Abstract
Staphylococcus aureus and Staphylococcus epidermidis are a frequent cause of biofilm-associated infections that are a tremendous burden on our healthcare system. Staphylococcal biofilms exhibit extraordinary resistance to antimicrobial killing, limiting the efficacy of antibiotic therapy and often require surgical intervention to remove infected tissues or implanted devices. Recent work has provided new insight into the molecular basis of biofilm development in these opportunistic pathogens. Extracellular bacterial products, environmental conditions, and polymicrobial interactions have all been shown to profoundly influence the ability of these bacteria to colonize and disperse from clinically relevant surfaces. In this article we review new developments in staphylococcal biofilm disassembly and set them in the context of potential strategies to control biofilm infections.
Introduction to staphylococcal biofilm development
Biofilms are complex microbial communities attached to a surface and embedded in an extracellular matrix. These communities can form on a diverse range of surface chemistries and numerous studies have investigated the factors required to carry out this intricate process. Bacteria of the genus Staphylococcus are thought to develop a biofilm in at least three stages: (i) cell attachment to a surface; (ii) assembly of these initial cells into a small clump, also called a microcolony; and (iii) growth of the biofilm into a mature structure. Once a biofilm is fully developed, it can disassemble (also called detachment or dispersion) through both mechanical and active processes. A number of excellent reviews cover the biofilm maturation process [1–3], and in this review, we will focus on biofilm disassembly.
Staphylococcal biofilm infections
In terms of bacterial infections, biofilms can manifest as growth on medical devices or a range of host tissues. The challenge presented by these infections is the recalcitrance to host defense mechanisms and antimicrobial therapy [4, 5], enabling the bacterial communities to persist and cause repeated waves of damage. Staphylococci have drawn attention as the dominant cause of biofilm-associated infections [2], with Staphylococcus epidermidis often cited as being associated with foreign body infections [6] and S. aureus with infections on host tissues (Figure 1), such as osteomyelitis [7], septic arthritis [8], and endocarditis [9]. The pronounced ability of staphylococci to develop biofilm-associated disease has drawn considerable interest over the past decade in understanding the complex mechanisms behind the formation of these persistent structures as well as developing strategies to target their disassembly.
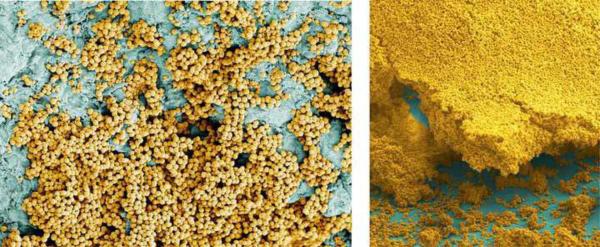
Figure 1.
Images of S. aureus biofilms on host surfaces. S. aureus cells (gold colored) attaching to and forming a biofilm on a heart valve (left) and an endotracheal tube (right).
Background on staphylococcal biofilm disassembly
The process of biofilm disassembly likely involves multiple steps that include degradation of the extracellular matrix and physiological changes that prepare cells for conditions outside the biofilm. One profound phenotype observed during biofilm disassembly is the conversion of bacteria existing in a state where they are resistant to antimicrobial chemotherapy to one where they regain sensitivity to these therapeutics [10, 11]. An in-depth understanding of staphylococcal molecular mechanisms and environmental conditions contributing to biofilm formation and disassembly could lead to innovative treatment options.
The benefits provided to bacteria through the biofilm mode of growth are impressive, but these advantages come at some cost and can trigger situations where biofilms disassemble. For example, matrix production imposes a synthetic burden on the bacteria and the nutrient gradients within biofilms can limit growth. Perhaps the most significant risk for biofilm bacteria occurs when local conditions deteriorate, such as nutrient depletion, the accumulation of wastes, the appearance of antimicrobial compounds, or other threats. Biofilm bacteria have a reduced ability to evade stresses because they are physically confined by the matrix. These costs associated with biofilm growth make it vital that bacteria possess mechanisms to separate from biofilms and assume a planktonic lifestyle for spreading to a more suitable habitat. Environmental conditions trigger active mechanisms that bring about bacterial separation [10]. The fact that disassembly can be triggered by several different cues could allow organisms to regulate their movement between the biofilm and planktonic growth states as the local environmental conditions change.
Biofilm disassembly likely plays an important role in most biofilm associated infections. A well-characterized example is the devastating embolic events of endocarditis caused by detachment of the biofilm growing on heart valves [12]. The dissemination of bacteria from biofilm infections can also result in severe acute infections such as sepsis [13]. In addition, many cases of hospital-acquired pneumonia are caused by bacteria detached from biofilms that form in a patient's endotracheal tube or oropharynx [14, 15].
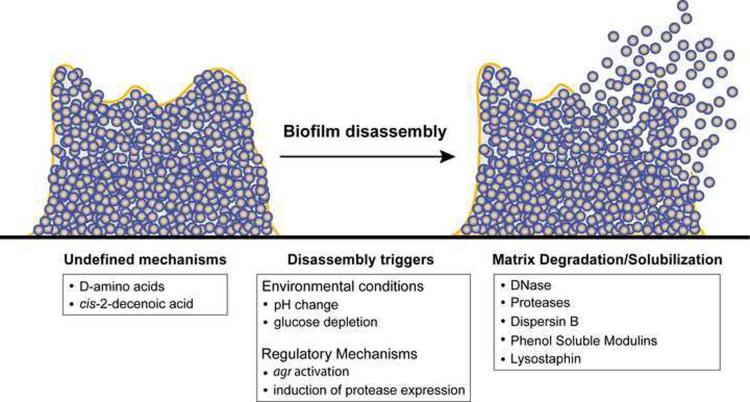
Because of their important role in human health and disease, considerable research effort has been aimed at defining the mechanistic basis of staphylococcal biofilm formation. This research effort has implicated numerous gene products in biofilm formation [1]. Many of these gene products have been identified by high-throughput screens accessing the ability of mutants to form biofilms [16, 17]. Finding genetic factors contributing to biofilm disassembly has lagged behind in large part because of the technical challenges in devising effective screening methods. However, in recent years progress toward uncovering the mechanisms of staphylococcal biofilm disassembly has been made and many of these mechanisms center around the breakdown or solubilization of the biofilm matrix. A summary of these disassembly mechanisms is outlined in Table 1 and Figure 2.
Table 1.
Biofilm disassembly mechanisms
| Species | Process or agent | Mechanism | Refs. |
|---|---|---|---|
| S. aureus & S. epidermidis | agr activation | Expression of agr regulated factors | [10, 13, 43] |
| S. aureus | Autoinducing peptide (AIP) | Activation of agr system | [10, 11] |
| S. aureus & S. epidermidis | Phenol-soluble modulins | Surfactant-mediated detachment | [13, 42] |
| S. aureus | Induction of extracellular protease expression | Cutting of matrix proteins | [10, 31] |
| S. aureus | pH change | Reactivation of agr or other regulatory systems | [10] |
| S. aureus & S. epidermidis | DNaseI addition | Degradation of eDNA matrix | [11, 18, 39, 40] |
| S. epidermidis | Dispersin B addition | Degradation of polysaccharide matrix | [18, 20] |
| S. aureus & S. epidermidis | Protease K, trypsin, V8, Esp, or other protease addition | Degradation of proteinaceous matrix | [10, 19, 20, 26, 44] |
| S. aureus & S. epidermidis | Lysostaphin addition | Degradation of cell wall | [52, 53] |
| S. aureus | cis-2-decenoic acid addition | Unknown | [54] |
| S. aureus | D-amino acids | Unknown | [56] |
Figure 2.
Model of known Staphylococcal biofilm disassembly mechanisms. See text and Table 1 for details.
The staphylococcal biofilm matrix
Staphylococcal biofilms are encased in an extracellular matrix composed of proteins, polysaccharides, extracellular DNA (eDNA), and presumably host factors. Compounds capable of dissolving matrix components can disrupt established biofilms or prevent the formation of a biofilm [10, 13, 18–20]. The matrix provides protection from a variety of insults such as attack by immune cells or exposure to antimicrobials. The precise composition of the biofilm matrix varies greatly depending on the Staphylococcus strain, its physiological status, the nutrients available and the prevailing physical conditions. Although much overlap exists between biofilm development in S. aureus and S. epidermidis, it should be noted that the biofilm matrix in these two species are not equivalent and variation has routinely been observed between strains of the same species.
The polysaccharide in Staphylococcus biofilms is a partially deacetylated β1–6 linked acetylglucosamine homopolymer [21]. This poly-N-acetyl glucosamine (PNAG) polysaccharide, which is also referred to as polysaccharide intercellular adhesion (PIA), is synthesized by enzymes encoded by the ica operon and deposited on the cell wall surface [22]. Environmental growth conditions that likely contribute to the role of PNAG in the Staphylococcus biofilm matrix are anaerobic growth, the presence of sub-inhibitory concentrations of antibiotics, high temperatures or osmolarity, and other environmental stresses [1]. PNAG has been shown to play a crucial role in several in vivo animal models of Staphylococcal biofilm infections [23–25]. While some strains rely more on polysaccharides for robust biofilm formation, others form polysaccharide-independent biofilms with the matrix composed primarily of protein and eDNA [1, 16, 18, 26]. Support for the non-essential role of PNAG in many strains comes from ica locus deletions that do not change the biofilm phenotypes [10, 27, 28], although these experiments were not conducted in animal models of biofilm infection. In cases of polysaccharide-independent biofilm formation, proteins and eDNA most likely substitute for PNAG as a structural matrix component [11, 16, 18].
Proteins make up the second major biofilm matrix component, as evidenced by the susceptibility of staphylococcal biofilms to proteases (Figure 3) [10, 11, 27, 29–31]. Most S. epidermidis isolates and S. aureus strains producing high levels of PNAG form biofilms that are not susceptible to protease activity [32], presumably because these biofilms rely more heavily on polysaccharides for structural integrity. Some surface proteins, such as the fibronectin binding proteins [19], protein A [33], SasG [34, 35], and biofilm associated protein (BAP) [31, 36], have been defined as being important in cell-cell and cell-surface interactions, although BAP has not been found in human isolates. One protein recently described to have a structural role in the S. aureus biofilm matrix is beta toxin [37]. Beta toxin is capable of binding eDNA and the authors suggest it forms covalent crosslinks to itself in the presence of DNA. This crosslink is protease susceptible, providing the first link between eDNA and proteins in forming the skeletal framework upon which staphylococcal biofilms are established. However, many clinical strains of S. aureus do not produce beta toxin due to the presence of a converting prophage [38], suggesting that other DNA-binding matrix components await identification.
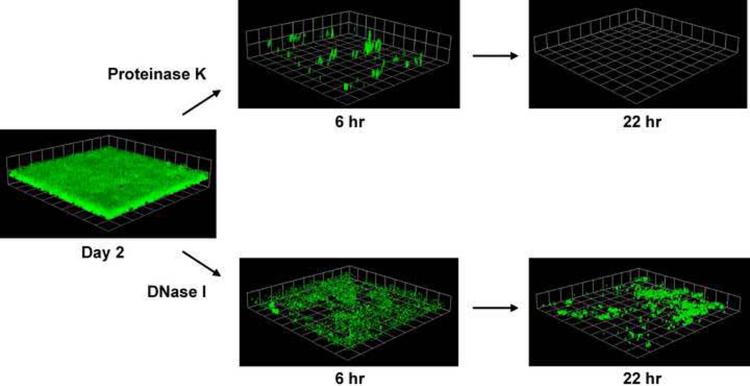
Figure 3.
Susceptibility of S. aureus biofilms to proteinase K and bovine DNaseI. Flow cell biofilms formed by a methicillin-resistant S. aureus (MRSA) isolate expressing GFP were treated with either proteinase K or bovine DNaseI and imaged at 6 and 22 hr post treatment. GFP-expressing biofilms were visualized with confocal laser scanning microscopy (CLSM) and each side of a grid square in the images represents 20 μM. This figure was adapted from a previous publication [11].
The most recently described staphylococcal biofilm matrix component is eDNA. Autolytic activity from a subpopulation of cells results in the release of genomic DNA that contributes to cell adhesion during biofilm maturation. eDNA is thought to serve a structural role in the S. aureus biofilm matrix and facilitate both cell-cell and cell-surface interactions [39, 40], whereas it is only a minor component of biofilms formed by S. epidermidis [18]. Taken together, our understanding of the complexities of the staphylococcal biofilm matrix remains incomplete, especially the emerging role for eDNA, and published findings to date are predominantly based on in vitro work, leaving considerable room for future development of this important research area.
Molecular mechanisms of staphylococcal biofilm disassembly
A primary mechanism of biofilm disassembly utilized by S. aureus and S. epidermidis is the production of extracellular enzymes or surfactants that degrade and solubilize adhesive components in the biofilm matrix. Since the biofilm matrix encases the bacterial cells within the biofilm colony, degradation of the matrix results in the detachment of cells from the colony and their release into the environment. Matrix-degrading gene products implicated in active staphylococcal biofilm dispersal include proteases, deoxyribonucleases (DNases), and surfactants.
One regulatory system controlling the production of matrix degrading enzymes is the accessory gene regulatory (agr) system. The agr system is controlled by a cyclic autoinducing peptide (AIP) that is synthesized and secreted into the environment. When the AIP concentration reaches an critical threshold concentration (in the low nanomolar range), it activates a two-component signal transduction cascade leading to the production of secreted virulence factors (for a recent review see [41]). The agr extracellular proteome includes multiple proteases and small pore-forming toxins called phenol-soluble modulins (PSMs). While the phenotypes of agr deficient strains in biofilms are variable depending on genetic background and assay conditions [10, 29, 42], activation of the agr system is generally accepted as being inhibitory towards biofilm maturation. In terms of species, S. aureus will not form a biofilm under conditions of high agr activity and reactivation of agr in a mature biofilm results in disassembly (Figure 4) [10]. Furthermore, the S. aureusagr system is more active in cells that have detached from a biofilm [43], and similar effects have been seen in S. epidermidis [13], providing further evidence that induction of the agr system results in biofilm disassembly.
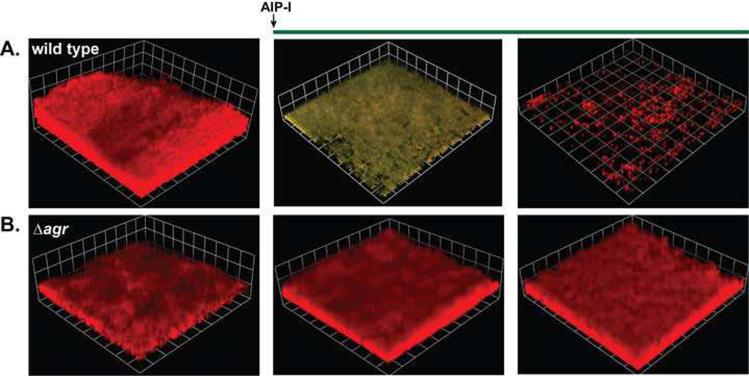
Figure 4.
AIP-mediated biofilm disassembly. Dual-labeled biofilms (PsarA-RFP, PagrP3-GFP) were grown for 2 days, and autoinducing peptide (AIP-I, 50 nM final) was added to the growth media. Biofilm integrity and RFP and GFP fluorescence were monitored with confocal laser scanning microscopy (CLSM) at Day 3 and 4. For the image reconstructions shown, AIP-I was added exogenously to either an agr type I wild type strain (A) or an agr deficient strain (B). The addition of AIP-I induces the agr system and dissembles the biofilm only in the wild type strain. Greenish yellow color indicates expression of the agr P3-GFP reporter and each side of a grid square in the images represents 20 μM. This figure was adapted from a previous publication [10].
The production of extracellular proteases has been implicated in the biofilm disassembly mechanism. In S. aureus, deletion of the genes encoding the proteases resulted in a significant increase in biofilm formation in flow cells, and a concomitant decrease in disassembly upon agr activation [10]. In addition, protease inhibitors have been shown to promote S. aureus biofilm formation under environmental conditions that normally accelerate disassembly [10, 16, 27]. Similarly, mutations that lead to strong upregulation of the extracellular proteases, such as sarA and sigB deficiencies, appear to lock S. aureus into a planktonic state [27, 29, 30], lending further support to the inverse correlation between protease expression and biofilm formation.
Recent work dissecting polymicrobial interactions in the nose uncovered that S. epidermidis is capable of producing a secreted protease named Esp that inhibits S. aureus biofilm formation and nasal colonization [44]. These findings support previous observations that S. aureus biofilm formation depends on the absence of extracellular protease activity and biofilm disassembly can target the proteinaceous matrix material, although the specific target(s) for Esp protease is not known. While the evidence of biofilm formation in the colonization state is limited, the fact that anti-biofilm treatments seem to also prevent colonization suggests there could be underappreciated parallels. However more work is needed to definitively determine if Esp protease activity inhibits nasal colonization via a biofilm disassembly mechanism or some other protease-dependent mechanism.
S. aureus secretes a potent DNase, also known as thermonuclease or micrococcal nuclease, that has been implicated in cell detachment from biofilms [40]. S. aureus biofilms are readily dispersed from microplate wells by the exogenous addition of DNases and restriction enzymes, indicating that eDNA is a major biofilm matrix adhesin in this species [18, 40]. It has been shown that a nuclease-deficient mutant strain of S. aureus exhibited significantly increased biofilm formation in flow cells compared with the biofilm capacity of a wild-type strain [40]. These findings suggest that nuclease may function as an endogenous mediator of biofilm disassembly in this species.
PSMs are surfactant-like peptides produced by both S. aureus and S. epidermidis and are capable of contributing to biofilm disassembly. PSMs are regulated by the agr quorum-sensing system and their amphiphilic α-helical structure lends them surfactant-like properties [45]. PSMs have been demonstrated to promote S. epidermidis biofilm disassembly in vitro and promote dissemination from colonized catheters in a mouse model of device-related infection [13]. In addition, the authors demonstrated that antibodies against PSMs inhibited bacterial spread from implanted catheters, showing that strategies to manipulate biofilm disassembly can prevent spread of infection and disease outcome. This important work is one of the only studies assessing the effect of biofilm disassembly in an animal model system, an area where more research is needed.
Environmental conditions promoting staphylococcal biofilm disassembly
In addition to the mechanistic complexity, the timing and extent of biofilm disassembly is likely to be under local regulation. Under favorable conditions, most wild-type biofilms release a small number of cells on a continual basis, but they also sporadically undergo major detachment events after prolonged periods of growth [43]. These events can be very heterogeneous in terms of timing, the regions of the biofilm affected, and perhaps the mechanism of detachment employed. This heterogeneity poses challenges to the investigation of biofilm disassembly mechanisms. One approach that has been used to overcome this challenge involves exposing biofilms to sudden changes in environmental conditions in order to induce a detachment event. This strategy has led to greater understanding of the process through which biofilm disassembly is triggered [10, 43]. One example of an environmental change that results in S. aureus biofilm disassembly is the removal of glucose from the growth medium. Under normal biofilm growth conditions, glucose represses the agr system through the non-maintained generation of low pH due to the excretion of acidic metabolites, and this common media supplement is used by many laboratories to induce staphylococcal biofilm formation. When glucose is depleted from the environment, the agr system reactivates and the production of matrix degrading enzymes and surfactants results in biofilm disassembly [10]. However, beyond glucose depletion, little is known about the contribution of other environmental conditions to the disassembly mechanism. Changes in nutrient levels are known to induce dispersion of Pseudomonas aeruginosa biofilms [46, 47], suggesting this is an area of research that warrants further investigation in staphylococcal strains.
Targeting biofilm disassembly
A directed approach to biofilm disassembly is possible through the targeting of biofilm matrix components. The diverse biofilm matrix chemistry, including proteinaceous material, eDNA, and polysaccharide, is susceptible to degradation by a range of exogenously added enzymes. A number of laboratories have observed that proteinase K and trypsin can readily disperse S. aureus and non-polysaccharide producing S. epidermidis biofilms [10, 11, 20, 26, 32]. Bovine DNAse I addition has also been successful at dispersing S. aureus biofilms (Figure 3) [18, 40]. These types of experiments contributed to the mounting evidence that proteins and eDNA are important structural components of the staphylococcal biofilm matrix. In a similar manner, enzymes capable of degrading PNAG should disassemble biofilms containing this polysaccharide as the primary matrix component. To date, no staphylococcal enzymes have been identified that possess PNAG-degrading activity, but it is possible that they simply remain to be discovered. An enzyme called dispersin B (DspB), produced by Actinobacillus actinomycetemcomitans [48], inhibits biofilm formation and promotes biofilm disassembly in many strains of S. epidermidis and S. aureus that utilize PNAG as a dominate component of their biofilm matrix [18, 20, 26, 49]. Dispersin B is a β-hexosaminidase that can hydrolyze the glycosidic linkages of PNAG [48, 50]. There is interest in utilizing this enzyme as an anti-biofilm agent [51], however the ability of S. aureus to form polysaccharide-independent biofilms suggests that such an application would have limitations. Finally, lysostaphin treatment disrupted established biofilms of S. aureus and S. epidermidis on abiotic surfaces [52]. Lysostaphin is a glycine endopeptidase produced by Staphylococcus simulans that degrades the pentaglycine bridge in the staphylococcal cell wall, perhaps indicating that cell wall material has an underappreciated role in the biofilm matrix. The treatment was also effective in a catheter mouse model of a S. aureus biofilm [53], suggesting lysostaphin could be a general therapy for staphylococcal biofilm infections.
Concluding remarks and future perspectives
Biofilm formation is an important mode of growth for S. aureus and S. epidermidis in many environments and is a significant contributor to the persistence of chronic infections. Staphylococcal biofilm disassembly likely plays a critical role in the transmission of these bacteria from host to host and in the spread of infection within a single host. Research on biofilm disassembly mechanisms is still in its infancy, but significant progress in uncovering molecular and biochemical mechanisms involved in the disassembly process has been made in the past five years. Despite these advances, there are still many open questions to be answered. For instance, in agr-mediated and Esp-mediated dispersal, the targets of the secreted proteases remain undetermined [10, 44]. Similarly, Staphylococcus strains secrete a potent thermonuclease enzyme, but its contribution to disassembly has not been investigated. In terms of environmental changes, little is known about conditions that trigger biofilm disassembly besides glucose removal. On the therapeutic side, the mechanism of action of cis-2-decenoic acid [54] and lysostaphin [52] on staphylococcal biofilms are unclear. At the same time, new important discoveries continue to be made, such as the recent determination that S. aureus produces D-amino acids in stationary phase [55] and that they have biofilm inhibitory properties [56], demonstrating that the disassembly field is rich with opportunities for future study. In biofilm pathogenesis models, investigators are only beginning to examine the consequences of targeting these disassembly mechanisms. One could imagine scenarios where addition of dispersal agents administered in combination with antibiotics could result in the elimination of biofilm infections. However, a major concern of utilizing biofilm dispersal agents clinically is the potential to spread the infection systemically or the generation of large detached biofilm chunks (also called `clumps' or `emboli') that have inherent resistance characteristics and could lead to embolism [57, 58]. Considering that most biofilm disassembly studies have been conducted using in vitro models of biofilm development, it is critical for future work to examine the consequences of the induction of biofilm disassembly in animal models of biofilm infection to address these concerns. Overall it is evident that our knowledge of the active mechanisms of disassembly are limited, and the more we gain insight on these mechanisms, the better we will be able to target the disassembly process for biofilm therapy.
Acknowledgements
BRB is supported by grant AI081748 and ARH by grants AI078921 and AI083211 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 2.Otto M. Staphylococcal biofilms. Current Topics Microbiol. Immun. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotz F. Staphylococcus and biofilms. Molecular Microbiology. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 4.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 5.del Pozo JL, Patel R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007;82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 6.Fey PD. Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections? Curr. Opin. Microbiol. 2010;13:610–615. doi: 10.1016/j.mib.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuluaga AF, Galvis W, Saldarriaga JG, Agudelo M, Salazar BE, Vesga O. Etiologic diagnosis of chronic osteomyelitis: a prospective study. Arch. Intern. Med. 2006;166:95–100. doi: 10.1001/archinte.166.1.95. [DOI] [PubMed] [Google Scholar]
- 8.Arnold SR, Elias D, Buckingham SC, Thomas ED, Novais E, Arkader A, Howard C. Changing patterns of acute hematogenous osteomyelitis and septic arthritis: emergence of community-associated methicillin-resistant Staphylococcus aureus. J. Pediatr. Orthop. 2006;26:703–708. doi: 10.1097/01.bpo.0000242431.91489.b4. [DOI] [PubMed] [Google Scholar]
- 9.Fowler VG, Jr., Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, Corey GR, Spelman D, Bradley SF, Barsic B, Pappas PA, Anstrom KJ, Wray D, Fortes CQ, Anguera I, Athan E, Jones P, van der Meer JT, Elliott TS, Levine DP, Bayer AS. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA. 2005;293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 10.Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathogens. 2008;4:e1000053. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J. Orthop. Res. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 12.Pitz AM, Yu F, Hermsen ED, Rupp ME, Fey PD, Olsen KM. Vancomycin susceptibility trends and prevalence of heterogeneous vancomycin-intermediate Staphylococcus aureus in clinical methicillin-resistant S. aureus isolates. J. Clin. Microbiol. 2011;49:269–274. doi: 10.1128/JCM.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 2011;121:238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adair CG, Gorman SP, Feron BM, Byers LM, Jones DS, Goldsmith CE, Moore JE, Kerr JR, Curran MD, Hogg G, Webb CH, McCarthy GJ, Milligan KR. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 1999;25:1072–1076. doi: 10.1007/s001340051014. [DOI] [PubMed] [Google Scholar]
- 15.Feldman C, Kassel M, Cantrell J, Kaka S, Morar R, Goolam Mahomed A, Philips JI. The presence and sequence of endotracheal tube colonization in patients undergoing mechanical ventilation. Eur. Resp. J. 1999;13:546–551. doi: 10.1183/09031936.99.13354699. [DOI] [PubMed] [Google Scholar]
- 16.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, Kelley WL. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 2007;75:1079–1088. doi: 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, Jabbouri S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 2007;75:125–132. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- 21.Maira-Litran T, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, Goldmann DA, Pier GB. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 24.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 1999;67:2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JK, Ragunath C, Kaplan JB, Mack D. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 27.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, Smeltzer MS. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS ONE. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marti M, Trotonda MP, Tormo-Mas MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penades JR. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 32.O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O'Gara JP. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penades JR, Lasa I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 36.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 2005;187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, Bayles KW, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006;188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem. Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 43.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 45.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, Deleo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 46.Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE. 2009;4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrobial Agents Chemo. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramasubbu N, Thomas LM, Ragunath C, Kaplan JB. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J Mol Biol. 2005;349:475–486. doi: 10.1016/j.jmb.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 51.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrobial Agents Chemo. 2003;47:3407–3414. doi: 10.1128/AAC.47.11.3407-3414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokai-Kun JF, Chanturiya T, Mond JJ. Lysostaphin eradicates established Staphylococcus aureus biofilms in jugular vein catheterized mice. J. Antimicrobial Chem. 2009;64:94–100. doi: 10.1093/jac/dkp145. [DOI] [PubMed] [Google Scholar]
- 54.Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang SJ, Nast CC, Mishra NN, Yeaman MR, Fey PD, Bayer AS. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob Agents Chemo. 2010;54:3079–3085. doi: 10.1128/AAC.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 2004;186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]