Abstract
Zebrafish you-too (yot) mutations interfere with Hedgehog (Hh) signaling during embryogenesis. Using a comparative synteny approach, we isolated yot as a zinc finger transcription factor homologous to the Hh target gli2. Two alleles of yot contain nonsense mutations resulting in carboxy-terminally truncated proteins. In addition to causing defects in midline development, muscle differentiation, and retinal axon guidance, yot mutations disrupt anterior pituitary and ventral forebrain differentiation. yot mutations also cause ectopic lens formation in the ventral diencephalon. These findings reveal that truncated zebrafish Gli2 proteins interfere with Hh signaling necessary for differentiation and axon guidance in the ventral forebrain.
Keywords: Zebrafish mutations, Hh signaling, gli2, ventral forebrain
Members of the Hedgehog (Hh) family of signaling molecules are secreted by midline cells that influence the differentiation of adjacent cells (Placzek 1995; Hammerschmidt et al. 1997; Dodd et al. 1998; Ingham 1998). Mutational analysis in mouse and zebrafish has established the essential role of sonic hedgehog (shh) in development. Mouse mutants lacking shh have ventral forebrain defects, lack a floor plate and motor neurons in the neural tube, and have reduced sclerotomal tissue in the somites (Chiang et al. 1996). Zebrafish sonic-you (syu) mutations disrupt the shh gene and lead to defects in the spinal cord, somites, and optic chiasm (Brand et al. 1996; van Eeden et al. 1996; Schauerte et al. 1998).
Genetic screens have identified several additional zebrafish mutations that have phenotypes similar to syu, and these may define additional components of the Hh signaling pathway (Brand et al. 1996; Karlstrom et al. 1996; van Eeden et al. 1996). In particular, you-too (yot) mutants lack an optic chiasm and a horizontal myoseptum in the somites. Overexpression of Shh fails to rescue the somite defects in yot (Schauerte et al. 1998). Furthermore, yot is required cell autonomously in the somites for proper muscle differentiation (van Eeden et al. 1996). These observations suggest that yot encodes a downstream component of the Shh signaling pathway.
We report here that yot mutations affect ventral forebrain development and disrupt zebrafish Gli2, a member of a family of zinc finger containing transcription factors implicated in mediating Hh signaling (Ding et al. 1998; Matise et al. 1998; Ruiz i Altaba 1998).
Results and Discussion
We mapped yot to linkage group 9 (LG9) of the zebrafish genetic map (Fig. 1A,B). Recent analysis revealed conserved synteny (gene linkage) between zebrafish LG9 and human chromosome 2 (Postlethwait et al. 1998). A survey of human chromosome 2 identified the zinc finger transcription factor GLI2, suggesting that a zebrafish gli2 gene might reside on LG9 (Fig. 1A). yot mutations affect tissues known to require Hh signaling for proper differentiation, and gli genes [Drosophila cubitus interruptus (ci), vertebrate gli1 and gli2] have been implicated in transducing Hh signals from the midline (Ruiz i Altaba 1997; Ding et al. 1998; Ingham 1998; Matise et al. 1998 and references therein). Thus, gli2 was a compelling candidate for the yot locus. We therefore cloned zebrafish gli2 and found that it resides on LG9 near yot (Fig. 1). The zebrafish gli2 cDNA contains a full-length open reading frame (ORF) of 1440 amino acids that is closely related to mouse Gli2 (Fig. 2). Sequencing revealed that both yot alleles contain nonsense mutations (Fig. 2B) predicted to result in carboxyl terminally truncated Gli2 proteins (Fig. 2C), confirming that the yot locus encodes Gli2.
Figure 1.
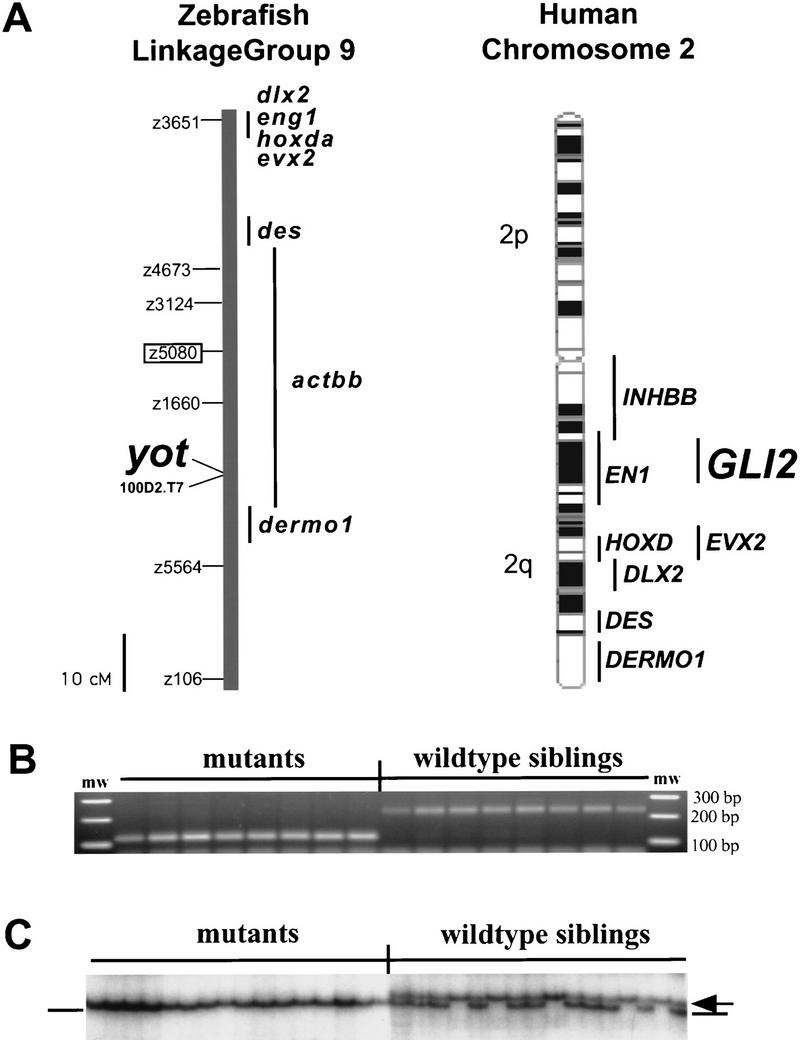
(A) Genetic map of linkage group 9 (LG9) showing position of yot/gli2 in relation to previously mapped genes (Postlethwait et al. 1998) and SSLP markers (Knapik et al. 1998). yotty17 was mapped to LG9 using centromere linkage analysis with centromeric marker z5080 (boxed) (see B and Materials and Methods), followed by mapping in a standard sexual intercross with marker z3124 (6 recombinants in 44 meioses). To determine the position of yot more precisely, we scored a marker that cosegregates with yot (BAC 100D2, see below) along with SSLP markers z3124, z5080, and z5564 on a haploid mapping panel (M.A. Gates, A.F. Schier, W.S. Talbot, in prep.). yot maps to the same linkage group as the zebrafish hoxda cluster (Amores et al. 1998), dlx2, engrailed1 (eng1), evx2, desmin (des), dermo1, and activinbb (actbb). The orthologs of these genes are located on human chromosome 2, where gli2 is also located. Human cytological map and locations were derived from Genome DataBase (http://gdbwww.gdb.org). Zebrafish actbb corresponds to human inhibinbb (INHBB). (B) Mapping to LG9 using SSLP marker z5080. z5080 primers amplified an ∼120-bp fragment from yotty17 DNA but an ∼220-bp band from wild-type siblings, indicating linkage to the marker. No recombinants were seen in 16 individuals. (C) SSCP analysis links BAC 100D2 DNA to yot. PCR amplification of DNA from individual mutant embryos produced a faster migrating band (lines), whereas DNA from wild-type siblings produced a slower migrating band (homozygotes, arrow) or both bands (heterozygotes). No recombinants were seen in 230 mutant embryos, indicating tight linkage.
Figure 2.
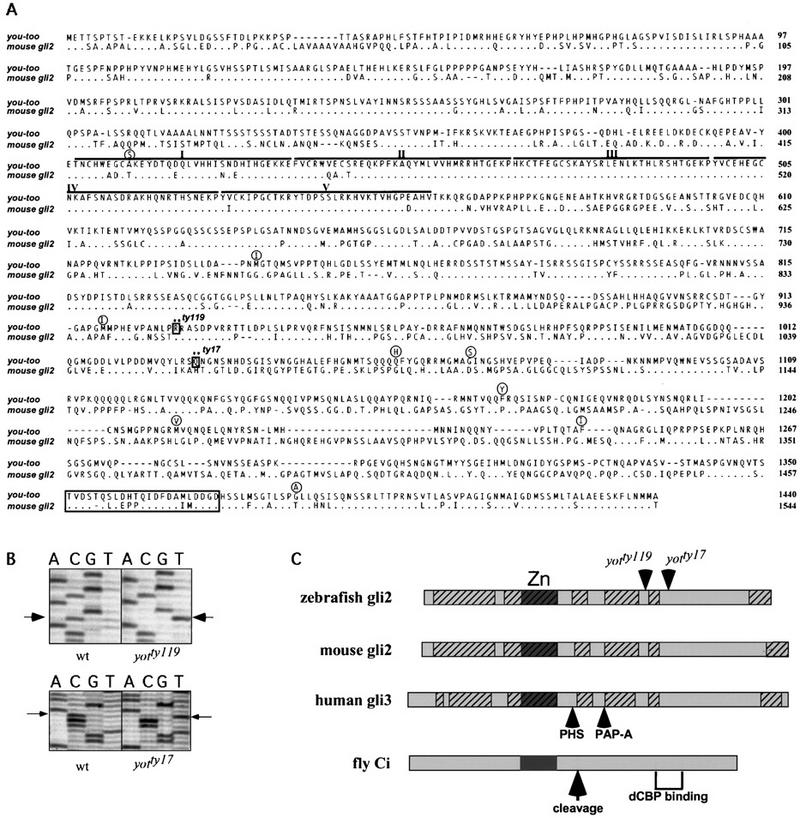
Sequence of zebrafish gli2 and identification of point mutations in yot alleles. (A) The deduced amino acid sequence of zebrafish gli2 (you-too) aligned with mouse gli2. The five zinc finger regions are indicated by lines and Roman numerals. The positions of stop codons in yotty119 and yotty17 are indicated by boxes and asterisks. Amino acid polymorphisms seen in wild-type strains are shown in circles over the zebrafish gli2 sequence. A putative VP16-like activation domain at the carboxyl terminus is boxed. (B) Sequencing gels showing point mutations in the two yot alleles. In yotty119 C3148 is mutated to U, changing Arg-929 to a stop codon. In yotty17 C3445 is mutated to U, changing Glu-1032 to a stop codon. (C) Schematic representation of the zebrafish gli2 protein sequence showing the positions of the stop codons (arrowheads) in the two mutant alleles. Alignment is shown with mouse Gli2, human GLI3, and fly Ci. Hatched regions have >70% sequence identity between the three Gli proteins. Overall amino acid sequence identities are zebrafish gli2 to mouse Gli2, 56%; zebrafish gli2 to mouse Gli1, 29%; zebrafish gli2 to zebrafish Gli1, 35%; zebrafish gli1 to mouse Gli1, 45%; zebrafish gli1 to mouse Gli2, 35% (R.O. Karlstrom, A. Kawakami, W.S. Talbot, A.F. Schier, unpubl.). Sites of stop codons resulting from nonsense mutations in Gli3 that lead to PAP-A and PHS are indicated by arrows. Site of cleavage that results in a repressor form of Ci is shown by an arrow. The carboxy-terminal region of Ci implicated in CBP binding is indicated by brackets. Zinc finger regions (Zn) are indicated by dark boxes.
The truncated Gli2 proteins retain the zinc finger DNA-binding domain but lack a region similar to a domain in Drosophila Ci implicated in binding dCBP, a transcriptional coactivator (Akimaru et al. 1997). They also lack a proposed VP16-like activation domain required for human Gli1 activity (Yoon et al. 1998). Because such carboxy-terminal deletions impair the ability of Ci and Gli1 to activate Hh targets (Alexandre et al. 1996; Yoon et al. 1998), the yot mutant proteins might not mediate full transcriptional activation in response to Hh signaling. The truncated Gli2 proteins share striking similarity to forms of Ci protein (Fig. 2C) that transcriptionally repress Hh target genes and form by post-translational processing in the absence of Hh signaling (Aza-Blanc et al. 1997). In addition, the yot truncations resemble potential repressor forms of Gli3 (Fig. 2C) that lead to dominant Pallister–Hall syndrome (PHS) (Kang et al. 1997) and postaxial polydactyly type A (PAP-A) (Radhakrishna et al. 1997). Taken together, these observations suggest that these proteins might act as repressors that form even in the presence of Hh and interfere with Hh signaling. Supporting this possibility, previous studies have shown that yot mutations block transmission of Hh signaling in target tissues (van Eeden et al. 1996; Schauerte et al. 1998). The weak dominant effects on somite patterning found in yot heterozygous embryos (van Eeden et al. 1996) are also consistent with the idea that yot mutations encode repressor forms of Gli2.
To determine if gli2 expression domains correspond to the regions affected by mutations in yot, we performed gli2 RNA in situ hybridization (Fig. 3). gli2 transcripts are first detected in the anterior neural plate and in somitic mesoderm precursors that are affected in yot mutants (Fig. 3A,B). As development proceeds, gli2 is expressed throughout the dorsal forebrain, midbrain, and hindbrain and is generally expressed dorsal and adjacent to cells that express shh. In the postoptic area of the ventral diencephalon, however, gli2 and shh expression overlap (Fig. 3E,G,H). This gli2-expression domain corresponds to the site of axon growth affected in yot mutants (see below) and is consistent with a direct role of gli2 in the formation of this area of the ventral diencephalon.
Figure 3.
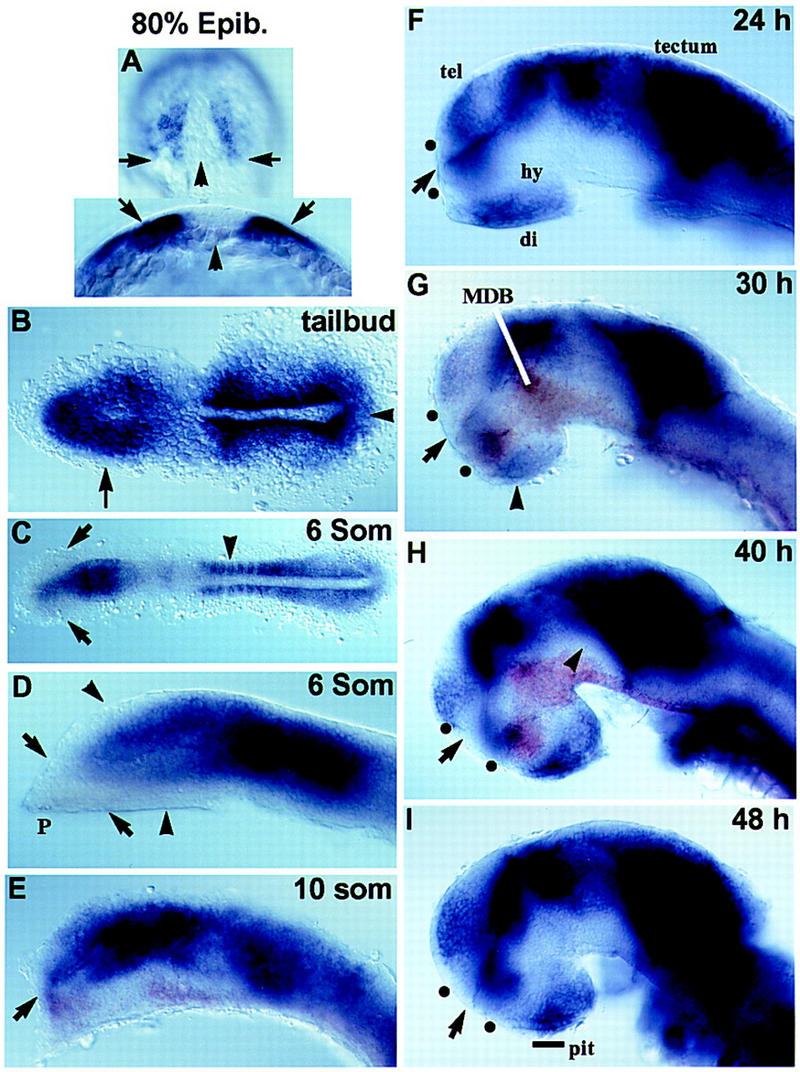
Zebrafish gli2 RNA expression (purple). Embryos in E, G, and H were double labeled with shh (brown). Anterior is to the left in all panels except A. Dots in F–I show positions of anterior and postoptic commissures. (A) 80% epiboly. (Top) Dorsal view, animal pole up, arrows point to the vegetal margin of the gastrula. (Bottom) Optical cross section, dorsal up, arrowhead marks midline. (Arrows) Lateral mesoderm. (B) Tailbud stage, dorsal view. (Arrow) Anterior neural plate; (arrowhead) tailbud. (C) Six-somite stage, dorsal view. (Arrows) Optic vesicles; (arrowhead) developing somites. (D–I) Lateral views of the brain, medial focal plane, eyes have been removed. (D) Six-somite stage. (Arrowheads) Most dorsal telencephalon and ventral diencephalon do not express gli2; (P) mesendodermal tissue of the polster. (Arrows) Border between polster and forebrain tissue. (E) 10-somite stage. (Arrow) Optic recess. (F) 24 hr; (G) 30 hr; (H) 40 hr. Expression is reduced in cells adjacent to shh-expressing cells in the forebrain (arrowhead). (I) 48 hr. Line, rostral portion of the anterior pituitary anlage. (di) Diencephalon; (hy) hypothalamus; (MDB) mid-diencephalon boundary, (pit) anterior pituitary; (tel) telencephalon; (dots) anterior and postoptic commissures.
Although gli genes have been implicated in forebrain development (Lee et al. 1997; Ruiz i Altaba 1998), the consequences of gli mutations on ventral forebrain patterning have not been analyzed. To define ventral forebrain defects in yot mutants more precisely, we first examined axon guidance and cell differentiation in this area. In the ventral diencephalon of yot mutant embryos, the formation of the optic chiasm (Brand et al. 1996; Karlstrom et al. 1996) and postoptic commissure are both affected (Fig. 4). Instead of crossing the midline in the postoptic area (Fig. 4C), retinal axons grow dorsally immediately after leaving the eye (Fig. 4D). The earlier forming postoptic commissure also fails to form in yot mutant embryos, and the anterior commissure in the telencephalon is reduced (Fig. 4B). Further, lens-like structures form in the anterior pituitary region of the ventral diencephalon (Fig. 4D) in nearly all yotty119 mutants and in 10%–50% of yotty17 mutants. These results establish that yot mutations lead to defects in axon guidance and cell differentiation in the ventral diencephalon.
Figure 4.
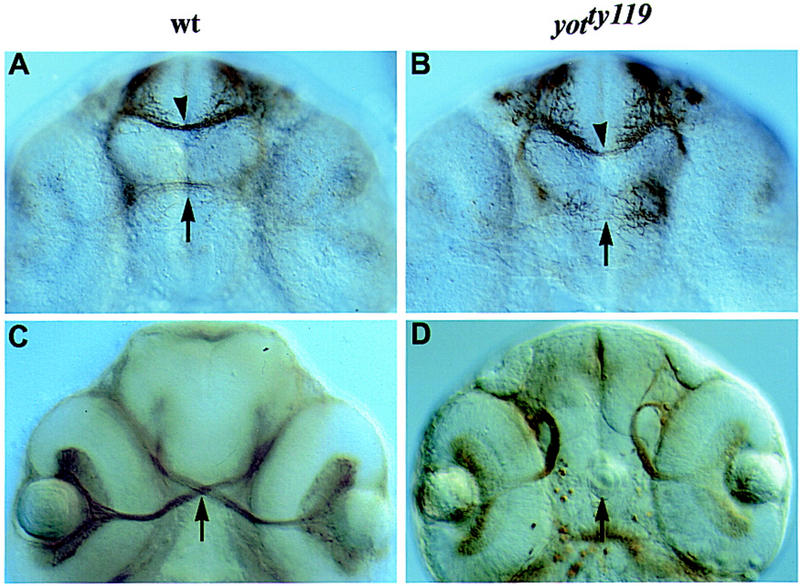
yot affects axon guidance in the ventral forebrain. (A–D) Ventral view, anterior up. (A,B) Anti-acetylated tubulin labeling. (A) In 36-hr wild-type embryos, commissural axons have crossed the forebrain to form the anterior (arrowhead) and postoptic (arrow) commissures. (B) In yot mutant embryos, the postoptic commissure fails to form (arrow) and the anterior commissure appears reduced (arrowhead). (C,D) ZN-8 antibody labeling. (C) In 48-hr embryos, retinal axons have crossed the midline to form the optic chiasm (arrow). (D) In yot mutant embryos, axons project ipsilaterally after leaving the eye and no chiasm forms. Lens-like structures form in yotty119 mutant embryos in the ventral diencephalon (arrow).
The mis-specification of ventral forebrain tissue, along with forebrain expression of gli2, suggests that axon guidance defects in yot mutants might be caused by abnormal development of ventral forebrain cells that serve as substrates or guidance cues for navigating axons. To test this idea, we examined the expression of ventral forebrain markers in yot mutant embryos. The homeobox gene nk2.2 is normally expressed by cells in the rostral part of the anterior pituitary anlage and in the postoptic area (Fig. 5A; Barth and Wilson 1995). In yot mutants, the rostral anterior pituitary expression of nk2.2 is absent (Fig. 5B). The homeobox gene lim3, a marker of the anterior pituitary anlage (Glasgow et al. 1997), is also reduced in the region of the anterior pituitary in yot mutants (not shown). six3, a homolog of Drosophila sine oculis, is a homeobox gene expressed in the anterior neural plate, anterior pituitary, and the hypothalamus (Fig. 5C; Kobayashi et al. 1998; Seo et al. 1998). In yot mutant embryos, six3 expression is absent from the rostral part of the anterior pituitary and the adjacent domain of the postoptic area but remains in the anterior postoptic area and caudal part of the anterior pituitary (Fig. 5D). The defects in the expression of nk2.2, lim3, and six3 indicate that yot mutations disrupt the formation of the rostral portion of the anterior pituitary as well as the adjacent region of the postoptic area. In contrast, markers for optic stalk (pax2), hypothalamus (dlx2), and telencephalon (dlx2, pax6) are expressed normally (not shown). These results are consistent with the idea that axon path-finding defects are caused by abnormal development of the postoptic area.
Figure 5.
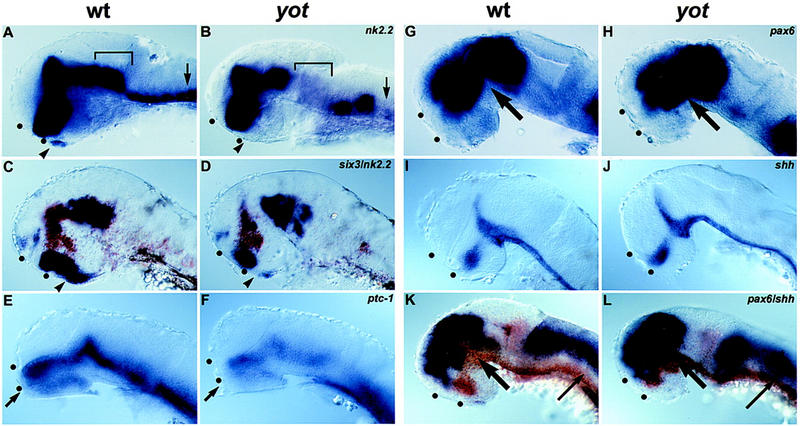
yot affects anterior pituitary formation and expression of Hh target genes in the brain. Expression of nk2.2 (A,B), six3 (C,D), ptc-1 (E,F), pax6 (G,H,K,L), and shh (I,J) in purple. Embryos in C, D, K, and L were double labeled with nk2.2 (C,D) or shh (K,L) (brown). Anterior is to the left in all panels; eyes have been removed. (A–D) 36 hr. (A,B) (Arrowheads) Anterior pituitary anlage; (square brackets) regions of the tegmentum; (arrows) ventral hindbrain. (C,D) In yot mutants, six3 expression (purple) is absent from the middle of its expression domain, which includes the rostral part of the anterior pituitary anlage (arrowheads). (E,F) 30 hr. (Arrow) Postoptic area. (G–L) 36 hr. Expansion of pax6 expression at mid-diencephalic boundary (thick arrows) and dorsal-to-shh–expressing cells (thin arrows). (K,L) (Dots) Anterior and postoptic commissures.
To further examine the role of Gli2 in mediating Hh signals during forebrain development, we analyzed the effect of yot on known Hh target genes. The putative Hh receptor patched-1 (ptc-1) is expressed in Hh-responsive cells adjacent to the source of Hh (Fig. 5E,F; Concordet et al. 1996) and has been shown to be up-regulated by Hh signaling in flies and vertebrates (Concordet et al. 1996; Ingham 1998). Consistent with defects in Hh signaling, we find that ptc-1 expression in yot mutant embryos is reduced throughout the brain and expression is absent in the postoptic area (arrow in Fig. 5F). The pax6 gene has been shown to be negatively regulated by shh in zebrafish (Ekker et al. 1995; Macdonald et al. 1995). Consistent with a role of gli2 in this repression, pax6 expression is expanded in yot mutants (Fig. 5H). The altered expression of pax6 and ptc-1 provides further evidence that yot disrupts Hh signaling in the forebrain. The relatively mild effects, however, also indicate that some Hh signaling remains in the ventral neural tube of yot mutants. It is conceivable that the carboxy-terminal truncations of Gli2 in yot mutants do not eliminate all Gli2 activity. In addition, other factors such as Gli1 or Gli3 might mediate Hh signaling in yot mutants. In support of this idea, we have found that zebrafish gli1 is expressed in the ventral neurectoderm of wild-type and yot mutant embryos (R.O. Karlstrom, W.S. Talbot, and AF. Schier, unpubl.).
shh has been implicated in the induction of floor plate and ventral motoneurons (Chiang et al. 1996; Dodd et al. 1998), but the role of gli2 in this process is less clear. Loss-of-function studies in the mouse demonstrate that gli2 is essential for floor plate, but not motoneuron, development (Ding et al. 1998; Matise et al. 1998). In contrast, overexpression studies in Xenopus have led to the proposal that gli2 represses floor plate development and mediates induction of motoneurons (Ruiz i Altaba 1998). Yet another result is observed in zebrafish. The Gli2 truncations lead to spinal cord defects but do not interfere with the induction of medial floor plate cells or motoneurons (Fig. 5I,J; Brand et al. 1996; Schauerte et al. 1998). The contrasting mouse and zebrafish mutant phenotypes might be due to differences in alleles, that is, the mouse gli2 mutation deletes zinc fingers 3–5 and more carboxy-terminal sequences, whereas the zinc finger region is unaffected in the yot mutations. As mentioned above, other factors that mediate Hh signaling in zebrafish, including gli1, might compensate for the mutations in gli2. It is interesting to note, however, that like yot, null mutations in zebrafish shh do not affect medial floor plate development (Schauerte et al. 1998). Thus, in contrast to mouse, medial floor-plate formation in zebrafish might be independent of Hh signaling (Schauerte et al. 1998). Alternatively, the presence of echidna hedgehog and tiggy-winkle hedgehog, as well as additional gli genes, might compensate for mutations in zebrafish shh and gli2, respectively (Dodd et al. 1998). The isolation of mutations in other gli and hh genes will be an important step toward determining the exact role of gli genes in mediating Hh signaling in zebrafish and will allow a more careful comparison of the developmental role of this signaling pathway in different vertebrates.
In summary, using comparative synteny in zebrafish for the first time, we have found that yot mutations encode carboxy-terminally truncated Gli2 proteins that affect Hh signaling. The use of synteny conservation, in combination with the rapid development of genomic resources in human, mouse, and zebrafish, is likely to facilitate the cloning of zebrafish mutations. Although ectopic expression experiments suggested a role for gli genes in ventral CNS specification (Hynes et al. 1997; Lee et al. 1997; Ruiz i Altaba 1998), it was previously unknown whether mutations in this gene family would affect ventral forebrain development or retinal axon pathfinding. The defects in yot mutant embryos underscore the importance of gli genes and the Hh signaling pathway in specifying cell fates in the ventral forebrain. We propose that these defects affect cellular cues that guide axons across the midline of the ventral forebrain and disrupt postoptic commissure formation and optic nerve crossing. Our phenotypic analysis also suggests a role for Hh signaling in differentiation of the anterior pituitary and inhibition of lens induction in this region. The defects in yot mutants and the involvement of shh in human holoprosencephaly (for review, see Ming and Muenke 1998) suggest that mutations in gli2 might underlie human diseases such as congenital malformations in facial and forebrain midline structures.
Materials and methods
Mutant and mapping strains
Two alleles of yot (yotty17 and yotty119) were identified previously in mutant screens (Brand et al. 1996; Karlstrom et al. 1996; van Eeden et al. 1996). Both alleles were kept in the Tübingen background and were also outcrossed to the AB background. For mapping, yotty17 was crossed to two polymorphic lines, the WIK line (Rauch et al. 1997) and the Leopard/Long Fin line. In situ and antibody analyses were performed with both the stronger (yotty119) and weaker (yotty17) alleles.
Genetic mapping by centromere linkage analysis
We determined the position of yot on the zebrafish genetic map using centromere linkage analysis (Johnson et al. 1996; Postlethwait and Talbot 1997). Gynogenetic diploid embryos were obtained from heterozygous females by early pressure treatment of eggs fertilized with inactivated sperm. Mutant and wild-type progeny were identified by visual inspection on day 1 or day 2. DNA prepared from individuals or from pools of eight mutant or wild-type individuals was assayed by PCR using polymorphic markers (simple sequence length polymorphisms; Knapik et al. 1998). This identified two genetic markers (z5080 and z3124) on LG9 that were linked to yot. Finer mapping was done using embryos obtained from pairwise matings of heterozygous yot parents in a WIK background.
Cloning zebrafish gli genes
Genomic clones were obtained by screening a gridded genomic bacterial artificial chromosome (BAC) library (Genome Systems) using radiolabeled probes for a mouse gli2 cDNA at low stringency hybridization conditions. BAC DNA was prepared for positive clones and the BAC ends were sequenced using vector primers. Sequence from the end of clone 100D2 had homology to gli2. Sequence from this clone was used to design primers that amplified a single strand conformation polymorphism (SSCP), which was used to map the BAC end to LG9 and to test cosegregation with the yot locus (Talbot and Schier 1999).
cDNA clones encoding gli genes were isolated from a 15- to 19-hr embryonic cDNA library (generously provided by Bruce Appel and Judith Eisen, University of Oregon, Eugene). BAC 100D2 sequence was used to design PCR primers that were used to make radiolabeled PCR probes. A low stringency screen using the mouse gli2 cDNA was also performed. Three clones encoding portions of the gli2 gene were identified. These clones were sequenced and their sequences assembled into the full gli2 coding region (GenBank accession no. AF085746) containing a putative translation initiation codon at position 364 and a stop codon at position 4681.
Sequencing mutant alleles
RT–PCR and cycle sequencing were used to sequence the two ENU-induced yot alleles (Talbot and Schier 1999). RNA was isolated from the following pools of 40 embryos: (1) yotty119 wild-type siblings; (2) yotty119 mutants; (3) yotty17 wild-type siblings; and (4) yotty17 mutants. First-strand cDNA was made using Superscript reverse transcriptase (GIBCO). Fragments (500–1000 bp) were amplified from first strand cDNA by the PCR using primers based on the deduced gli2 cDNA sequence. DNA fragments were then gel purified and cycle sequenced (Stratagene Cyclist). Sequence was compared between pools and to the gli2 cDNA sequence. The fragments containing the yot point mutations were also subcloned using the TA cloning system (Invitrogen). DNA from two separately isolated clones was purified and the mutant sequence was verified.
In situ hybridization and antibody labeling
Single-color in situ labeling was performed as described (Schier et al. 1997). Antibody labeling (Karlstrom et al. 1996) was performed using the anti-acetylated tubulin antibody (Sigma) to label axon tracts and ZN-8 to label retinal ganglion cells and their axons. Two-color in situ labeling (Jowett and Yan 1996) was performed using the INT substrate (Boehringer Mannheim) for the second phosphatase reaction. A 3′ gli2 probe lacking the conserved zinc finger region was made using pBS–gli2-4.9 linearized with Bst98-1 and T7 RNA polymerase. Other probes used were nk2.2 (Barth and Wilson 1995), lim3 (Glasgow et al. 1997), shh (Krauss et al. 1993), ptc-1 (Concordet et al. 1996), and pax6 (Krauss et al. 1991). Our six3 probe corresponds to that of one group (Kobayashi et al. 1998) but is called six6 by another (Seo et al. 1998).
Acknowledgments
We thank members of the Talbot and Schier laboratories for helpful discussions; C. Nüsslein-Volhard, P. Haffter, H.-G. Frohnhoefer for zebrafish strains; M. Gates for map data; B. Appel, J. Eisen, K. Platt, and A. Joyner for reagents; our colleagues in the zebrafish field for in situ probes; S. McManus and R. Feeney for fish care; and R. Burdine, S. Dougan, B. Feldman, G. Fishell, A. Joyner and H. Sirotkin for critical comments on the manuscript. This work was supported by National Institutes of Health (NIH) fellowship 1F32NS10185 to R.O.K., NIH grant 1RO1RR12349 to W.S.T., and Skirball Institute start-up funds to A.F.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL karlstrom@bio.umass.edu; schier@saturn.med.nyu.edu; FAX (212) 263-7760.
References
- Akimaru H, Chen Y, Dai P, Hou DX, Nonaka M, Smolik SM, Armstrong S, Goodman RH, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signaling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- Alexandre C, Jacinto A, Ingham PW. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the Cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes & Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho R, Langeland J, Prince V, Wang Y-L, Westerfield M, Ekker M, Postlethwait J. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber F-A, Laget M-P, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Barth KA, Wilson SW. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development. 1995;121:1755–1768. doi: 10.1242/dev.121.6.1755. [DOI] [PubMed] [Google Scholar]
- Brand M, Heisenberg C-P, Warga R, Pelegri F, Karlstrom R, Beuchle D, Picker A, Jiang Y-J, Furutani-Seiki M, van Eeden F, Granato M, Haffter P, Hammerschmidt M, Kane D, Kelsh R, Mullins M, Odenthal J, Nüsslein-Volhard C. Mutations affecting development of the midline and general body shape during zebrafish embryogenesis. Development. 1996;123:129–142. doi: 10.1242/dev.123.1.129. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, Scott MP, Ingham PW. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Rong M, Sasaki H, Rossant J, Hui C-C. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- Dodd J, Jessell T, Placzek M. The when and where of floor plate induction. Science. 1998;282:1654–1657. doi: 10.1126/science.282.5394.1654. [DOI] [PubMed] [Google Scholar]
- Ekker SC, Ungar AR, Greenstein P, vonKessler DP, Porter JA, Moon RT, Beachy PA. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr Biol. 1995;5:944–955. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- Glasgow E, Karavanov AA, Dawid IB. Neuronal and neuroendocrine expression of lim3, a LIM class homeobox gene, is altered in mutant zebrafish with axial signaling defects. Dev Biol. 1997;192:405–419. doi: 10.1006/dbio.1997.8761. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Brook A, McMahon AP. The world according to hedgehog. Trends Genet. 1997;13:14–21. doi: 10.1016/s0168-9525(96)10051-2. [DOI] [PubMed] [Google Scholar]
- Hynes M, Stone DM, Dowd M, Pitts-Meek S, Goddard A, Gurney A, Rosenthal A. Control of cell pattern in the neural tube by the zinc finger transcription factor and oncogene gli-1. Neuron. 1997;19:15–26. doi: 10.1016/s0896-6273(00)80344-x. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Transducing Hedgehog: The story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Gates MA, Johnson M, Talbot WS, Horne S, Baik K, Rude S, Wong JR, Postlethwait JH. Centromere-linkage analysis and consolidation of the zebrafish genetic map. Genetics. 1996;142:1277–1288. doi: 10.1093/genetics/142.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T, Yan Y-L. Two colour whole-mount in situ hybridisation. Trends Genet. 1996;12:387–389. doi: 10.1016/s0168-9525(96)90091-8. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JMJ, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Karlstrom R, Trowe T, Klostermann S, Baier H, Brand M, Crawford A, Grunewald B, Haffter P, Hoffman H, Meyer SU, Müller B, Richter S, van Eeden F, Nüsslein-Volhard C, Bonhoeffer F. Zebrafish mutations affecting retinotectal axon pathfinding. Development. 1996;123:427–438. doi: 10.1242/dev.123.1.427. [DOI] [PubMed] [Google Scholar]
- Knapik EW, Goodman A, Ekker M, Chevrette M, Delgado J, Neuhauss S, Shimoda N, Driever W, Fishman MC, Jacob HJ. A microsatellite genetic linkage map for zebrafish (Danio rerio) Nat Genet. 1998;18:338–343. doi: 10.1038/ng0498-338. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Toyama R, Takeda H, Dawid IB, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish pax genes suggests a role in early brain regionalization. Nature. 1991;353:267–270. doi: 10.1038/353267a0. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet J-P, Ingham PH. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. gli1 is a target of sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Macdonald R, Barth KA, Xu Q, Holder N, Mikkola I, Wilson SW. Midline signaling is required for Pax gene regulation and patterning of the eyes. Development. 1995;121:3267–3278. doi: 10.1242/dev.121.10.3267. [DOI] [PubMed] [Google Scholar]
- Matise M, Epstein D, Park H, Platt K, Joyner A. gli2 is required for the induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Holoprosencephaly: From Homer to Hedgehog. Clin Genet. 1998;53:155–163. doi: 10.1111/j.1399-0004.1998.tb02666.x. [DOI] [PubMed] [Google Scholar]
- Placzek M. The role of the notochord and floor plate in inductive interactions. Curr Opin Genet Dev. 1995;5:499–506. doi: 10.1016/0959-437x(95)90055-l. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Talbot WS. Zebrafish genomics: From mutants to genes. Trends Genet. 1997;13:183–190. doi: 10.1016/s0168-9525(97)01129-3. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Wild A, Grzeschik K-H, Antonarakis SE. Mutation in Gli3 in postaxial polydactyly type A. Nat Genet. 1997;17:269–271. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- Rauch, G.J., M. Granato, and P. Haffter. 1997. A polymorphic zebrafish line for genetic mapping using SSLPs on high percentage agarose gels. Trends Tech. Tips Online. TO1208
- Ruiz i Altaba A. Catching a Gli-mpse of hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- ————— Combinatorial Gli function in floor plate and neuronal inductions by Sonic hedgehog. Development. 1998;125:2203–2212. doi: 10.1242/dev.125.12.2203. [DOI] [PubMed] [Google Scholar]
- Schauerte H, van Eeden F, Fricke C, Odenthal J, Strähle U, Haffter P. Sonic hedgehog is not required for the induction of medial floor plate cells in zebrafish. Development. 1998;125:2983–2993. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SCF, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Seo HC, Drivenes O, Ellingsen S, Fjose A. Expression of two zebrafish homologues of the murine Six3 gene demarcates the initial eye primordia. Mech Dev. 1998;73:45–57. doi: 10.1016/s0925-4773(98)00028-8. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Schier AF. Positional cloning of mutated genes in zebrafish. Methods Cell Biol. 1999;60:259–286. doi: 10.1016/s0091-679x(08)61905-6. [DOI] [PubMed] [Google Scholar]
- van Eeden F, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg C-P, Jiang Y-J, Kane D, Kelsh R, Mullins M, Odenthal J, Warga R, Allende M, Weinberg E, Nüsslein-Volhard C. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- Yoon J, Liu C, Yang J, Swart R, Iannaccone P, Walterhouse D. GLI activates transcription through a herpes simplex viral protein 16-like activation domain. J Biol Chem. 1998;273:3496–3501. doi: 10.1074/jbc.273.6.3496. [DOI] [PubMed] [Google Scholar]


