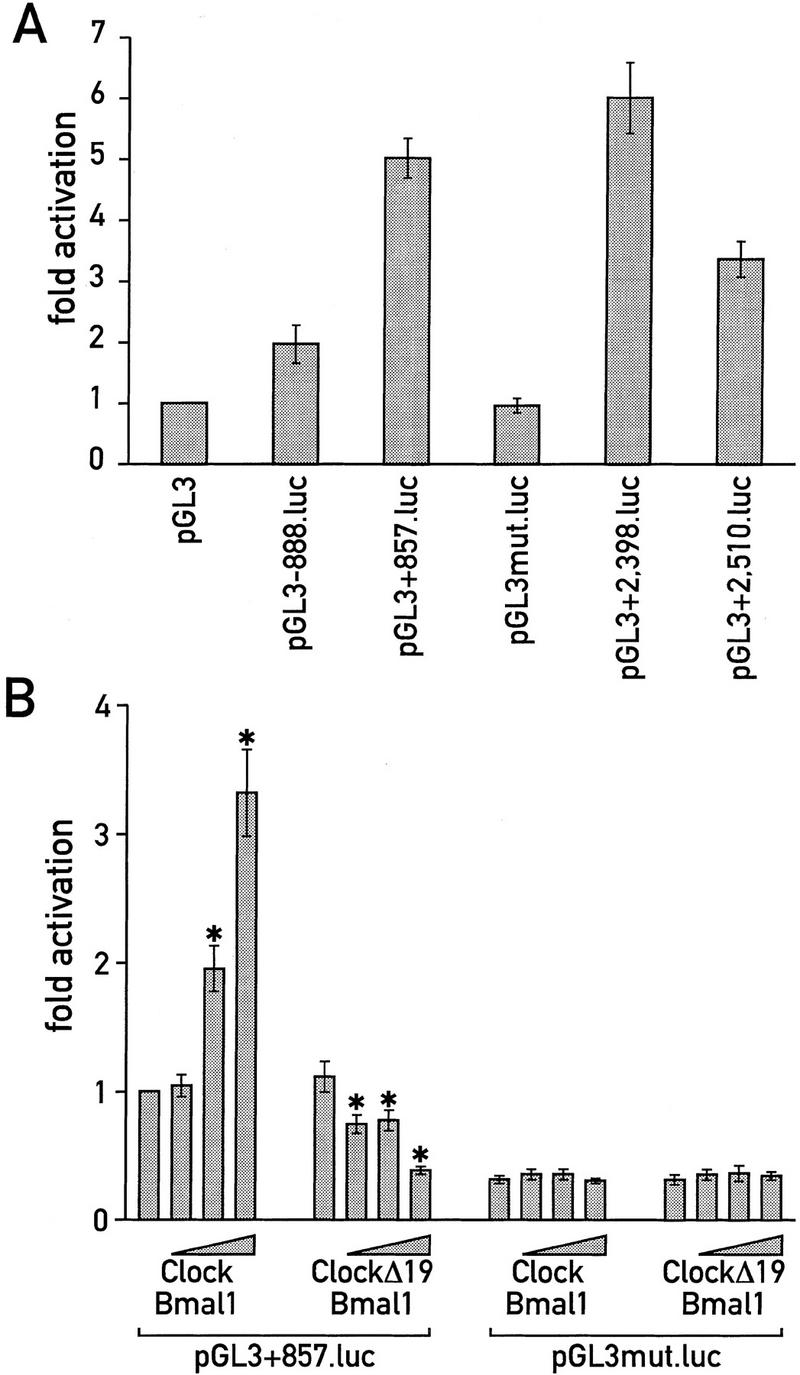
CLOCK activates transcription from the intronic Dbp E box. (A) Comparison of basal reporter gene activities of luciferase reporter gene constructs containing four different E-box motifs identified in Dbp. (B) Mouse LTK− fibroblasts were cotransfected with either pGL3+857.luc or pGL3mut.luc and increasing amounts of expression vectors for CLOCK and BMAL1 or CLOCKΔ19 and BMAL1 (80 ng, 400 ng, or 2 μg each). Mean ± s.d. of six experiments. The transfection experiment with pGL3+857.luc and no expression vectors was set to onefold. An asterisk marks highly significant differences (P < 0.001) within the pGL3+857.luc series. There were no such differences within the pGL3mut.luc series.
The Dbp mRNA expression is severely down-regulated in the SCN and the liver of Clock mutant mice
On the basis of the observations described above, we considered the possibility that circadian Dbp expression involves CLOCK. To examine this conjecture, the daily accumulation of Dbp mRNA was determined in the SCN of Clock/Clock mutant mice that express the CLOCKΔ19 protein (King et al. 1998b) by in situ hybridization and in the liver by RNase protection experiments. In the SCN of Clock/Clock mutant mice Dbp mRNA accumulation still appears to be rhythmic (ANOVA, P = 0.0018), but the amplitude is significantly reduced as compared with wild-type mice (ANOVA, P = 0.0001; Fig. 5). In liver, the difference between mutant and wild-type mice is even more dramatic. As seen in Figure 6A, circadian Dbp mRNA accumulation is completely abolished in homozygous Clock mutant animals. Surprisingly, the Dbp mRNA peak levels are very low even in heterozygous Clock mutant mice (Fig 6B). These experiments indicate that the CLOCKΔ19 mutant protein, which lacks a glutamine-rich segment of its supposed transactivation domain, interferes with its wild-type counterpart in a dominant-negative fashion, conceivably by competing for its DNA recognition sequences.
Figure 5.
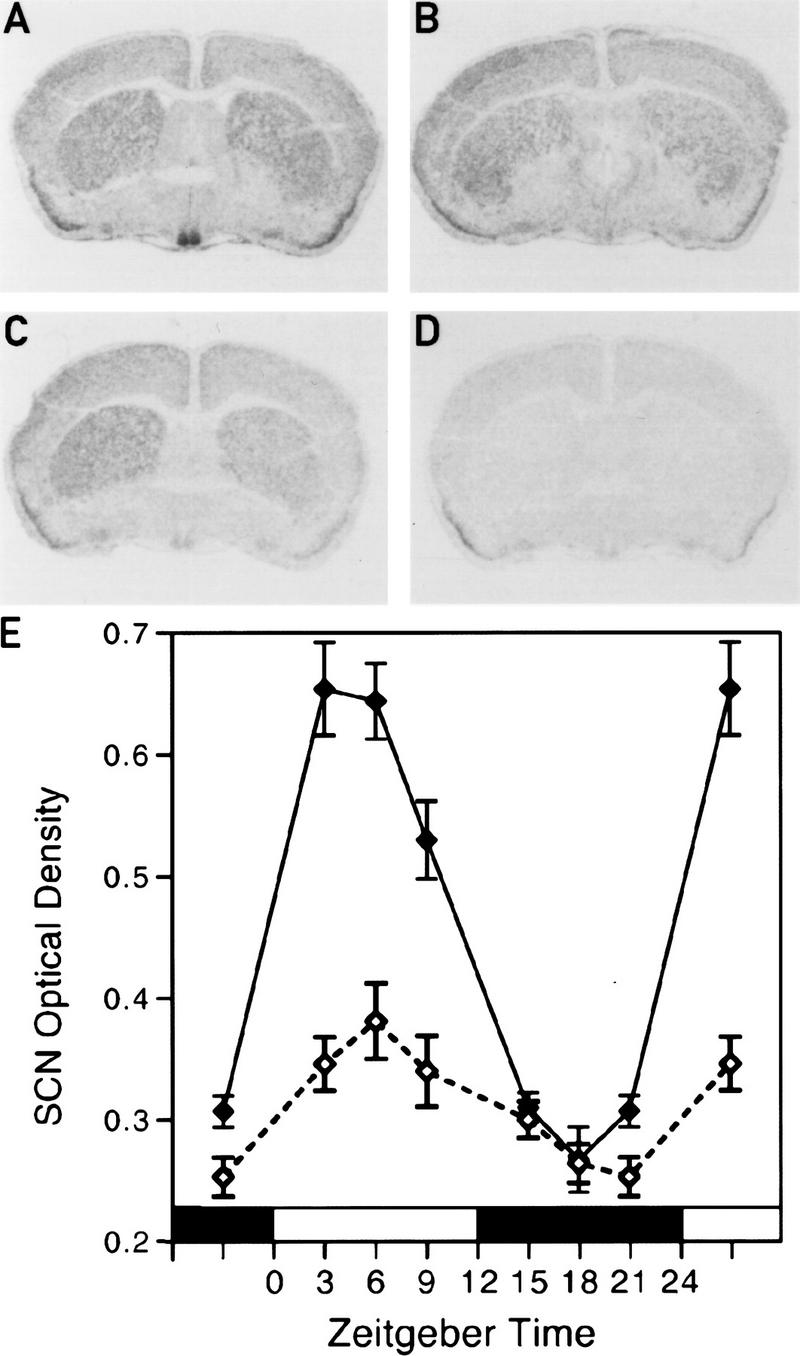
Dbp mRNA levels are reduced in the SCN of Clock/Clock mice. Representative autoradiographs of in situ hybridizations with coronal brain sections containing the SCN from wild-type mice at ZT3 (A), wild-type mice at ZT15 (B), and Clock/Clock mutant mice at ZT3 (C). A Dbp antisense cRNA was used as a hybridization probe to detect Dbp RNA. (D) A section adjacent to that of the one shown in A hybridized with a Dbp sense cRNA probe (negative control). (E) The temporal accumulation profiles of Dbp mRNA in the SCN of wild-type mice (solid line) and Clock/Clock mutant mice (broken line) in a 12-hr light/dark cycle. Each value is the mean ± s.e.m. of 5–6 animals.
Figure 6.
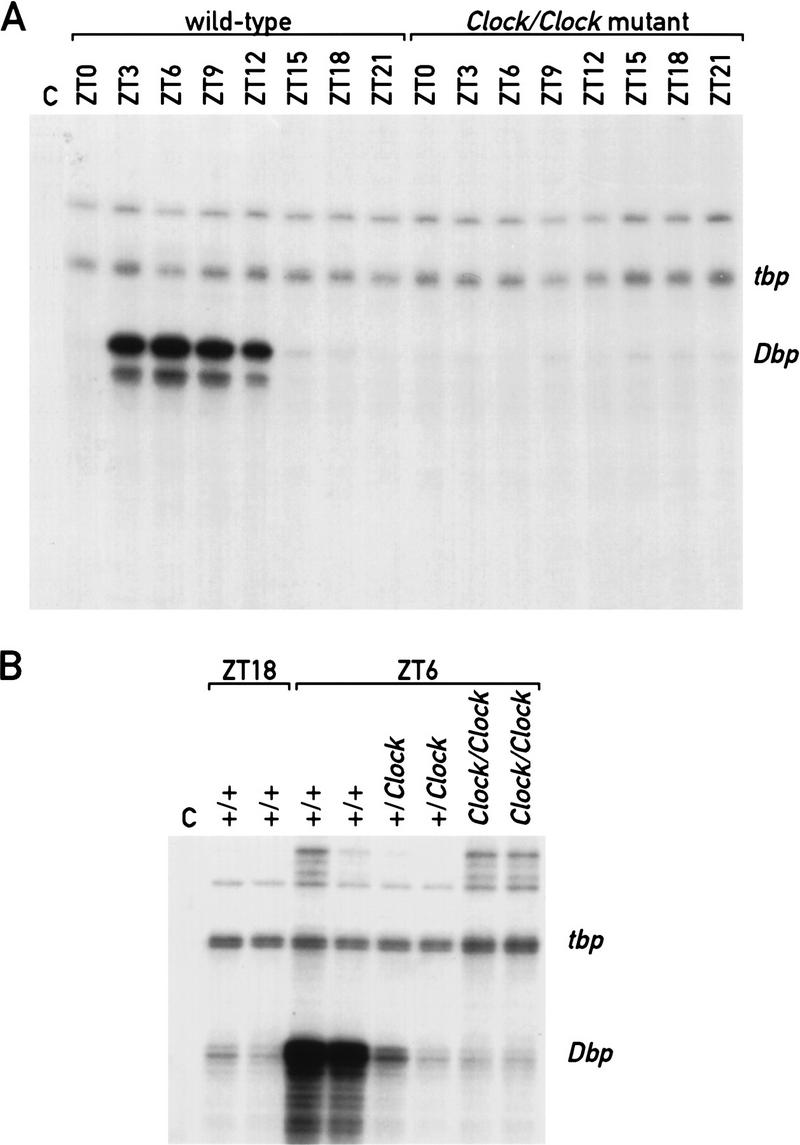
Circadian Dbp expression is obliterated in the liver of Clock mutant mice. (A) RNase protection assays with an antisense Dbp cRNA probe and whole-cell liver RNAs prepared from wild-type and Clock/Clock mutant mice at the indicated times (ZT0–ZT21). A tbp antisense cRNA was included as an internal control for a constitutively expressed mRNA (see Fig. 1). (Lane C) Negative control in which liver RNA was substituted with yeast tRNA. (B) RNase protection assays with an antisense Dbp cRNA probe and whole-cell liver RNAs prepared from mice of different genotypes. The ZT at which the animals were sacrificed and the genotypes of the mice are indicated at top. (Lane C) Negative control in which liver RNA was substituted with yeast tRNA.
Discussion
Dbp contains multiple putative enhancer regions
We have studied the transcriptional regulation of circadian DBP expression by using a variety of genetic and biochemical tools. Cis-acting DNA regulatory elements occupied by their cognate transcription factors frequently render the neighboring chromatin hypersensitive to nucleolytic cleavage by DNase I and other nucleases. As a consequence, the mapping of DNase I hypersensitive chromatin regions can be used as a reliable tool to identify transcriptional control elements such as promoters, enhancers, locus control regions, and silencing sequences (Weintraub and Groudine 1976; Thomas and Elgin 1988; Fraser et al. 1990; Felsenfeld et al. 1996; Kingston et al. 1996; Sippel et al. 1996). We have employed this technique to locate putative cis-acting regulatory regions in the Dbp gene. The robust amplitude in circadian Dbp transcription makes this search particularly attractive, as it should allow the temporal correlation of hypersensitive sites with the transcription status of Dbp. At times of maximal Dbp transcription rates [ZT or circadian time (CT) 7 and 11] seven DNase I hypersensitive regions could be identified, of which one maps to the cap site at position +1. The cleavage at the cap site is strictly correlated with transcription efficiency and probably reflects the occupancy of this site by components of the RNA polymerase II transcription machinery. Hence, formation of the transcription preinitiation complex may be limited to the time window during which Dbp is actively transcribed. The susceptibility to nucleolytic attack of four of the six remaining DNase I hypersensitive regions (2, 4, 6 and 7, see Fig. 2) also varies significantly with circadian time, albeit with an amplitude lower than that observed for the site mapping to +1. The hypersensitive region 6 displays a somewhat more complex pattern. Whereas some of the cleavages oscillate with the same phase as transcription, others appear to be anticyclic. Conceivably, the DNA-binding proteins responsible for the anticyclic cleavages are transcriptional repressors rather than activators. Interestingly, the hypersensitive sites 5 and 7 (see Fig. 2) were completely absent from the 5′-flanking region of the Dbp–lacZ fusion gene, whereas DNase I hypersensitive region 6 appeared as a single band that was constitutively present around the clock (Fig. 2F). This suggests that some of the transcription factors binding to DNA sequences within 5′-flanking sequences can do so only in concert with transcription factors binding to intragenic enhancer elements. Therefore, transcriptional regulatory proteins occupying upstream and intragenic DNA elements are likely to stimulate Dbp transcription synergistically, for example by cooperative binding.
The poor transcriptional activity of the Dbp–lacZ allele indicates that intragenic regulatory sequences are essential for high-amplitude circadian Dbp transcription. However, given the possibly synergistic action of upstream and intragenic regulatory sequences, the same may be true for enhancers located within the 5′-flanking region. In fact, cis-acting elements conferring circadian transcription are likely to exist within this region as well, as the few transcripts issued by the Dbp–lacZ allele still exhibit circadian accumulation (Fig. 1B). Clearly, the examination of the role that upstream sequences may play in rhythmic Dbp transcription will require further studies.
CLOCK is an upstream regulator of Dbp
In Clock/Clock mutant mice Dbp circadian expression is dramatically dampened in the SCN and nearly extinguished in the liver. However, this genetic analysis does not establish whether CLOCK is a direct or indirect regulator of Dbp. Based on two observations we favor a direct interaction between Clock and Dbp. First, the circadian expression of Dbp and mPer1, another putative CLOCK target gene (Gekakis et al. 1998; Jin et al. 1999), display peak expression at the same time of day. Secondly, we could identify CLOCK recognition sequences within circadian hypersensitive chromatin regions of Dbp that in cotransfection assays are capable of conferring CLOCK/BMAL1-mediated transcriptional stimulation (Fig. 3B; data not shown). Another related E-box motif and potential binding site for CLOCK is located within the circadian DNase I hypersensitive region 7 at position −888. Conceivably, this E box is responsible for the rhythmic accumulation of the few transcripts issued from the Dbp–lacZ fusion allele observed in liver (Fig. 1B). In accordance with this speculation, the trace amounts of Dbp mRNA detected in the livers of Clock/Clock mutant mice, in contrast to those in Dbp+/− mice, are not subject to circadian variation (Figs. 1 and 6A).
Wild-type CLOCK protein and its DNA-binding sites within extra- and/or intragenic Dbp enhancer sequences are likely to participate in both the positive and negative limbs of the circadian feedback loop. This has been suggested for the circadian regulation of both Drosophila and mammalian pacemaker genes (Allada et al. 1998; Darlington et al. 1998; Gekakis et al. 1998; Rutila et al. 1998; Jin et al. 1999; Kume et al. 1999). We wish to emphasize, however, that not all E box-containing DNase I hypersensitive regions displayed a strong circadian pattern. In fact, the E box motif centered around +2510 within DNase I hypersensitive region 2 shows less dramatic circadian accessibility than the E boxes at positions +857 and +2398 to nuclease digestion throughout the day, yet binds CLOCK avidly in vitro (data not shown). We can thus only speculate on the molecular mechanisms leading to the different behavior of the various E box-containing motifs in nuclease digestion experiments. One possibility to account for the differential nuclease sensitivity of different E boxes in Dbp chromatin would be that only the circadian E-box elements participate in the negative limb of the circadian autoregulatory loop, whereas the other E box acts more as a constitutive enhancer element. Given the multitude of different bHLH proteins present in liver nuclei that bind E-box motifs in vitro (see Fig. 3A), it is conceivable that the circadian and constitutive E-box motifs bind different proteins in vivo. In fact, the occupancy of a given cis-acting element in vivo is determined not only by the intrinsic sequence specificity of a transcription factor but also by the cooperative interactions this factor establishes with other proteins binding to nearby or distant promoter and enhancer elements. Therefore, although in vitro binding studies with short DNA recognition sequences can establish a repertoire of possible cognate proteins present in a cellular extract, they are insufficient to positively identify the proteins that occupy these sites in vivo. Nevertheless, together with the genetic evidence obtained with Clock and Dbp mutant mice, the biochemical experiments presented above suggest that CLOCK protein may interact directly with cognate E boxes in Dbp to regulate circadian transcription of this gene.
Dbp transcription may be linked directly to the circadian clock
CLOCK was found to be an essential upstream regulator of Dbp, and this observation links the expression of this gene directly to the molecular oscillator. Interestingly, we observed some minor differences regarding the effect of CLOCKΔ19 on the expression of Dbp in the SCN and the liver. Although in the SCN neurons some rhythmic expression was still observed, in the periphery circadian expression was completely abolished, even in Clock heterozygous mice (Figs. 5 and 6). Conceivably, the CLOCKΔ19 protein accumulates to higher levels in liver nuclei as compared with nuclei of SCN neurons.
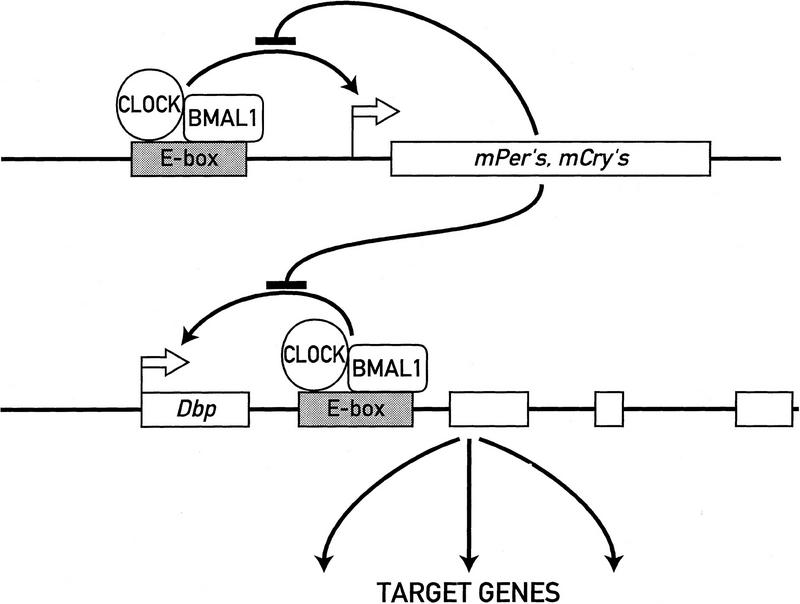
As a transcription factor expressed in most cell types, DBP can contribute to the cyclic transcription of many target genes both in the SCN and in peripheral tissues. In liver, for example, DBP participates in the circadian expression of coumarin 7 hydroxylase, steroid 15α hydroxylase, and several other cytochrome P-450 enzymes (Lavery et al. 1999; B. Kornmann and U. Schibler, unpubl.). The way by which the molecular feedback loop of cellular pacemakers can drive circadian outputs are illustrated in Figure 7. Whereas some output genes such as the neuropeptide vasopressin (Jin et al. 1999) may be directly controlled by the molecular oscillator, others may be regulated by circadian transcription factors that themselves are controlled by the molecular pacemaker.
Figure 7.
Dbp expression may be hardwired to the circadian feedback loop circuitry. The cartoon depicts a speculative model according to which circadian oscillations are generated by a negative-feedback loop of gene expression. The CLOCK/BMAL1 heterodimer activates transcription of essential pacemaker genes such as mPers and mCrys through an E-box motif. The protein products of these pacemaker genes then form complexes that, once they reach a critical threshold level, attenuate CLOCK/BMAL1-mediated activation. The same mechanism may be employed to regulate circadian transcription of Dbp. The cyclic accumulation of DBP may then drive circadian transcription of downstream target genes.
Although the experimental data on Dbp are in strong favor of a model in which the expression of this output gene is directly hardwired to the molecular oscillator, they cannot be regarded as direct proof for such a scenario. In fact, the unequivocal demonstration of a direct functional interaction between a transcription factor and its target gene requires experiments with specificity-shift mutations in both the DNA-binding domain of the transacting regulatory protein and its DNA recognition sequence(s). Owing to the technical difficulty of such an approach in metazoan organisms, this has been accomplished only in very rare cases. One very elegant example is the study by Schier and Gehring (1992) on the negative autoregulatory loop established by the Drosophila segmentation gene fushi tarazu.
In conclusion, our work has identified CLOCK, a central component of the circadian pacemaker, as a regulator of Dbp transcription. The presence of CLOCK-binding sites within DNase I hypersensitive regions, which oscillate during the day in phase with Dbp transcription, strongly suggests that CLOCK controls rhythmic hepatic Dbp expression in a direct manner. Hence, we propose that the regulation of Dbp transcription is directly hardwired to the circadian feed back circuitry, as has been suggested for another output gene, vasopressin (Jin et al. 1999). In contrast to vasopressin, however, Dbp is rhythmically expressed in both SCN neurons and most peripheral cell types and, as a transcription factor, can control the circadian expression of many target genes. The observation that the same transcriptional hierarchies can be observed in SCN neurons and hepatocytes supports the idea that central and peripheral circadian systems are similar in their molecular makeup.
Materials and methods
Animal care and handling
The Dbp−/− mouse strain with a substitution of the ORF of the Dbp gene by a lacZ cassette (Lopez-Molina et al. 1997) and the Clock/Clock mutant mouse strain bearing a deletion of exon 19 have been described previously (Antoch et al. 1997; King et al. 1997b). Mice were housed in a strict 12:12 hour light/dark regimen (lights on 7 a.m.; lights off 7 p.m.). Experiments under constant conditions were performed with animals kept in constant darkness for at least one day before the first animal was sacrificed (CT). For animals housed 10 days under constant conditions the period lengths of locomoter activity and their individual time schedules were calculated from their running-wheel activities as described (Lopez-Molina et al. 1997).
Analysis of whole-cell liver RNA
RNA was purified from mouse livers and analyzed by ribonuclease protection assays using probes for Dbp and tata box-binding protein (tbp) described previously (Lopez-Molina et al. 1997). The probe for Dbp–lacZ, overlapping the 5′ insertion site of the lacZ cassette in the Dbp gene, was obtained by PCR using a sense primer from Dbp, 5′-TTCTTTGCGAGAAGTGC-3′, an antisense primer from lacZ, 5′-AAACCAGGCAAAGCGCAT-3′, and genomic DNA from a Dbp−/− mouse as a template.
Mapping of DNase I hypersensitive sites
Nuclei from mouse liver were isolated as described (Lichtsteiner et al. 1987). The accessibility of the chromatin to limited DNase I digestion was tested as reported by Boyes and Felsenfeld (1996). Briefly, nuclei were treated with 1 U/μl of DNase I for five min on ice. Isolated genomic DNA (20 μg per lane) was digested to completion with the indicated restriction enzyme(s), separated on 0.7% agarose gels, blotted to nylon (Nitran) membranes, and hybridized with a BamHI/HindIII fragment (2680 to 3447, GenBank accession no. U29762) or a PvuII/EcoRV fragment (4333 to 5010, GenBank accession no. U29762) of the mouse Dbp gene. For the probing of C/ebpα a NcoI/HindIII fragment of the ORF from pMSV-C/EBP (Friedman et al. 1989) was used. The mapping in Dbp+/− animals was performed with EcoRI/HindIII-digested DNA (the EcoRI/HindIII fragment of Dbp–lacZ has about the same length as the HindIII/HindIII fragment of Dbp). The probe for mapping of the Dbp–lacZ locus was obtained as an EcoRI/SacI fragment from the targeting vector pTK-B.A.-LacZ-NEO-UMS-D (Lopez-Molina et al. 1997). For a fine mapping of hypersensitive sites we obtained restriction fragments of a defined length from the Dbp locus. These fragments were adjusted in their quantity to match the hybridization signals derived from DNase I hypersensitive sites, and they were run together with DNase I-digested genomic DNA and detected with the appropriate probes. The resolution of these experiments should be ±100 bp.
Two-dimensional gel mobility shift and Western blot experiments
Briefly, an oligonucleotide encompassing the E box whose thymine residues were substituted with highly photoreactive azido-uracil residues was used as a DNA probe (Ossipow et al. 1993). The protein–DNA complexes formed with liver nuclear extracts were separated by electrophoresis on a composite agarose–polyacrylamide gel. The gel was irradiated with UV light, and the region containing the specific protein–DNA complexes was excised, placed horizontally onto a SDS–polyacrylamide gel, and the photo-cross-linked protein–DNA complexes were size-fractionated by electrophoresis. Because the UV dose chosen for these experiments resulted in the covalent protein–DNA cross-linking of only a small proportion of the complexes, these contain mainly protein monomers.
Proteins from liver nuclei were obtained as described (Ripperger et al. 1995). A double-stranded oligonucleotide encompassing the intronic E-box motif of Dbp (+857) was generated according to Ossipow et al. (1993) with Klenow polymerase us-ing the 5′-primer 5′-CCTCGCAGGG-3′, 5-azido-dUTP, [α-32P]dATP, and the template 5′-CCTAGTTTCCATGTGACCCTGCGAGG-3′. The gel mobility shift reactions were performed in a 20 μl final volume of 10% glycerol, 50 mm KCl, 10 mm HEPES (pH 7.6), 1 mm dithiothreitol, 0.1 mm EDTA, 5 mm MgCl2, 50 ng/μl poly[d(I-C)], 0.4 μg/μl sheared salmon sperm DNA, 20 μg of protein, and 1 × 106 cpm of the oligonucleotide (1 × 107 cpm/pmole). After an incubation of 15 min at room temperature the protein–DNA complexes were separated on a 4% polyacrylamide/0.5% agarose gel in 36 mm Tris-borate (pH 8.3)/0.8 mm EDTA, cross-linked in situ for 7 min with UV302nm, and layered on top of a 7% SDS-polyacrylamide gel (Laemmli 1970). After electrophoretic separation, the protein–DNA complexes were electroblotted to a nitrocellulose membrane, which was exposed for autoradiography for 92 hr. Rat antibodies against the amino terminus of mCLOCK (amino acids 1–424) and full-length hBMAL1 were raised and purified using standard techniques. Nuclear proteins (30 μg/lane) were separated on a 7% SDS-polyacrylamide gel, transferred to nitrocellulose, and specific antigen/antibody complexes were visualized using a horseradish peroxidase-conjugated secondary antibody and an ECL kit (Pierce).
Cotransfection experiments
Oligomerized E-box motifs (n = 4) were ligated into the BglII restriction site of pGL3 (Promega). The oligonucleotides used were: +888, 5′-GATCCACGTCCCATGTGGCCTTCT-3′; −857, 5′-GATCTAGTTTCCATGTGACCCTGC-3′; mut, 5′-GATCTAGTTTCACTGGTACCCTGC-3′; +2,398, 5′-GATCCCCTCGCCACGTGAGTCCGC-3′; +2510, 5′-GATCTAGGCCACACGTGATGCGGC-3′; and their corresponding complementary sequences containing a 5′-GATC overhang to facilitate cloning. Expression vectors for CLOCK, CLOCKΔ19, and BMAL1 have been described previously (Jin et al. 1999). Mouse LTK− fibroblasts of 60%–80% confluency were transfected with 150 ng of reporter plasmids and the indicated amounts of either CLOCK expression vector made up to a total of 4 μg with an empty plasmid (pcDNA3.1, Invitrogen), using Superfect (Qiagen). After 48 hr of recovery the reporter gene activity was measured using a luciferase kit (Boehringer), and normalized for the transfection efficiency with a cotransfected lacZ plasmid using a β-galactosidase kit (Tropix).
In situ hybridization
The in situ hybridizations of mouse coronal brain sections have been described previously (Jin et al. 1999; Shearman et al. 1999). The cRNA probes were obtained from a 1065-bp fragment of the rat Dbp gene (357–1422, GenBank accession no. J03179) in vitro synthesized in sense or antisense orientation.
Acknowledgments
We are most grateful to Joseph S. Takahashi and Martha H. Vitaterna for supplying us with Clock/Clock founder mice; Steven L. McKnight for his generous gifts of C/ebpα and hBmal1/JAP3 recombinant plasmids; Charles J. Weitz for providing expression constructs; David R. Weaver for his help with in situ hybridizations; Steven A. Brown for valuable discussions; and Nicolas Roggli for expert preparation of the figures. This work was supported by the Canton of Geneva, grant 3147314.96 of the Swiss National Science Foundation to U.S., grant R37 HD14427 to S.M.R., and a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft to J.A.R. (Ri 910/1-1).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ueli.schibler@molbio.unige.ch; FAX 41 22 702 68 68.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Schibler U. The ins and outs of circadian timekeeping. Curr Opin Genet Dev. 1999;9:588–594. doi: 10.1016/s0959-437x(99)00009-x. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Drolet DW, Scully KM, Simmons DM, Wegner M, Chu KT, Swanson LW, Rosenfeld MG. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes & Dev. 1991;5:1739–1753. doi: 10.1101/gad.5.10.1739. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Falvey E, Fleury-Olela F, Schibler U. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- Franken P, Lopes-Molina L, Marcacci L, Schibler U, Tafti M. The transcription factor DBP affects circadian sleep consolidation and rhythmic EEG activity. J Neurosci. 2000;20:617–625. doi: 10.1523/JNEUROSCI.20-02-00617.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Hurst J, Collis P, Grosveld F. DNase1 hypersensitive sites 1, 2 and 3 of the human beta-globin dominant control region direct position-independent expression. Nucleic Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD, Landschulz WH, McKnight SL. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes & Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the Clock protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Hao H, Allen DL, Hardin P. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Glossop NRJ, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Förster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates PER-like developmental, spatial, and circadian expression expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Ohyashiki K, Toyama K, Cleary ML. HLF, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes & Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- Inaba T, Roberts WM, Shapiro LH, Jolly KW, Raimondi SC, Smith SD, Look AT. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- Iyer SV, Davis DL, Seal SN, Burch JB. Chicken vitellogenin gene-binding protein, a leucine zipper transcription factor that binds to an important control element in the chicken vitellogenin II promoter, is related to rat DBP. Mol Cell Biol. 1991;11:4863–4875. doi: 10.1128/mcb.11.10.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- King DP, Vitaterna MH, Chang AM, Dove WF, Pinto LH, Turek FW, Takahashi JS. The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics. 1997a;146:1049–1060. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian Clock gene. Cell. 1997b;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Schibler U. Circadian transcription of the cholesterol 7 α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes & Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C. Circadian expression of the steroid 15 alpha hydroxylase (cyp2a4) and coumarin 7 hydroxylase (cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S, Wuarin J, Schibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987;51:963–973. doi: 10.1016/0092-8674(87)90583-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CR, Maire P, Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990;61:279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter JJ, Cheneval D, Dang CV, Resar LM, Mezey E, Yang VW. The upstream stimulatory factor binds to and activates the promoter of the rat class I alcohol dehydrogenase gene. J Biol Chem. 1991;266:15457–15463. [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Fritz S, Richter K, Hocke GM, Lottspeich F, Fey GH. Transcription factors Stat3 and Stat5b are present in rat liver nuclei late in an acute phase response and bind interleukin-6 response elements. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- Rosbash M, Allada R, Dembinska M, Guo WQ, Le M, Marrus S, Qian Z, Rutila J, Yaglom J, Zeng H. A Drosophila circadian clock. Cold Spring Harb Symp Quant Biol. 1996;61:265–278. [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Schibler U. Circadian rhythms. New cogwheels in the clockworks. Nature. 1998;393:620–621. doi: 10.1038/31337. [DOI] [PubMed] [Google Scholar]
- Schibler U, Lavery DJ. Circadian timing in animals. In: Russo E, Cove D, Edgar L, Jaenisch R, Salamini F, editors. Develoment—Genetics, epigenetcs, and environmental regulation. Heidelberg, Germany: Springer Verlag; 1999. pp. 487–505. [Google Scholar]
- Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Reppert SM, Weaver DR. Expression of basic-helix-loop-helix-PAS genes in the mouse suprachiasmatic nucleus. Neuroscience. 1999;89:387–397. doi: 10.1016/s0306-4522(98)00325-x. [DOI] [PubMed] [Google Scholar]
- Sippel AE, Saueressig H, Huber MC, Hoefer HC, Stief A, Borgmeyer U, Bonifer C. Identification of cis-acting elements as DNase1 hypersensitive sites in lysozyme gene chromatin. Methods Enzymol. 1996;274:233–246. doi: 10.1016/s0076-6879(96)74020-x. [DOI] [PubMed] [Google Scholar]
- Thomas GH, Elgin SC. Protein/DNA architecture of the DNase1 hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988;7:2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Viollet B, Lefrançois-Martinez A-M, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- ————— Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW. Molecular control of circadian behavioral rhythms. Recent Prog Horm Res. 1999;54:87–94. [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts in the outside of the brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]