Abstract
Although the suprachiasmatic nucleus (SCN) is the major pacemaker in mammals, the peripheral cells or immortalized cells also contain a circadian clock. The SCN and the periphery may use different entraining signals—light and some humoral factors, respectively. We show that induction of the circadian oscillation of gene expression is triggered by TPA treatment of NIH-3T3 fibroblasts, which is inhibited by a MEK inhibitor, and that prolonged activation of the MAPK cascade is sufficient to trigger circadian gene expression. Therefore, such prolonged activation of MAPK by entraining cues may be involved in the resetting of the circadian clock.
Keywords: Circadian clock, entrainment, gene expression, MAP kinase, signal transduction
Daily rhythms of biological activity are driven by self-sustaining, endogenous oscillators called circadian clocks, which typically run with an intrinsic period that is close to, but not exactly, 24 hr. Under natural conditions, circadian clocks become precisely entrained to the 24-hr light/dark cycle because exposure to light at certain times induces a phase shift of the clock. Entrainment to light/dark cycles ensures that the clock adopts a specific and stable phase relation to the natural day, setting the clock to local time. The molecular mechanisms of entrainment are not well understood, but photic induction of immediate-early genes, such as c-fos, FosB, JunB, and Per (a homolog of the Drosophila clock gene period), in the suprachiasmatic nucleus (SCN) is thought to have a role (Rusak et al. 1990; Kornhauser et al. 1996; Albrecht et al. 1997; Shigeyoshi et al. 1997; Morris et al. 1998; Takumi et al. 1998). Although the SCN is the major pacemaker in mammals (Rusak and Zucker 1979), recent studies have indicated that the peripheral cells or immortalized cells also contain a circadian clock (Plautz et al. 1997; Balsalobre et al. 1998; Rosbash 1998; Sakamoto et al. 1998; Whitmore et al. 1998; Zylka et al. 1998; Dunlap 1999; Ishida et al. 1999; Krishnan et al. 1999) and suggested that the SCN and the periphery use different entraining signals: light for the former and some humoral factors for the latter (Silver et al. 1996; Balsalobre et al. 1998; Oishi et al. 1998; Rosbash 1998; Sakamoto et al. 1998; Zylka et al. 1998; Dunlap 1999; Ishida et al. 1999). The serum shock of rat fibroblasts, which induces the circadian expression of various genes whose transcription also oscillates in living animals, also results in transient stimulation of c-fos and rPer (rat homolog of period) expression and thus mimics light-induced immediate-early gene expression in the SCN (Balsalobre et al. 1998). Although the circadian time-keeping properties of the SCN require gene expression, little is known about the signal transduction pathways that initiate transcription. It has been reported that a brief exposure to light during the subjective night, but not during the subjective day, activates the p44/42 mitogen-activated protein kinase (MAPK) signaling cascade in the SCN (Obrietan et al. 1998). In addition, the stimulation of MAPK activates CREB (cAMP response element binding protein), suggesting that potential downstream transcription factors are stimulated by the MAPK pathway in the SCN (Obrietan et al. 1998). Here, we show first that induction of the circadian oscillation of expression of various clock and clock-related genes is triggered by treatment of NIH-3T3 cells with TPA, and that this triggering of the induction is inhibited by a MEK (MAPKK) inhibitor, U0126. We then show evidence that sustained activation of the MAPK cascade alone is sufficient to trigger the induction of circadian gene expression. These results strongly suggest that the MAPK cascade is involved in the resetting of circadian gene expression.
Results and Discussion
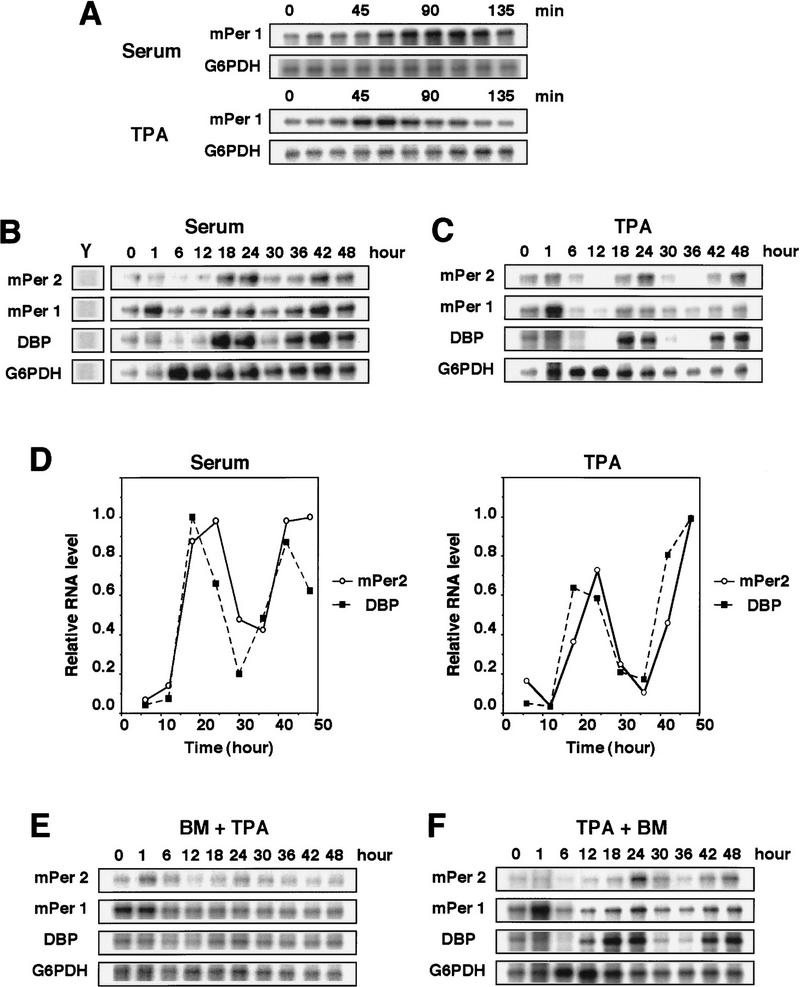
The light-induced entrainment of the circadian clock is accompanied by the induction of some immediate-early genes in the SCN (Rusak et al. 1990; Kornhauser et al. 1996; Albrecht et al. 1997; Shigeyoshi et al. 1997; Morris et al. 1998; Takumi et al. 1998), and the serum shock, which triggers the induction of the circadian gene expression in cultured cells (Balsalobre et al. 1998), also results in a transient and immediate induction of some genes such as mPer1, a mammalian homolog of the Drosophila clock gene period (Sun et al. 1997; Tei et al. 1997). The acute induction of mPer1 mRNA in the SCN after light exposure is thought to be involved in light-induced phase shifting of the overt rhythm (Akiyama et al. 1999). Then, we searched for stimuli that could induce the transient expression of mPer1 in mouse fibroblast NIH-3T3 cells and found that TPA treatment as well as a serum shock (50% serum) is able to induce the transient and strong expression of mPer1 (Fig. 1A); as little as 10 nm TPA was effective. We then monitored the mRNA expression levels of mPer1 and mPer2, which is also a period homolog (Albrecht et al. 1997; Shearman et al. 1997; Takumi et al. 1998) whose homozygous mutation in a PAS domain results in a shorter circadian period followed by a loss of circadian rhythmicity in constant darkness (Zheng et al. 1999), and albumin site D-binding protein (DBP), which is a clock-related gene encoding transcription factor (Lopez-Molina et al. 1997) during 2 days by RNase protection assays. As demonstrated previously for Rat-1 fibroblasts (Balsalobre et al. 1998), after the transient exposure to 50% serum expression levels of all the three mRNAs oscillated with an approximate period length of 24 hr in confluently grown NIH-3T3 cells in the absence of serum (Fig. 1B,D). A control gene, glucose-6-phosphate dehydrogenase (G6PDH) did not oscillate in the serum-shocked cells (Fig. 1B). Thus, the serum shock is able to trigger the induction of a circadian oscillation of expression of clock and clock-related genes in NIH-3T3 cells as well as in Rat-1 cells. Remarkably, TPA treatment (50 nm for 2 hr) without serum also triggered the induction of a circadian oscillation of expression of the three genes, mPer1, mPer2, and DBP, with essentially the same period length as seen in serum-shocked cells (Fig. 1C,D). The TPA treatment was as effective as the serum shock in triggering the induction of circadian gene expression (Fig. 1D). Pretreatment with a specific inhibitor of protein kinase C (PKC), bisindolylmaleimide I (BM), abolished the TPA-induced circadian oscillation of gene expressions (Fig. 1E), confirming that TPA exerted its effect through activation of PKC. In contrast, the addition of the PKC inhibitor after TPA treatment failed to inhibit the triggering of the induction of circadian gene expression (Fig. 1F), suggesting that the inhibitor does not have a toxic effect. These results suggest that PKC activation is able to entrain the circadian rhythm of the gene expression.
Figure 1.
TPA-triggered induction of circadian gene expression in NIH-3T3 fibroblasts. (A) Cells were shifted to medium containing 50% serum or 50 nm TPA (t = 0). The levels of mPer1 mRNA were determined by RNase protection assays. G6PDH is a loading control. Cells were shifted to medium containing 50% serum (B) or 50 nm TPA (C) (t = 0), and incubated for 2 hr, after which the medium was replaced with serum-free medium. Whole-cell RNA was prepared at the indicated times, and the relative levels of the mRNAs were determined by RNase protection assays. Yeast RNA (Y) was used as negative control. Three independent experiments gave similar results. (D) The signals obtained the RNase protection assays in B and C for mPer2(○) and DBP(█) mRNAs were quantified and normalized to signals obtained for G6PDH mRNA. The maximum value was set to 1.0. (E,F) Bisindolylmaleimide I (BM, 5 μm final concentration), a PKC inhibitor, was added 30 min before the addition of TPA (E) or 8 hr after the addition of TPA (F).
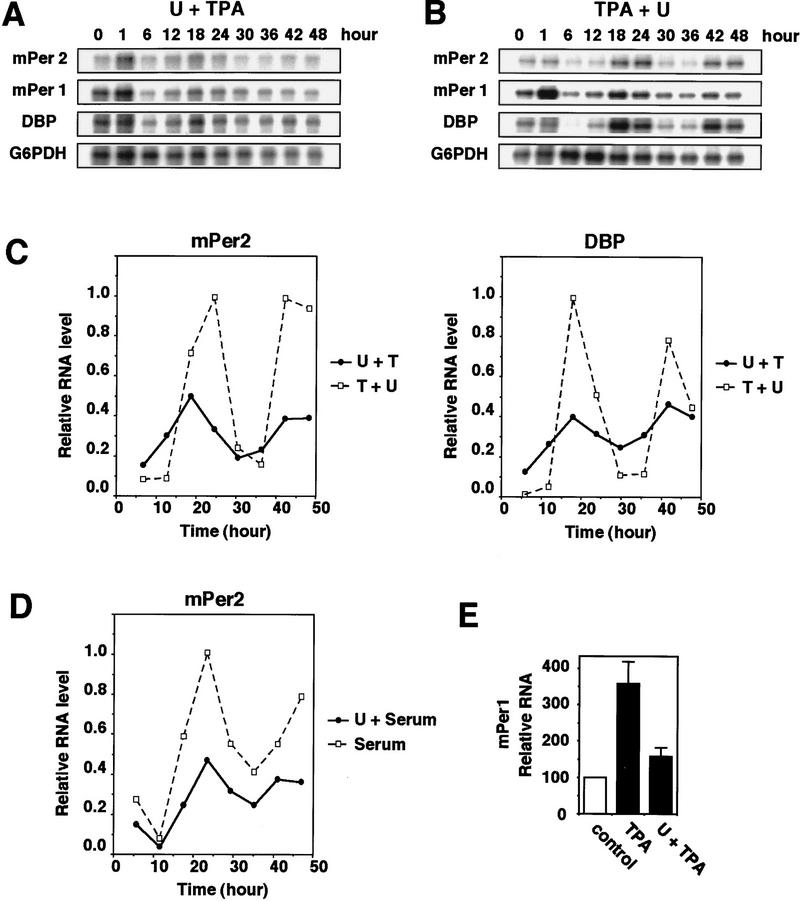
Both serum stimulation and TPA treatment (PKC activation) induce the activation of the classical MAPK cascade (the MEK/ERK cascade) (Cobb et al. 1991; Nishida and Gotoh 1993), which is known to result in the induction of immediate-early genes (Treisman 1996). Then we hypothesized that the MAPK cascade might be involved in triggering the induction of the circadian oscillation of gene expression. To test this idea, we used U0126, a specific inhibitor of MEK (Favata et al. 1998). Pretreatment with U0126 significantly inhibited both the TPA-induced immediate expression of mPer1 (Fig. 2A,E) and the TPA-triggered induction of a circadian oscillation of expression of the three genes (mPer1, mPer2, and DBP) (Fig. 2A,C). U0124, an inactive derivative of U0126, had no effect on them (data not shown). Therefore, activation of the MAPK cascade is required for TPA treatment to trigger the induction of circadian gene expression. In SCN, the activity of ERK/MAPK is shown to oscillate in a circadian manner (Obrietan et al. 1998, 1999). In TPA-treated NIH-3T3 cells, however, the activity of ERK/MAPK (both ERK1 and ERK2) was only transiently activated and did not oscillate (data not shown). Moreover, the addition of U0126 8 hr after TPA treatment did not inhibit the circadian oscillation of gene expression (Fig. 2B,C), indicating that although activation of the MAPK cascade is required for triggering the induction of circadian gene expression, it is not required for a run of circadian oscillation itself. The serum shock-triggered induction of circadian gene expression was also significantly inhibited by pretreatment with U0126 (Fig. 2D). Thus, it is likely that activation of the MAPK cascade is required for entraining the circadian oscillation of gene expression.
Figure 2.
Inhibition of TPA-triggered induction of circadian gene expression by pretreatment with MEK inhibitor U0126. (A) 10 μm U0126 was added 30 min before the addition of 50 nm TPA (A) or 8 hr after the addition of TPA (B). Three independent experiments gave similar results. (C) The signals obtained in the RNase protection assays shown in A (●, U + TPA) and B (□, TPA + U) for mPer2 and DBP mRNAs were quantified and normalized as in Fig. 1D. (D) Inhibition of serum shock-triggered induction of circadian gene expression by pretreatment with U0126. The signals obtained in the RNase protection assays for mPer2 mRNA were quantified and normalized. (●); 20 μm U0126 before 50% serum treatment; (□) 50% serum treatment. (E) Relative levels of mPer1 mRNA induction at t = 70 min (means ± s.d.; n = 3).
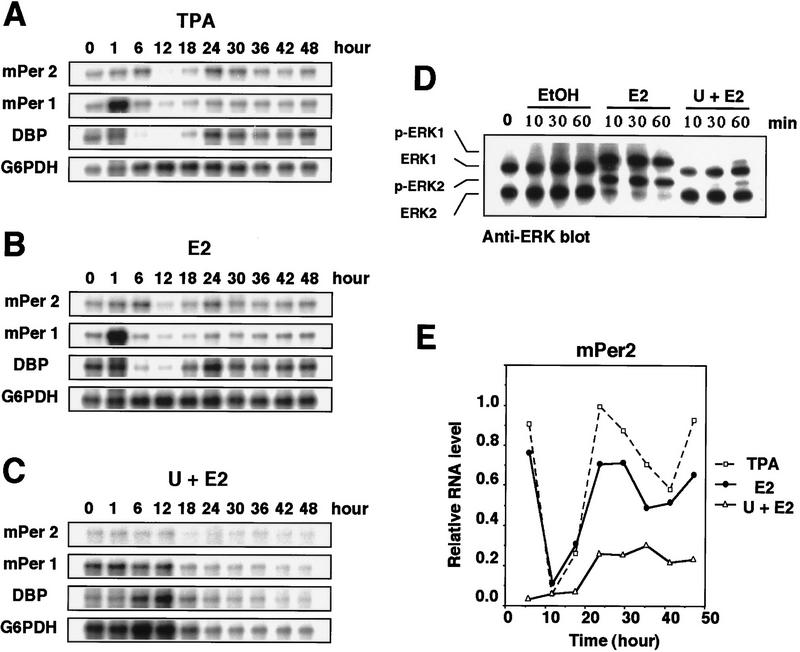
We then examined whether the activation of the MAPK cascade is able to trigger the induction of circadian gene expression. We made use of the ΔB-Raf:ER (estrogen receptor) NIH-3T3 cells in which ΔB-Raf-conjugated with ER is stably transfected (Pritchard et al. 1995). B-Raf is known to function as a specific and direct activator of MEK; in ΔB-Raf:ER cells the addition of estrogen results in immediate activation of ERK/MAPK and the removal of estrogen induces gradual inactivation of ERK/MAPK (Pritchard et al. 1995, Fig. 3D; data not shown). As a control, we treated ΔB-Raf:ER NIH-3T3 cells with TPA (50 nm for 2 hr). This treatment induced immediate expression of mPer1 and triggered the induction of circadian oscillation of expression of the three genes (Fig. 3A,E), the pattern of which was slightly irregular and incomplete, as compared with the oscillation in TPA-treated parental NIH-3T3 cells (Fig. 1C,D). This incompleteness may be intrinsic to the ΔB-Raf:ER cell line. Exposure of the ΔB-Raf:ER cells to estrogen for 1 hr resulted in both the immediate induction of mPer1 and the triggering of the induction of circadian oscillation of the three genes (Fig. 3B,E). The extent of the induction and the pattern of the oscillation were almost identical to those in TPA-treated cells (Fig. 3A,B,E). Pretreatment with the MEK inhibitor U0126 almost completely abolished the estrogen-induced expression of mPer1 and oscillation of the three genes (Fig. 3C,E). It was confirmed that pretreatment with U0126 inhibited the activation of ERK MAPKs (Fig. 3D). These results therefore suggest that activation of the MAPK cascade is sufficient for triggering the induction of the circadian oscillation of gene expression in cultured cells.
Figure 3.
Induction of circadian gene expression triggered by activation of the Raf/MEK/ERK cascade in ΔB-Raf:ER NIH-3T3 cells. TPA (A, 50 nm) or estradiol-17β (E2) (B, 1 μm) was added to the medium (t = 0) and incubated for 2 hr (TPA) or 1 hr (E2), as described. Thirty minutes before the addition of 1 μm E2 (C) 10 μm U0126 was added. The relative levels of the mRNAs were determined by RNase protection assays. Three independent experiments gave similar results. (D) Cells were pretreated with or without 10 μm U0126 for 30 min prior to E2 treatment, and the protein extracts prepared at the indicated times were subjected to immunoblotting with anti-ERK/MAPK antibody. The electrophoretically retarded bands represent active forms. (E) Signals obtained in the RNase protection assays shown in A (□), B (●), and C (▵) for mPer2 mRNAs were quantified and normalized.
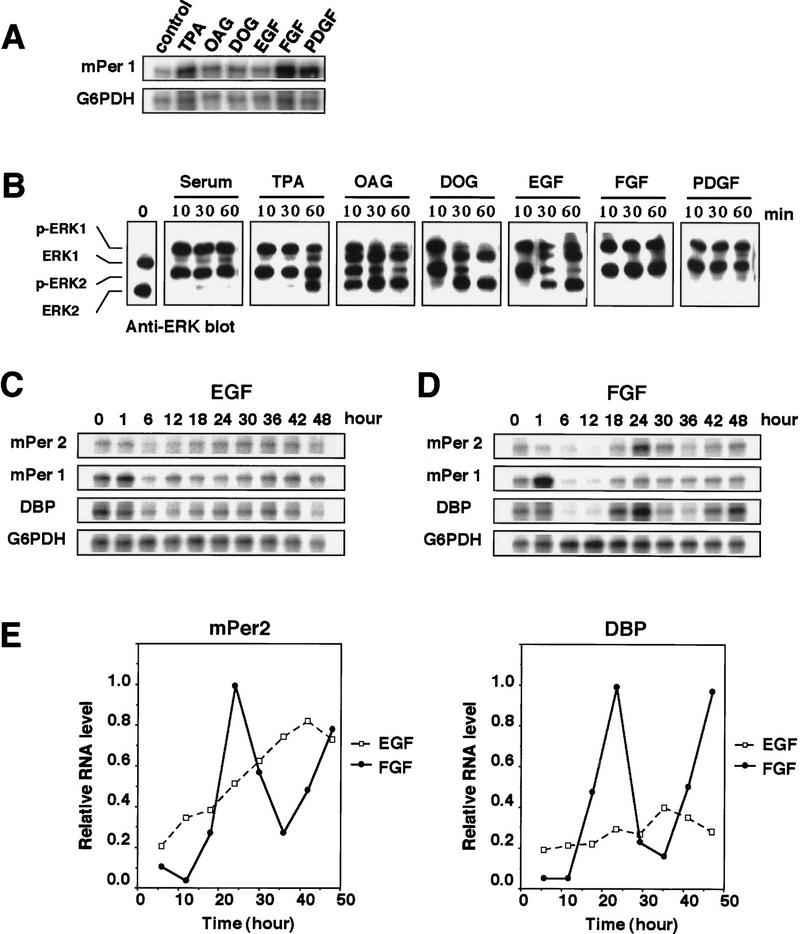
ERK MAPKs are activated in response to various extracellular stimuli including growth factors (Cobb et al. 1991; Nishida and Gotoh 1993; Treisman 1996). Then, we finally tested whether these extracellular stimuli are able to trigger the induction of circadian gene expression. It was reported that treatment of SCN with nerve growth factor (NGF) induced phase shift of circadian rhythm (Bina and Rusak 1996). The ability of several stimuli to induce immediate expression of mPer1 was first examined. Although fibroblast growth factor (FGF) and platelet-derived growth factor (PDGF), in addition to serum (50% serum shock) and TPA, induced the immediate expression of mPer1 strongly, epidermal growth factor (EGF) and membrane-permeable diacylglycerol analogs such as OAG and DOG induced very weakly (Fig. 4A). We then examined their ability to activate ERK MAPKs. Interestingly, serum, TPA, FGF, and PDGF induced prolonged activation of both ERK1 and ERK2, whereas OAG, DOG, and EGF induced their transient and shorter activation; Both ERK1 and ERK2 remained strongly activated 60 min after the treatment in the case of the former, whereas both were mostly inactivated 60 min after in the case of the latter (Fig. 4B). We then examined the expression pattern of the three genes, mPer1, mPer2, and DBP, after treatment with FGF or EGF. The result showed that FGF, but not EGF, triggered the induction of the circadian oscillation of gene expressions (Fig. 4C, D, E). Thus, both the immediate induction of mPer1 and the triggering of the induction of the circadian expression of the three genes correlate well with the prolonged activation of ERK MAPKs.
Figure 4.
Prolonged activation of ERK MAPK is able to trigger the induction of robust circadian gene expression. NIH-3T3 cells were treated with 50% serum, 50 nm TPA, 200 μm OAG, 200 μm DOG, 30 nm EGF, 25 ng/ml FGF, or 30 ng/ml PDGF. (A) Whole-cell RNA was prepared at 70 min after treatment, and the relative levels of the mRNAs were determined by RNase protection assays. (B) Protein extracts prepared at the indicated times were subjected to immunoblotting with anti-ERK/MAPK antibody. EGF (C, 30 nm) or FGF (D, 25 ng/ml) was added to the medium (t = 0) and incubated for 2 hr as described. Relative levels of the mRNAs were determined by RNase protection assays. Three independent experiments gave similar results. (E) Signals obtained in the RNase protection assays shown in C and D (□, EGF; ●, FGF) for mPer2 and DBP mRNAs were quantified and normalized.
The serum-, TPA- or FGF-triggered induction of the circadian oscillation of gene expression observed here is independent of the cell cycle. Balsalobre et al. (1998) also demonstrated clearly that the circadian oscillation of gene expression after serum shock is independent of the cell cycle. The oscillation of circadian gene expression proceeded for 2 days under serum-free conditions after the treatment, whereas the number of the cells did not increase at all (data not shown). Moreover, an increase of cyclin A, which is associated with the progression of the cell cycle, was not detected in the cells (data not shown).
Our results indicate that prolonged activation of the classic MAPK cascade (MEK/ERK) is able to induce immediate expression of mPer1 and trigger the induction of the circadian oscillation of expression of clock and clock-related genes in mammalian cultured cells and therefore suggest that the MAPK cascade has a key role in entrainment of the circadian rhythm in cultured cells. Previous studies have suggested that not only the SCN but also the periphery has a circadian clock (Balsalobre et al. 1998; Rosbash 1998; Sakamoto et al. 1998; Zylka et al. 1998; Dunlap 1999; Ishida et al. 1999), and light and some humoral factor(s) may act as an entraining cue in the SCN and the periphery, respectively (Silver et al. 1996; Oishi et al. 1998; Sakamoto et al. 1998; Dunlap 1999; Ishida et al. 1999); transcriptional activation is an essential event linking the cue and the circadian entrainment as well (Rusak et al. 1990; Kornhauser et al. 1996; Albrecht et al. 1997; Shigeyoshi et al. 1997; Morris et al. 1998; Reppert 1998; Takumi et al. 1998; Dunlap 1999). In the SCN, light induces both ERK activation and immediate-early gene expression (Rusak et al. 1990; Kornhauser et al. 1996; Albrecht et al. 1997; Shigeyoshi et al. 1997; Morris et al. 1998; Obrietan et al. 1998, 1999; Takumi et al. 1998), which in general is mediated by the ERK pathway (Treisman 1996). It has also been reported that PKC activation in the SCN may have a role in rodent circadian rhythm (Biello et al. 1997; McArthur et al. 1997; Schak and Harrington 1999) and that treatment of the SCN with NGF induced the phase shift of circadian rhythm (Bina and Rusak 1996). Taken together, these results suggest that the MAPK cascade may function as a key mediator in common in the signal transduction pathways for entrainment of circadian rhythm in the SCN, the periphery, and immortalized cultured cells and that circadian entrainment by light and humoral factors may employ similar signal transduction mechanisms. Our results also suggest that an unidentified humoral factor(s) that functions as an entraining cue in the periphery may induce the prolonged activation of the MAPK cascade in peripheral cells.
Materials and methods
Cell culture and preparation of RNA samples
NIH-3T3 fibroblasts were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FCS. NIH-3T3 cells stably transfected with ΔB-Raf:ER were grown in DMEM supplemented with 10% FCS. Under these conditions, the cells reached confluence after ∼4 days and were kept for 2 days in medium containing 1% serum. At t = 0, reagents were added to the medium, which was replaced with serum-free DMEM after 2 hr. At the indicated times, total RNA was extracted using an RNeasy mini kit according to the manufacturer's instructions (Qiagen).
Cloning cDNA fragments
Whole-cell RNA from NIH-3T3 cells was reverse-transcribed into cDNA using random hexamers. Fragments of mPer1 (position 412–631 of mPer1 cDNA), mPer2 (position 455–779 of mPer2 cDNA), DBP (position 606–780 of DBP cDNA), and G6PDH (position 309–428 of G6PDH cDNA) were amplified by PCR cloned into pCR2.1-TOPO vector (Invitrogen), and sequenced to verify their identity and orientation.
RNase protection assay
RNase protection assays were performed using a Hybspeed RPA kit according to the manufacturer's instructions (Ambion). The probes were produced from the cDNA fragments described above. The plasmids were linearized with a suitable restriction enzyme, and antisense RNA probes were prepared by in vitro transcription of the linearized templates with T7 RNA polymerase using [32P]UTP. The signals were quantified using a Bio-Rad PhosphorImager. Data were analyzed using Molecular Analyst software (Bio-Rad).
Immunoblotting analysis
After cell lysates were subjected to SDS-PAGE (12%), proteins were transferred to PVDF membrane (Immobilon P, Millipore). Membranes were incubated with anti-ERK2 polyclonal antibody (Santa Cruz Biotechnology) in Tris-buffered saline (20 mm Tris-Cl at pH 7.5, 500 mm NaCl) containing 3% BSA and, subsequently, with HRP-conjugated anti-goat IgG (Santa Cruz Biotechnology). Immunoreactive bands were detected by the ECL Western blotting detection system (Amersham Corp.).
Acknowledgments
We thank M. McMahon for kindly providing ΔB-Raf:ER cells. We also thank Y. Tsuchiya for technical support and members of our laboratory for stimulating discussion. This work was supported by grants from the Ministry of Education, Science, and Culture of Japan (to E.N.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL L50174@sakura.kudpc.kyoto-u.ac.jp; FAX 81-75-753-4235.
References
- Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Biello SM, Golombek DA, Schak KM, Harrington ME. Circadian phase shifts to neuropeptide Y In vitro: Cellular communication and signal transduction. J Neurosci. 1997;17:8468–8475. doi: 10.1523/JNEUROSCI.17-21-08468.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina KG, Rusak B. Nerve growth factor phase shifts circadian activity rhythms in Syrian hamsters. Neurosci Lett. 1996;206:97–100. doi: 10.1016/s0304-3940(96)12432-0. [DOI] [PubMed] [Google Scholar]
- Cobb MH, Robbins DJ, Boulton TG. ERKs, extracellular signal-regulated MAP-2 kinases. Curr Opin Cell Biol. 1991;3:1025–1032. doi: 10.1016/0955-0674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Ishida N, Kaneko M, Allada R. Biological clocks. Proc Natl Acad Sci. 1999;96:8819–8820. doi: 10.1073/pnas.96.16.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhauser JM, Mayo KE, Takahashi JS. Light, immediate-early genes, and circadian rhythms. Behav Genet. 1996;26:221–240. doi: 10.1007/BF02359382. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AJ, Hunt AE, Gillette MU. Melatonin action and signal transduction in the rat suprachiasmatic circadian clock: Activation of protein kinase C at dusk and dawn. Endocrinology. 1997;138:627–634. doi: 10.1210/endo.138.2.4925. [DOI] [PubMed] [Google Scholar]
- Morris ME, Viswanathan N, Kuhlman S, Davis FC, Weitz CJ. A screen for genes induced in the suprachiasmatic nucleus by light. Science. 1998;279:1544–1547. doi: 10.1126/science.279.5356.1544. [DOI] [PubMed] [Google Scholar]
- Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–131. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Humoral signals mediate the circadian expression of rat period homologue (rPer2) mRNA in peripheral tissues. Neurosci Lett. 1998;256:117–119. doi: 10.1016/s0304-3940(98)00765-4. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Pritchard CA, Samuels ML, Bosch E, McMahon M. Conditionally oncogenic forms of the A-Raf and B-Raf protein kinases display different biological and biochemical properties in NIH 3T3 cells. Mol Cell Biol. 1995;15:6430–6442. doi: 10.1128/mcb.15.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM. A clockwork explosion. Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Rosbash M. Why the rat-1 fibroblast should replace the SCN as the in vitro model of choice. Cell. 1998;93:917–919. doi: 10.1016/s0092-8674(00)81197-6. [DOI] [PubMed] [Google Scholar]
- Rusak B, Zucker I. Neural regulation of circadian rhythms. Physiol Rev. 1979;59:449–526. doi: 10.1152/physrev.1979.59.3.449. [DOI] [PubMed] [Google Scholar]
- Rusak B, Robertson HA, Wisden W, Hunt SP. Light pulses that shift rhythms induce gene expression in the suprachiasmatic nucleus. Science. 1990;248:1237–1240. doi: 10.1126/science.2112267. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, et al. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- Schak KM, Harrington ME. Protein kinase C inhibition and activation phase advances the hamster circadian clock. Brain Res. 1999;840:158–161. doi: 10.1016/s0006-8993(99)01787-4. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]