Abstract
Splicing of certain pre-mRNA introns is dependent on an enhancer element, which is typically purine-rich. It is generally thought that enhancers increase the use of suboptimal splicing signals, and one specific proposal is that enhancers stabilize binding of U2AF65 to weak polypyrimidine (Py) tracts. Here, we test this model using an IgM pre-mRNA substrate, which contains a well-characterized enhancer. Although the enhancer was required for in vitro splicing, we found it had no effect on U2AF65 binding. Unexpectedly, replacement of the natural IgM Py tract, branchpoint, and 5′ splice site with consensus splicing signals did not circumvent the enhancer requirement. These observations led us to identify a novel regulatory element within the IgM M2 exon that acts as a splicing inhibitor; removal of the inhibitor enabled splicing to occur in the absence of the enhancer. The IgM M2 splicing inhibitor is evolutionarily conserved, can inhibit the activity of an unrelated, constitutively spliced pre-mRNA, and acts by repressing splicing complex assembly. Interestingly, the inhibitor itself forms an ATP-dependent complex that contains U2 snRNP. We conclude that splicing of IgM exons M1 and M2 is directed by two juxtaposed regulatory elements—an enhancer and an inhibitor—and that a primary function of the enhancer is to counteract the inhibitor.
Keywords: Pre-mRNA splicing, U2AF, splicing enhancer, splicing inhibitor
A typical metazoan precursor mRNA (pre-mRNA) contains multiple exons that must be joined precisely by splicing, a process that can occur constitutively and in some cases is regulated in a tissue and developmental-specific fashion. Cis-acting elements required for splicing of all higher eukaryotic introns include the 5′ and 3′ splice sites, and the highly variable polypyrimidine (Py) tract and branchpoint immediately upstream of the 3′ splice site (Krämer 1996; Reed 1996).
Pre-mRNA splicing involves the stepwise assembly of both RNA and protein components to form an active spliceosome. In higher eukaryotes, spliceosome assembly initiates through recognition of the 5′ splice site by U1 snRNP and the Py tract by the U2 small nuclear riboncleoprotein particle (snRNP) auxiliary factor U2AF (Reed 1996). U2AF is a heterodimer comprising a large subunit, U2AF65, and a small subunit, U2AF35 (Zamore and Green 1989). U2AF65 directly contacts the Py tract and has been shown to be required for splicing in several in vitro systems. U2AF65 has a bipartite structure comprising an RNA-binding domain (RBD) and an arginine–serine-rich (RS) region. The RS region is dispensable for binding of U2AF65 to the Py tract but is required for the ability of U2AF65 to promote the U2 snRNP–branchpoint interaction, the first ATP-dependent event in spliceosome assembly (Zamore et al. 1992). Although the small U2AF subunit is dispensable for splicing in vitro, it is required for Drosophila viability, implying a critical in vivo role for at least a subset of pre-mRNAs (Rudner et al. 1996, 1998b).
Some higher eukaryotic pre-mRNAs contain additional cis-acting elements that can regulate splicing activity. The best-characterized of these regulatory elements are so-called splicing enhancers, which are typically purine-rich and function by binding one or more SR proteins, a conserved family of essential splicing factors (Zahler et al. 1992; Fu 1995; Manley and Tacke 1996; Valcárcel and Green 1996; Hertel et al. 1997). Enhancers are often associated with introns that contain apparently weak splicing signals. It has been proposed that enhancers function by promoting binding of U2AF65 to weak Py tracts (Wang et al. 1995; Zuo and Maniatis 1996; Bouck et al. 1998). The increase in U2AF65 binding is thought to occur through formation of a network of protein–protein interactions, ultimately involving direct contact of U2AF through its small subunit U2AF35 (Wu and Maniatis 1993; Zuo and Maniatis 1996).
In this report we test the generality of this model using the mouse immunoglobulin (IgM) model exon enhancer substrate. Splicing of IgM exons M1 and M2 is dependent on a purine-rich enhancer element located within the M2 exon (Watakabe et al. 1993). The data presented below indicate that the IgM M2 enhancer does not function by stabilizing U2AF65 binding but, rather, reveals a new mechanism by which an enhancer can promote splicing.
Results
The IgM M2 enhancer does not function by increasing U2AF65 binding
To determine whether the IgM M2 splicing enhancer functions by increasing U2AF65 binding, we measured splicing and binding of U2AF65 to the Py tract under identical conditions. U2AF65 binding was measured in a standard splicing reaction mixture using a UV cross-linking/immunoprecipitation assay with the α-U2AF65 monoclonal antibody MC3 (Gama-Carvalho et al. 1997).
Figure 1A shows that following a 90-min incubation in a HeLa nuclear extract, an enhancer-containing IgM substrate (μM) was spliced >75%, whereas splicing of the substrate lacking the enhancer (μMΔ) was undetectable. Because binding of U2AF65 to the Py tract is an initial step of spliceosome assembly and splicing (Reed 1996), we analyzed early time points. Figure 1B shows that splicing of the enhancer-containing substrate could be detected as early as 20 min (top). Unexpectedly, at all time points, binding of U2AF65 to the substrate containing or lacking the enhancer was equivalent (bottom).
Figure 1.
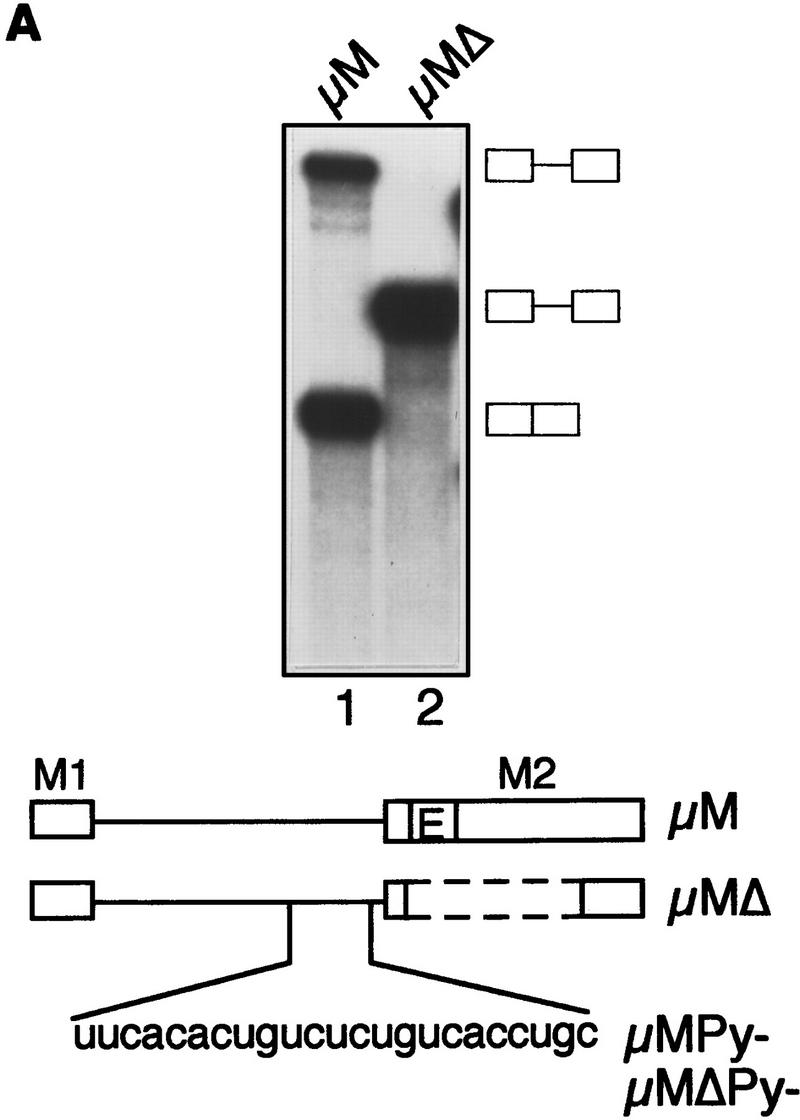
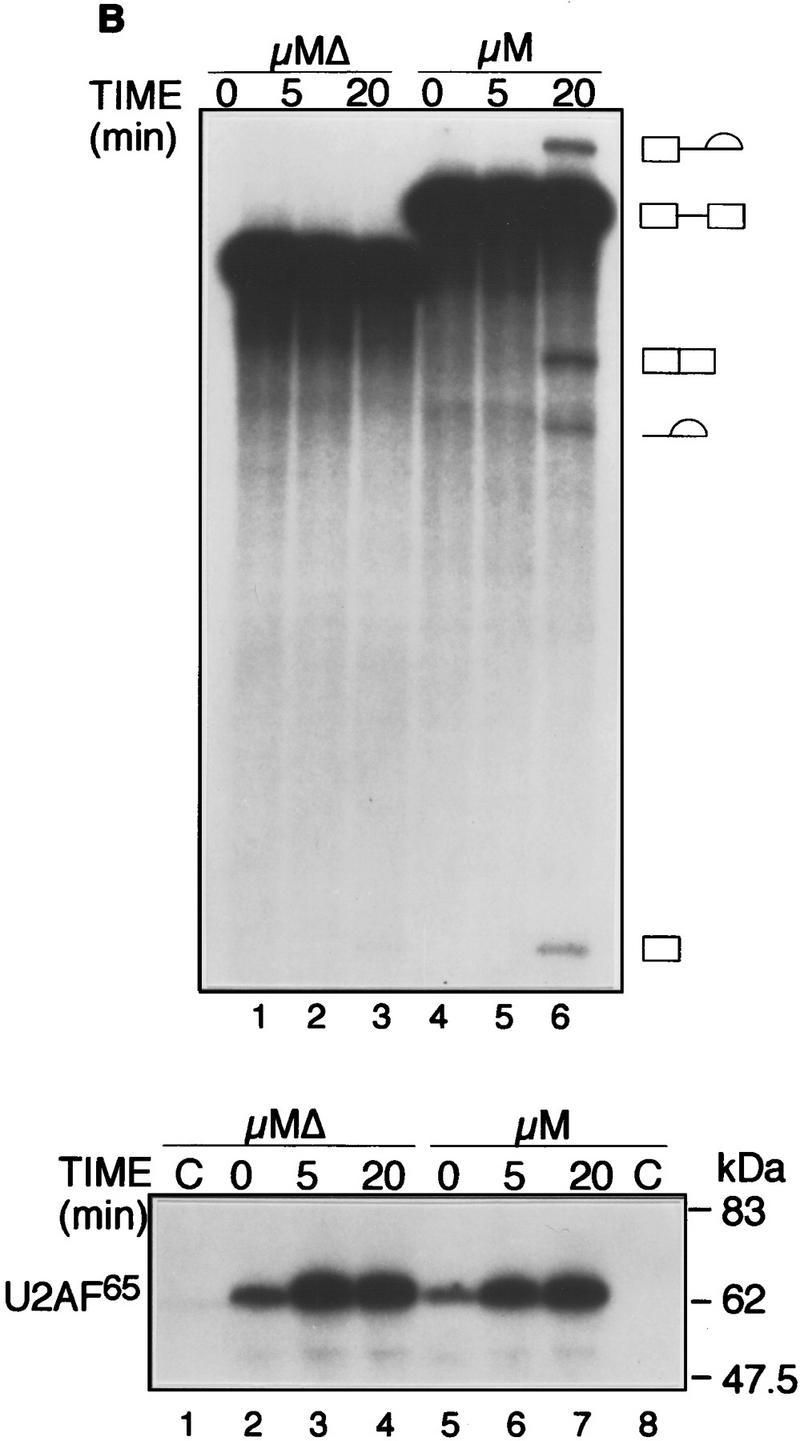
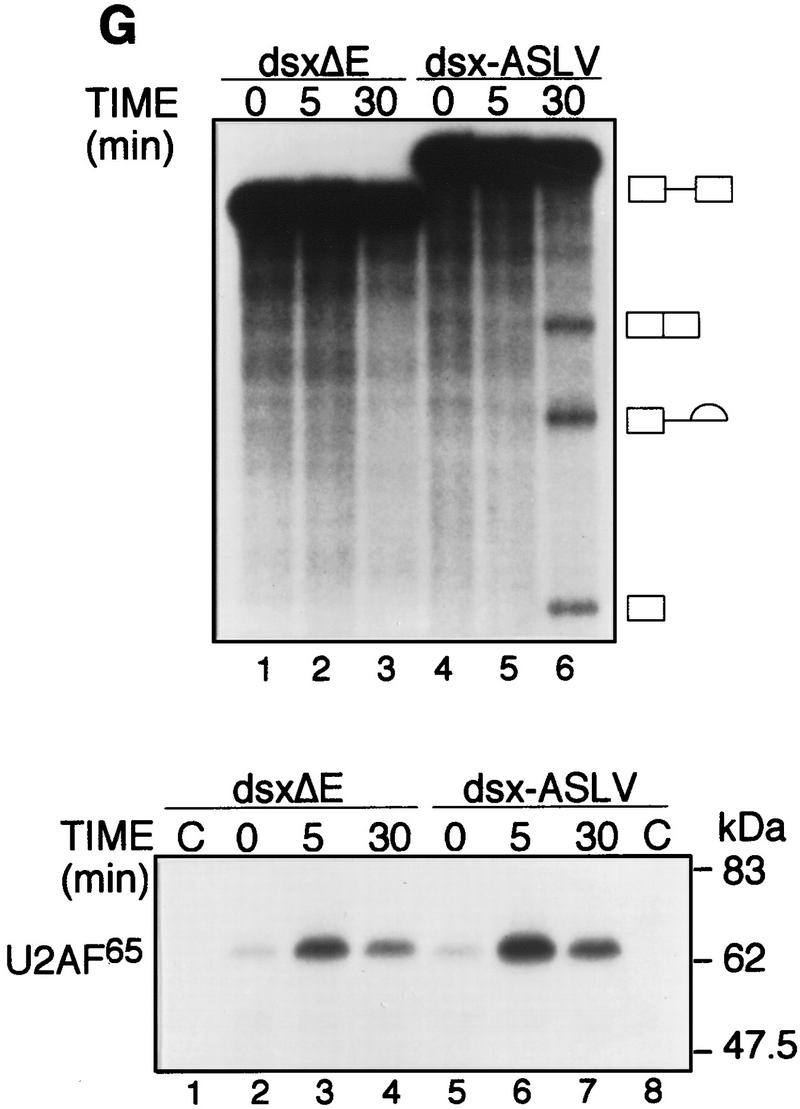
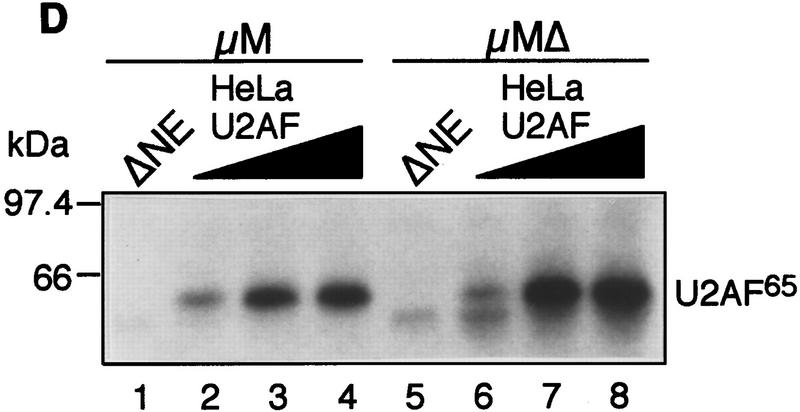
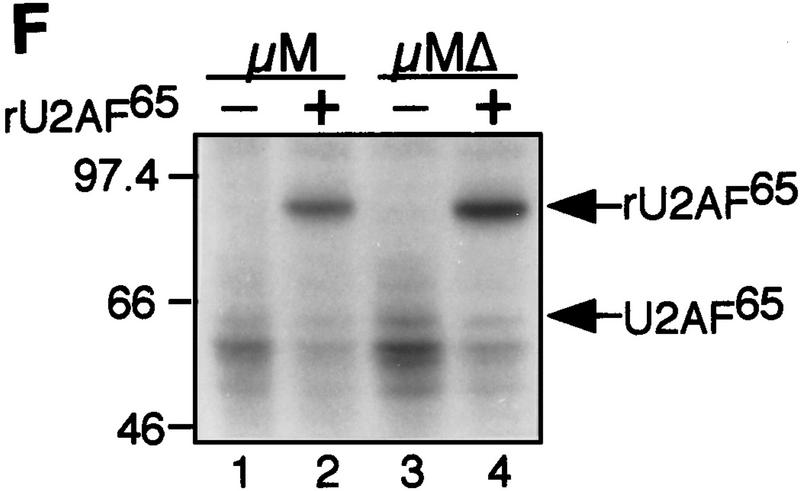
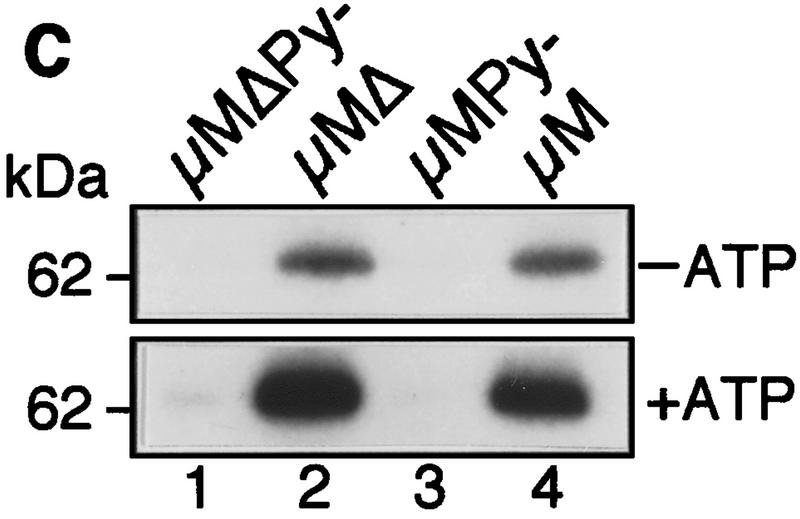
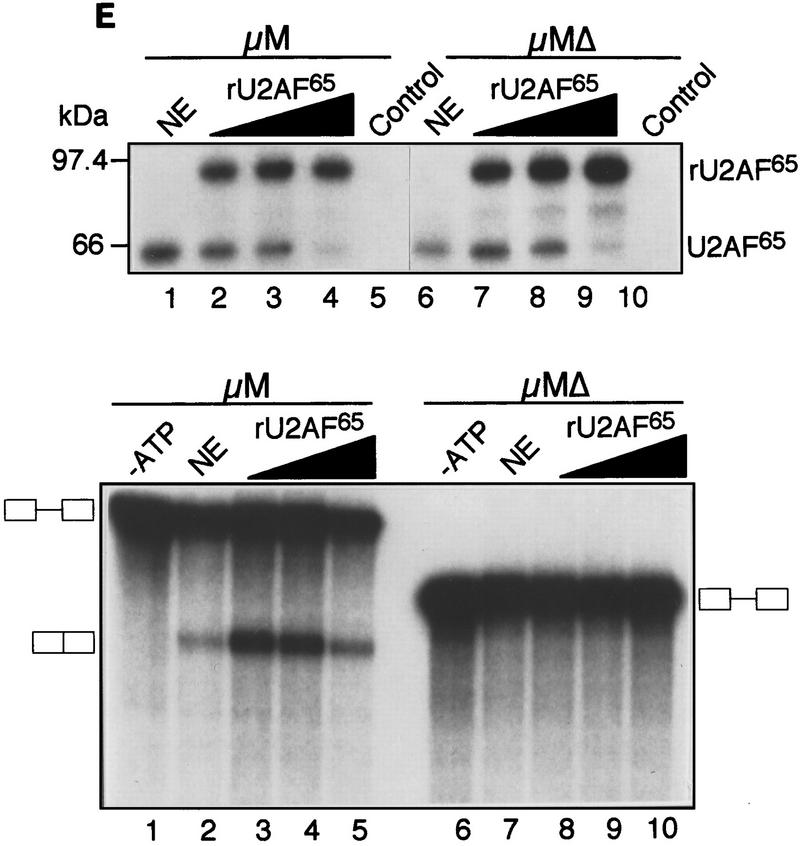
The IgM M2 enhancer does not function by increasing U2AF65 binding. (A) In vitro splicing of IgM substrates. (Bottom) A diagram of the RNA substrates μM and μMΔ (formerly named pμM1-2 and pμMΔ; Watakabe et al. 1993). The intron sequence deleted to generate RNA substrates lacking a Py tract, μMPy- and μMΔPy- is shown. (Top) Splicing was performed at 30°C for 90 min using standard conditions for μM (lane 1) and μMΔ (lane 2). The unspliced RNA substrates and spliced product are indicated. (B) Early splicing time course. In vitro splicing of RNA substrates was performed at 30°C for the times indicated. A portion of the reaction mixture was used to assess splicing by electrophoresis on a 13% denaturing gel. (Bottom) The remainder was UV cross-linked and immunoprecipitated as described in Materials and Methods and electrophoresed on a 10% SDS–polyacrylamide gel. (C) Specificity of U2AF65 binding. In vitro splicing reactions were performed as described above for 20 min in the presence or absence of ATP, cross-linked, and immunoprecipitated with the MC3 antibody. RNA substrates lacking (lanes 1,2) or containing (lanes 3,4) the enhancer or lacking the pyrimidine tract (lanes 1,3) are shown. (D) HeLa U2AF add-back. Cross-linking was performed as described for C. UV–RNA cross-linking/immunoprecipitation was performed in U2AF-depleted nuclear extract (NE) (lanes 1,5) and with increasing addition of U2AF purified from HeLa cells (lanes 2–4,6–8). (E) rU2AF65 add-back. (Top) UV–RNA cross-linking/immunoprecipitation in the presence of NE (lanes 1,6), with increasing amounts of rU2AF65 (lanes 2–4, 7–9), and immunoprecipition performed in the absence of the primary monoclonal antibody (lanes 5,10). (Bottom) In vitro splicing was performed in parallel and is shown. (F) Total UV–RNA cross-linking of IgM substrates. In vitro splicing and UV–RNA cross-linking was performed as described in C in the presence of NE (lanes 1,3) and with the addition of rU2AF65 (lanes 2,4). RNA substrates containing (lanes 1,2) or lacking (lanes 3,4) the enhancer are shown. (G) Splicing time course and UV–RNA cross-linking/immunoprecipitation of dsx RNA. (Top) In vitro splicing of RNA substrates, dsx–ASLV and dsxΔE (formerly named dsx–SO; Tanaka et al. 1994), was performed at 30°C for the times indicated. A portion of the reaction mixture was used to assess splicing by electrophoresis on an 8% denaturing gel. (Bottom) The remainder was UV cross-linked and immunoprecipitated as described in Materials and Methods and electrophoresed on a 10% SDS–polyacrylamide gel. Splicing substrates, intermediates, and products are indicated. Control UV–RNA cross-linking/immunoprecipitation (lanes 1,8) were performed in the absence of U2AF65 monoclonal antibody.
To verify that U2AF65 bound to the Py tract, we constructed and analyzed Py tract deletion mutants μMPy and μMΔPy (see Fig. 1A). The results of Figure 1C show that deletion of the Py tract eliminated U2AF65 binding, which confirmed that the data of Figure 1B measured sequence-specific binding of U2AF65 to the Py tract. On the basis of these combined data, we conclude that under conditions in which the enhancer promoted splicing it did not increase binding of U2AF65 to the Py tract.
To confirm and extend this conclusion, we analyzed U2AF65 binding at different protein concentrations. Varying amounts of purified HeLa U2AF (large and small subunit) (Figure 1D) or recombinant U2AF65 (Fig. 1E) were added to nuclear extracts immunodepleted of U2AF using an α-U2AF65 monoclonal antibody (Gama-Carvalho et al. 1997). In both experiments, at all concentrations of U2AF65 tested, binding was comparable in the presence or absence of the enhancer. Moreover, Figure 1E (top) shows that as the binding of rU2AF65 increased there was a concomitant loss of endogenous U2AF65 binding, and at the highest rU2AF65 concentration, binding of endogenous U2AF65 became undetectable. We interpret this latter result to mean that the highest U2AF concentration was saturating and thus the Py tract was fully occupied by rU2AF65. However, despite such full occupancy, in the absence of the enhancer, splicing still did not occur (Fig. 1E, bottom). Collectively, these results indicate that under a variety of conditions in which the enhancer promoted splicing, binding of U2AF65 to the Py tract did not increase.
To verify that U2AF65 binding detected in the UV cross-linking/immunoprecipitation experiments was representative of the total cross-linked product, we performed UV cross-linking in the absence of immunoprecipitation. Figure 1F shows that the amount of total cross-linked U2AF65 and rU2AF65 was equivalent in the presence or absence of the enhancer, in agreement with the results of the UV cross-linking/immunoprecipitation assay.
Finally, to determine how the results with the IgM M2 splicing enhancer compared to the well-studied Drosophila doublesex (dsx) pre-mRNA, we analyzed U2AF65 binding to a chimeric dsx pre-mRNA containing the enhancer from avian sarcoma-leukosis virus, dsx–ASLV (Tanaka et al. 1994). Figure 1G shows that under splicing conditions, binding of U2AF65 was comparable (less than twofold difference) in the presence or absence of the ASLV enhancer, whereas splicing was detectable only in the presence of the enhancer. Deletion analysis confirmed that U2AF65 bound to the Py tract (data not shown).
The IgM M2 enhancer can function in the absence of U2AF35
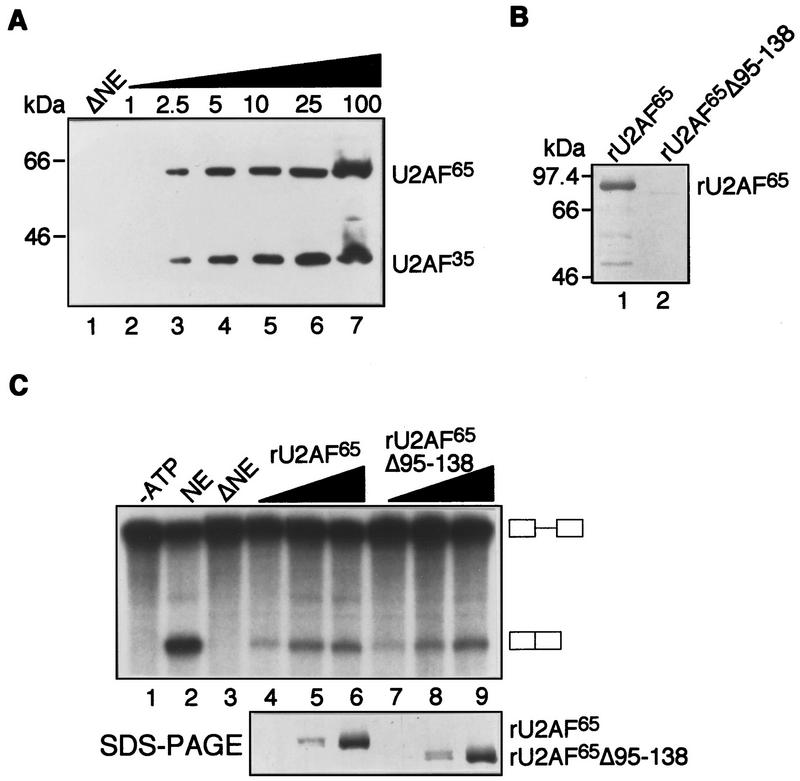
A central feature of models invoking an enhancer-dependent increase in U2AF65 binding is the formation of a network of protein–protein interactions involving U2AF35 (Wu and Maniatis 1993; Wang et al. 1995; Zuo and Maniatis 1996). We therefore tested whether U2AF35 was required for IgM M2 enhancer-dependent splicing function. U2AF65 was immunodepleted from a HeLa nuclear extract using the MC3 monoclonal antibody. The quantitative immunoblotting data of Figure 2A show that both U2AF subunits were depleted equally and present <2.5% of that in the standard HeLa nuclear extract—our limit of detection. Figure 2C shows that the immunodepleted extract failed to splice and that addition of U2AF65 restored splicing in a dose-dependent fashion. To rule out the unlikely possibility that the function of U2AF65 was due to interaction with a small amount of putative residual U2AF35, we repeated the experiment with rU2AF65Δ95–138, a U2AF65 derivative lacking the U2AF35 binding site (Fleckner et al. 1997). The Far Western analysis of Figure 2B confirmed the previous two-hybrid result (Fleckner et al. 1997) that rU2AF65Δ95–138 is severely compromised for interaction with rU2AF35. The addition of rU2AF65Δ95–138 also rescued splicing of this enhancer-dependent substrate at concentrations equivalent to that observed with wild-type rU2AF65. rU2AF65Δ95–138 bound equally to substrates containing or lacking the enhancer (data not shown). On the basis of these combined results, we conclude that U2AF35 is dispensable for the enhancer-dependent splicing of the IgM pre-mRNA substrate.
Figure 2.
The IgM M2 enhancer can function in the absence of U2AF35. (A) Quantitative immunoblotting of U2AF-depleted HeLa nuclear extract (NE). Immunoblot analysis of HeLa NE immunodepleted with the MC3 monoclonal antibody to U2AF65. Depleted NE (ΔNE, lane 1) and a concentration curve of varying amounts of HeLa NE (lanes 2–7) were probed with the U2AF65 and U2AF35 antibodies. The numbers above each lane represent the percentage of HeLa NE in the sample. (B) Far Western analysis of the rU2AF65Δ95–138–U2AF35 interaction. Far Western analysis with 35S-labeled in vitro-translated U2AF35 against rU2AF65 (lane 1) and rU2AF65Δ95–138 (lane 2). (C) In vitro splicing of IgM M2 splicing enhancer substrate in ΔNE. Splicing of IgM substrate in the absence of ATP (lane 1, control), in NE (lane 2), in ΔNE (lane 3), with increasing rU2AF65 (lanes 4–6), with rU2AF65Δ95–138 (lanes 7–9). The amount of recombinant protein used in these splicing assays is the same as those used for UV–RNA cross-linking in Fig. 1E. Equivalent levels of rU2AF65 and rU2AF65Δ95–138 were used in this experiment, as indicated by silver staining of protein added (bottom).
Substitution of IgM splicing signals with consensus elements does not relieve the enhancer requirement
To confirm the above conclusions, we replaced the natural IgM Py tract with a consensus U2AF65 binding site. We reasoned that if the enhancer functioned by promoting U2AF65 binding to a weak site, changing the Py tract to the consensus sequence should relieve the enhancer requirement. Figure 3 shows that like the original substrate, an IgM substrate containing a consensus U2AF65 binding site was not detectably spliced unless the enhancer was present (Figure 3, cf. lanes 1 and 2). Next, we tested whether changes in any of the other splicing signals could restore splicing in the absence of an enhancer. The results indicate that changing the Py tract, branchpoint, 5′ splice site, combinations of any two signals, or all three splicing signals also failed to restore splicing in the absence of the enhancer. Moreover, the enhancer promoted splicing of each pre-mRNA derivative. Taken together, these results indicate that the inability of IgM pre-mRNA to splice in the absence of the enhancer is not due to suboptimal splicing signals.
Figure 3.
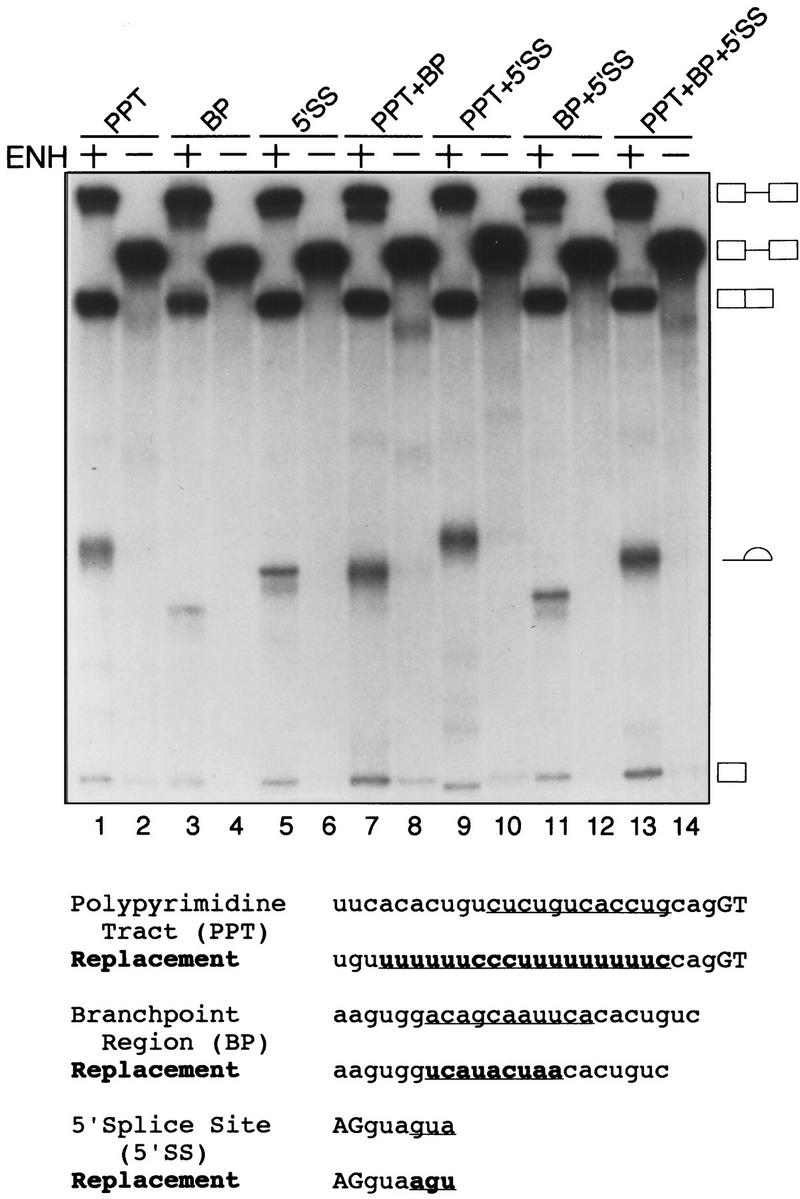
Substitution of IgM splicing signals with consensus elements does not relieve the enhancer requirement. (Bottom) Schematic representation of the replacements made to the IgM RNA substrate Py tract, branchpoint, and 5′ splice site. The sequence removed from the RNA substrate is underlined and the replacement sequence is in boldface type and underlined. (Top) In vitro splicing with different compensatory replacements of splice signals is shown. The presence or absence of the enhancer and the compensatory change made is indicated above the lanes.
Identification of a region within IgM exon M2 that inhibits splicing
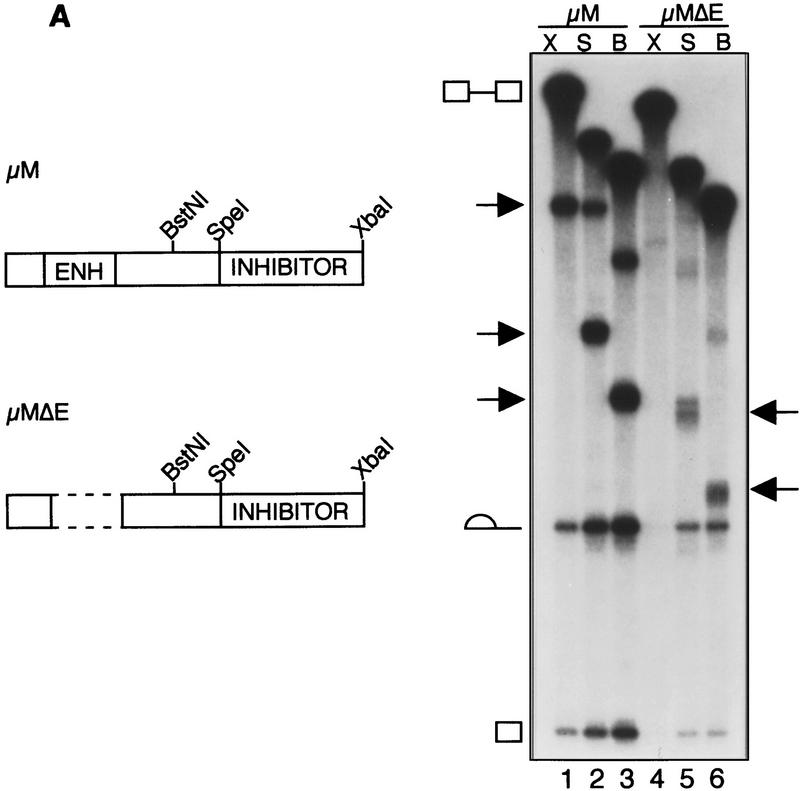
To investigate regulation of the IgM substrate in further detail, we constructed and analyzed a series of M2 exon deletion mutants. Figure 4A shows that pre-mRNA substrates in which exon M2 is truncated to the SpeI or BstN1 site gave rise to spliced products in the absence of the enhancer. Furthermore, even in the presence of the enhancer, these truncations increased splicing. These results indicate that the SpeI–XbaI fragment contains a region that inhibits splicing.
Figure 4.
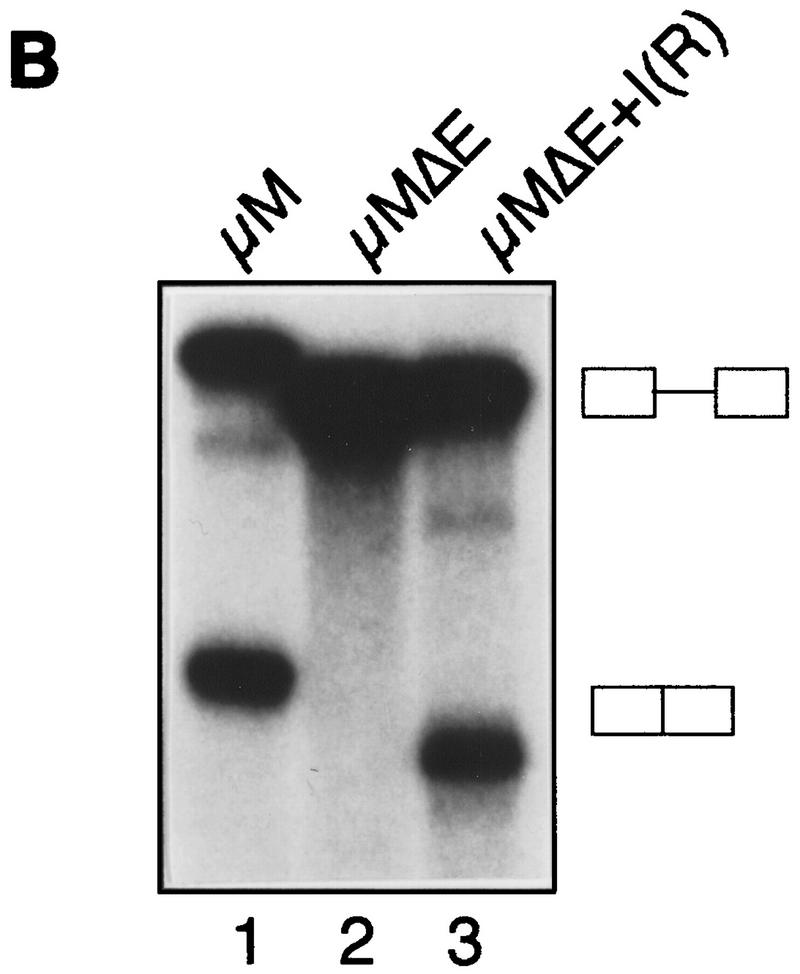
Identification of a region within IgM exon M2 that inhibits splicing. (A)(Left) Schematic diagram of IgM substrates. (ENH) The purine-rich element; (inhibitor) the region encompassing the inhibitory element. (Right) RNA substrates for in vitro splicing were generated by use of the restriction endonuclease shown above each lane: (X) XbaI; (B) BstNI; and (S) SpeI for μM (lanes 1–3) and μMΔE (lanes 4–6). Spliced products are indicated by arrows. The splicing substrate and intermediates are indicated. (B) The splicing inhibitor function is orientation-dependent. In vitro splicing of RNA substrates are μM (lane 1), μMΔE (lane 2), and with the inhibitor in reverse orientation, μMΔE+I(r) (lane 3). For in vitro transcription, μM and μMΔE were linearized with XbaI, and μMΔE+I(r) was linearized with HincII. (C) U2AF65 binding. IgM substrate in the absence of the inhibitor with (lane 1) and without the enhancer (lane 2).
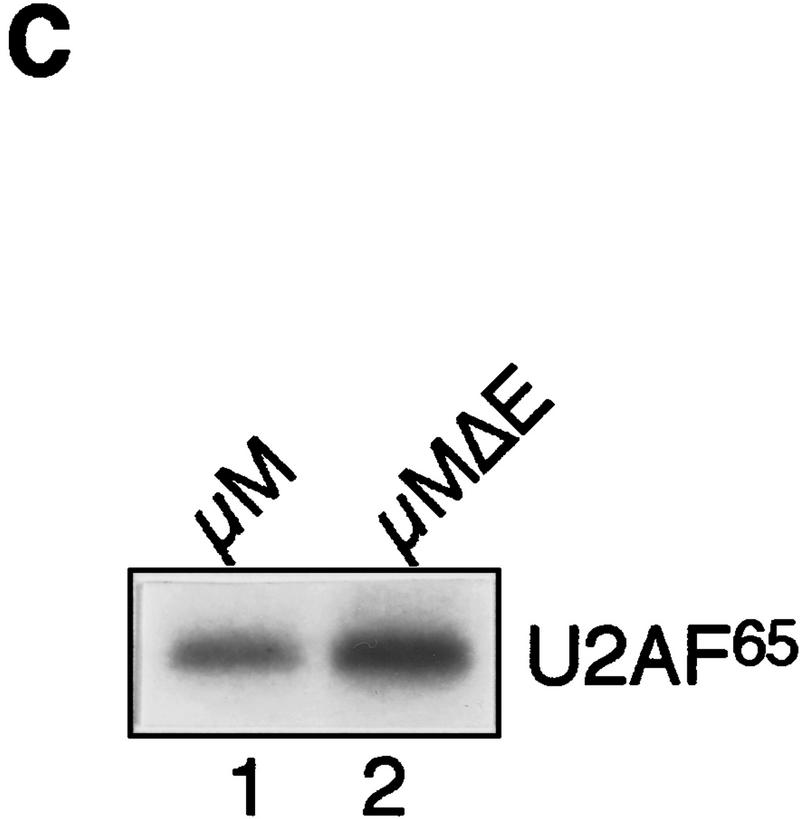
To confirm and extend this conclusion, we tested the orientation dependence of the putative IgM M2 inhibitory element. Figure 4B shows that following reversal of the SpeI–XbaI fragment (lane 3), splicing occurred in the absence of the enhancer, providing further evidence for the presence of a splicing inhibitor. Finally, on the basis of the results of Figure 1, we tested the possibility that the IgM M2 splicing inhibitor might influence U2AF65 binding. Figure 4C shows that in the absence of the inhibitory region, there was essentially equal binding of U2AF65 in the presence (lane 1) or absence (lane 2) of the enhancer.
Function of IgM M2 splicing inhibitor and enhancer in a heterologous RNA substrate
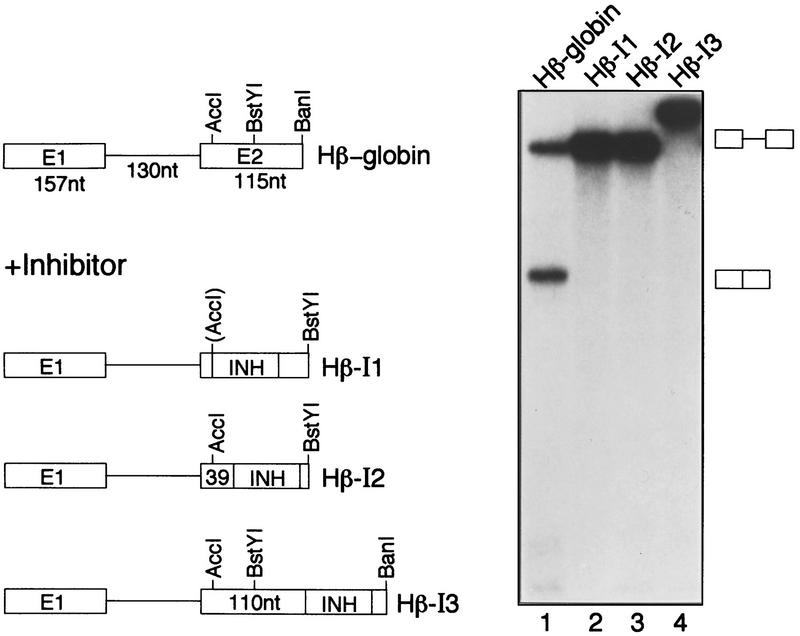
To characterize further the IgM M2 splicing inhibitor, we asked whether it could act upon a heterologous pre-mRNA. The SpeI–XbaI fragment was inserted into the second exon of a human β-globin pre-mRNA, a well-characterized efficiently spliced substrate. Figure 5 shows that placement of the IgM M2 splicing inhibitor within the second exon 14, 39, or 104 nucleotides from the 3′ splice site inhibited splicing completely.
Figure 5.
Function of the IgM M2 splicing inhibitor in a heterologous substrate. (Left) Schematic diagram of human β-globin constructs. (Right) In vitro splicing of human β-globin (lane 1) or human β-globin containing the IgM M2 splicing inhibitor in various positions (lanes 2–4). Spliced products and intermediates are indicated.
Next, we tested whether the IgM M2 splicing enhancer would restore splicing of the human β-globin derivative containing the inhibitor. To mimic the organization within the natural IgM M2 exon, the enhancer was placed upstream of the inhibitor, creating a substrate with an enhancer 14 nucleotides from the 3′ splice site followed by the IgM M2 splicing inhibitor positioned 25 nucleotides farther downstream. Figure 6 (lane 3) shows that this β-globin derivative was spliced, indicating that enhancer-dependent splicing was recapitulated in a heterologous, constitutively spliced pre-mRNA with consensus splicing signals. On the basis of these results and those presented above, we conclude that the IgM M2 splicing enhancer functions, at least in part, by counteracting the IgM M2 splicing inhibitor.
Figure 6.
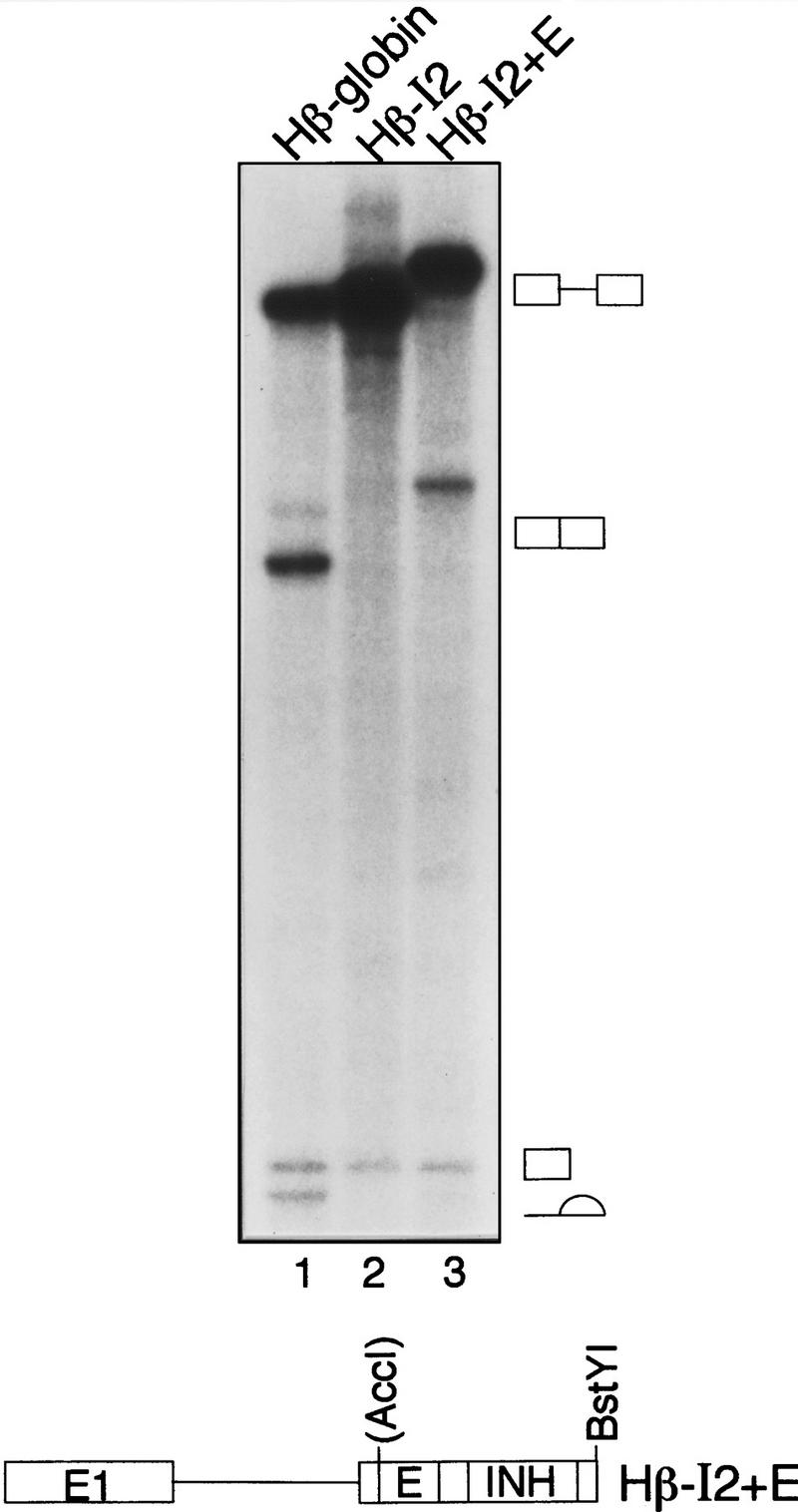
Enhancer-dependent splicing of a human β-globin pre-mRNA derivative. (Top) In vitro splicing of human β-globin pre-mRNA (lane 1), human β-globin pre-mRNA containing the IgM M2 splicing inhibitor (lane 2), and human β-globin pre-mRNA containing both splicing inhibitor and enhancer (lane 3). Spliced products and intermediates are indicated. (Bottom) For in vitro transcription, RNA substrates were linerized with BstYI.
The IgM M2 splicing inhibitor represses splicing complex assembly
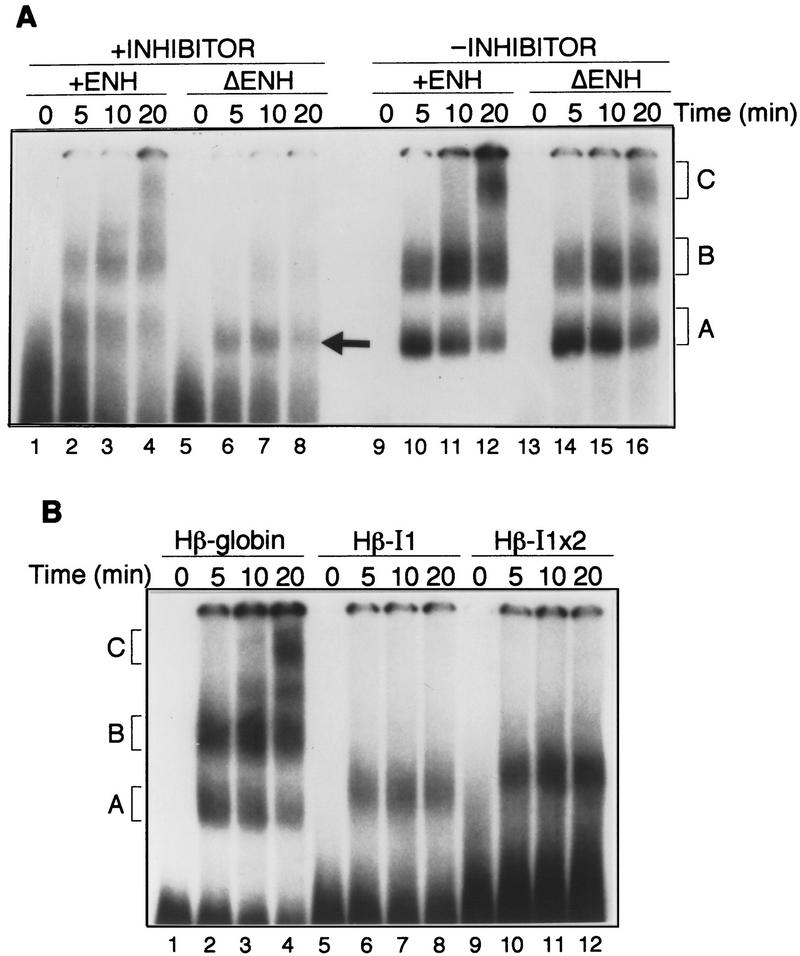
To understand how the IgM M2 splicing inhibitor functions, we analyzed splicing complex assembly. Figure 7A shows that in the wild-type substrate, which contains both enhancer and inhibitor, formation of the standard splicing complexes A, B, and C was readily detected (lanes 2–4). However, upon removal of the enhancer, formation of the B and C complexes was diminished greatly (lanes 6–8). Interestingly, and as discussed further below, the substrate lacking the enhancer formed a complex with an electrophoretic mobility similar to but distinct from that of the normal A complex (arrow). Following removal of the IgM M2 splicing inhibitor from the enhancer-less substrate, complexes B and C were again observed (lanes 14–16). Removal, of the IgM M2 splicing inhibitor also increased complex formation from the enhancer-containing substrate (cf. lanes 1–4 and 9–12), reminiscent of the splicing data of Figure 4A. We conclude that the inhibitor acts at the level of splicing complex assembly.
Figure 7.
The IgM M2 splicing inhibitor represses splicing complex assembly. (A) Complex assembly of IgM substrates. Time course of splicing complex assembly for IgM pre-mRNA substrates with enhancer and inhibitor, μM/XbaI (lanes 1–4), without enhancer and with inhibitor, μMΔE/XbaI (lanes 5–8), with enhancer and without inhibitor, μM/BstNI (lanes 9–12), or without enhancer and inhibitor, μMΔE/BstNI (lanes 13–16). The identities of the complexes are indicated. (B) Complex assembly of human β-globin substrates. Time course of complex formation for human β-globin (lanes 1–4), for human β-globin with one copy of the inhibitor (lanes 5–8), and for human β-globin with two tandem copies of the inhibitor (lanes 9–12).
We also analyzed splicing complex assembly in the heterologous human β-globin substrate. Figure 7B shows that in the presence of the IgM M2 splicing inhibitor, formation of B and C complexes did not occur, although a complex with an electrophoretic mobility similar to but clearly distinct from the normal complex A was evident (lanes 6–8). A human β-globin substrate containing two tandem copies of the IgM M2 splicing inhibitor gave rise to an increased amount of this A-like complex with a slightly reduced electrophoretic mobility (lanes 10–12).
The IgM M2 splicing inhibitor forms an ATP-dependent complex that contains U2 snRNA
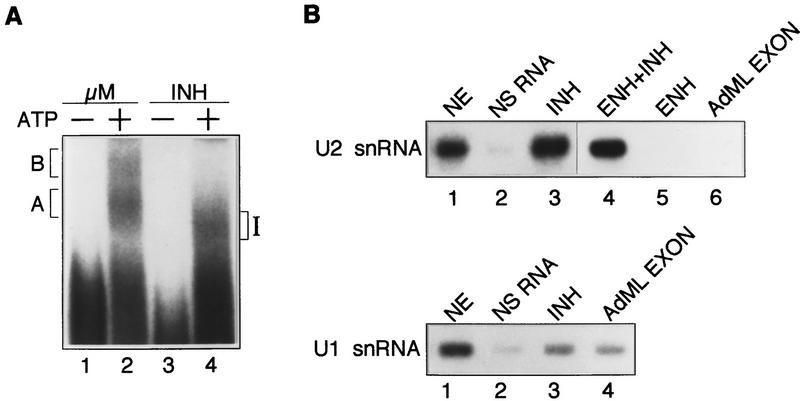
The data of Figure 7 suggests that the IgM M2 splicing inhibitor may itself be assembled into a complex. To test this possibilty, we analyzed complex formation using an RNA substrate that contains the inhibitory region of the M2 exon but lacks splicing signals. Figure 8A shows that this substrate (INH) formed an ATP-dependent complex that we define as inhibitor complex (complex I).
Figure 8.
The IgM M2 splicing inhibitor forms an ATP-dependent complex that contains U2 snRNA. (A) Complex assembly of IgM M2 splicing inhibitor. μM RNA substrate with enhancer (lanes 1,2) and the inhibitor only (INH; lanes 3,4) incubated in the absence (lanes 1,3) or presence (lanes 2,4) of ATP. (B) Northern blot analysis of inhibitor complex. Biotinylated RNA substrates were incubated in standard in vitro splicing reaction mixtures and purified on streptavidin beads. Samples were treated with proteinase K and electrophoresed on a 10% urea–polyacrylamide gel. (Top) RNAs purified from nuclear extract (NE; lane 1), nonspecific RNA (lane 2), splicing inhibitor (lane 3), splicing enhancer and inhibitor (lane 4), splicing enhancer (lane 5), or exon sequence from AdML (lane 6) were probed with a 32P-end-labeled α–U2 snRNA oligonucleotide. (Bottom) RNAs purified from NE (lane 1), nonspecific RNA (lane 2), splicing inhibitor (lane 3), or exon sequence from AdML (lane 4) were probed with a 32P-end-labeled α-U1 snRNA oligonucleotide.
Because of the similiarity in electrophoretic mobility to complex A, we analyzed whether complex I contained U2 snRNA. Biotinylated RNA substrates of the inhibitor and several control RNAs were synthesized, incubated under splicing conditions, affinity purified, and analyzed for U2 snRNA by Northern blotting. The results of Figure 8B show that the inhibitor contained stably bound U2 snRNA (lane 3). Binding was specific, as evidenced by the fact that U2 snRNA did not bind to RNA substrates comprising irrelevant polylinker sequences (lane 2), Adenovirus major late (AdML) exon sequences (lane 6), or the IgM M2 splicing enhancer (lane 5). The results also show that binding of U2 snRNA to the IgM inhibitor was not affected by the enhancer (cf. lanes 3 and 4). In contrast to the results with U2 snRNA, binding of U1 snRNA to the inhibitor was not above the background level of this assay, as evidenced by comparison to the nonspecific RNA control and the AdML exon RNA (cf. lane 3 and lanes 2 and 4).
Discussion
In this study we have identified a novel splicing inhibitor in the IgM exon M2, located near the previously defined enhancer. The IgM M2 splicing inhibitor forms a complex that contains U2 snRNP and represses normal spliceosome assembly and splicing. The IgM M2 splicing enhancer antagonizes the inhibitor, enabling splicing to occur.
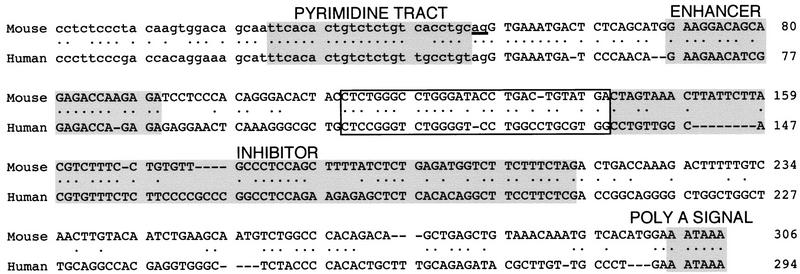
Figure 9 compares IgM M2 from mouse and human. Clearly, several regions are conserved, including known processing signals such as the Py tract/3′ splice site, the purine-rich enhancer, and the polyadenylation signal. Also conserved are portions of the region containing the inhibitor, corresponding to the mouse SpeI–XbaI fragment, consistent with an important regulatory function. Finally, we note that an ∼32-nucleotide region immediately upstream of the inhibitor is also highly conserved.
Figure 9.
Sequence comparison of mouse and human IgM exon M2. Conserved elements, Py tract , enhancer, inhibitor, and polyadenylation signal are shaded and conserved sequence immediately upstream of the inhibitor is boxed.
Splicing enhancers and U2AF
Although it seems clear that splicing enhancers function by binding one or more members of the SR protein family (for review, see Fu 1995; Manley and Tacke 1996; Valcárcel and Green 1996), their detailed mechanism of action remains to be elucidated. Previous studies have reported that binding of U2AF65 to the Py tract was increased by an enhancer (Wang et al. 1995; Zuo and Maniatis 1996; Bouck et al. 1998). Increased U2AF65 binding was proposed to result from the formation of a network of protein–protein interactions that requires the RS domain of U2AF35 (Wu and Maniatis 1993; Zuo and Maniatis 1996).
We have found that under splicing conditions, the enhancer did not significantly affect binding of U2AF65 to IgM or dsx substrates. Zuo and Maniatis (1996), measured U2AF binding in reaction mixtures containing purified U2AF and a single SR protein, whereas our experiments were performed in crude HeLa nuclear extract under in vitro splicing conditions. It seems likely that the disparate conclusions reached in these studies is related to the significant differences in the biochemical systems.
A prediction of the U2AF65 binding model is that raising the U2AF concentration should circumvent the enhancer requirement, a prediction not met for IgM pre-mRNA (Fig. 1E) or, to our knowledge, in any other instance. Finally, inconsistent with the proposed model are recent in vivo studies demonstrating that the RS domain of the Drosophila U2AF35 homolog dU2AF38 is dispensable both for proper regulation of dsx splicing and for viability (Rudner et al. 1998a).
Splicing inhibitors
A complete elucidation of IgM pre-mRNA splicing will now require an understanding of how both the enhancer and the inhibitor function. Although many splicing enhancers conform to a purine-rich consensus sequence, the splicing inhibitors identified to date appear remarkably diverse (Nemeroff et al. 1992; Siebel et al. 1992; Amendt et al. 1995; Del Gatto and Breathnach 1995; Staffa and Cochrane 1995; Staffa et al. 1997; Valcárcel and Gebauer 1997; Grabowski 1998). For example, there is no obvious splicing inhibitor consensus sequence, perhaps reflecting the specificity and complexity of these elements for tissue-specific and developmentally regulated splicing. Some splicing inhibitors appear to bind the Py tract binding protein (PTB), a known inhibitor of splicing (for review, see Valcárcel and Gebauer 1997). In this regard, both the mouse and human IgM M2 sequences contain the core consensus sequence for PTB binding (UCUU) (Pérez et al. 1997), raising the possibility that the function of the IgM M2 splicing inhibitor may involve PTB binding.
Although diverse in sequence, several splicing inhibitors have been found to bind snRNPs. For example, splicing regulation of the Drosophila P element is mediated by binding of U1 snRNP to a pseudo 5′ splice site (Siebel et al. 1992); a negative regulator in Rous sarcoma virus binds U1 and U11 snRNP (Gontarek et al. 1993; Cook and McNally 1998; McNally and McNally 1998); and we have found the IgM M2 splicing inhibitor associates with U2 snRNP.
Mechanism of IgM M2 splicing
The IgM M1–M2 RNA substrate has been used extensively as a model for splicing enhancer function. It was first identified based on the ability of exon M2 sequences to stimulate splicing of the preceding intron (Watakabe et al. 1991). Further characterization identified a purine-rich element responsible for splicing enhancement (Watakabe et al. 1993). Here, we identify a novel splicing inhibitor located near the purine-rich enhancer. This organization of nearby positive and negative splicing elements is reminiscent of several other pre-mRNAs, for example, the terminal HIV-1 tat/rev exon (Amendt et al. 1995; Staffa and Cochrane 1995) and the human fibronectin EDA exon (Caputi et al. 1994; Staffa et al. 1997).
Watakabe et al. (1993) reported that several IgM pre-mRNA derivatives lacking both the enhancer and the inhibitor were not spliced. Because these substrates lacked the inhibitor, our results would have predicted that splicing should occur. We suggest that the lack of splicing may be a consequence of the relatively short incubation (20 min) used in the study by Watakabe et al. (1993). We note that even in the one substrate that was spliced (μMΔ+U1), there was only a very small amount of spliced product. In our experiments, substrates lacking both the enhancer and the inhibitor were spliced significantly more efficiently following long incubation times (90 min). Moreover, although substrates lacking the inhibitor were spliced, the enhancer still increased the amount of spliced product (see Fig. 4).
Unexpectedly, we found that the IgM M2 splicing inhibitor bound the essential splicing factor, U2 snRNP. It will be important to determine whether the U2 snRNP–inhibitor interaction requires U2AF65 and the other factors involved in the normal U2 snRNP–branchpoint interaction. Of particular interest is how the U2 snRNP complex formed on the IgM M2 splicing inhibitor resembles the normal U2 snRNP–branchpoint complex.
Although U2AF65 is required for an efficient U2 snRNP–branchpoint interaction, following deletion of the Py tract U2 snRNP binds the branchpoint at a low level (Nelson and Green 1989). Thus, U2AF65 may be required for the U2 snRNP–inhibitor interaction, even in the absence of a high affinity binding site. Alternatively, another factor may substitute for U2AF in promoting binding of U2 snRNP to the inhibitor.
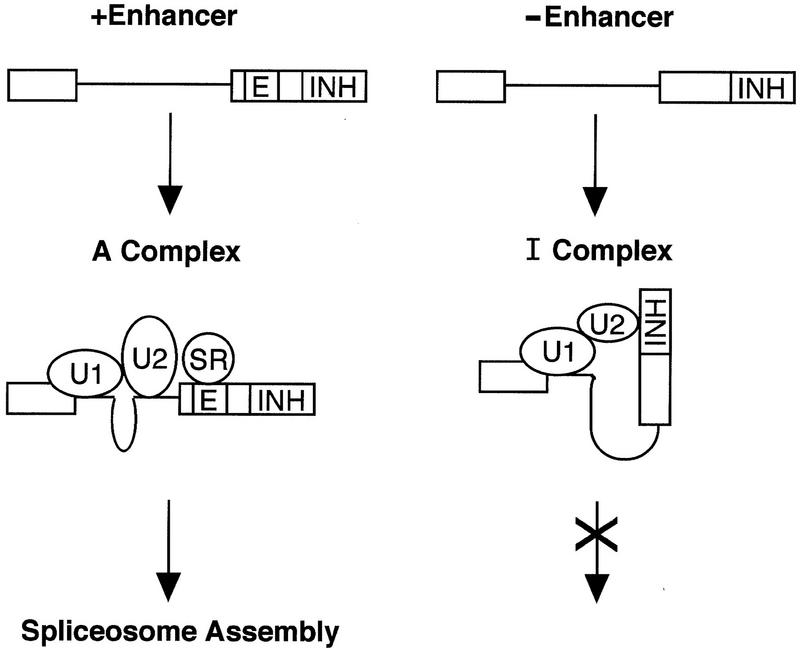
We speculate that the U2 snRNP–inhibitor complex interacts physically and/or functionally with the U1 snRNP-containing complex at the 5′ splice site, forming a ‘dead-end’ complex (see Fig. 10). The IgM M2 splicing enhancer could then function to counteract the inhibitor in several ways, including: increasing formation of the bona fide U2 snRNP-branchpoint complex; promoting pairing of the U2 snRNP-branchpoint complex with the 5′ splice site complex; or directly antagonizing formation or activity of the inhibitor complex.
Figure 10.
Model of splicing enhancer function in IgM pre-mRNA. In the absence of the enhancer (right), a U2 snRNP-inhibitor complex pairs with the 5′ splice site complex resulting in a dead-end complex. In the presence of the enhancer (left), a functional U2 snRNP–branchpoint complex productively pairs with the 5′ splice site complex, which is subsequently assembled into an active spliceosome.
Materials and methods
Plasmid construction
Plasmids μMPy, μMΔPy, μMΔE, and all plasmids containing splice site replacements (Fig. 3) were constructed by PCR using appropriate primer pairs starting with μM and μMΔ (formerly called pμM1-2 and pμMΔ, respectively; Watakabe et al. 1993). To obtain the RNA substrate that contained the RNA splicing inhibitor, the SpeI–XbaI fragment of μM was inserted into the XbaI site of pSP73. The RNA substrate with the IgM M2 splicing enhancer only was constructed by insertion of annealed oligo nucleotides into the HindIII–EcoRI site of pSP72. To obtain μMΔE(r), μMΔE was digested with SpeI–XbaI and religated with itself. To generate the Hβ-I constructs, first exon 3 and a portion of exon 2 was removed from pSP64-HβΔ6 (Krainer et al. 1984) by digestion with BamHI to create SP64–HβT. Hβ-I1 was constructed by insertion of the blunted SpeI–XbaI fragment from pμM into the AccI site of SP64–HβT. Hβ-I2 and Hβ-I3 were generated by PCR of SP64–HβT and insertion of the blunted SpeI–XbaI fragment from μM. Hβ–I2+E was generated by digesting Hβ–I2 with AccI and insertion of annealed oligonucleotides corresponding to the enhancer of μM.
Expression and purification of U2AF65
U2AF65 and U2AF65Δ95–138 were expressed in Escherichia coli as GST fusion proteins and purified by standard procedures. Plasmids used for protein expression were described previously (Zamore et al. 1992; Fleckner et al. 1997). Protein gels for assessing recombinant protein addition in splicing assays were stained with Plusone silver stain (Pharmacia).
Splicing and spliceosome assembly assays
Splicing assays were performed in 10μl reaction mixtures containing 50% HeLa nuclear extract or 30% HeLa nuclear extract depleted of U2AF with 2 mm MgCl2, 0.6 mm ATP, 20 mm creatine phosphate, and 5 units of RNasin. Capped RNAs were prepared using either SP6 or T7 RNA polymerase. Nuclear extracts were depleted of U2AF using the monoclonal antibody MC3 to U2AF65. In brief, 350 μl of monoclonal antibody was incubated for 4 hr at 4°C with 100 μl of anti-mouse IgG agarose beads (Sigma). Antibody was removed, and the beads were washed twice with Bfr D/0.1 m KCl (20 mm HEPES at pH 8.0, 20% glycerol, 0.2 mm EDTA, 1 mm DTT, and 100 mm KCl) followed by addition of 160 μl of HeLa nuclear extract and incubation for 3 hr at 4°C. The levels of U2AF depletion achieved were assessed by immunoblotting with the MC3 and U2AF35 antibodies (Zuo and Maniatis 1996; Gama-Carvalho et al. 1997).
Splicing complex assembly assays were performed as described above in a 10-μl reaction mixture, treated with 0.5 mg/ml heparin at room temperature for 10 min, followed by addition of native gel loading dye. An aliquot of the reaction mixture was loaded on a 4% polyacrylamide (37.5:1) gel containing 0.5% agarose in Tris–glycine buffer (95 mm glycine, 12 mm Tris at pH 8.5). All complex assembly gels were electrophoresed for 6–8 hr at 200 V and 4°C.
UV–RNA cross-linking and immunoprecipitation assays
UV–RNA crosslinking was performed essentially as described (Wang et al. 1995). In brief, splicing reactions were performed as described above for the times indicated in a total volume of 60 μl. UV cross-linking was for a total of 1.2 J using a Stratagene cross-linker. The UV–RNA cross-linked reaction mixtures were treated with 0.2 mg/ml RNase A at room temperature for 15 min, followed by addition of SDS–sample buffer and electrophoresed on a 10% SDS–polyacrylamide gel. For immunoprecipitation, the RNase A-treated splicing reaction mixtures were incubated with 8 μl of MC3 monoclonal antibody for 2 hr at 4°C. Anti-mouse IgG agarose beads (15-μl bead volume) were added and incubated for an additional 2–3 hr with continuous mixing on a rotator device at 4°C. The samples were spun at 1000g for 2–3 min to pellet the beads, and the supernatant was removed. Washes were performed by addition of buffer followed by gentle mixing and subsequently allowing the beads to settle on ice for 10 min. The beads were washed four times with high salt buffer (500 mm NaCl, 1% NP-40, 50 mm Tris-Cl at pH 8.0) and once with 50 mm Tris-Cl at pH 8.0. SDS–sample buffer was added to the beads and boiled to release the immunoprecipitated protein, and the supernatant was electrophoresed on a 10% SDS–polyacrylamide gel. The gel was subsequently incubated in a solution containing 50% methanol and 10% acetic acid for 8–12 hr. This was followed by incubation in 10% methanol and 5% acetic acid for 1 hr. The gel was dried before autoradiography.
Far Western blot analysis
In vitro-translated 35S-labeled U2AF35 was produced using the coupled in vitro transcription/translation system (Promega). U2AF65 and U2AF65Δ95–138 were electrophoresed on a 10% SDS–polyacrylamilde gel, transferred to poly(vinylidene difluoride) membrane (Immobolin-P, Millipore) by electroblotting. Far Western analysis was performed as described in Zhang et al. (1992).
Northern blot analysis
In vitro splicing was performed with a biotinylated (biotin-14-CTP, GIBCO-BRL) RNA substrate (50-μl reaction mixture) and subsequently treated with 0.4 mg/ml heparin at room temperature for 10 min. The reaction mixture was incubated with streptavidin beads at 4°C for 2 hr. Beads were washed four times using 0.2 m KCl bead wash buffer (50 mm Tris-Cl at pH 7.9, 0.2 m KCl, 0.05% NP-40, 0.5 mm DTT). A solution containing 3 mg/ml proteinase K in PK buffer (50 mm Tris-Cl at pH 7.6, 100 mm NaCl, 10 mm EDTA, 1% SDS) was added to the beads, followed by incubation at 55°C for 30 min. The supernatant was transferred to a new tube, extracted with phenol/chloroform and chloroform, and ethanol precipitated. The RNA was electrophoresed on a 10% denaturing polyacrylamide gel and transferred to Zeta probe GT membrane (Bio-Rad) by electroblotting in 0.5× TBE buffer for 30 min at 60 mA. The membrane was probed with a 32P-end-labeled anti-U1 or anti-U2 snRNA oligonucleotide.
Acknowledgments
We thank Y. Shimura for the IgM constructs, pμM1-2 and pμMΔ, and the dsx constructs, dsx–ASLV and dsx–SO, P. Zuo and T. Maniatis for the polyclonal antibody to U2AF35, and M. Carmo-Fonsenca for the MC3 monoclonal antibody to U2AF65. We thank A.T. Dang and J. Zhu for preparation of nuclear extracts and members of the Green laboratory for helpful discussions. J.L.C.K. was supported by a postdoctoral fellowship from the National Institutes of Health (NIH). This work was supported by a grant from the NIH to M.R.G.. M.R.G. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL michael.green@ummed.edu; FAX (508) 856-5473.
References
- Amendt BA, Si A, Stoltzfus CM. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: Evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck J, Fu X-D, Skalka AM, Katz RA. Role of the constitutive splicing factors U2AF65 and SAP49 in suboptimal RNA splicing of novel retroviral mutants. J Biol Chem. 1998;273:15169–15176. doi: 10.1074/jbc.273.24.15169. [DOI] [PubMed] [Google Scholar]
- Caputi M, Casari G, Guenzi S, Tagliabue R, Melo CA, Baralle FE. A novel bipartite splicing enhancer modualtes the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CR, McNally MT. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;9:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcárcel J, Green MR. U2AF65 recruits a novel human dead box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splciing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcárcel J, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontarek RR, McNally MT, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes & Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- Grabowski PJ. Splicing regulation in neurons: Tinkering with cell-specific control. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Lynch KW, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- McNally LM, McNally MT. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol Cell Biol. 1998;18:3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KK, Green MR. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes & Dev. 1989;3:1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Utans U, Krämer A, Krug RM. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting prespliceosome complexes. Mol Cell Biol. 1992;12:962–970. doi: 10.1128/mcb.12.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez I, Lin C-H, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Reed R. Initial splice-site recogniton and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci. 1996;93:10333–10337. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Breger KS, Rio DC. Molecular genetic analysis of the heterodimeric splicing factor U2AF: The RS domain on either the large or small Drosophila subunit is dispensable in vivo. Genes & Dev. 1998a;12:1010–1021. doi: 10.1101/gad.12.7.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Interactions between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol. 1998b;18:1765–1773. doi: 10.1128/mcb.18.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Fresco LD, Rio DC. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: Multiprotein complexes at an exon pesudo-5′ splice site control U1 snRNP binding. Genes & Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements withing the terminal tat-rev exon of human immunodefiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffa A, Acheson NH, Cochrane A. Novel exonic elements that modulate splicing of the human fibronectin EDA exon. J Biol Chem. 1997;272:33394–33401. doi: 10.1074/jbc.272.52.33394. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J, Gebauer F. Post-transcriptional regulation: The dawn of PTB. Curr Biol. 1997;7:705–708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Wang Z, Hoffmann HM, Grabowski PJ. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Sakamoto H, Shimura Y. Repositioning of an alternative exon sequence of mouse IgM pre-mRNA activates splicing of the preceding intron. Gene Expression. 1991;1:175–184. [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes & Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splciing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicng factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]