Abstract
The ordered progression through the cell cycle depends on regulating the abundance of several proteins through ubiquitin-mediated proteolysis. Degradation is precisely timed and specific. One key component of the degradation system, the anaphase promoting complex (APC), is a ubiquitin protein ligase. It is activated both during mitosis and late in mitosis/G1, by the WD repeat proteins Cdc20 and Cdh1, respectively. These activators target distinct sets of substrates. Cdc20–APC requires a well-defined destruction box (D box), whereas Cdh1–APC confers a different and as yet unidentified specificity. We have determined the sequence specificity for Cdh1–APC using two assays, ubiquitination in a completely defined and purified system and degradation promoted by Cdh1–APC in Xenopus extracts. Cdc20 is itself a Cdh1–APC substrate. Vertebrate Cdc20 lacks a D box and therefore is recognized by Cdh1–APC through a different sequence. By analysis of Cdc20 as a substrate, we have identified a new recognition signal. This signal, composed of K-E-N, serves as a general targeting signal for Cdh1–APC. Like the D box, it is transposable to other proteins. Using the KEN box as a template, we have identified cell cycle genes Nek2 and B99 as additional Cdh1–APC substrates. Mutation in the KEN box stabilizes all three proteins against ubiquitination and degradation.
Keywords: APC, Cdh1, ubiquitin, proteolysis, specificity, KEN box
Ubiquitin-mediated protein degradation plays a key role at several places in the cell cycle. Attachment of ubiquitin to a lysine residue on a substrate protein is mediated by a ubiquitin-conjugating enzyme (an E2 or UBC) linked to ubiquitin via a thioester. This can occur directly or with the participation of a ubiquitin protein ligase (an E3) (for review, see Ciechanover 1994; Hochstrasser 1996). Several types of ligases have been identified including the anaphase promoting complex (APC) (for review, see Peters 1998; Wolf and Jackson 1998; Zachariae and Nasmyth 1999), the HECT domain proteins (Scheffner et al. 1993, 1995; Kumar et al. 1997), and the SCF complex (for review, see Peters 1998; Zachariae and Nasmyth 1999). Target substrates carrying polyubiquitinated high molecular weight conjugates are recognized and degraded by the 26S proteasome (for review, see Goldberg and Rock 1992; Coux et al. 1996; Zachariae and Nasmyth 1999). The proteasome is not specific to substrates and is active throughout the cell cycle (Mahaffey et al. 1993). The timing and specificity of degradation is regulated at the level of the conjugation of ubiquitin to the substrates.
The ubiquitin ligase, APC, regulates exit from mitosis, late mitotic events, and events in G1 (Irniger and Nasmyth 1997; Fang et al. 1998; for review, see Zachariae and Nasmyth 1999). Recent reports have shown that WD-containing proteins Cdc20 (Fizzy in Drosophila) and a related protein Cdh1 (Fizzy-related in Drosophila) can activate APC (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998). Each activator associates with APC in a cell cycle-dependent manner and is thought to target a distinct set of substrates. Cdc20-activated APC corresponds to the mitotic form of APC. All known Cdc20–APC substrates contain a destruction box, composed of the sequence R-X-X-L-X-X-X-X-N (Glotzer et al. 1991). In metaphase, APC targets the anaphase inhibitors now called securins (Pds1 in Saccharomyces cerevisiae, Cut2 in Schizosaccharomyces pombe, PTTG in human) for degradation, allowing for sister chromatid separation (Cohen-Fix et al. 1996; Funabiki et al. 1996; Yamamoto et al. 1996; Zou et al. 1999). Subsequent targeting of mitotic cyclins and Ase1 allow cells to exit mitosis (Glotzer et al. 1991; Juang et al. 1997).
In late mitosis Cdh1 activates APC. Cdh1–APC recognizes both D-box and non-D-box-containing substrates (Fang et al. 1998), including the non-D-box (in vertebrates) substrate Cdc20 itself. Cdc20 protein levels fluctuate with the cell cycle (Weinstein 1997; Prinz et al. 1998), rising in G2, peaking in mitosis, and declining as cells exit mitosis and enter G1. The rapid loss of Cdc20 at the end of mitosis is thought to be due to protein degradation targeted by Cdh1–APC (Fang et al. 1998; Prinz et al. 1998; Shirayama et al. 1998). Human Cdc20 lacks a D box (even though the budding yeast Cdc20 contains two D boxes) but is recognized by Cdh1–APC, making it a good candidate for analyzing Cdh1-dependent degradation.
We investigated the specificity in protein substrates that are recognized by Cdh1–APC but lack an obvious D box. We show that Cdh1–APC acts in a sequence-specific manner and is not generally a permissive for all proteins. The lack of a D-box requirement reflects not a relaxed specificity, but a strict requirement for specific sequences including the D box. We identify a new targeting signal, the KEN box, which directs the recognition of Cdc20 by Cdh1 and is transposable like the B-type cyclin D box to other proteins. Using this information about target specificity, we also identify the cell cycle-regulated proteins Nek2 and B99 as Cdh1 substrates, and show that the KEN box directs their ubiquitination and subsequent degradation.
Results
Cdh1 and Cdc20 confer on APC different substrate preferences for ubiquitination and for degradation
Xenopus egg extracts represent a complete system for regulated cyclin degradation (Murray and Kirschner 1989; Murray 1991). Interphase extracts supplemented with a nondegradable form of sea urchin cyclin B (Δ90) enter and remain in a mitotic state (Glotzer et al. 1991), in which they degrade mitotic APC substrates, such as securin/Pds1 (Zou et al. 1999) and cyclin B (Glotzer et al. 1991). Cdh1 is absent in the egg and does not appear during embryonic development until the mid-blastula transition (Lorca et al. 1998), so Xenopus egg extracts do not show degradation of known Cdh1 substrates. However, by supplementing interphase extracts with baculovirus-purified hCdh1 protein, we were able to reconstitute Cdh1-dependent degradation (Fig. 1).
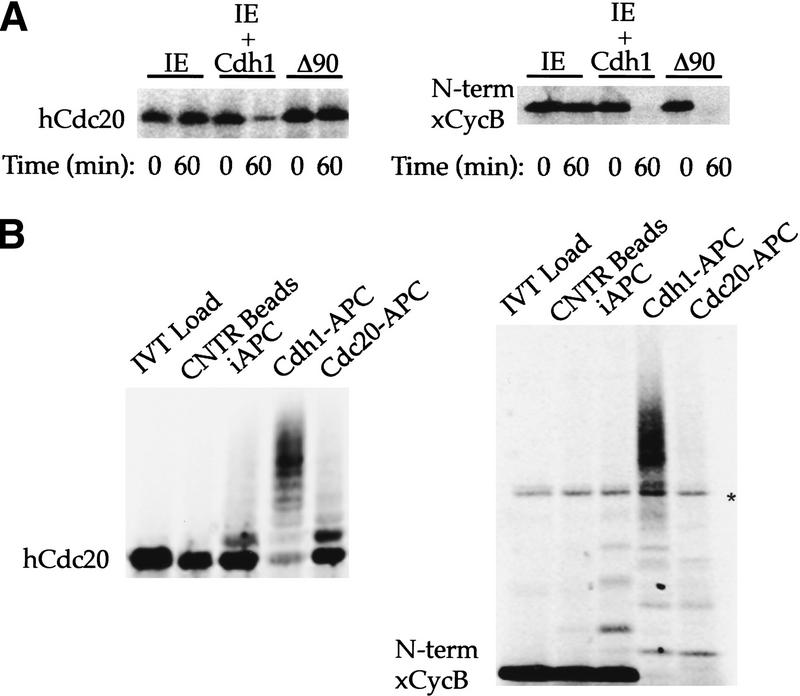
Figure 1.
Recognition of Cdc20 as a substrate by Cdh1–APC but not Cdc20–APC. (A) Incubation of 35S-labeled in vitro-translated hCdc20 and the amino terminus of Xenopus cyclin B (xCycB) in interphase extract supplemented with buffer (IE), interphase extract supplemented with Cdh1 protein (IE + Cdh1), or interphase extract pushed into mitosis by sea urchin cyclin B Δ90 (Δ90). Both hCdc20 and xCycB are stable in IE, but degrade rapidly in IE + Cdh1. xCycB, and not hCdc20, disappears in the mitotic extract. (B) Incubation of hCdc20 and amino-terminal xCycB in a purified system ubiquitination assay using control beads (Cntr Beads), immunopurified interphase APC on beads (iAPC), or iAPC on beads activated by Cdh1 (Cdh1–APC) or Cdc20 (Cdc20–APC). Neither substrate forms high molecular weight conjugates in the presence of control beads or iAPC, but both form significant higher molecular weight conjugates in the presence of Cdh1–APC. Only xCycB forms higher molecular weight conjugates in the presence of Cdc20–APC. Polyubiquitinated species of xCyc B spread out more in the presence of Cdc20–APC than in the presence of Cdh1–APC, creating a much lighter smearing not always easily visualized for the 35S-labeled cyclin fragment as with iodinated substrate in previous reports. A darker exposure (not shown) makes visualization of the conjugates more obvious. An asterisk indicates the presence of a background band present in the in vitro translation products.
hCdc20 and the amino-terminal fragment of Xenopus cyclin B are both stable in interphase extracts in which APC, presumably, is inactive because of the absence of a bound activator. Both proteins become unstable upon the addition of Cdh1 to the interphase extracts (Fig. 1A). When Δ90 cyclin B is added to an interphase extract, the extract enters mitosis, in which the active form of APC is the form associated with Cdc20. hCdc20 remains stable, but amino-terminal Xenopus cyclin B degrades rapidly. This demonstrates a clear difference in specificity between the Cdc20- and Cdh1-activated forms of APC in the complete extract system.
The direct recognition of hCdc20 by Cdh1–APC can be studied by generating ubiquitin conjugates in a purified system. In this system, APC is immunopurifed from interphase extracts with anti-Cdc27 antibody coupled to beads and activated with baculovirus-expressed hCdh1 or hCdc20 protein. The subsequent addition of purified E1, Ubcx, ubiquitin, and an energy-regenerating system results in ubiquitination of [35S]-methionine-labeled in vitro translation products of APC substrates (Fang et al. 1998). As substrates become conjugated to ubiquitin, the signal corresponding to the unmodified substrate disappears and higher molecular weight species are produced. In the purified system there is always some very low level of ubiquitin conjugation of APC substrates. Hence, it is the quantitative loss of the unmodified protein and its conversion of the input peptide into high molecular weight conjugates that we consider evidence for significant activity on a given substrate.
Few conjugates of ubiquitin to hCdc20 or the amino terminus of xcyclin B occur with control beads (beads incubated with extract, but lacking Cdc27 antibodies), or with unactivated interphase APC on beads (Fig. 1B). In the presence of Cdc20–APC, the pattern of conjugation to hCdc20 and to xcyclin B is strikingly different. Except for a few monoubiquitinated species, there is no evidence of conjugation of Cdc20 to ubiquitin. In contrast, the cyclin B signal is converted completely to conjugated species. With Cdh1-activated APC, there is conjugation of ubiquitin to both substrates. The difference in specificity between Cdc20–APC and Cdh1–APC for degradation, therefore, is preserved in the purified system for ubiquitination and does not require additional elements from the extract. In the presence of unactivated interphase APC, there is a small background of monoubiquitination, but little quantitative conversion of the unmodified substrate.
Specificity versus permissivity of Cdh1–APC
The budding yeast homolog of Cdc20 contains two destruction boxes (shown schematically in Fig. 2A), but the vertebrate homologs lack any motif similar to the R-L-N of the D box. Furthermore, previous work (Fang et al. 1998; Gieffers et al. 1999) reports the ability of Cdh1–APC in the purified system to recognize the amino terminus of Xenopus cyclin B with the D box deleted. To determine how Cdh1–APC recognizes substrates lacking a D box, whereas apparently both WD-repeat-containing proteins confer on APC the capacity to recognize D box-containing substrates, we first tested the less interesting possibility that Cdh1–APC simply shows little substrate selectivity. Previous work (Fang et al. 1998) had indicated that Cdh1 was not completely permissive in the purified system. In the Cdh1–APC ubiquitination assay on a random sampling of in vitro-translated individual proteins as well as in vitro-translation products from cDNA pools, they showed that Cdh1did not recognize any of the proteins chosen. We performed a similar random sampling of 10 cDNA pools from a Xenopus-dorsalized gastrula library containing ∼30 clones each. We tested the in-vitro translation products of each pool in the purified system Cdh1–APC ubiquitination assay as well as the degradation assay of Cdh1-supplemented interphase extracts. Of the 300 clones assayed in both systems, we found only 2 positives. The positives were isolated from the pools, sequenced, and identified to be xcyclin B1 and xcyclin B3. Because the only two clones to respond positively in these assays are well-documented substrates containing the specific targeting signal, the D box, we conclude that Cdh1 is not permissive in its activation of APC, but clearly targets substrates with some unknown degree of specificity.
Figure 2.
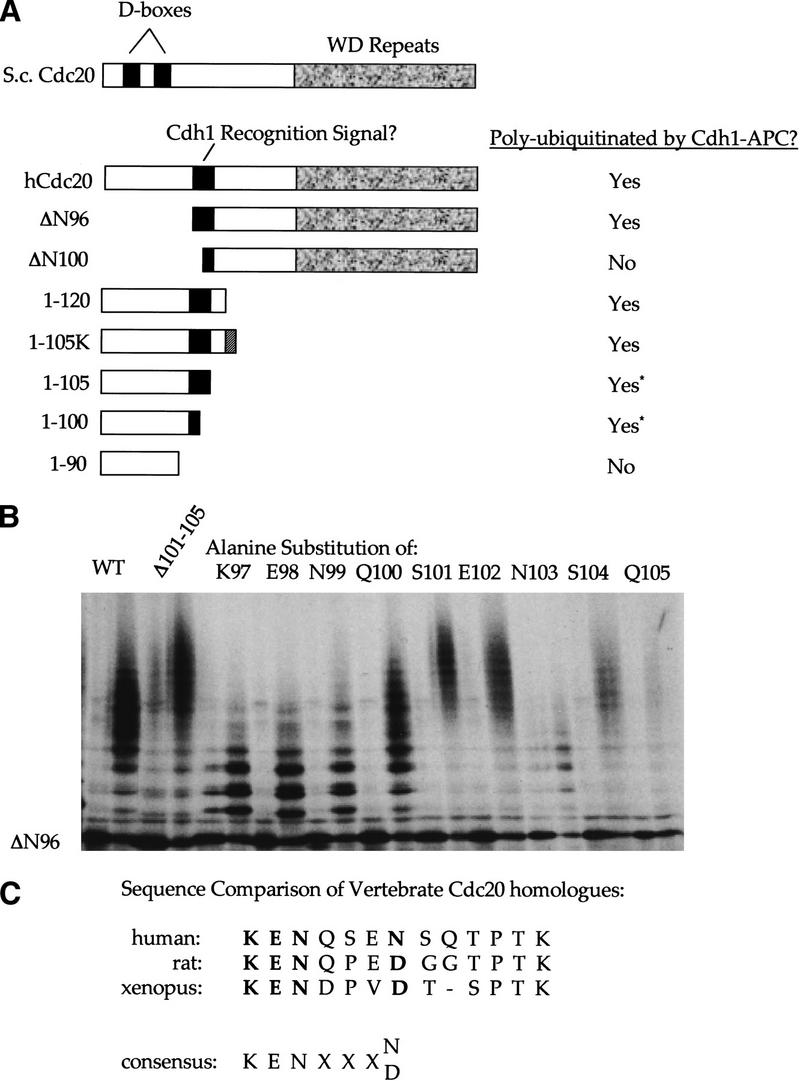
Isolation of the region in hCdc20 recognized by Cdh1–APC. (A) Schematic representation comparing yeast and human Cdc20. D boxes are indicated by black boxes. The WD repeat region is indicated by a textured gray box. Amino- and carboxy-terminal deletion mutants of hCdc20 were incubated in the purified system ubiquitination assay in the presence of Cdh1–APC, and the results are summarized at right. Deletion mutants narrowed the region recognized by Cdh1–APC, shown as a solid gray box. (B) Ubiquitination assays of hCdc20 point mutants using Cdh1–APC. Each pair represents an IVT load and the Cdh1–APC ubiquitination reaction. (WT) An internal deletion of amino acids 101–105, individual alanine substitution mutants of amino acids 97–105 within the Δ96 context are shown. All alanine mutants produced a ubiquitination profile showing high molecular weight conjugates except for K97A, E98A, N99A, and N103A. These four substitution mutants formed only lower molecular weight conjugates representing monoubiquitinated species. (C) Comparison of various vertebrate Cdc20 sequences of this region. Residues shown to play a role in recognition by the alanine scanning mutagenesis are shown in bold. Below the sequence comparisons, a putative consensus for Cdh1 recognition on the basis of sequence comparison and experimental results is suggested. The Cdc2 site is highly conserved among species (even in clam, not shown) even though it is not required for recognition by Cdh1.
Identifying the KEN box
Given the absence of an obvious D box in vertebrate Cdc20, we systematically deleted sequences from the amino and carboxyl termini of human Cdc20 (represented schematically in Fig. 2A) and assayed ubiquitination using Cdh1-activated APC (results summarized to the right of the schematic). Deletion of the amino-terminal 96 amino acids did not alter the ubiquitination profile in terms of amount of full-length protein converted to polyubiquitinated species, or the average number of ubiquitins added per molecule, but deletion of the amino-terminal 100 amino acids blocked ubiquitination almost completely. A few conjugates formed, but most of the original protein remained unconjugated. Those conjugates that did form were probably monoubiquitinated on, at most three sites. The amino-terminal 120 amino acids showed a wild-type ubiquitination profile, and the amino-terminal 100 and 105 amino acids were each sufficient for a ubiquitin laddering of up to nine ubiquitins added per molecule, even though we did not observe the smearing associated with branched polyubiquitinated chains. The amino-terminal 90 amino acids remained largely unconjugated or formed only monoubiquitinated species.
Despite the lower stoichiometry of ubiquitination, the 100 and 105 amino acid amino-terminal fragments formed conjugates with almost complete modification of the initial input peptide. The reduced level of conjugation as compared with that of the amino-terminal 120 amino acid fragment might have been due to the presence of additional lysines and not specific sequence elements involved in recognition. To test this idea, we tagged the amino-terminal 105 mutant with four additional lysines at its carboxyl terminus. This restored the branched polyubiquitin smearing seen with the 120 amino acid mutant, confirming that the smearing profile resulted from availability of lysines, not from specific sequence recognition elements.
Given the identification of the region between amino acids 97–105 as required for ubiquitination, we created a series of alanine substitutions in this region within the Δ96 construct. Substitution of four amino acids produced a marked drop in ubiquitination (Fig. 2B) compared with the Δ96 construct. The K97, E98, N99, and N103 alanine substitutions showed a collapse from the polyubiquitin smearing to a monoubiquitin laddering. Because we had seen significant ubiquitination of an amino-terminal 100 amino acid fragment, we were puzzled by the effect of alanine substitution at position N103, outside of the amino-terminal domain. To study the role of the N103 position, we created a Δ96 mutant containing an internal deletion of amino acids 101–105. This mutant formed polyubiquitinated chains of significantly greater average length than the Δ96 mutant (Fig. 2B). N103, in the context of the amino-terminal fragment of Cdc20, therefore, is not required for recognition, but it might contribute to recognition, partially in the context of surrounding residues.
PEST sequences are sequences enriched for the amino acids proline, glutamic acid, serine, and threonine and have been shown previously to contribute to protein instability (Rogers et al. 1986). Amino acids 97–108 of Cdc20 qualify as a PEST sequence (http://www.at.embnet.org/embnet/tools/bio/PESTfind/). Changing S101, E102, S104, T106, or P107 to alanine should reduce ubiquitination of Cdc20 if recognition depends on this sequence serving as a PEST sequence. However, these point mutants within the Δ96 context did not reduce recognition. The average length of the polyubiquitin chains formed actually increased following substitution of alanine for the PEST residues S101, E102, and S104.
The human, rat, and Xenopus homologs of Cdc20 in this region (Fig. 2C) are conserved for the required K-E-N residues. The fourth position, shown to contribute to recognition, although not being required, is an asparagine in the human sequence, and an aspartic acid in the other vertebrate homologs shown. Replacement of N103 in the Δ96 context with aspartic acid has no effect on the ubiquitination profile (data not shown), confirming that a similar structure that carries a negative charge is acceptable at this position.
Mutation of the KEN box blocks recognition by Cdh1–APC
To confirm the importance of the K-E-N-X-X-X-N sequence for ubiquitination and subsequent degradation of Cdc20, we constructed a mutant in which all four residues shown to be important by alanine scanning (K97, E98, N99, and N103) were changed to alanine within the full-length protein. This mutant (hCdc20KENN-A) showed decreased ubiquitination compared with wild-type Cdc20, both in terms of the extent and rate of conjugation (Fig. 3A). Furthermore, hCdc20KENN-A was much more stable than wild-type protein in Cdh1-supplemented extracts (Fig. 3B). To rule out gross misfolding of the mutant protein, we compared the ability of hCdc20KENN-A with wild-type hCdc20 to activate APC. Mutant protein showed similar activity to wild-type protein in stimulating cyclin ubiquitination in the purified system (Fig. 3C).
Figure 3.
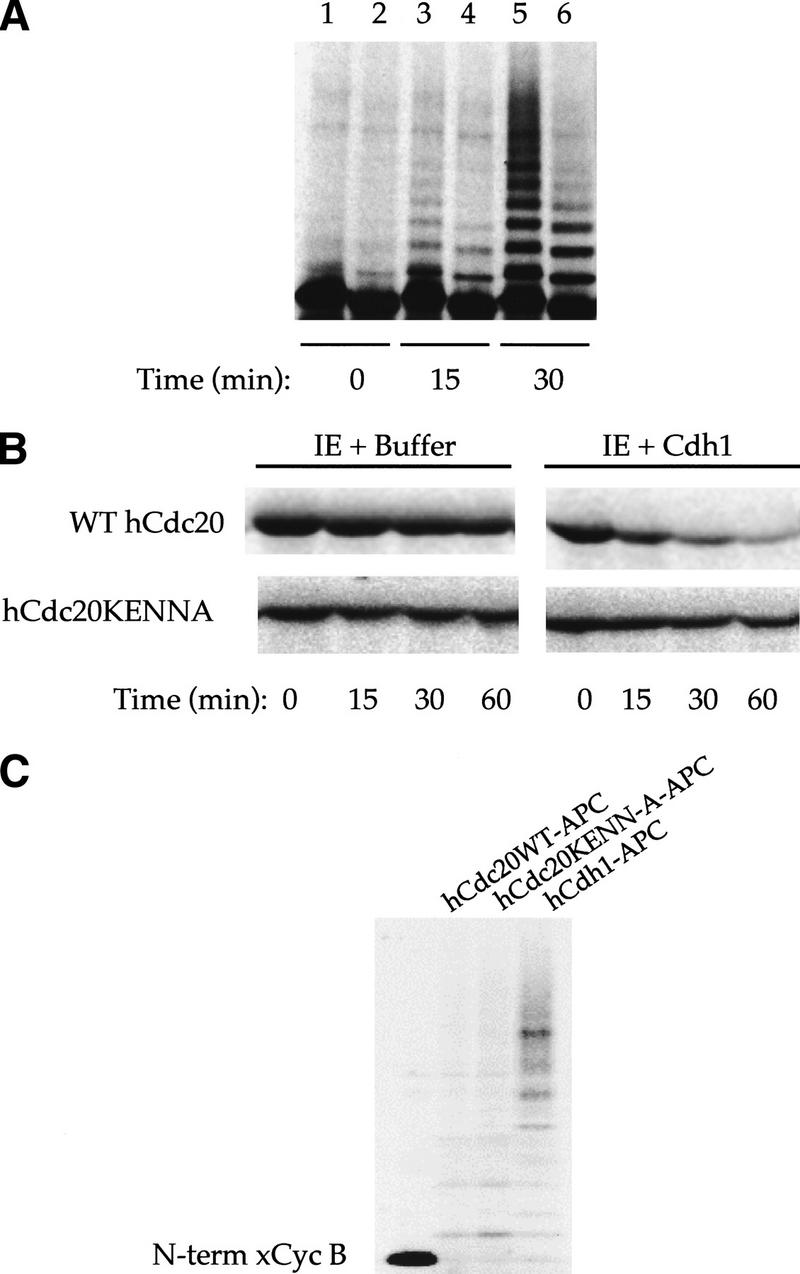
Cdh1–APC does not recognize hCdc20 with mutant KEN-box recognition signal. (A) Time series of ubiquitination of wild-type hCdc20 (lanes 1,3,5) and mutant hCdc20KENN-A (key recognition residues changed to alanine—97AAAQSEA103, lanes 2,4,6). The side-by-side comparison shows reduced and slower ubiquitination of the mutant by Cdh1–APC. After 30 min, significant polyubiquitinated species have formed for wild-type hCdc20, whereas only monoubiquitinated species exist for the mutant. (B) Degradation assays using interphase extracts supplemented with buffer or Cdh1 show rapid degradation of wild-type but not mutant hCdc20. (C) Ubiquitination assay of Xenopus amino-terminal cyclin B using similar amounts of cold in vitro translated wild-type hCdc20–APC, hCdc20KENN-A–APC, and hCdh1–APC to activate iAPC on beads. Wild-type and mutant Cdc20 show similar ability to recognize amino-terminal xcyclin B.
Recognition of the R-L-N D box motif by Cdh1–APC: a cryptic D box in Xenopus cyclin B
Previous work showed that Cdh1–APC can promote the ubiquitination of an amino-terminal fragment of Xenopus cyclin B lacking the D box at position 36–44 (Fang et al. 1998; Gieffers et al. 1999). Although there were no KEN-box sequences in this fragment, we did find another R-L-N motif at the extreme amino terminus, positions 7–15 that had been overlooked previously (Fig. 4A). This potential D box is present only in Xenopus cyclin B, and is not found in other species. Because deletion of the conserved 36–44 D box blocks degradation in mitotic extracts (King et al. 1996), the 7–15 D box of cyclin B cannot in itself promote mitotic degradation. To test whether this additional R-L-N motif was responsible for the recognition of cyclin ΔD box by Cdh1 in the purified system, we constructed mutants of the amino-terminal fragment that lacked this second D box or double mutants lacking both D boxes. Ubiquitination assays revealed that deletion of the 7–15 D box alone resulted in a slight drop in ubiquitination by Cdh1 as compared with wild type. However, deletion of both D boxes eliminates ubiquitination completely (summarized quantitatively in Fig. 4B). Thus, Cdh1–APC recognizes D boxes or KEN boxes in target proteins.
Figure 4.
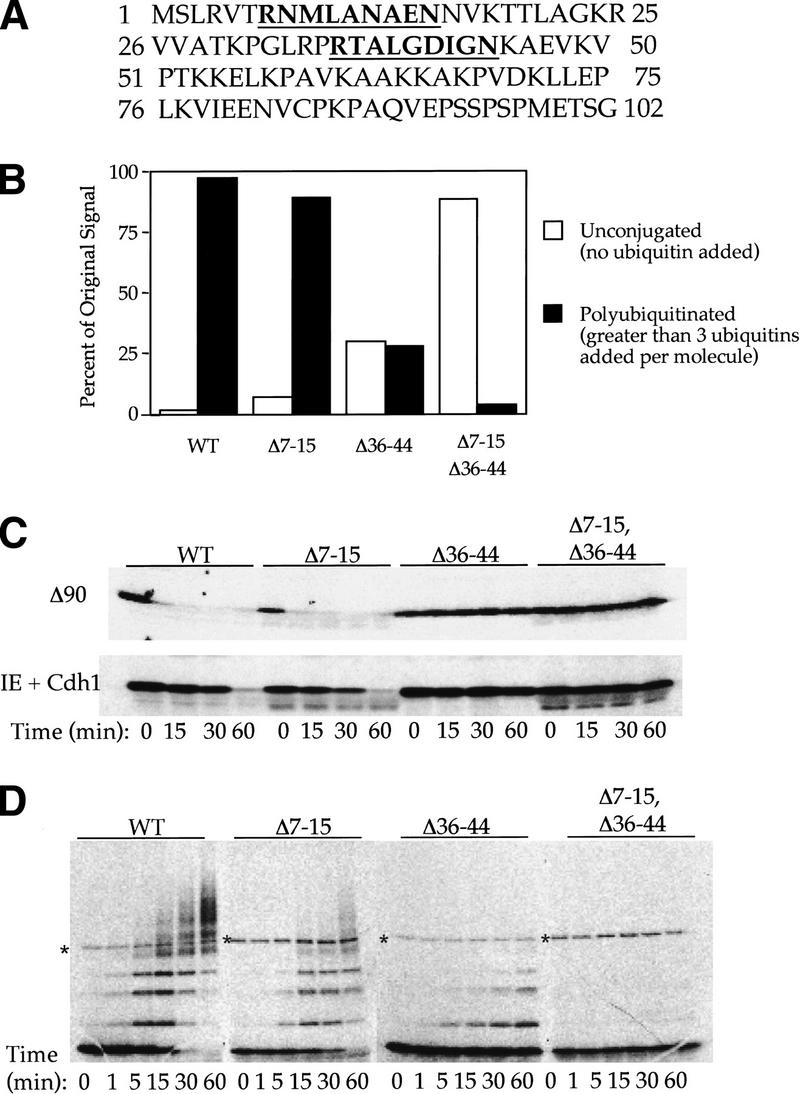
Cdh1 recognizes the R-L-N destruction box motif. (A) Sequence of Xenopus cyclin B amino terminus. Both R-L-N motifs are underlined and shown in bold. The first R-L-N motif, at position 7–15 is not conserved between species, whereas the second, at position 36–44, is well conserved and well characterized as the B-type cyclin D box. (B) Quantitation of ubiquitination assays in the purified system using Cdh1–APC in the presence of 35S-labeled in vitro translated substrate. Bar graph depicting unconjugated signal remaining after a 1-hr incubation with Cdh1–APC (open bar) and polyubiquitinated (greater than three ubiquitins added per molecule) species formed for each mutant (solid bar). Polyubiquitinated species form for wild-type Xenopus amino-terminal cyclin B as well as the Δ7–15 and Δ36–44 mutants. Only monoubiquitination occurs for the double mutant, and most of the signal remains unconjugated. The unconjugated and polyubiquitinated percentages do not add up to 100%; the remaining signal reflects monoubiquitinated species not shown in the graph. (C) Degradation assays of cyclin and D-box mutants in the extract system Cdh1-supplemented interphase extracts and Δ90 extracts. Both mutants that lack the 36–44 D box are stable in the Δ90 and Cdh1-supplemented extracts. (D) Time series of Cdh1–APC ubiquitination assay of xCycB and D-box deletion mutants using 20% of ubiquitin in the other assays. By limiting the addition of ubiquitin, the specificity of the extract system from C is restored in the purified system. The Δ36–44 mutant no longer forms significant polyubiquitin chains. An asterisk indicates a background band from the in vitro translation products.
Both in degradation assays and in ubiquitination assays, we have found quantitative differences in target sequence recognition. For example, in Figure 4b, Cdh1–APC recognizes the standard D-box deletion (Δ36–44), by promoting nearly 30% of the original protein into polyubiquitinated species; wild type show nearly 100% conversion. Even though there is some ubiquitination of this deletion in the purified system, that particular deletion is completely inactive for degradation in the Cdh1-supplemented extracts (Fig. 4C). In contrast, both the wild-type and Δ7–15 constructs were rapidly degraded under similar conditions. The difference between ubiquitination with purified components and degradation in extracts may be due to the optimized conditions for ubiquitination in the purified system, in which there is no isopeptidase activity. Alternatively, certain crucial components may be limiting or be subject to competition in the cell and in extracts, so that recognition occurs only for optimal substrates.
To restore artificially some of the selectivity to the purified system, we reduced the concentration of ubiquitin. At a ubiquitin concentration of 20% of the original amount, selectivity at the level of ubiquitination was much more apparent (Fig. 4D). Conjugation of the Δ36–44 D-box mutant was reduced to monoubiquitination with no higher molecular weight conjugates, whereas the Δ7–15 mutant was ultimately fully conjugated to ubiquitin, although there was a significant decrease in the rate and extent of conjugation compared with wild type. Thus, under less optimal conditions, substrate selectivity was enhanced and different D boxes were differentially active.
D-box and KEN-box-containing peptides completely inhibit degradation of full-length Cdc20
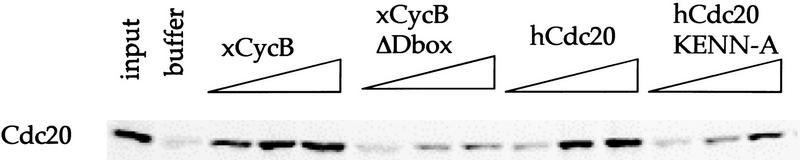
The amino-terminal peptides of Xenopus cyclin B (McGarry and Kirschner 1998; Zou et al. 1999) and sea urchin cyclin B (Holloway et al. 1993) containing the conserved D box, have been well characterized as competitive substrates for Cdc20–APC degradation in mitotic extracts. Because Cdh1–APC recognizes D-box substrates, we asked whether the amino-terminal peptide of cyclin B can compete for Cdh1-dependent degradation. We find that the amino-terminal cyclin B peptide competed for hCdc20 degradation in Cdh1-supplemented, whereas the corresponding deletion mutant of the D box did not compete (Fig. 5). Because the amino-terminal 120 amino acids of hCdc20 is an excellent Cdh1 substrate, we tested the ability of this peptide also to inhibit Cdh1-dependent degradation. As shown in Figure 5, wild-type amino-terminal Cdc20 blocked degradation of full-length hCdc20 5 μm. The KEN-box mutant amino-terminal peptide shows partial inhibition at 10 μm and none at 5 μm. At 5 μm, the mutant caused no increase in inhibition over a buffer control, whereas 5 μm of wild-type KEN-box peptide stabilized initial substrate protein to >75%.
Figure 5.
Peptides containing, but not lacking, the D box or KEN box inhibit degradation of Cdc20 in Cdh1-supplemented extracts. Degradation of full-length hCdc20 in Cdh1-supplemented interphase extracts. The left-most lane shows the initial input of substrate for each condition. Subsequent lanes indicate 1-hr incubations of buffer, followed by incubation with 1, 5, and 10 μm amounts of amino-terminal xcyclin B, Δbox amino-terminal xcyclin B, amino-terminal hCdc20, and amino-terminal hCdc20KENN-A.
The KEN box acts as a transposable degradation signal
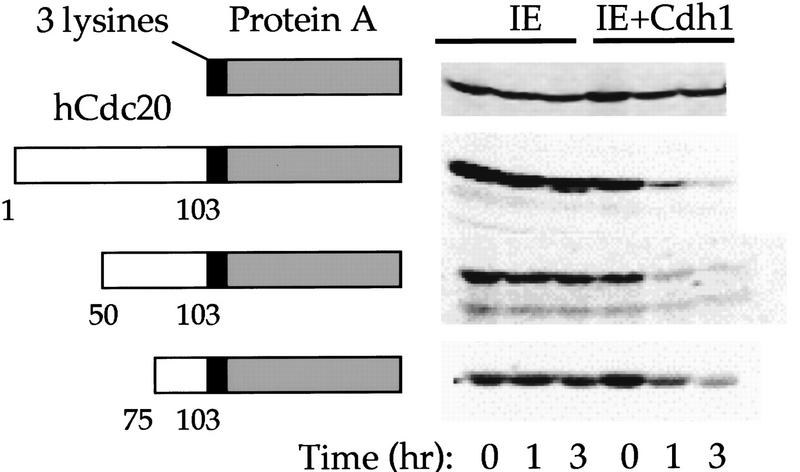
A key property of the cyclin B D box is that the degradation signal is transposable. Fusion of a small region containing the cyclin B D box with the immunoglobulin binding region of protein A (a protein stable in mitotic extracts, therefore not recognized by Cdc20–APC) resulted in rapid degradation of the fusion protein in mitotic extracts (Glotzer et al. 1991; King et al. 1996). The immunoglobulin-binding region of protein A is not recognized by Cdh1–APC in ubiquitination or degradation assays. We constructed a series of fusions of the region containing 97–103 to protein A and assayed these fusion proteins for recognition by Cdh1–APC using both the ubiquitination and degradation assays. Fusion of the amino-terminal 103 amino acids, or even the amino-terminal 100 amino acids (containing KENQ, but lacking the second asparagine residue shown to contribute to recognition in the point mutant analysis) to protein A resulted in a ubiquitination profile similar to full-length wild-type Cdc20 protein (not shown). Smaller fusions reduced the ubiquitination profile but still yielded visible ubiquitin conjugates, whereas protein A alone was not recognized at all. These fusion proteins were stable in interphase extracts supplemented with buffer, but degraded in the presence of Cdh1 (Fig. 6). Fusion of the KEN box alone did not result in ubiquitination or degradation (not shown), but fusion of the 28 amino acid region from 75–103 was sufficient to allow ubiquitination and degradation, although the degradation occurred somewhat more slowly than full-length hCdc20 or the larger fusion proteins.
Figure 6.
The Cdc20 KEN box acts as a transposable ubiquitination and degradation signal. (Open box) Schematic representation of fusion proteins containing the hCdc20 KEN box; (shaded box) and the immunoglobulin-binding region of protein A. Three lysines have been inserted at the hinge region (small black box). The schematic is not scaled to size. Degradation assays using in vitro translated proteins in IE or IE + Cdh1 are shown at right.
The KEN-box sequence can be used to predict additional Cdh1 substrates
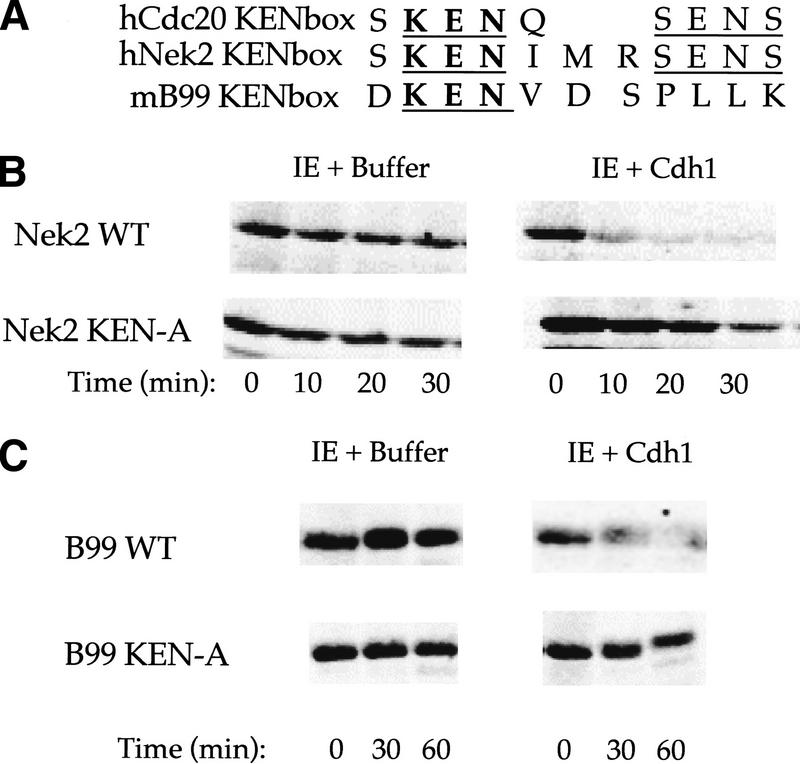
Because the KEN box directs the ubiquitination of Cdc20 by Cdh1 and can be transferred to another protein, we investigated to see whether this sequence serves a general targeting signal or is specific to Cdc20. Database searching for other KEN-box-containing proteins revealed that the serine–threonine kinase Nek2, a NimA related kinase, contains a region extremely similar to the recognition signal in Cdc20 (Fig. 7A). NimA has been identified recently as an APC substrate in Aspergillis nidulans (for review, see Zachariae and Nasmyth 1999) making Nek2 a likely Cdh1–APC candidate. Furthermore, Nek2 has been reported to vary with the cell cycle, peaking in protein expression level and activity in S and G2 and disappearing during mitosis and G1 (Schultz et al. 1994). The sequence of B99, a p53-inducible gene that peaks in expression during G2 (Utera et al. 1998), also contains a putative KEN box (Fig. 7A).
Figure 7.
Cdh1–APC targets cell cycle proteins Nek2 and B99 and recognizes the KEN box as a general targeting signal. (A) Sequence comparison of the Cdc20 KEN box with the KEN-containing regions of human Nek2 and mouse B99. KEN residues are in bold and conserved residues are underlined. Incubation of Nek2 (B) and B99 (C) and corresponding alanine substitution KEN mutants IE and IE + Cdh1. Both Nek2 and B99 form polyubiquitin chains in the presence of Cdh1–APC in the purified system and degrade rapidly in the presence of Cdh1 in the extracts. Alanine substitution of the KEN box stabilizes these mutants in the extract.
In the purified system, both Nek2 and B99 show significant ubiquitin conjugation by Cdh1–APC (not shown) but not with antibody control beads. Nek2, but not B99, shows some degree of recognition by unactivated interphase APC, but conjugation is still not as great as that achieved by Cdh1–APC. Nek2, in particular, forms significant branched chains of varying lengths that extend over a much broader range than for other substrates. Because the conjugates are more spread out as they run on a gel, we do not visualize the dark smear commonly seen for hCdc20.
Nek2 is degraded in interphase extracts with a half-life of 50–60 min (not shown). Addition of Cdh1 to these extracts results in complete degradation of Nek2 by 10 min (Fig. 7B), a 12-fold increase in rate. Alanine substitution of the KEN box in Nek2 significantly stabilizes the protein against both ubiquitination in the purified system (not shown) as well as degradation in extracts (Fig. 7B). B99 is completely stable in interphase extracts but is degraded with a half-life of 25–30 min when the extracts are supplemented with Cdh1. Alanine substitution of the B99 KEN box completely stabilizes the protein to degradation (Fig. 7C).
Discussion
Genetic and biochemical studies suggest that the WD repeat proteins, Cdc20 and Cdh1, sequentially activate the ubiquitin ligase APC during mitosis to achieve a temporal order of substrate degradation. Cdc20 itself is a substrate for APC when APC is activated by Cdh1, and therefore, Cdc20 is no longer present at the end of mitosis; Cdh1 is a stable protein present throughout the cell cycle, whose interaction with APC is most likely regulated by phosphorylation (Kotani et al. 1999). Although it has been suggested that Cdc20 and Cdh1 might confer different substrate specificities on APC, the nature of substrate recognition is still unclear. In the SCF complex, the corresponding WD proteins such as HOS or βTrCP (Margottin et al. 1998; Yaron et al. 1998; Fuchs et al. 1999) bind directly to the substrate, whereas the affinity of Cdc20 and Cdh1 with potential substrates has not been shown, possibly because it is too low to measure. Mutagenesis experiments in many species defines a target sequence recognized by APC that has been activated by Cdc20. This sequence, which includes the destruction box plus some nonconserved lysine containing sequences, has been identified on all species of cyclin B plus several other proteins degraded at metaphase or soon after, such as Geminin, Pds1 (securin), Ase1, and others. The targeting sequence to APC activated by Cdh1 has, up to now, been undetermined. Its characterization, the subject of this work, would allow a better understanding of specificity and temporal order of protein degradation during mitosis and in G1.
We have used Cdc20 itself as a substrate for Cdh1–APC to define recognition sequences within Cdh1–APC substrates. Cdc20 has been shown to be degraded late in mitosis (Weinstein 1997) in an APC-dependent manner (Prinz et al. 1998; Shirayama et al. 1998) and to be a substrate of Cdh1–APC in vitro. Using ubiquitination assays in a completely defined system and degradation assays in complete extracts, we have examined mutants of Cdc20 to learn the requirements for ubiquitination and degradation by Cdh1–APC. Deletion mutagenesis narrowed down the recognition region to amino acids 97–105, and point mutants in this region identified four residues that decreased recognition when changed to alanine, K-E-N-X-X-X-N, (in which aspartic acid in the final position supported similar polyubiquitination as the asparagine). However, within a truncated fragment of Cdc20, deletion of residues 101–105, containing the second crucial asparagine, showed a greater than wild-type response. This indicates to us that the second asparagine is not required for recognition overall, but that it may be required within the context of the surrounding residues, perhaps modulating a negative regulatory region.
Sequence alignment between the vertebrate Cdc20 sequences and the Cdc20 sequence of S. cerevisiae show no KEN-similar motif in the same region of the protein. However, there is a potential KEN box at position 586 near the carboxyl terminus of S. cerevisiae Cdc20 (protein YGL116w), K-E-N-X-X-X-N (pointed out to us by E. Hildebrant and A. Hoyt, pers. comm.). The carboxy-terminal KEN boxes of hNek2 and mB99 support ubiquitination and degradation, suggesting the possibility that the KEN box in any part of the protein could be functional. We do not know if recognition of the KEN box extends to the yeast system or is limited to vertebrate systems, or whether there are undiscovered Cdh1–APC recognition signals in other proteins.
Previous reports already suggested that Cdh1–APC differed from Cdc20–APC in that it could mediate the ubiquitination of substrates like hCdc20 that lack a D box and substrates like amino-terminal xcyclin B from which the conserved D box had been deleted (deletion of residues 36–44). A possible interpretation of these results is that Cdh1–APC is promiscuous in substrate recognition, particularly since there were no known conserved sequence elements in the amino-terminus of cyclin B other than the D box that could serve as a targeting sequence. Yet, countering this conclusion is the lack of activity of Cdh1–APC against 300 random substrates. By mutagenesis of Cdc20 and Xenopus cyclin B, we could demonstrate that Cdh1–APC shows specificity for two classes of sequences, those that contain the R-L-N motif of the D box and those that show a new targeting sequence, the KEN box. The paradoxical observation that Cdh1–APC recognizes a cyclin fragment lacking both a D box and a KEN box was explained when an additional D box sequence in Xenopus cyclin B was found upstream of the conserved D box. Deletion of this second, nonconserved motif blocks polyubiquitination completely.
There are notable quantitative differences between ubiquitination in the purified system and degradation in extracts. For example, there is significant polyubiquitination of cyclin B from which the conserved D box has been deleted (Δ36–44 N), whereas the protein is very stable in Cdh1-supplemented extracts. The degree and rate of ubiquitination must be a very complex function of many factors in extracts and in living cells. It may depend on competition among substrates, the activity of isopeptidases, and other factors that could modify any of the several steps in the process. In the purified in vitro system, ubiquitination, is optimized and inhibitory factors are absent. Dilution of ubiquitin reduced the efficiency of ubiquitination, but most importantly, it also elicited a difference between the behavior of the two D boxes in Xenopus cyclin B in the purified system, a difference reflected in the aggregate rates of degradation in extracts.
Like the D box, the KEN-box domain was transposable to other proteins. A fusion of only 28 amino acids around the KEN-box sequence (residues 75–103 of hCdc20) to protein A resulted in ubiquitination in the purified system and degradation in the extract system. Degradation was slower than larger fusion constructs and wild-type protein. Fusion of the KEN box alone did not confer degradation, suggesting that there may be additional structural elements required for recognition. Alternatively, there could be separate signals mediating recognition and degradation, as suggested by work with cyclin protein A fusions (Klotzbücher et al. 1996). There also may be a minimum size for proper degradation.
In the initial mutagenesis studies of KEN box, we found unexpectedly that alanine substitution of the two serines and one of the glutamic acids in a region carboxy-terminal to the KEN sequence noticeably increased polyubiquitination. A similar increase in the average chain length of the polyubiquitin chain was seen upon deletion of five amino acids including the previously mentioned serines and glutamate. This increased activity in the mutants suggested that negative charge in that region could inhibit ubiquitination and that phosphorylation might be used physiologically to regulate the process. It may be significant that there is a conserved Cdc2 site just carboxy-terminal to the KEN box, making it a likely target for cell cycle regulated phosphorylation. Although phosphorylation has been postulated to control the ubiquitination machinery in APC-dependent processes, it has not yet been implicated in controlling substrate selectivity. For the SCF complex the situation is reversed, phosphorylation regulates substrate selectivity positively, although there is no known regulation of the ubiquitination machinery. These mutagenesis results suggest that it would be useful to examine APC substrates for phosphorylation regulation, which might further specify the timing of protein degradation at the end of mitosis.
The explanation for the partially overlapping specificities of Cdc20–APC and Cdh1–APC for substrates will ultimately require studies at an atomic level resolution. The amino-terminal peptide of cyclin B containing the D box shows no regular structure in solution by NMR (H. Yu and M. Kirschner, unpubl.), so there is little hope that the structure of the peptide independent of APC would be informative. One way of thinking of the structural basis for the overlap in specificity mediated by Cdc20 and Cdh1 is a bump and hole model (assuming binding of both sequences to the same site). In this model, the KEN box peptide contains a bump that is accommodated by a hole in Cdh1 when bound to APC. It would not be accommodated by Cdc20; hence, KEN peptides would not be able to bind to Cdc20–APC. The D-box sequence would not have this feature and would bind to both Cdc20–APC and Cdh1–APC.
Knowledge of the respective roles of the D box and KEN box has provided us with a new understanding of how APC manages to achieve temporal, spatial, and substrate specificity throughout the cell cycle. In addition, it has given us new predictive tools to identify Cdh1 substrates. Given the low stringency of the KEN sequence, each candidate substrate needs to be validated by other means. However, using this information, we were able to identify two new substrates, Nek2 and B99, which show a pattern of abundance in vivo expected for a Cdh1 substrate. Both have functional KEN boxes. In addition, the Aurora kinase family all have the conserved KEN residues (for review, see Bischoff and Plowman 1999), making them likely candidates to test for KEN-box-dependent degradation. Recently, we carried out an unbiased screen by in vitro expression cloning for Cdh1–APC substrates and identified two novel proteins, which on sequencing, have KEN sequences which, by mutagenesis, are essential for degradation (S. Rankin, N. Ayad, and C. Pfleger, unpubl.). Because Cdh1–APC activity is found to be high in nondividing cell populations, such as brain (Gieffers et al. 1999), we anticipate finding substrates that are involved in cellular processes beyond cell cycle events.
Materials and methods
Preparation of low speed supernatant interphase extracts
Extracts were prepared as described previously (Murray 1991; Fang et al. 1998). In brief, eggs from HCG-injected PMSG-primed frogs were collected and washed in 1XMMR. Eggs were dejellied with 2% cysteine and activated by calcium ionophore, and then washed into XB and XB containing protease inhibitors. A 1-min packing spin at 1000 rpm was performed, followed by a crushing spin at 10,000 rpm. The cytoplasmic layer was collected and subjected to a clarifying spin also at 10,000 rpm. The clarified cytoplasmic layer was collected. After addition of an energy-regenerating system, protease inhibitors, cytochalasin B, and glycerol to 4%, extracts were frozen in liquid nitrogen, and stored at −80°C.
Construction of Cdc20 mutants
Mutants were generated by PCR with primers designed for the desired mutations. Construction of deletion mutants was achieved by designing primers with a restriction site, a start ATG (for amino-terminal deletions) or stop codon (for carboxy-terminal deletions) and sequence for at least five amino acids at the position of the desired deletion. Point mutations within the 96 amino acid deletion mutant were achieved using an amino-terminal primer encoding the altered codon for the desired amino acid change. Insertion of an alanine cassette in place of the KEN box in the full-length Cdc20 was achieved by designing both up and down primers introducing a NotI restriction site and sequence for amino acids on either side of the deletion. Two separate fragments were PCRed, digested with NotI, and ligated together and into a pCS2+ based vector, in position for in vitro translation with the Sp6 primer. For more information on primer sequences used for construction of mutants of Cdc20 and other proteins used in this study, please contact C. Pfleger or M. Kirschner.
Construction of Cdc20–protein A fusions
Insertion of a StuI site at the beginning of the IgG-binding region of protein A allowed ligation to small regions of Cdc20 PCRed with a carboxy-terminal primer lacking a stop site and containing a StuI site in the same frame. Coding sequence for three additional lysines was added just amino-terminal to the StuI site to ensure the close proximity of lysines to the KEN box in the fusion protein. Inserts were cloned into a pCS2+ vector for in vitro translation with the Sp6 primer.
Expression and purification of hCdc20 and hCdh1
His-tagged hCdc20 and hCdh1 were expressed in insect cells by the baculovirus system (virus obtained from G. Fang, Stanford University, Palo Alto, CA). Sf9 cells were infected with the appropriate virus, and harvested 2 days later. Protein was purified over a nickel column and eluted with high imidazole. The resulting eluate was run over a sizing column with PBS and containing 1 mm DTT. The appropriate fractions were collected and reconcentrated. Glycerol was added to 10%, then protein was frozen in liquid nitrogen and stored at −80°C.
Expression and purification of Cdc20 and cyclin B amino-terminal peptides
Amino-terminal Cdc20 peptides corresponding to the first 120 amino acids of either human wild-type sequence or KENN-A mutant were cloned into a pET 28 bacterial expression vector tagging them with an amino-terminal His tag. Protein was expressed in bacteria and purified on nickel beads. Previously described Xenopus cyclin B amino-terminal peptides (amino acids 1–102) containing or lacking the Δ36–44 D box (Fang et al. 1998) were expressed in bacteria, purified on nickel columns, and eluted with high imidazole. Peptides were dialyzed into XB-sucrose containing 1 mm DTT, reconcentrated, and glycerol was added to 10%. Peptides were frozen in liquid nitrogen and stored at −80°C.
In vitro ubiquitination assays
Ubiquitination assays were described previously (Fang et al. 1998). In brief, interphase APC (iAPC) was immunopurified from interphase extract with anti-Cdc27 antibodies covalently coupled to protein A beads and washed 5 times with XB (minus sucrose) containing 500 mm NaCl and 0.5% NP-40 detergent and two times with XB (minus sucrose). iAPC beads were incubated for 1 hr with baculovirus-expressed and purified Cdc20 or Cdh1, unless otherwise stated. Activator-bound APC on beads was then washed three times with XB (minus sucrose) followed by addition of 50 μg/ml Ubcx, 1.25 mg/ml ubiquitin, 200 μg/ml recombinant E1, 0.1 mg/ml cycloheximide, 1–5 μm ubiquitin aldehyde, and one-fifth the volume of 35S-labeled in vitro-translated substrate (typically, 2 μl of beads would be added to a 5-μl reaction). Ubiquitination assays were scored either visually, by observing the decrease in original signal with the appearance of polyubiquitinated (greater than three ubiquitins added per molecule) conjugates, or quantitated with NIH Image 1.61.
Competition experiments
In vitro degradation assays were performed in the presence of increasing amounts of bacterially expressed his-tagged peptides of amino-terminal Cdc20 wild-type or KEN-box mutant or amino-terminal cyclin B wild type or ΔD box.
Construction of Nek2 and B99 KEN-box mutants
Nek2 was cloned by PCR using the marathon cDNA library and the advantage polymerase from Clontech. B99 was obtained from L. Collavin in Claudio Schneider's laboratory (Trieste) and cloned into a pCS2+ vector. Mutants of Nek2 were made by PCR by introducing an alanine cassette containing a NotI site in the manner described for the Cdc20 KENN-A mutant. The B99 mutant was also made by PCR, using a carboxy-terminal primer encoding the alanine substitution.
Acknowledgments
We thank Guowei Fang and Hongtao Yu for their assistance in the initiation of this project and we thank Hui Zou, Ethan Lee, and Adrian Salic for their suggestions and advice throughout the course of this work. We also thank Teresita Bernal and Tanya Civco for their assistance. We thank the National Institute of General Medical Science (GM26875 and GM39023) for their support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marc@hms.harvard.edu; FAX (617) 432-0420.
References
- Bischoff J, Plowman GD. The Auror/Ip1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in Mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Chen A, Xiong Y, Pan ZQ, Ronai Z. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IΚB and beta-catenin. Oncogene. 1999;18:2039–2046. doi: 10.1038/sj.onc.1202760. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters J-M. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–137. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Goldberg A, Rock KL. Proteolysis, proteasomes, and antigen presentation. Nature. 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;74:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters J-M, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzbücher A, Stewart E, Harrison D, Hunt T. The 'destruction box' of cyclin A allows B-type cyclins to be ubiquitinated, but not efficiently destroyed. EMBO J. 1996;15:3053–3064. [PMC free article] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kumar S, Kao WH, Howley PM. Physical interaction between specific E2 and HECT E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- Lorca T, Castro A, Martinez A-M, Vigneron S, Morin N, Sigrist S, Lehner S, Dorée M, Labbé J-C. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey D, Yoo Y, Rechsteiner M. Ubiquitin metabolism in cycling Xenopus egg extracts. J Biol Chem. 1993;268:21205–21211. [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-βTrCP, that interacts with the HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell-cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Peters J-M. SCF and APC:The Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for the APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schultz SJ, Fry AM, Sutterlin C, Ried T, Nigg EA. Cell cycle-dependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 1994;5:625–635. [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 Cyclin Proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utera R, Collavin L, Lazarevic D, Delia D, Schneider C. A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J. 1998;17:5015–5025. doi: 10.1093/emboj/17.17.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. Cdc20 and Cdh1: A Family of Substrate-Specific Activators of APC-Dependent Proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Weinstein J. Cell cycle-regulated expression, phosphorylation, and degradation of p55Cdc. A mammalian homolog of CDC20/Fizzy/slp1. J Biol Chem. 1997;272:28501–28511. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- Wolf DA, Jackson PK. Cell cycle: Oiling the gears of anaphase. Curr Biol. 1998;8:R636–R639. doi: 10.1016/s0960-9822(07)00410-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]