Abstract
Pitx1 is a Bicoid-related homeodomain factor that exhibits preferential expression in the hindlimb, as well as expression in the developing anterior pituitary gland and first branchial arch. Here, we report that Pitx1 gene-deleted mice exhibit striking abnormalities in morphogenesis and growth of the hindlimb, resulting in a limb that exhibits structural changes in tibia and fibula as well as patterning alterations in patella and proximal tarsus, to more closely resemble the corresponding forelimb structures. Deletion of the Pitx1 locus results in decreased distal expression of the hindlimb-specific marker, the T-box factor, Tbx4. On the basis of similar expression patterns in chick, targeted misexpression of chick Pitx1 in the developing wing bud causes the resulting limb to assume altered digit number and morphogenesis, with Tbx4 induction. We hypothesize that Pitx1 serves to critically modulate morphogenesis, growth, and potential patterning of a specific hindlimb region, serving as a component of the morphological and growth distinctions in forelimb and hindlimb identity. Pitx1 gene-deleted mice also exhibit reciprocal abnormalities of two ventral and one dorsal anterior pituitary cell types, presumably on the basis of its synergistic functions with other transcription factors, and defects in the derivatives of the first branchial arch, including cleft palate, suggesting a proliferative defect in these organs analogous to that observed in the hindlimb.
Keywords: Pitx1, morphogenesis, hindlimb development, pituitary
The role of homeodomain factors in early and late development is genetically well established (Gehring et al. 1994; Scott 1997), with factors of the Hox gene cluster exhibiting specific domains of expression not only along the anterior–posterior axis but also in the developing appendages (Krumlauf 1994; Maconochie et al. 1996). For example, the most posterior members of the vertebrate, Hox a and Hox d gene clusters are expressed in a fashion that presage the subdivision of the limb along the anterior, posterior, and proximodistal axes (Dollé et al. 1989; Izpisua-Belmonte et al. 1991; Yokouchi et al. 1991; Nelson et al. 1996). In this paper we investigate the function of a member of a second family of homeodomain factors that exert critical regulatory roles during development, the bicoid-related family of homeobox genes, which includes goosecoid (gsc), Otx1, and Otx2. Goosecoid in vertebrates, Orthodenticle in Drosophila, and the vertebrate homologs Otx1 and Otx2 are critical in determination of head structures (Cho et al. 1991; Simeone et al. 1992, 1993). Deletion of Otx1 results in loss of all head structures (Acampora et al. 1998), whereas forebrain and midbrain regions are deleted in Otx2−/− mice (Acampora et al. 1995, 1996, 1998; Ang et al. 1996; Rhinn et al. 1998). Recently, a search for factors interacting with the pituitary-specific transcription factor Pit-1 (Szeto et al. 1996), or for interactions with a cis-acting element in the POMC promoter (Lamonerie et al. 1996), led to the cloning of a novel member of this bicoid-related gene family, P-Otx/Ptx1. The human homolog, Backfoot, was found in a screen for novel homeodomain factors (Shang et al. 1997). This factor, now referred to as Pitx1, has been shown to be expressed in the pituitary gland (Lamonerie et al. 1996; Szeto et al. 1996), in the first branchial arch, and its derivatives, and in the lateral mesenchyme and developing hindlimb, but only at very low levels in forelimb (Szeto et al. 1996; Lanctôt et al. 1997; Shang et al. 1997). Although considerable insight has been obtained in the molecular basis underlying the establishment of the different limb axes (Johnson and Tabin 1997; Martin 1998; Schwabe et al. 1998), the intriguing question of the molecular mechanisms that underlie distinctions between forelimb and hindlimb are less well studied.
Pitx1 is one of the few known transcription factors that exhibit a striking hindlimb/forelimb difference in their expression. Its preferential expression in the hindlimb suggests that this transcription factor may exert a critical role in distinguishing hindlimb from forelimb identity. Two other genes, members of the T-box family (Tbx) of transcriptional activators, exhibit differential expression in limbs. Tbx4 is expressed primarily in the developing hindlimb, whereas Tbx5 is initially selectively expressed in the forelimb, although Tbx5 later exhibits some expression in the hindlimb (Chapman et al. 1996; Gibson-Brown et al. 1996; Li et al. 1997).
Pitx1 is also expressed in the pituitary throughout its development (Lamoneier et al. 1996; Szeto et al. 1996), being uniformly expressed in oral ectoderm during the period of exclusion of Sonic hedgehog (Shh) from the invaginating Rathke’s pouch (Treier et al. 1998), as well as dorsal–ventral Fgf8 gradient (Erickson et al. 1998; Takuma et al. 1998; Treier et al. 1998). Pitx1 expression continues during the subsequent ventral–dorsal emergence of distinct cell types including gonadotropes, expressing luteinizing hormone β, and follicle-stimulating hormone β (LHβ, FSHβ); thyrotropes, expressing thyroid-stimulating hormone β, (TSHβ); somatotropes, expressing growth hormone (GH); lactotropes, expressing prolactin; and corticotropes, producing adrenocorticotropic hormone (ACTH). Early in pituitary development, Pitx1 is coexpressed with a second, highly related gene, Pitx2/RIEG (Semina et al. 1996; Gage and Camper 1997), which was initially identified by positional cloning of the gene responsible for the Rieger Syndrome in humans. This autosomal dominant disease is characterized by anterior chamber ocular abnormalities, dental hypoplasia, mild craniofacial dysmorphism, and occasionally decreased levels of growth hormone. Pitx2 is asymmetrically expressed in lateral plate mesoderm and appears to exert critical roles in left–right situs (Logan et al. 1998a; Meno et al. 1998; Piedra et al. 1998; Ryan et al. 1998; St. Amand et al. 1998; Yoshioka et al. 1998).
In this paper we report evidence, on the basis of gene deletion in mice, that Pitx1 exerts critical roles in the hindlimb, pituitary, and first branchial arch development. The most striking phenomena in the Pitx1 gene-deleted mouse are alterations of specific skeletal structures within a specific region of the hindlimb, which assume many morphological and growth features of the corresponding bones in the forelimb, suggesting they are dependent on Pitx1 expression in hindlimb mesenchymal populations. Misexpression of Pitx1 in the chicken wing bud further supports the role of Pitx1 in limb growth and morphogenesis. Further, Pitx1, probably on the basis of its synergistic actions with other transcription factors, is important for proliferation and differentiated function of specific pituitary cell phenotypes, as well as for closure of the palate and mandibular development.
Results
Deletion of the Pitx1 genomic locus
Pitx1 and the highly-related gene Ptx2/RIEG referred to as Pitx2, which is linked to a human genetic disease (Semina et al. 1996) and to determination of left–right situs (Logan et al. 1998a; Meno et al. 1998; Piedra et al. 1998; Ryan et al. 1998; St. Amand et al. 1998; Yoshioka et al. 1998) are expressed from mouse embryonic day 7 (E7) onward, in distinct, yet highly overlapping patterns (e.g., Fig.1A) (Szeto et al. 1996; Gage and Camper 1997; Lanctôt et al. 1997; Shang et al. 1997). Both Pitx1 and Pitx2 are robustly expressed in early development in specific mesenchymal populations and in the ectodermal primordium of the pituitary gland and derivatives of the first branchial arch (Fig. 1A). Most strikingly, Pitx1 is selectively expressed in the mesenchyme of the developing hindlimb bud (Fig. 1B), where it is initially detected in the lateral plate mesenchyme at the level at which the hindlimb will emerge (E9–E10) (Szeto et al. 1996; Shang et al. 1997; Lanctôt et al. 1997). Pitx1 transcripts are detected by whole mount in situ hybridization by E10.5–E11, and Pitx1 remains robustly and widely expressed in the hindlimb mesenchyme through E12.5–E13.5. At later stages, Pitx1 transcripts are absent in the centers of chondrogenesis, becoming confined to the perichondral regions and soft tissues of the hindlimb. Loss of Pitx1 transcripts occurs in a proximal to distal fashion in the developing limb (Fig. 1B; data not shown). Pitx1 is expressed in a very restricted fashion and only at later stages in the forelimb (Fig. 1B,D).
Figure 1.
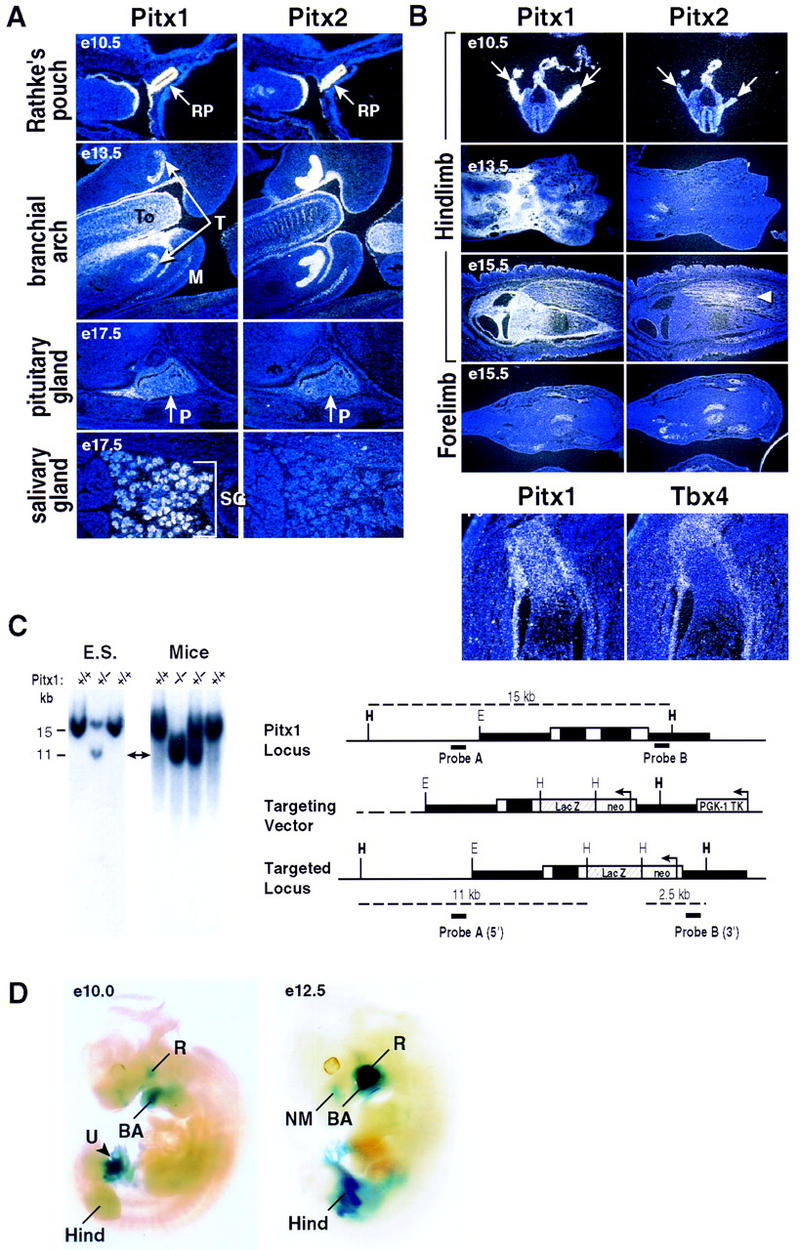
Pitx1 and Pitx2 expression patterns and targeted disruption of the Pitx1 locus. (A) Expression of Pitx1 and Pitx2 analyzed by in situ hybridization. The distinct and overlapping expression patterns of Pitx1 and Pitx2 Rathke’s pouch (RP) (E10.5) and branchial arch structures (E13.5) (T) tooth; (To) tongue; (M) mandible; salivary gland (SG); (E17.5) and pituitary gland (P). (B) Expression of Pitx1, Pitx2, and Tbx4. Pitx1 is highly expressed in early limb bud, decreasing distally by E12.5; there is a differential expression of Pitx1, in the sheath and growth plate of the long bones, whereas Pitx2 is expressed in muscle (arrow). Note the similar, limited expression of Pitx1 and Pitx2 in forelimb. Pitx1 is robustly expressed in the developing hindlimb (arrows, top), throughout development with serial restriction from proximal, and then distal regions (second panel), finally in a pattern in perichondral regions, at the end of long bones, whereas Pitx2 is selectively expressed in muscle (arrow) as well as transiently in developing pelvis (not shown), Hind (hindlimb) and Fore (forelimb). Pitx1 expression overlaps with the Tbx4 expression in limb development, and is shown in the perichondral region and growth plate of the femur (P0, bottom right). (C) Targeted deletion of the Pitx1 genomic locus. Schematic representation of the Pitx1 locus (top), the targeting vector (middle), and the Pitx1 targeted allele (bottom). White and black boxes represent exons and introns, respectively, and restriction enzymes: HindIII (H); EcoRI (E). Analysis of transfected ES cells by genomic Southern blot analysis with the 5′-external probe A identifies a 11-kb HindIII fragment in the mutant allele and a 15-kb HindIII fragment in the wild-type allele. A 3′-external probe B was used to identify a 2.5-kb HindIII fragment in the mutant allele, while hybridizing to the 11-kb HindIII fragment in the wild-type mouse (not shown). Homozygous and heterozygous mice were identified by Southern blot analysis with probes A and B. (D) The normal Pitx1 expression patterns analyzed by whole-mount lacZ staining at E10–E12.5. (R) Rathke’s pouch; (NM) nasal mesenchyme; (BA) branchial arch; (U) umbilical cord; (Hind) hindlimb.
In contrast, Pitx2 is expressed in the population of mesenchymal cells that migrate from the somite into both limb buds and eventually will differentiate into the limb musculature (Fig. 1B). Pitx1 transcripts are transiently present in the pelvis (data not shown) and ultimately expressed in the most distal of the overlapping domains at the end of the long bones that express parathyroid hormone related peptide (PTHrP), parathyroid hormone receptor (PTHR) and Indian hedgehog (Ihh) (Lanske et al. 1996; Vortkamp et al. 1996). Pitx1 mesenchymal expression overlaps with that of a member of the Tbx of transcriptional activators, Tbx4, and is later localized in the long bones (Fig. 1B). Tbx4 provides a marker exhibiting hindlimb, but not forelimb, expression (Gibson-Brown et al. 1996), until late in development. In contrast, a second member of the family, Tbx5, is initially selectively expressed in forelimb, but later is also detected in the hindlimb (Chapman et al. 1996; Gibson-Brown et al. 1996; Li et al. 1997). Both Pitx1 and Pitx2 are also transiently expressed late in development in a few restricted regions of forelimb, and subsequently decline to practically undetectable levels in the mature limbs (Fig. 1B; data not shown). Pitx1 appears to be selectively expressed in the olfactory pit, submandibular gland, ventral body wall mesenchyme, and Pitx1 and Pitx2 exhibit distinct expression patterns in the gastrointestinal tract and urogenital sinus (Fig. 1A; data not shown).
To examine the potential roles of Pitx1 in development of the hindlimb and other tissues in which it is developmentally expressed, a targeting vector was designed to delete virtually the entire coding sequence region, except for the amino-terminal sequence 80 amino acids to which lacZ was fused in-frame (Fig. 1C). This targeting construct was used to obtain homologous recombinants in ES cells, which were injected into blastocysts to generate chimeric mice. Gene-deleted murine lines were generated by appropriate breeding, and homologous recombination was confirmed by genomic Southern blot analysis with both 5′- and 3′-specific probes (Fig. 1C), as well as lack of Pitx1 transcripts in the first branchial arch, Rathke’s pouch, and hindlimb (data not shown). In these mice, Pitx1-directed lacZ expression is identical to the characteristic distribution of endogenous Pitx1 transcripts (e.g., Fig. 1D). Pitx1−/− mice, which die immediately or shortly after birth, and are born at levels statistically slightly below expected Mendelian ratios (∼20 per 100 births). This could be explained by the finding that a small subset of null mice exhibit embryonic lethality after E11.5.
Pitx1 role in hindlimb morphogenesis
Although most distinctions between the mammalian hindlimb and forelimb are morphological, including joint articulations, shape, and size of radius/ulna versus tibia/fibula, there are apparently patterning differences that are evolutionarily conserved in mammals that include the patella in the hindlimb, and a particular proximal element of the forelimb carpus, referred to as the pisiform element, which is not present in the hindlimb tarsus (Romer 1986). Thus, the pisiform is a characteristic forelimb-specific structure arising from a specific condensation center. The hindlimb of Pitx1−/− mice is significantly shorter than in wild-type littermates (Fig. 2A). Examination of the skeletal structure reveals that the long bones of the hindlimb, including the femur, tibia, and fibula are altered in length, width, and overall structure (Figs. 2B and 3A). The size of the pelvis, a structure in which Pitx1 is also transiently expressed, is also remarkably reduced (Fig. 2B). However, the overall structure of pelvis and femur in Pitx1−/− mice appears to retain the morphological features of the wild-type counterparts. In contrast, the tibia and fibula of the distal hindlimb in Pitx1−/− mice both exhibit striking alterations in morphology, relative size, and shape. This includes alterations in the cnemial crest of the tibia (Romer 1986), altered angles of articulation of fibula both proximally and distally, and a striking alteration in relative size and diameters of tibia and fibula. Thus, the tibia and fibula of the Pitx1−/− hindlimb are now morphologically more similar, although not identical, to the radius and ulna of the forelimb, exhibiting equivalent cross sectional diameters (Fig. 3A). Further, the patella is now absent (Fig. 2B) and there is a loss of the Zucker nodes (Fig. 3C), both characteristic features of the hindlimb (Fig. 3C).
Figure 2.
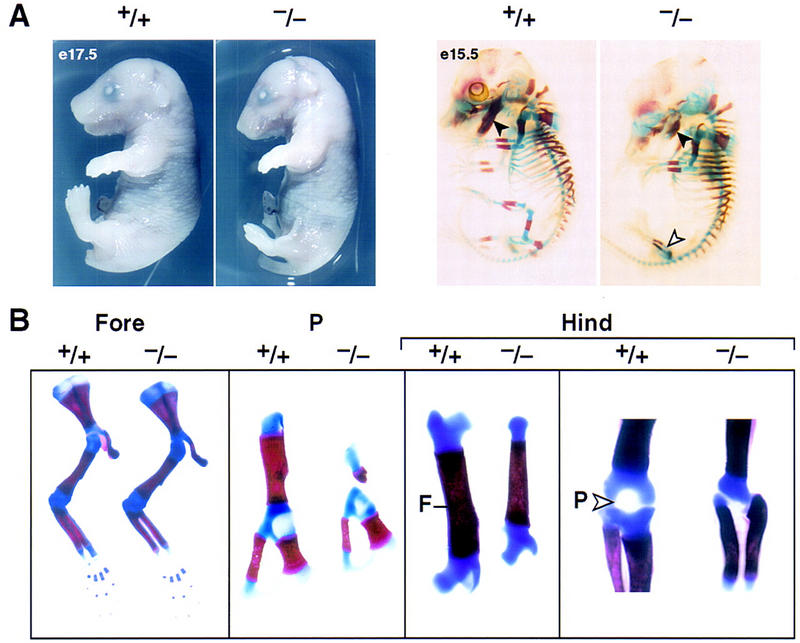
Morphological alterations of Pitx1+/+ and Pitx1−/− mice at different stages of development. (A) Pitx1+/+ and Pitx1−/− mice at E17.5; and E15.5 skeletal structures. Note the foreshortening of the mandible (solid arrowhead) and altered hindlimb (open arrowhead). (B) Details of skeletal structures of hindlimb and forelimb of Pitx1+/+ and Pitx1−/− mice at P0. The Forelimb (Fore) is unaltered, Pelvis (P) is abnormal with particular loss of the femur (F). The hindlimb (Hind) is altered in overall length and size. The patella (P) is absent (open arrowhead).
Figure 3.
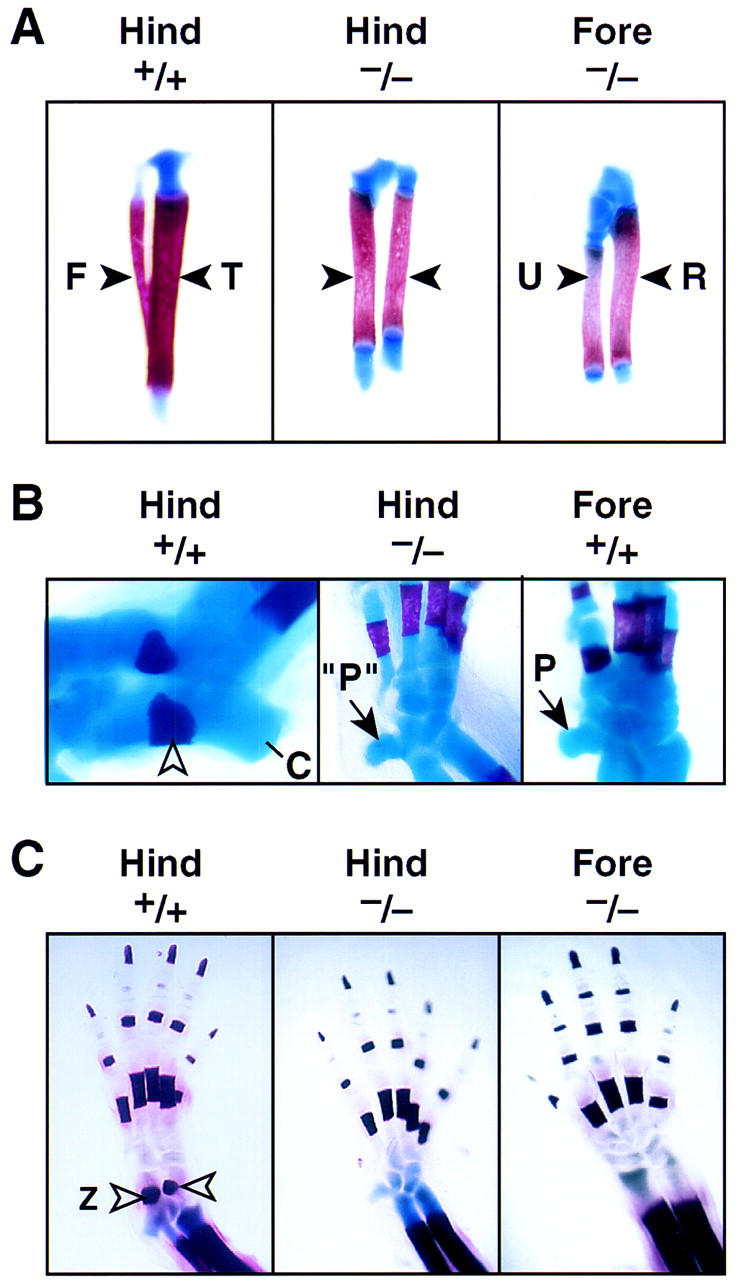
Analysis of the structural components of hindlimb development of Pitx1+/+ and Pitx1−/− at P0. (A) Skeletal structures of the tibia (T) and fibula (F) in P0 Pitx1+/+ and Pitx1−/− mice. There is alteration in size of the femur, whereas the growth and relative size of the tibia and fibula (solid arrowheads) now more closely resembles the morphological resemblance of the relationship between radius (R) and ulna (U) of the forelimb. (B) Tarsal development of the Pitx1+/+ and Pitx1−/− mice. Note the normal development of the Zucker nodes (open arrowhead) and calcaneous (C) in Pitx1+/+ mouse. The hindlimb (Hind) proximal tarsus of the Pitx1−/− mouse now contains an element that resembles the pisiform (P) seen in forelimb (Fore) carpus development of Pitx1+/+ littermate, with a very small calcaneous. (C) The development of the hindlimb distal components of Pitx1+/+ and Pitx1−/− mice at P0. Digit morphology is not reproducibly altered. Zucker nodes (open arrowhead) are absent in the hindlimb (Hind) of the Pitx1−/− mouse.
There is also a striking alteration in the tarsal structure of the ankle (Fig. 3B), with the appearance of an additional proximal tarsal element that appears analogous to the pisiform, which is an evolutionarily conserved characteristic element in the mammalian forelimb carpus (Romer 1986). However, the adjacent proximal tarsus does not assume a carpal-like morphology. In parallel, there is a marked alteration in morphology and size of the calcaneus, no longer characteristic of the wild-type hindlimb structure. The distal tarsus of the Pitx1−/− mouse is not clearly distinct from the tarsus of the wild-type littermates. Whereas the size of digits is somewhat smaller, there do not appear to be clear structural alterations; consistent with the similarity of the forelimb and hindlimb digits in the wild-type mouse. Together, these alterations in morphology and bone patterning properties of the distal hindlimb of the Pitx1−/− mouse, cause it to have bone structures, including the tibia/fibula, patella, and the appearance of a pisiform element-like structure in the proximal tarsus, that are quite distinct from that of the wild-type hindlimb. The mutant hindlimb has assumed several morphological features that highly resemble those of the corresponding structures in the forelimb.
The regions of the cartilaginous growth plates, in which neither Pitx1 nor Pitx2 are expressed, appear to be normally maintained. In the Pitx1−/− mice, expression of the hindlimb-specific marker gene, Tbx4, is reduced compared with normal, and is more strikingly reduced in the hindlimb region in which the morphological alterations are most dramatic, as shown both by in situ hybridization of sectioned embryos and whole-mount staining (Fig. 4A,B). No alterations in the Tbx5 expression pattern are observed (Fig. 4A). Because Pitx1 expression patterns correspond to structures adjacent to the growth plates, we evaluated expression Ihh, PTHR, and PTHrP, and found no alterations in their expression (data not shown). No effects are observed on genes that are normally expressed at similar levels in hindlimb and forelimb including Wnt5a, Fgf8, Bmp4, gsc, Hoxd10, d11, or d13 expression (data not shown).
Figure 4.
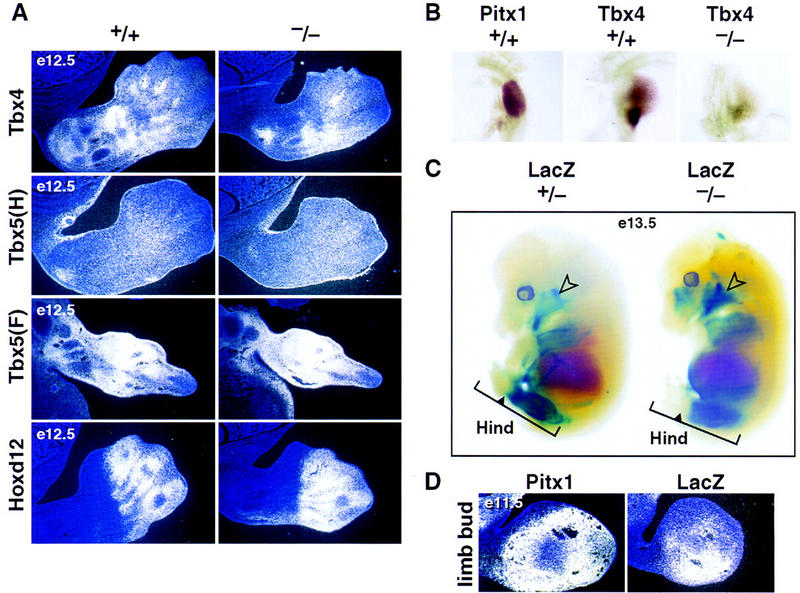
In situ hybridization and whole-mount analysis of limbs in Pitx1+/+ and Pitx1−/− mice. (A) In situ hybridization analysis of markers known to be expressed during limb development. Tbx4 expression is diminished distally, and no alteration in Tbx5 or Hoxd12 expression is noted in the hindlimb [Tbx5 (H)] or forelimb [Tbx5 (F)]. The expression of Hoxd12, PTHrP, PTHR, and Ihh are also similar in +/+ and −/− hindlimb at E12.5 and P0 (data not shown). (B) The expression patterns of Pitx1 and Tbx4 in the hindlimb by whole-mount in situ hybridization at E10.5. The Tbx4 expression overlaps with that of Pitx1 in the hindlimb of Pitx1+/+ mice and is reduced in the hindlimb of Pitx1−/− mice. (C) Whole-mount lacZ staining analysis. lacZ (marker of targeted gene) expression is present in the mandibular structures (open arrow) and is subtly but reproducibly restricted in an anterior/distal portion of the hindlimb (Hind) of Pitx1−/− mice (solid arrow in the bracket). (D) In situ hybridization analysis of Pitx1 and lacZ expression in hindlimb at E11.5, showing anterior/distal restriction of Pitx1/lacZ expression, consistent with findings in the whole-mount LacZ staining (C).
To determine the effects of Pitx1 gene deletion on the population of mesenchyme that normally expresses Pitx1, we examined lacZ expression in these mice. Surprisingly, lacZ expression is slightly diminished in the proximal developing mesenchyme (E11.5) compared with the levels in Pitx1+/− mouse, and is reproducibly diminished in anterior, distal mesenchyme, as determined by the linear portion of limb in which lacZ staining can be detected (Fig. 4C,D). We therefore suggest that a population of Pitx1-expressing mesenchyme promotes a Pitx1-dependent hindlimb-specific morphogenesis program, modulating growth and exerting specific effects on the tibia, fibula, patella, and tarsal morphology of ankle.
Effects of cPitx1 misexpression
On the basis of the apparent requirement for Pitx1 to achieve certain hindlimb-specific characteristics, it became of particular interest to investigate whether expressing Pitx1 in forelimb would modify its developmental program. A chick Pitx1 (cPitx1) cDNA was isolated, encoding a protein 80% identical to murine Pitx1, as has been recently reported (Logan et al. 1998b; Lanctôt et al. 1997). The cPitx1 gene is expressed in a very similar pattern to the murine Pitx1. Initially, cPitx1 transcripts are present almost exclusively in the limb bud that will develop into the leg but not in the limb bud that will give rise to the wing (Fig. 5A). As in the mouse, cPitx1 is first detected in the lateral plate mesoderm before the limb bud emerges and as limb outgrowth proceeds, cPitx1 is expressed throughout the entire limb mesenchyme (Fig. 5A). With progressive development of the limb bud, cPitx1 becomes differentially expressed. Between stages 23 and 30, cPitx1 transcripts are gradually excluded from the most proximal aspect of the developing limb bud and the adjacent flank. By stage 25, cPitx1 transcripts begin to disappear from the distal region of the developing leg in which the digits are beginning to form, although low levels are observed in the interdigit region (Fig. 5A). Chick Pitx1 transcripts start to be weakly detected in the developing wing at stage 26 outside of the prechondrogenic regions. Chick Pitx1 transcripts are never detected in the limb ectoderm nor in the apical ectodermal ridge. This expression pattern as well as cell-grafting experiments led Logan et al. (1998b) to hypothesize that Pitx1 could be involved in specifying chick leg identity. If cPitx1 is involved in encoding limb identity, its expression should be stable when leg tissue is grafted into wing tissue. When small pieces of leg mesoderm grafts are implanted beneath the apical ectodermal ridge of host wing buds, the original cPitx1 expression is retained (Logan et al. 1998b).
Figure 5.
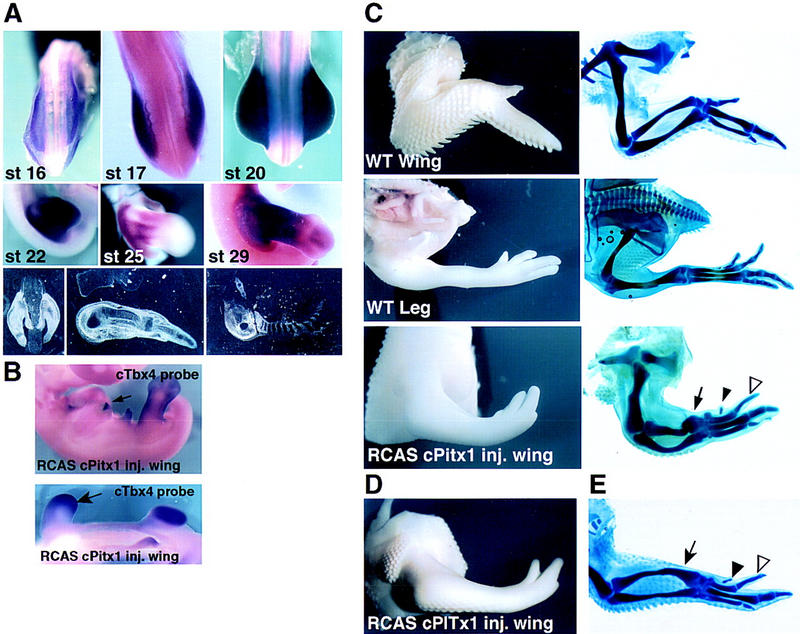
(A) Expression of cPitx1 in the developing chick hindlimb. Whole-mount and radioactive in situ hybridizations at different stages of hindlimb development. cPitx1 transcripts start to be detected in the presumptive leg bud cells. At later stages, transcripts are detected throughout the hindlimb bud cells until stage 24 where they start to disappear from the condensing cartilage (panels 4–8). The bottom right panel shows the transient expression of cPitx1 in the pelvis. (B) As seen by the ectopic patches (top) or broader expression (bottom) of Tbx4, retroviral infection of cPitx1 in the presumptive wing cells can, later on, induce the expression of the hindlimb-specific gene, Tbx4. (C) Misexpression of cPitx1 perturbs the outgrowth and patterning of the chick wing. Shown are whole-mount and skeletal preparations of control, and not injected, chick wing and leg. The chick wing has three digits of variable length, is covered by feathers, and displays a characteristic downward flexure at the wrist level. Instead, the chick leg is straight in its most distal part, not covered by feathers but by small scales, and has three digits of similar length and an additional small four digits located toward the back of the foot. Ectopic expression of cPitx1 in the wing transforms the curvature of the wing into an almost straight position, increases the size of digit 2 (open arrowhead) and induces the formation of a fourth digit (solid arrowhead). All of these changes in growth and patterning, together with the disappearance of feathers from the distal side of the wing, induce the infected wing to resemble a leg. Some of the infected embryos showed alterations in the size and morphology of the radius and ulna (solid arrow). (D,E) Results of a second experiment, to illustrate the range of phenotypic variations.
On the basis of the hypothesis that the wing also contains the population of mesenchymal cells that would, if they expressed Pitx1, modulate a leg-like pattern, the retrovirus encoding full-length Pitx1 was injected into the forelimb bud. Misexpression of Pitx1 in the developing forelimb causes several morphological changes in the wing that are noticeably more striking at the distal aspect of the limb (Fig. 5E). One of the unique features of the chick wing is the posterior flexure of the most distal segment, the autopod, with respect to the middle segment, the zeugopod. This flexure is not observed in the leg in which the distal elements are placed in a straight orientation. Fifty-seven percent of the Pitx1-infected wings show a loss in the downward flexure of the wrist joint, thus giving them a leg-like appearance. When the skeletal pattern was examined, we observed a striking change in the relative size of the infected wing digits. The three wild-type wing digits have a variable length, with digit IV being the longest and II the shortest (Fig. 5C–E). In the leg, on the contrary, digits II, III, and IV are very similar in length. In addition, the leg has an extra smaller digit positioned toward the back of the foot. In 61% of the Pitx1 infected wings, we observed an increase in the size of digit II (Fig. 5C–E). In addition, an extra short digit appeared at the anterior side of the infected wings in 58% of the cases (Fig. 5C–E). This uniformity in digit length, as well as the appearance of an extra digit, has some resemblance to the digital patterning of the leg. The fact that only minimal abnormalities in the zeugopodal segment were observed after Pitx1 misexpression, is likely to reflect a loss in developmental plasticity by the time the Pitx1 virus is active in the regions fated to become the adult chick radius and ulna. The most distal cells of the limb bud, which will give rise to the digits, are, however, heavily infected by stage 20, a stage in which they are still plastic and competent to change cell fate (data not shown). This could explain the fact that the wing alterations are mainly restricted to the distal part of the limb. Finally, another effect of Pitx1 misexpression was on the integument. The distal part of the chick wing, contrary to the leg, is normally covered by feathers. In 38% of the injected wings, we observed a reduction in the number of distal feathers, suggesting that Pitx1 could act as a suppressor of feather formation during the developing leg integument.
In situ hybridization for various markers expressed during normal limb development in both leg and wing buds (including Shh, Bmp2, Bmp4, Hoxd11, or Hoxd13) showed no changes in their expression pattern. The limb alterations were preceded by ectopic patches of expression of the hindlimb-specific Tbx4 gene (15% of the injected limbs) (Fig. 5B). However, no change is observed for the forelimb-specific Tbx5 gene (data not shown). Taken together, these results suggest that overexpression of Pitx1 induces cell proliferation of a certain population of wing mesenchyme cells, complementing the lack of proliferation and morphological alterations observed after ablation of Pitx1 in the mouse.
Role of Pitx1 in pituitary and branchial arch development
Consistent with the pattern of Pitx1 gene expression, the Pitx1−/− mice also exhibit developmental defects in the anterior pituitary gland. Throughout the entire period in which cell phenotypes are established during pituitary organogenesis, Pitx1 is continuously expressed. All known early events, including invagination of the Rathke’s pouch, exclusion of Shh from invaginating epithelium, activation of Fgf8, P-Lim, Msx-1, Lhx3, αGSU, Bmp2, the ventral marker Isl-1 (Treier et al. 1998) and Prop-1 on E12.5 and Pit-1 on E13.5, (Sornson et al. 1996) are normally maintained (Fig. 6A; data not shown). Analysis of expression of the trophic hormones that are characteristic of the differentiated pituitary cell types at E15.5 through postpartum day 0 (P0) indicates a selective decrease in the most ventral cell type populations. Examination of thyroid-stimulating hormone β (TSHβ), luteinizing hormone β (LHβ), follicle stimulating hormone β (FSHβ), and the common glycoprotein α subunit (αGSU) expression suggests that both the number of gonadotropes and thyrotropes, as well as the level of LHβ and TSHβ transcripts and protein within the individual cells, are diminished (Fig. 6B,C; data not shown). Interestingly, the level of TSHβ transcripts is most severely reduced in the rostral tip thyrotrope population, which does not require Pit-1 for TSHβ gene activation (Lin et al. 1994). Growth hormone expression in somatotropes appears unchanged (Fig. 6B,C), whereas the number and expression levels of the POMC gene in the intermediate lobe melanotropes appears normal between E15.5 and P0. There is a consistent increase in the levels of both number of, and ACTH transcripts and peptide levels in the anterior pituitary corticotropes (Fig. 6B,C).
Figure 6.
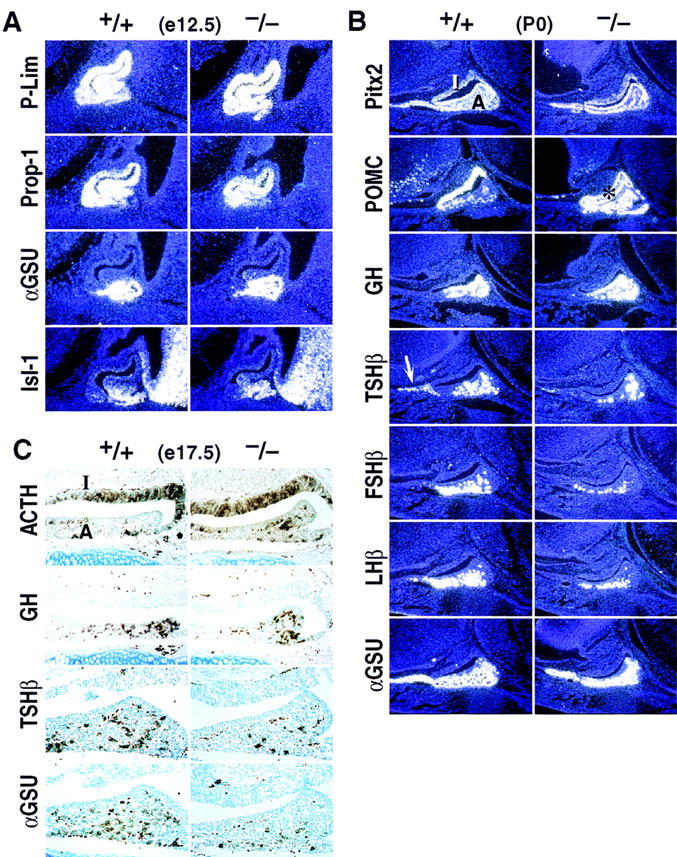
Effects of Pitx1 deletion on development of the anterior pituitary gland. (A) Effects of Pitx1 gene deletion on P-Lim, Prop-1, αGSU, and Isl-1 expression in the pituitary gland. No differences were observed in the expression of these factors in Pitx1−/− embryos as compared with that of the wild-type at E12.5. (B,C) Pitx1 gene deletion alters expression of ventral pituitary-specific cell types at P0 in mice analyzed by in situ hybridization (B) or by immunohistochemical analysis at E17.5 (C). The expression of TSHβ, FSHβ, and LHβ are markedly decreased, with some reduction of αGSU. Note particularly the loss of TSHβ in the rostral tip (arrow). Pitx2 and growth hormone (GH) gene expression are unchanged. POMC transcripts and ACTH immuno activity in anterior lobe are consistently increased. Because of saturation of the film with the POMC probe, the region is artificially black in a portion of the intermediate lobe (asterisk, B). Note that ACTH staining is similar on wild-type and −/− glands in the intermediate lobe (I), while clearly increased in the anterior lobe (A) (C).
Development of the palate and derivatives of the first branchial arch are invariably severely affected in Pitx1−/− mice (Fig. 7), probably accounting for the early postnatal death of the homozygous null mice. In addition to cleft palate (Fig. 7A), the distal mandible and the tongue are significantly foreshortened, the ventral sublingual mesenchyme is hypoplastic, and the submandibular gland does not form (Fig. 7B). However, the spatial relationships between most of the components of lower jaw and mouth are apparently normally maintained. The expression of a number of markers expressed early in the first branchial arch, including Msx1, Msx2, gsc, Shh, Bmp2/4, Wnt5a, and Pitx2, are unaltered in Pitx1 gene-deleted mice (Fig. 7B; data not shown). The craniofacial defects in Pitx1−/− mice, are particularly intriguing in light of the observation that the human Pitx1 gene maps to 5q31 (Crawford et al. 1997), which the investigators suggest could indicate that mutant Pitx1 alleles might be responsible for a subset of patients with Treacher–Collins syndrome (Rogers 1964; Fazen et al. 1967; McDonald and Gorski 1993).
Figure 7.
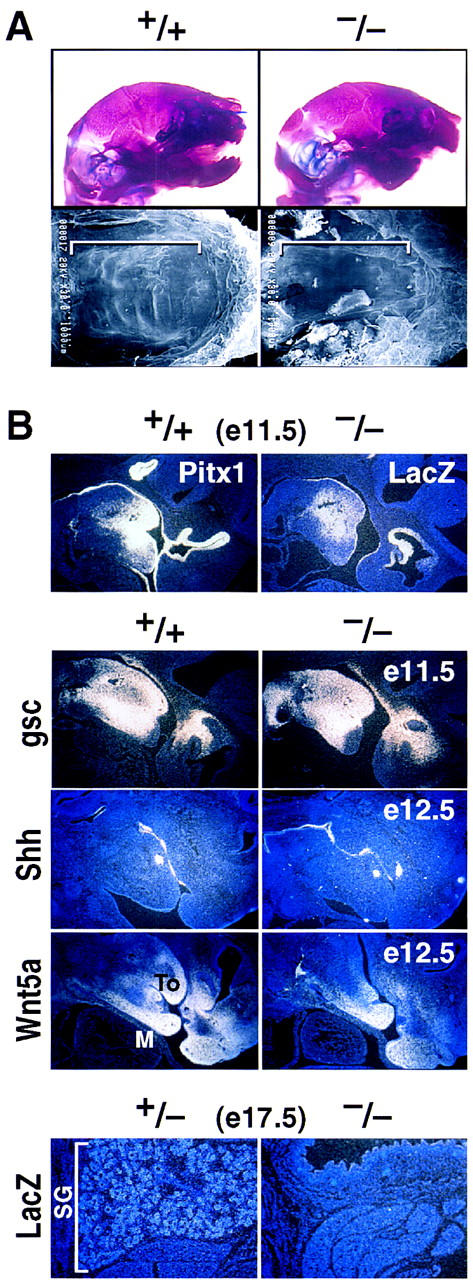
Effects of Pitx1 gene deletion on mandible, palate, and submandibular gland development at E17.5 and P0. (A) Marked reduction in size of mandible analyzed by skeletal staining (top) and failure of palatal closure (see brackets) analyzed by scanning electron microscopy in the Pitx1−/− mouse (bottom). (B) Mandibular expression of Pitx1 and lacZ as a marker in Pitx1+/+ and Pitx1−/− mice, respectively at E11.5. Markers of mandibular development analyzed at E11.5 and E12.5 by in situ hybridization included gsc, Shh, and Wnt5a. Submandibular gland (SG) development in Pitx1+/− and Pitx1−/− mice at E17.5 marked by the lacZ probe. (To) tongue; (M) mandible.
Discussion
On the basis of these data, the bicoid-related gene, Pitx1, appears to be a critical transcriptional component of limb development, as well as exerting roles in development of a derivative of the midline stomadeum, the anterior pituitary, and on derivatives of the first branchial arch. Our data suggest a model in which expression of Pitx1 in hindlimb mesenchyme is required for correct hindlimb morphogenesis and growth, particularly in the region encompassing the tibia, fibula, patella, and proximal tarsus. Thus, in the absence of Pitx1, tibia and fibula are morphologically more similar to the forelimb radius and ulna, and evolutionarily conserved hindlimb patterning features are altered. These include loss of the hindlimb-specific patella and Zucker’s nodes, and appearance of a potential pisiform-like element, an evolutionary-conserved characteristic of the forelimb carpus, in the proximal tarsus, with altered growth and morphology of the calcaneus, but without clear alterations in other elements of the proximal tarsus. Because both of these morphological and potential patterning features are well-recognized aspects of the distinction between forelimb and hindlimb, we suggest that the presence of Pitx1 is required for a transcriptional program required for the characteristic growth and morphological alterations that are a component of the distinctions between hindlimb and forelimb. The molecular basis of this morphological change in the Pitx1−/− mouse hindlimb could reflect either altered patterning, and/or altered formation of, or response to, an anterior–posterior growth gradient in the limb bud, resulting in normalization of the size of the tibia and fibula, loss of the patella and Zucker’s nodes, and altered proximal tarsus. Consistent with this model, targeted misexpression of Pitx1 in the chicken wing bud causes distinct proliferative alterations of the digits, as well as altered morphological features, suggesting that the appropriate population of mesenchyme is present in both limbs to mediate Pitx1-dependent morphological and proliferative alterations. The presence of Pitx1 therefore appears to be required for a transcriptional program required for the characteristic growth and morphological alterations that are a component of the distinctions between hindlimb and forelimb. It is therefore tempting to speculate that Pitx1 exerts either a patterning function or/and dictates the formation of the response to an anterior–posterior gradient-mediating proliferation in the limb bud.
Another family of genes that have been suggested as being involved in determining identity are the Tbx family (Chapman et al. 1996; Gibson-Brown et al. 1998; Isaac et al. 1998; Logan et al. 1998b, Ohuchi et al. 1998), with Tbx4 specifically expressed in the hindlimb. Expression of Tbx4 in the Pitx1−/− hindlimb is clearly diminished, especially in its distal aspect. Expression of Tbx4 is also induced in the chick forelimb after Pitx1 misexpression. These results suggest that Pitx1 is epistatic to at least a portion of the Tbx4 program and required for the remainder of the gene inductions for segmental morphogenesis and patterning in a hindlimb-specific fashion. In the absence of Pitx1, the forelimb-specific gene Tbx5 is not induced in the hindlimb: This may account for a portion of the distinctions that remain between the hindlimb of the Pitx1−/− mouse and the wild-type forelimb.
The actions of Pitx1 in pituitary development may provide several clues to the molecular basis of its actions in limb development. In pituitary, any potential early roles of Pitx1 may be redundant with those of Pitx2/RIEG (Crawford et al. 1997), as there is no defect in early pituitary organogenesis; however, there is a consistent, late, pituitary developmental phenotype, which involves decreased proliferation and levels of distal target gene expression in two ventral pituitary cell types—gonadotropes and thyrotropes—and an element of a distal cell type, the corticotrope, expressing ACTH, in which Pitx1 protein is expressed (Lamonerie et al. 1996). On the basis of identification of synergistic interactions between Pitx1 and other transcription factors (Szeto et al. 1996; Tremblay et al. 1998), it is likely that a major aspect of the phenotype reflects the synergistic role of Pitx1 in target gene induction. Furthermore, as there are likely to be proliferative roles of the Fgf8, Shh, Bmp, and Wnt signaling factors in these cell types (Treier et al. 1998), Pitx1 may alter sensitivity to critical trophic factors in a cell-autonomous fashion, and may exert similar effects in the limb. A similar explanation is likely to account for Pitx1 effects in closure of the palatal bone structure. Thus, in the affected targets, Pitx1 could alter expression of, or response to, trophic factors thereby exerting its effects on growth, as well as morphology. In this regard, the mechanism of Pitx1 actions may be analogous in the various affected target organs.
On the basis of our in vivo data, we speculate that the growth, and morphogenetic roles of Pitx1 in hindlimb actually contribute a critical component of the differential hindlimb and forelimb developmental programs that generate limb identity.
Materials and methods
Generation of Pitx1-gene-deleted mice
A mouse Pitx1 genomic clone was isolated from a J1 129/Sv mouse genomic library with the complete Pitx1 cDNA probe. The 5′-flanking region comprising a 4.0-kb EcoR1–HindIII fragment and a 3.0-kb PmlI 3′-flanking fragment were subcloned into the corresponding cloning sites of the lacZ/neomycin containing vector, in which expression of lacZ is driven by the Pitx1 promoter and a neomycin gene is driven by the mouse phosphoglycerate kinase (PGK) promoter (Bermingham et al. 1996). The targeting vector was completed by subcloning into PGK-thymidine kinase plasmid. The entire homeodomain and virtually all of the 3′-coding region is replaced by the lacZ/neomycin gene. The R1 cell line was cultured in DMEM high glucose medium containing 15% FCS and supplemented with leukemia inhibitory factor (LIF). Targeting vector DNA was linearized (12 μg) and electroporated into 2 × 107 ES cells in 0.8 ml of electroporation buffer at 250 V and 500 μF with a Genepulser. Cells were grown for 7–9 days in 150 μg/ml G418 and 2 mm Gancyclovir and 500 double drug-selected clones were grown for an additional 3 days. Cell lines that had undergone homologous recombination were identified with the 5′ external (1.0 kb) probe that hybridizes to a 15-kb HindIII wild-type Pitx1 locus fragment and a 11-kb HindIII fragment in the Pitx1−/− allele. A 3′ internal (0.5 kb) probe, which recognizes a 2.5-kb HindIII fragment in both wild-type and Pitx1−/− allele, was used to identify homologous recombination in the 3′-flanking region. Three ES cell lines, that met the requirement for homologous recombination at the Pitx1 locus, were microinjected into C57BL/6 blastocysts that were then transferred to pseudopregnant females. Chimeric male mice were backcrossed to C57BL/6 females and germ-line transmission was scored by the presence of the agouti coat color. Heterozygotes and homozygotes were identified by Southern blot analysis (Bermingham et al. 1996). Three lines were generated and analyzed.
In situ hybridization, whole-mount hybridization, lacZ staining, immunohistochemistry, and differential bone/cartilage staining
Isolation, fixation, and hybridization with 35S-labeled antisense RNA probes and exposure were performed as described previously (Ryan et al. 1998; Simmons et al. 1989). Chick embryos were staged according to Hamburger and Hamilton (1951). Whole-mount in situ hybridization of chick embryos and sectioning was performed as described (Ryan et al. 1998). After fixation, whole-mount in situ hybridization was performed on mouse embryos, which were dehydrated with methanol and treated with hydrogen peroxide and proteinase K. Transcripts were then detected with AP-conjugated DIG antibodies and stained with NBT/BCIP. Whole-mount lacZ staining was performed in the presence of 1 mg/ml X-gal substrate. Immunohistochemistry was done on 5/7-mm-thick paraffin sections stained by indirect immunoperoxidase method. Peroxidase activity was visualized with DAB/metal enhancer (Pierce, Rockford, Il). Sections were counterstained with methyl green and mounted in Permount (Fisher). Antibodies were obtained and used diluted as follows: ACTH (Chemicon, Temecula, CA) 1:1000; GH and TSHβ (DAKO, Carpinteria, CA) 1:1000; αGSU (National Hormone and Pituitary Program) 1:1000. Anti-rabbit horseradish peroxidase-conjugated antibodies were from Chemicon and used at a 1:500 dilution. For bone and cartilage staining, embryos were isolated by cesarean section and the abdomens of embryos were immediately cut open prior to being placed into 95% ethanol for 24 hr. After embryos were skinned and eviscerated, they were fixed in 95% ethanol for 72 hr, and placed in 0.3% Alcian Blue/0.1% Alizarin Red S staining solution for at least 72 hr. After staining, each embryo was washed in tap water to remove excess dye, and then placed in 0.75% potassium hydroxide solution for maceration. After 24 hr, the embryos were cleared by successive washes in 20% and 50% glycerin solution.
Retroviral infection
The replication competent retroviral vectors RCASBP(A)-containing cDNAs encoding full-length cPitx1 were generated as described (Ryan et al. 1998). Chicken embryos (MacIntrye Poultry, San Diego) were infected by injecting virus into the presumptive wing region at stages 10–12 (Hamburger and Hamilton staging table). After appropriate periods of incubation, chick embryos were fixed in 4% paraformaldehyde overnight, dehydrated in methanol, evaluated under a dissecting microscope, and stored at −20°C prior to in situ hybridization.
Acknowledgments
We thank Mario Capecchi, Mitch Kronenberg, and Andy McMahon for generously providing reagents, and Andy McMahon, Cliff Tabin, and Mathias Treier for discussions and sharing of results prior to publication. We thank Peggy Meyer for her expertise and generous assistance in preparation of illustrations and Marie Fisher for assistance in manuscript preparation. M.G.R. is an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants to M.G.R. and J.C.I.B., and the Leila Y. Mathers Charitable Foundation Grant to J.C.I.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mrosenfeld@ucsd.edu; FAX (619) 534-8180.
E-MAIL belmonte@salk.edu; FAX (619) 455-1349.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brûlet P. Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brûlet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat Genet. 1996;14:218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Tuorto F, Avantaggiato V, Tremblay JJ, Lazzaro D, di Carlo A, Mariano A, Macchia PE, Corte G, Macchia V, Drouin J, Brulet P, Simeone A. Transient dwarfism and hypogonadism in mice lacking Otx-1 reveal prepubescent stage-specific control of pituitary levels of Gh, FSH and LH. Development. 1998;125:1229–1239. doi: 10.1242/dev.125.7.1229. [DOI] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- Bermingham JR, Jr, Scherer SS, O’Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes & Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dynam. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Cho KW, Blumberg B, Steinbeisser H, De Robertis EM. Molecular nature of Spemann’s organizer: The role of the Xenopus homeobox gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford MJ, Lanctôt C, Tremblay JJ, Jenkins N, Gilbert D, Copeland N, Beatty B, Drouin J. Human and murine PTX1/Ptx1 gene maps to the region for Treacher Collins syndrome. Mamm Genome. 1997;8:841–845. doi: 10.1007/s003359900589. [DOI] [PubMed] [Google Scholar]
- Dolle P, Izpisúa-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Erickson J, Norlin S, Jessel TM, Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998;125:1005–1015. doi: 10.1242/dev.125.6.1005. [DOI] [PubMed] [Google Scholar]
- Fazen LE, Elmore J, Nadler HL. Mandibulo-facial dysostosis (Treacher-Collins syndrome) Am J Dis Child. 1967;113:405–410. doi: 10.1001/archpedi.1967.02090190051001. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Camper SA. Pituitary homeobox 2, a novel member of the bicoid-related family of homeobox genes, is a potential regulator of anterior structure formation. Hum Mol Genet. 1997;6:457–464. doi: 10.1093/hmg/6.3.457. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Silver LM, Papaioannou VE. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech Dev. 1996;56:93–101. doi: 10.1016/0925-4773(96)00514-x. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. Involvement in T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125:2499–2509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton H. A series of normal stages in the development of the chick embryos. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Isaac A, Rodriguex-Estaban C, Ryan A, Altabef M, Tsukui T, Patel K, Tickle C, Izpisúa-Belmonte JC. Tbx genes and limb identity in chick embryo development. Development. 1998;125:1867–1875. doi: 10.1242/dev.125.10.1867. [DOI] [PubMed] [Google Scholar]
- Izpisúa-Belmonte JC, Tickle C, Dolle P, Wolpert L, Duboule D. Expression of the homeobox Hox-4 genes and the specification of position in chick wing development. Nature. 1991;50:585–589. doi: 10.1038/350585a0. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Tabin CJ. Molecular models for vertebrate limb development. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lamonerie T, Tremblay JJ, Lanctôt C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeobox transcription factor involved in transcription of the proopiomelanocortin gene. Genes & Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- Lanctôt C, Lamolet B, Drouin J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defize LHK, Ho C, Mulligan RC, Abou-Samra AB, Jüppner H, Segre GV, Kronenberg HM. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- Li H, Tierney C, Wen L, Wu JY, Rao Y. A single morphogenetic field gives rise to two retina primordia under the influence of the prechordal plate. Development. 1997;124:603–615. doi: 10.1242/dev.124.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Li S, Drolet DW, Rosenfeld MG. Pituitary ontogeny of the Snell dwarf mouse reveals Pit-1-independent and Pit-1-dependent origins of the thyrotrope. Development. 1994;120:515–522. doi: 10.1242/dev.120.3.515. [DOI] [PubMed] [Google Scholar]
- Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor of Ptx2 mediates situs-specific morphogenesis in response to left-right asymmetric signaling. Cell. 1998a;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- Logan M, Simon H, Tabin C. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development. 1998b;125:2825–2835. doi: 10.1242/dev.125.15.2825. [DOI] [PubMed] [Google Scholar]
- Maconochie MS, Nonchev S, Morrison A, Krumlauf R. Paralogous Hox genes: function and regulation. Annu Rev Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- Martin G. The roles of Fgf in the early development of vertebrate limbs. Genes & Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- McDonald MT, Gorski JL. Nager acrofacial dysostosis. J Med Genet. 1993;30(9):779–782. doi: 10.1136/jmg.30.9.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Takenchi J, Yoshioka H, Ishimaru Y, Ogura K, Takahashi N, Ogura T, Noji S. Correlation of wing-leg identity in ectopic FgF-induced chimeric limbs with the differential expression of chick Tbx5 and Tbx4. Development. 1998;125:51–60. doi: 10.1242/dev.125.1.51. [DOI] [PubMed] [Google Scholar]
- Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dierich A, Shawlot W, Behringer RR, Le Meur M, Ang SL. Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development. 1998;125:845–856. doi: 10.1242/dev.125.5.845. [DOI] [PubMed] [Google Scholar]
- Rogers B. Barry-Treacher Collins syndrome: A review of 200 cases. Br J Plast Surg. 1964;17:109–136. doi: 10.1016/s0007-1226(64)80020-5. [DOI] [PubMed] [Google Scholar]
- Romer AS. In: The vertebrate body. Parsons TS, editor. Philadelphia, PA: Saunders; 1986. [Google Scholar]
- Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, Norris DP, Robertson EJ, Evans RM, Rosenfeld MG, Izpisúa Belmonte JC. Ptx2 determines asymmetries in vertebrate. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- St. Armand TR, Ra J, Zhong Y, Hu Y, Baber SI, Qiu M, Chen Y. Cloning and expression pattern of chicken Pitx2: A new component in the SHH signaling pathway controlling embryonic heart looping. Biochem Biophys Res Commun. 1998;247:100–105. doi: 10.1006/bbrc.1998.8740. [DOI] [PubMed] [Google Scholar]
- Scott MP. Hox genes, arms and the man. Nat Genet. 1997;15:117–118. doi: 10.1038/ng0297-117. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Rodriguez-Esteban C, Izpisúa Belmonte JC. Limbs are moving: Where are they going? Trends Genet. 1998;14:229–235. doi: 10.1016/s0168-9525(98)01477-2. [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Shang J, Luo Y, Clayton DA. Backfoot is a novel homeobox gene expressed in the mesenchyme of the development hind limb. Dev Dynam. 1997;209:242–253. doi: 10.1002/(SICI)1097-0177(199706)209:2<242::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DM. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabeled single stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- Sornson MW, Wu W, Dasen JS, Flynn S, Norman DJ, O’Connell SM, Gukovsky I, Carrière C, Ryan AK, Miller AP, Zuo L, Gleiberman AS, Andersen B, Beamer WG, Rosenfeld MG. Pituitary lineage determination by the Prop-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Ryan AK, O’Connell SM, Rosenfeld MG. P-OTX: A Pit-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuma N, Sheng HZ, Furuta Y, Ward JM, Sharma K, Hogan BL, Pfaff SL, Westphal H, Kimura S, Mahon KA. Formation of Rathke’s pouch requires dual induction from the diencephalon. Development. 1998;125:4835–4840. doi: 10.1242/dev.125.23.4835. [DOI] [PubMed] [Google Scholar]
- Treier M, Gleiberman AS, O’Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. Multistep signaling requirements for pituitary organogenesis in vivo. Genes & Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit 1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;112:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Sasaki H, Kuroiwa A. Homeobox gene expression correlated with the bifurcation process of limb cartilage development. Nature. 1991;353:443–445. doi: 10.1038/353443a0. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, Hamada H, Noji S. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]


