Abstract
The ability to attach to host cells is one of the main determinants of the host range of influenza A viruses. By using virus histochemistry, we investigate the pattern of virus attachment of both a human and an avian influenza virus in colon and trachea sections from 12 wild bird species. We show that significant variations exist, even between closely related avian species, which suggests that the ability of wild birds to serve as hosts for influenza viruses strongly varies among species. These results will prove valuable to assess the possibilities of interspecies transmission of influenza viruses in natural environments and better understand the ecology of influenza.
Introduction
Wild waterbirds are considered to be the fundamental reservoir of influenza A viruses [1]. However, the role played by different waterbird species in the ecology of influenza viruses remains unclear, and important parameters such as the frequency of interspecies transmission are unknown [1]. The host range of influenza viruses is determined by, among other factors, the ability of the virus to attach to sialic acid (SA) residues on the surface of host cells [2]. Because avian viruses preferentially attach to α-2,3-linked SAs whereas human viruses attach to α-2,6-linked SAs, the distribution of α-2,3- and α-2,6- linked SAs on surface epithelia is usually studied by using plant lectins, such as the Maackia amurensis agglutinin (MAA) and the Sambucus nigra agglutinin (SNA), which respectively recognize α-2,3- and α-2,6-linked SAs [2], [3], [4]. Lectin studies conducted on bird tissues suggest that SA expression varies depending on species and tissues [5], [6], [7], [8], [9].
However, lectin histochemistry has inherent limitations in predicting host-virus interactions. Indeed, MAA and SNA lectins do not detect differences involving the inner chain of SAs [10], [11] and MAA binding specificity may differ depending on isotypes, with unspecific binding to unsialylated glycoconjugates as well as binding to α2,3-sialylated structures of limited/unknown role in the attachment of influenza viruses [3], [4], [12], [13]. Therefore, the use of labeled influenza virus particles to study the pattern of virus attachment (PVA) in target tissues is a better tool to determine whether a species is receptive to influenza virus infection, and to assess the cell and tissue tropism of viruses [6], [14], [15].
A previous study, comparing human and avian influenza virus PVAs on the upper and lower respiratory tract of humans and several other mammal species [15], identified differences in cell and tissue tropism between human and avian influenza viruses and showed that the PVA in the human respiratory tract corresponds to the main presentation of the disease [15]. The objective of the present study was to determine the PVA of avian and human influenza A viruses in the colon and trachea of wild bird species. Our hypothesis was that differences in PVA could help classify avian species with regard to their roles in the ecology of low pathogenic avian influenza viruses (LPAIVs).
As representatives of influenza viruses with an avian-like and a human-like receptor specificity, we respectively used a LPAIV (H6N1 A/Mallard/Sweden/81/02), which does not attach to human trachea [15], and a seasonal human influenza virus (H3N2 A/Netherlands/213/03), which attaches abundantly to human and pig trachea [15]. We chose the colon and trachea as target tissues because LPAIVs are known to replicate in these tissues in mallards and related domestic ducks [16], [17]. We selected six waterbird species (Table 1) that share similar habitats and from which LPAIVs have been isolated [1]. For comparison, we examined tissues from domestic chickens and six terrestrial species, including three species (rock pigeon, hooded crow, house sparrow) suspected of acting as bridge species for the transmission of avian influenza viruses between wild waterfowl and domestic poultry [18], and three insectivorous species (European robin, goldcrest, blue tit) unlikely exposed to influenza viruses.
Table 1. Pattern of virus attachment of an avian influenza virus and a human seasonal influenza virus in the trachea and colon of 12 wild bird species and domestic chicken.
| Habitat type | Taxonomic order | Species | Latin name | Prevalencea (%)[95% binomial confidence interval] | Avian virus | Human virus | ||
| Colon | Trachea | Colon | Trachea | |||||
| Aquatic | Anseriformes | Mallard | Anas plathyrhynchos | 12.9 [12.4–13.4] | +++ | +++ | − | − |
| Eurasian wigeon | Anas penelope | 0.8 [0.4–1.5] | ± | +++ | +c | − | ||
| Greylag goose | Anser anser | 1.1 [0.6–2.0] | ++ | +++ | ++c | − | ||
| Tufted duck | Aythya fuligula | 1.1 [0.2–3.3] | − | +++ | − | + | ||
| Charadriiformes | Herring gull | Larus argentatus | 1.4 [0.7–2.5] | + | + | − | ++ | |
| Pelecaniformes | Great cormorant | Phalacrocorax carbo | 0.4 [0.2–0.6] | ±b | +++ | ± | − | |
| Terrestrial | Galliformes | Domestic chicken | Gallus gallus | ++ | +++ | +c | ± | |
| Columbiformes | Rock pigeon | Columba livia domesticus | − | − | − | +++ | ||
| Passeriformes | Hooded crow | Corvus corone cornix | + | +++ | − | − | ||
| House sparrow | Passer domesticus | + | −d | ±e | − | |||
| European robin | Erithacus rubecula | ++ | +++ | ± | +++ | |||
| Goldcrest | Regulus regulus | + | ± | − | −d | |||
| Blue tit | Cyanystes caeruleus | ± | + | ±e | −d | |||
Scoring indicates attachment to ciliated cells for the trachea and intestinal epithelial cells from the crypts or villae for the colon.
−: no significant attachment.
±: attachment to rare or few cells.
+: attachment to a moderate number of cells.
++: attachment to many cells.
+++: attachment to a large majority of cells.
: prevalence data are extracted from Olsen et al. 2006 [1] and are calculated considering altogether all low pathogenic avian influenza subtypes.
: attachment to intestinal epithelial cells in 1 of the 3 cormorants.
: attachment to intestinal epithelial cells in the crypts.
: attachment to tracheal goblet cells.
: attachment to colon goblet cells.
Results and Discussion
Attachment of the avian virus
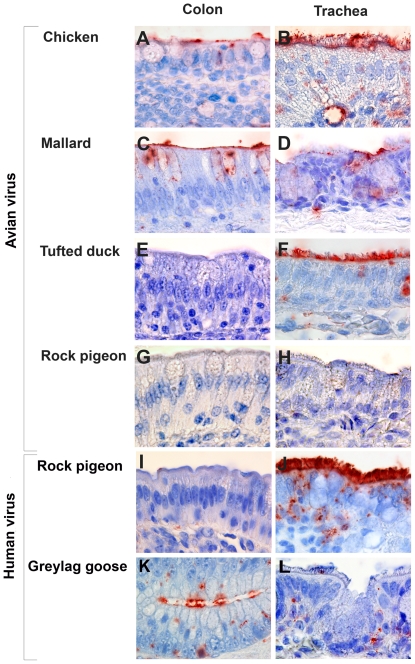
The overall PVA was consistent among individuals of the same bird species. As expected, the avian virus attached to tissues from wild birds more frequently than the human virus (Table 1). The avian virus attached abundantly to both the trachea and colon of mallards and chickens (Figure 1), which corroborates earlier observations of α2,3-linked SAs and avian virus attachment in tracheal and intestinal cells of chickens and mallards [6], [7], [19] and fits with the observation that LPAIV infection results in virus replication in both the respiratory and digestive tracts of these species [16], [20].
Figure 1. Colon and trachea sections from selected wild bird species showing that the pattern of attachment of avian and human influenza viruses varies between bird species.
(a) chicken colon with avian virus; (b) chicken trachea with avian virus; (c) mallard colon with avian virus; (d) mallard trachea with avian virus; (e) tufted duck colon with avian virus; (f) tufted duck trachea with avian virus; (g) rock pigeon colon with avian virus; (h) rock pigeon trachea with avian virus; (i) rock pigeon colon with human virus; (j) rock pigeon trachea with human virus; (k) greylag goose colon crypt with human virus; (l) greylag goose trachea with human virus.
The main purpose of this study was to investigate wild bird species other than mallards. Because all waterbird species investigated were reported to carry influenza viruses [1], we expected the avian influenza virus to attach to their colon surface epithelium. Attachment to many cells was observed for geese and European robin, and attachment to a moderate number of cells was observed for gulls and several terrestrial species. However, attachment to only a few goblet cells was detected in wigeons and no attachment was observed in the colon of tufted ducks (Figure 1). This result suggests that the susceptibility of tufted ducks to intestinal infection with this avian virus is relatively low. Given that epidemiological surveys have shown that tufted ducks occasionally shed LPAIVs [1], experimental infections combined with PVA studies should be conducted to assess whether other avian viruses can replicate in their digestive tract.
The detection of significant virus attachment to tracheal ciliated cells of all bird species, except pigeons and sparrows, suggests that the upper respiratory tract may be another major entry route for influenza viruses in wild bird species. These results are consistent with surveillance [21], [22], [23] and experimental [24], [25] studies showing that LPAIVs are frequently detected in oropharyngeal swabs from wild waterfowl as well as lectin studies suggesting that α-2,3-linked SAs are expressed on domestic bird trachea and bronchus [6], [9].
The fact that no avian virus attachment was detected on the colon and trachea of domestic pigeons (Figure 1) suggests that pigeons have low susceptibility to infection by this avian influenza virus. This result fits with observations that H5N1 highly pathogenic avian influenza virus (HPAIV) replication in pigeons only is observed with high inoculation doses [26] and that α-2,3-linked SAs are expressed poorly in pigeons [27].
Attachment of the human virus
The overall lack of attachment of the human influenza A virus is consistent with a lack of evidence of human influenza virus infection in birds. Exceptions were observed for intestinal epithelial cells from the colon crypts of chickens, wigeons, and geese (Figure 1), and for tracheal ciliated cells of tufted ducks, gulls, robins, and pigeons (Figure 1). Experimental infection is needed to determine whether these avian species are susceptible to human influenza virus infection. If infection by both a human and an avian influenza virus proved possible in these species, then they could be considered, along with domestic pig (Sus scrofa) [28] and Japanese quail (Coturnix coturnix) [7], as potential mixing vessels for the generation of human-avian reassortants. However, one would expect that, if such infections with human viruses were possible, they already would have been detected, at least for species in very close contact with humans such as chickens, pigeons and geese. A recent experimental inoculation of chickens with human H1N1 influenza A viruses showed very limited infection [29].
Conclusions
Our results suggest that cell and tissue tropisms are important in determining the host species range of influenza viruses in birds. We found that the PVA of LPAIVs differs between wild bird species, even between species from the same taxonomic family (i.e. with shared phylogenetic origins), and between species with similar feeding behavior (i.e. with a similar risk of ingesting virus particles while feeding). The screening for virus attachment was more discriminating than lectin histochemistry, which has been used to detect the α-2,6-linked SA receptor pattern on wild bird tracheas [5], and proved useful in identifying species with unexpected attachment patterns.
Because the ability of a virus to replicate in cells does not depend solely on attachment, experimental inoculation studies are needed to determine whether viruses can replicate. However, determining differences in PVA between different viruses and bird species is a valuable preliminary screening method. Because we cannot exclude the possibility that other avian and human influenza viruses show a different PVA than those used in this study, we encourage further PVA studies using various influenza A virus isolates and tissues from a broader range of bird species including gulls, which are known to host specific virus subtypes [30], [31]. Knowledge about the PVA of HPAI H5N1 virus in wild bird tissues would be helpful in identifying wild bird species that could potentially spread this zoonotic virus. In the human respiratory tract, the PVA of two LPAIVs (H5N9 and H6N1) and HPAI H5N1 virus did not differ [15]. Whether similar results would be observed with avian tissues remains to be determined.
Materials and Methods
Tissue sampling procedures were approved by the Swedish Environmental Protection Agency (permits number 412-6267-08NV and 412-5977-08NV) and the Swedish Board of Agriculture (permits number 74-08 and 43-09).
For each species, three individual birds were euthanized humanely and both trachea and colon were sampled within three minutes after death. Tissues were fixed in formalin for 48 h and paraffin-embedded. For each bird and tissue, three transverse sections were incubated with the human or avian virus labeled with fluorescein isothiocyanate (FITC; Sigma-Aldrich, Stockholm, Sweden) [15]. For each tissue tested, in each run an omission control was included to check for unspecific staining and sections from pig trachea (displaying α2,6-linked SAs) and mallard colon (displaying α2,3-linked SAs) were included as positive control tissues, respectively for the human and the avian virus.
We used H6N1 A/Mallard/Sweden/81/02 and H3N2 A/Netherlands/213/03 as representatives of LPAIV and seasonal human influenza virus, respectively. The specificity of the avian virus for α2,3-linked SAs was confirmed by the hemagglutination of horse erythrocytes expressing α2,3-linked SAs only. The specificity of the human virus for α2,6-linked SAs was confirmed by the hemagglutination of modified turkey erythrocytes expressing α2,6-linked SAs only, as described by Chutinimitkul et al. [32].
Virus attachment on tissue sections was detected by virus histochemistry as described previously [14], [15]. Briefly, formalin-fixed paraffin-embedded tissues were deparaffinized with xylene and rehydrated with graded alcohol. FITC-labeled influenza viruses (50 hemagglutination units) were incubated with tissues overnight at 4°C. The FITC label was detected with a peroxidase-labeled rabbit anti-FITC antibody (ab 19492-500, Abcam, Cambridge, UK), and the signal was amplified with a tyramide signal amplification system (Perkin-Elmer, Upplands Väsby, Sweden) according to the manufacturer's instructions. Peroxidase was revealed with 3-amino-9-ethyl-carbazole (Sigma-Aldrich), and tissues were counterstained with hematoxylin and embedded in glycerol-gelatin (Merck, Darmstadt, Germany). Attachment of influenza virus to tissues was visible as granular to diffuse red staining on the apical surface of epithelial cells. We considered that the most important cells for virus attachment were ciliated cells in the trachea and intestinal epithelial cells from crypts and villi in the colon [16], [17]. The proportion of cells to which the virus attached was scored as follows: - no significant attachment, ± attachment to rare or few cells, + attachment to a moderate number of cells, ++ attachment to many cells, +++ attachment to a large majority of cells.
Acknowledgments
We thank the people involved in collecting tissues; Charlotte Bergström from Kalmar hospital, Eva Westergren from SVA Stockholm and Joakim Palovaara from Linnaeus University for helpful practical advice. This is contribution no. 247 from Ottenby Bird Observatory.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was financially supported by the Swedish Research Council (VR) and the Swedish Research Council Formas, the French Fondation pour la Recherche Médicale (reference SPE20070709608) and the Intramural Research Program of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, et al. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki Y. Sialobiology of influenza molecular mechanism of host range variation of influenza viruses. Biological & Pharmaceutical Bulletin. 2005;28:399–408. doi: 10.1248/bpb.28.399. [DOI] [PubMed] [Google Scholar]
- 3.Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Laboratory Investigation. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008;16:149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Ellström P, Jourdain E, Gunnarsson O, Waldenstrom J, Olsen B. The “human influenza receptor” Neu5Ac alpha 2,6Gal is expressed among different taxa of wild birds. Arch Virol. 2009;154:1533–1537. doi: 10.1007/s00705-009-0476-8. [DOI] [PubMed] [Google Scholar]
- 6.Kimble B, Nieto GR, Perez DR. Characterization of influenza virus sialic acid receptors in minor poultry species. Virology Journal. 2010;7:365. doi: 10.1186/1743-422X-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan H, Perez DR. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology. 2006;346:278–286. doi: 10.1016/j.virol.2005.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo CT, Takahashi N, Yagi H, Kato K, Takahashi T, et al. The quail and chicken intestine have sialyl-galactose sugar chains responsible for the binding of influenza A viruses to human type receptors. Glycobiology. 2007;17:713–724. doi: 10.1093/glycob/cwm038. [DOI] [PubMed] [Google Scholar]
- 9.Kuchipudi SV, Nelli R, White GA, Bain M, Chang KC, et al. Differences in influenza virus receptors in chickens and ducks: implications for interspecies transmission. Journal of Molecular and Genetic Medicine. 2009;3:143–151. doi: 10.4172/1747-0862.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambaryan A, Yamnikova S, Lvov D, Tuzikov A, Chinarev A, et al. Receptor specificity of influenza viruses from birds and mammals: New data on involvement of the inner fragments of the carbohydrate chain. Virology. 2005;334:276–283. doi: 10.1016/j.virol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Mansfield KG. Viral tropism and the pathogenesis of influenza in the Mammalian host. Am J Pathol. 2007;171:1089–1092. doi: 10.2353/ajpath.2007.070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JS. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinkman-Van der Linden ECM, Sonnenburg JL, Varki A. Effects of sialic acid substitutions on recognition by Sambucus nigra agglutinin and Maackia amurensis hemagglutinin. Analytical Biochemistry. 2002;303:98–104. doi: 10.1006/abio.2001.5539. [DOI] [PubMed] [Google Scholar]
- 14.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 15.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. Intestinal influenza - Replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kida H, Yanagawa R, Matsuoka Y. Duck influenza lacking evidence of disease signs and immune response. Infect Immun. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitworth D, Newman SH, Mundkur T, Harris P, editors. FAO. Wild birds and avian influenza: an introduction to applied field research and disease sampling techniques. 2007. FAO Animal Production and Health Manual No. 5 Whitworth D, Newman SH, Mundkur T, Harris P, editors. Rome.
- 19.Gambaryan A, Webster R, Matrosovich M. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol. 2002;147:1197–1208. doi: 10.1007/s00705-002-0796-4. [DOI] [PubMed] [Google Scholar]
- 20.Munster VJ, Schrauwen EJA, de Wit E, van den Brand JMA, Bestebroer TM, et al. Insertion of a multibasic cleavage motif into the hemagglutinin of a low-pathogenic avian influenza H6N1 virus induces a highly pathogenic phenotype. J Virol. 2010;84:7953–7960. doi: 10.1128/JVI.00449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellström P, Latorre-Margalef N, Griekspoor P, Waldenström J, Olofsson J, et al. Sampling for low-pathogenic avian influenza A virus in wild Mallard ducks: oropharyngeal versus cloacal swabbing. Vaccine. 2008;26:4414–4416. doi: 10.1016/j.vaccine.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 22.Hoye BJ, Munster VJ, Nishiura H, Klaassen M, Ron Fouchier AM. Surveillance of wild birds for avian influenza virus. Emerg Infect Dis. 2010;16:1827–1834. doi: 10.3201/eid1612.100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleijn D, Munster VJ, Ebbinge BS, Jonkers DA, Muskens G, et al. Dynamics and ecological consequences of avian influenza virus infection in greater white-fronted geese in their winter staging areas. Proceedings of the Royal Society B-Biological Sciences. 2010;277:2041–2048. doi: 10.1098/rspb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jourdain E, Gunnarsson G, Wahlgren J, Latorre-Margalef N, Brojer C, et al. Influenza Virus in a Natural Host, the Mallard: Experimental Infection Data. Plos One. 2010;5(1):e8935. doi: 10.1371/journal.pone.0008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JD, Stallknecht DE, Berghaus RD, Swayne DE. Infectious and lethal doses of H5N1 highly pathogenic avian influenza virus for house sparrows (Passer domesticus) and rock pigeons (Columbia livia). Journal of Veterinary Diagnostic Investigation. 2009;21:437–445. doi: 10.1177/104063870902100404. [DOI] [PubMed] [Google Scholar]
- 27.Liu YH, Han CH, Wang XQ, Lin J, Ma M, et al. Influenza A virus receptors in the respiratory and intestinal tracts of pigeons. Avian Pathol. 2009;38:263–266. doi: 10.1080/03079450903055363. [DOI] [PubMed] [Google Scholar]
- 28.Ito T, Couceiro J, Kelm S, Baum LG, Krauss S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babiuk S, Albrecht R, Berhane Y, Marszal P, Richt JA, et al. 1918 and 2009 H1N1 influenza viruses are not pathogenic in birds. J Gen Virol. 2010;91:339–342. doi: 10.1099/vir.0.016246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamnikova SS, Gambaryan AS, Tuzikov AB, Bovin NV, Matrosovich MN, et al. Differences between HA receptor-binding sites of avian influenza viruses isolated from Laridae and Anatidae. Avian Dis. 2003;47:1164–1168. doi: 10.1637/0005-2086-47.s3.1164. [DOI] [PubMed] [Google Scholar]
- 32.Chutinimitkul S, van Riel D, Munster VJ, van den Brand JMA, Rimmelzwaan GF, et al. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 2010;84:6825–6833. doi: 10.1128/JVI.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]