Abstract
Despite many years of dedicated efforts, high-resolution structural determination of membrane proteins lags far behind that of soluble proteins. Computational methods in general, and molecular dynamics (MD) simulations in particular, have represented important alternative resources over the years to advance understanding of membrane protein structure and function. However, it is only recently that much progress has been achieved owing to new high-resolution membrane protein structures, specialized parallel computer architectures, and efficient simulation algorithms. This has definitely been the case for G protein-coupled receptors (GPCRs), which have assumed a leading role in the area of structural biology with several new structures appearing in the literature during the past five years. We provide here a concise overview of recent developments in computational biophysics of membrane proteins, using GPCRs as an example to showcase important information that can be derived from modern MD simulations.
Introduction
The paucity of structural data for membrane proteins at atomic resolution, combined with the realization that the dynamics of such systems involve large conformational changes occurring usually over long time scales, limits the extent of our current mechanistic understanding of their physiological function. Information regarding the specific motions associated with certain functions can be derived from various spectroscopic techniques, albeit only indirectly and often uncertainly. In light of this, molecular dynamics (MD) simulations have become indispensable tools to obtain more detailed information on membrane protein dynamics and have proven to be particularly valuable, especially when they go hand in hand with experiments.
Since several excellent surveys of MD simulations of membrane proteins with an emphasis on transporters and channels have appeared in the literature recently [1-4], we focus this review on showcasing state-of-the-art MD simulations that have been carried out to provide insights into biological processes of another major class of membrane proteins, the superfamily of G protein-coupled receptors (GPCRs). GPCRs are a key component in signal transduction across the cell membrane [5] and represent about 25 % of current drug targets [6]. The large number of GPCR crystal structures that have appeared in the literature in the past few years [7] has rendered this class of membrane proteins an ideal target for current investigation by MD. After a concise introduction to the different types of cutting-edge MD techniques that have recently been used to explore the structure and dynamics of GPCRs, we survey a few applications that have contributed to an increase in our understanding of their function.
State-of-the-art MD Simulations
Although there have been important developments in experimental techniques to study membrane proteins in dynamical motion, MD is the method of choice to obtain information about unstable species that is impossible or difficult to retrieve experimentally. Keeping the focus of this review on the latest revolutionary MD simulation studies of GPCRs, we briefly introduce below the various combinations of all-atom, coarse-grained, and biased MD simulation strategies that have been used by these studies. We direct the reader elsewhere for a recent review offering a longer history of MD applied to GPCRs [8], as well as for recent state-of-the-art quantum mechanics/molecular mechanics (QM/MM) studies on these receptors [9-12].
All-Atom Standard MD Simulations
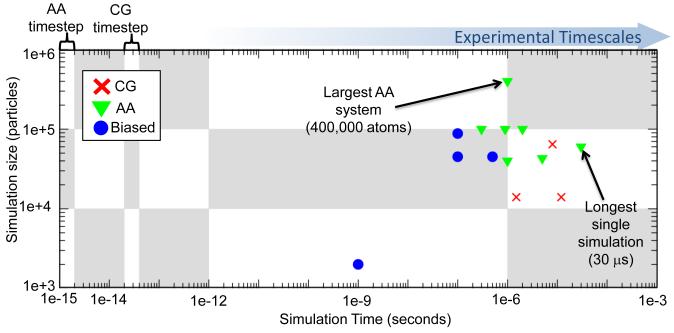
A complete overview of atomistic classical MD simulations, even if focused on GPCRs only, would not be practical within the framework of this review. With apologies to the many authors who contributed significantly to the GPCR field through their MD studies, here we will only report on the longest published MD simulations of explicitly represented GPCR/lipid/water systems, i.e. those breaching the microsecond threshold [13]. A laudable outcome of recent advances in computer hardware (e.g., Anton [14] and Blue Gene [15,16] built at DE Shaw Research and IBM, respectively) and software (e.g., Desmond [17] and Blue Matter [18]), these simulations allow us to reach timescales that still fall far short of biologically relevant ones for GPCRs, not to mention the fact that they are a prerogative of a restricted group of investigators. In addition to known limitations of standard MD simulations (e.g., 1-2 femtoseconds integration time step, simplified energy functions, etc.), the stochastic nature of GPCR-mediated biological processes adds an extra level of complexity, since more than one single long simulation might be expected to be required to obtain statistically accurate thermodynamic estimates from such simulations [19]. Figure 1 illustrates the timescales and system sizes of the MD simulations of GPCRs reviewed herein.
Figure 1.
Scatter plot of simulation length vs. simulation size for studies reviewed in this manuscript. The blue arrow indicates the direction of increasing experimental timescales and a comparison of the time step for atomistic and for CG simulations is highlighted. Atomistic simulations reviewed here are represented by green triangles, CG simulations by red crosses and biased simulations by blue circles. Where multiple simulations were performed in the same publication, the total time used for the analysis of the single system is reported. For instance, for the biased umbrella sampling, the total time is the sum of the lengths of all windows, whereas for multiple simulations of different systems only the longest is reported in this graph.
Coarse-Grained MD Simulations
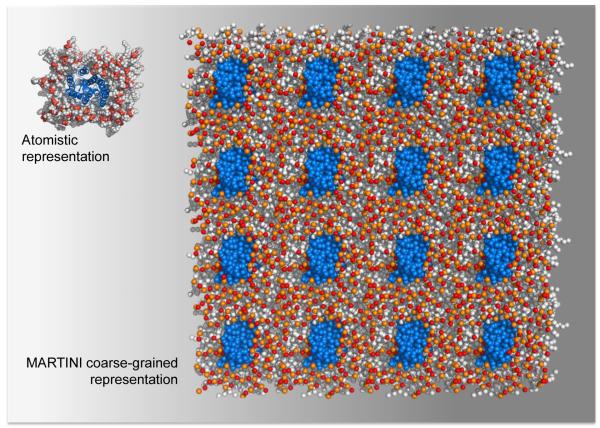
While not a substitute for all-atom simulations, coarse-grained (CG) MD simulations have rendered lengthy MD simulations of several large, explicitly represented, membrane protein systems more tractable [1]. Offering high CG resolution (i.e., four-to-one mapping of heavy atoms, including water molecules) of proteins and lipid molecules, the GROMACS [20] compatible MARTINI force field [21] has enabled time steps approximately 10 times those possible with an all-atom classical MD simulation, resulting in a four-fold scaling factor for the effective time sampled for simulations of GPCRs in an explicit lipid-water environment. The major drawback of this force field arises from the necessity of using restraints to maintain the integrity of the secondary structure of the receptor, thus limiting the capacity to study protein conformational changes. Figure 2 shows a comparison of the smallest explicit atomistic and the largest CG simulation setups used in the studies reviewed here.
Figure 2.
A comparison between approximations of the smallest explicit atomistic system setup of a GPCR protomer (e.g., [13]) and the largest CG setup of the simulations [48] surveyed in this manuscript. The protomers are shown in blue, and the lipids are shown in grey (hydrophobic tails) and red/orange (head groups).
Enhanced MD Simulations
An alternative way to bridge the gap between computational and experimental timescales is through enhanced sampling of so-called “rare events” in a given system [22]. A key aspect for the successful application of enhanced MD methods is that the collective variables (CVs) or reaction coordinates that are chosen to represent the process of interest are adequate to describe it appropriately.
Umbrella sampling [23] is one of several enhanced sampling methods that have very recently been applied to GPCRs. This prototypic biased MD technique improves sampling of a potential of mean force (PMF) along a predefined reaction coordinate divided into sections (windows). The windows are then combined, most often using the weighted histogram analysis method (WHAM) [24], and the unbiased distribution function is used to obtain the PMF along the reaction coordinate. Another of these techniques is well-tempered metadynamics (WTM) [25]. This method enables reconstruction of the free-energy surface (FES) of a system as a function of a small set of CVs, the exploration of each of which is enhanced by a history-dependent potential. Although the bias potential does not fully compensate the FES, the free-energy of a complex system can be estimated using this method. We also report below results obtained by a recent application of random acceleration MD (RAMD) [26] to GPCRs, which has enabled efficient exploration of ligand binding pathways by addition of an artificial, randomly directed force imposed upon the ligand.
Insights into Biological Processes of GPCRs from MD Simulations
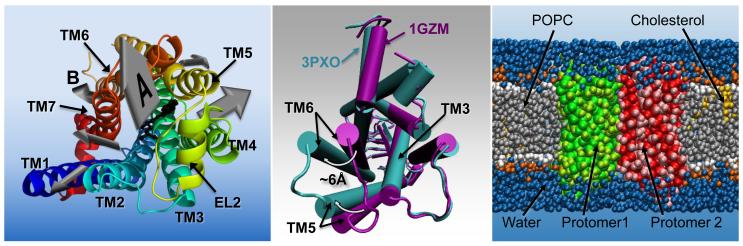
In response to stimulation by an external signal (e.g., ligand binding), an inactive GPCR undergoes a series of conformational rearrangements usually occurring on a microsecond or millisecond timescale [27], leading to active conformations. These activated conformations, in turn, trigger activation of intracellular proteins (e.g., G-proteins, arrestin), leading to specific biological responses. Receptor activation may or may not occur in the context of a dimeric or oligomeric arrangement, recently suggested to be transient in nature (on timescales of hundreds of milliseconds to seconds) at least for a few GPCRs [28-31]. Here we survey recent MD simulations aimed at a molecular-level understanding of ligand recognition, activation, and dimerization/oligomerization events of GPCRs, all illustrated in Figure 3.
Figure 3.
Three key events in the life of a GPCR. The leftmost panel illustrates ligand recognition, showing the exit pathways of carazolol (black, stick representation) from the crystallographically found binding site in the β2AR (rainbow blue to red by residue number, cartoon representation, viewed from the extracellular side) as determined by the RAMD study [34] reviewed here. The size of the arrow denotes the approximate relative frequency of occurrence of each of the exit pathways during the RAMD. Arrow A is the major exit pathway found by RAMD, and is highlighted as the closest equivalent to the entry pathway observed for NLX into the binding pocket of the DOR, in the cited WTM study [33]. Arrow B is highlighted as the exit pathway found by RAMD that is the closest equivalent to the entry pathway of 2-AG into the cannabinoid receptor, via a cleft between TM6 and TM7, suggested in [32]. The central panel illustrates the largest conformational changes occurring activation using an intracellular view of the aligned structures of inactive rhodopsin (PDBid: 1GZM) [53] and metarhodopsin II (PDBid: 3PXO) [46]. Changes highlighted in the activated structure are the movement of TM6 by ~ 6Å, movement and torsion of TM5. The right-most panel shows a system setup for a coarse-grained simulation of a GPCR homodimer, in an explicit lipid environment. Water is shown in blue, the phosphocholine bilayer is shown in grey (hydrophobic tails) and red/orange (head groups), cholesterol is yellow and the two protomers are shown in red and green.
Ligand Recognition
Both unbiased and biased MD techniques have recently been employed to reveal details of ligand binding pathways in GPCRs. A total of four microseconds of unbiased MD simulations [32] were shown to be sufficient for the endogenous cannabinoid sn-2-arachidonoylglycerol (2-AG) ligand to access the binding pocket of a rhodopsin-based model of the inactive cannabinoid CB2 receptor from the lipid bilayer, along a pathway that is in agreement with inferences from isothiocyanate covalent labeling studies. Whilst the conformational changes associated with ligand-induced activation were all observed, the authors acknowledged manual intervention to force the steps that are rate-limiting in vivo/in vitro (e.g., protonation of the receptor) to occur during the simulation.
We recently applied [33] WTM to study the binding pathway(s) of the non-selective antagonist naloxone (NLX) from the water environment into the well-accepted alkaloid-binding pocket of δ-opioid receptor (DOR). Using only two CVs, we were able to reconstruct a FES based on which, our calculated equilibrium binding constant for NLX was remarkably close to experimental values [33]. Our simulations suggested a preferential entry of NLX from the extracellular region of the receptor, through a cleft formed by EL2 and EL3, causing breaking of hydrogen bonds between residues in these regions. Notably, although not directly comparable, results obtained by an application of the RAMD technique to the β2 adrenergic receptor (β2AR) crystal structure [34], both in the presence and absence of the inverse agonist carazolol, suggested that the main exit route of the ligand from the binding pocket caused the breakage of a hydrogen bond between residues within EL2 and EL3/top TM7, while the ligand main entry route to the binding pocket was through a cleft at the receptor extracellular side formed by TM2, TM3, TM7, and a hydrophobic patch bridging EL2, EL3, and the top of TM7. A ligand entry pathway similar to that observed for 2-AG into the cannabinoid receptor, via a cleft between TM7 and TM6, was also observed in the RAMD studies, but with lower frequency of occurrence. While interesting and in line with experiments, to the best of our knowledge, the novel hypotheses of ligand-receptor interactions along the ligand entry or exit pathways predicted by these studies have not yet been verified.
Activation
Several groups have employed exceptionally long time-scale MD simulations to study distinct aspects of the activation process of GPCRs. In addition to large computational investments (>1.5 microseconds) to assess the role of the membrane environment [35], or of the so-called ‘ionic lock’ in relation to activation [36,37], long-scale atomistic MD simulations have also been employed to explore the conformational space of activated conformations of GPCRs. DE Shaw Research recently produced the longest atomistic ‘classical’ MD simulations (30 microseconds), by one order of magnitude, performed on a membrane protein of any kind [13]. The overall conclusion drawn from this extraordinary amount of simulation data was that agonist binding alone is insufficient to maintain the active state of a GPCR over a long time scale, and stabilizing interactions from a G protein or a G protein-mimic (e.g., the nanobody used to facilitate crystallization of the activated β2AR) are a necessity. Also impressive is the recently published microsecond-scale 400,000 atom MD simulation of the rhodopsin-transducin complex in an explicit lipid bilayer [38]. Although based on a docking model of the complex rather than a crystal structure (currently unavailable), these simulations reveal a dynamic interface between the C-termini of both protein domains, suggested to be in overall agreement with experimental studies.
Since long time scale simulations have not been able yet to produce fully activated conformations of GPCRs starting from an inactive one, biased MD techniques have been used to simulate this transition. In line with previous studies involving activation models derived from biophysical data (e.g. [39]), Tikhonova and colleagues [40] sequentially applied two sets of experimentally derived distance restraints to the LUMI rhodopsin crystal structure [41,42] and performed biased MD with mass-weighted RMSD restraints to obtain putative activated models. From comparison of the results of these calculations on wild-type and several mutants, a clear correlation to known phenotypes was revealed.
Taking an alternative approach, we [43] steered the crystal structure of a photoactivated deprotonated intermediate of rhodopsin (PDBid: 2I37) [42] towards the alleged active low pH crystal structure of opsin (PDBid: 3CAP) [44], using adiabatic biased molecular dynamics (ABMD) simulations. Path CVs derived from these ABMD trajectories were used for subsequent WTM simulations from which the reconstructed FES revealed common metastable states characterized by a different degree of opening between TM3 and TM6. The predicted largest separation between TM3 and TM6 was in line with data obtained for Meta IIb from spectroscopy [45], as well as for both opsin [44] and, more recently, the active Meta II state from crystallography [46]. It still remains to be proven (and we have suggested mutants to shift the conformational equilibrium [43]) whether a conformational state with a reduced opening between TM3 and TM6 exists, and can still activate the G-protein.
Oligomerization
The main problems in simulating dimers/oligomers of GPCRs with a view to characterizing their lifetime, inducing forces, aggregation specificity, allosteric modulation, etc., arise from the increased size of the systems and the long timescales required, not to mention the absence of any crystal structures of physiologically relevant dimeric complexes. Although an impressive study of a microsecond simulation of an atomistic resolution rhodopsin dimer was recently published [47], this is far from ‘the norm’, and CG representations have proven more effective for investigating multi-protomer GPCR systems. To date, the most expansive computational study of GPCR oligomerization (16 rhodopsin protomers in four different CG lipid environments, simulated for up to 8 μs each) involved application of the MARTINI CG model to simulate non-biased self-aggregation of the receptors [48]. Not only was this a series of very long simulations, but also it is remarkable in terms of simulated system size. The results of these simulations suggested a localized membrane adaptation to the presence of the receptor possibly due to hydrophobic mismatch between the length of the hydrophobic part of the monomeric receptor and the equilibrium hydrophobic bilayer thickness for the different lipid types. An implicit solvation model might not have revealed some of these effects, and an atomistic simulation of this magnitude would be unfeasible on even the most efficient current computer architectures, rendering the CG representation the current most effective technique for addressing self-aggregation phenomena on this scale.
We have recently combined a CG representation with both umbrella sampling [49] and metadynamics [50] to estimate a possible lifetime for a GPCR dimer in a lipid bilayer. We described the relative position of two interacting DOR protomers in terms of their relative orientation and separation. The resulting umbrella sampling FES as a function of the separation between receptors was combined with a diffusion limited model of dimerization to calculate a half-life of DOR TM4-TM4 dimers in the lipid bilayer of the order of seconds, approximating the kinetics of M1 muscarinic receptors recently assessed by single-molecule studies [28]. When we compared these results with those from a similar calculation for a DOR homodimer with both TM4 and TM5 at the interface [50], we found the half-life of this dimer to be still of the order of seconds, but slightly shorter than that of the TM4 dimer. We look forward to further integration between computation and single-molecule imaging for a more comprehensive characterization of the GPCR monomer–dimer dynamic equilibrium in live cells, and a better understanding of their inducing and stabilizing forces.
Conclusions
MD simulations, both classical MD at very long timescales and simulations involving biasing techniques or reduced representations have been an invaluable tool to aid in the rationalization of biophysical phenomena observed from crystallization, fluorescence and mutation studies of membrane proteins in general, and GPCRs in particular. The capacity for MD simulations as a route to calculating measurable quantities that can be verified ex silico is greater than ever, with the advent of faster hardware, increasingly efficient parallelizable algorithms, and ever more realistic force field descriptions. We look forward to more crystallographic information, and the applications of the newest graphical processing unit (GPU) technologies [51] and polarizable force fields [52] to GPCRs, as well as other membrane proteins, in the hope of obtaining more accurate descriptions of the dynamics of these systems, while bridging the gap between the timescales of biological processes observed in vivo and those accessible in computer simulation.
Highlights.
-
-
GPCRs as an example to showcase cutting-edge molecular dynamics simulations.
-
-
Brief introduction to all-atom, coarse-grained, and biased MD simulation strategies.
-
-
Focus on studies addressing GPCR ligand recognition, activation and oligomerization.
Acknowledgements
The authors’ work on membrane proteins is supported by National Institutes of Health grants DA020032, DA026434, and MH091360. Computations are supported in part by the National Science Foundation through TeraGrid advanced computing resources (TRAC MCB080077). We wish to thank Dr. Alan Grossfield for comments on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindahl E. Sansom MSP: Membrane proteins: molecular dynamics simulations. Current Opinion in Structural Biology. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Khalili-Araghi F, Gumbart J, Wen PC, Sotomayor M, Tajkhorshid E, Schulten K. Molecular Dynamics Simulations of Membrane Channels and Transporters. Current Opinion in Structural Biology. 2009;12:128–137. doi: 10.1016/j.sbi.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discovery Today. 2009;14:1130–1135. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Shaikh SA, Tajkhorshid E. Exploring Transmembrane Diffusion Pathways With Molecular Dynamics. Physiology. 2010;25:142–154. doi: 10.1152/physiol.00046.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldo J, Pin J-P. G protein-coupled receptors: From structure to function. The Royal Society of Chemistry; Cambridge, UK: 2011. in press. [Google Scholar]
- 6.Overington JP, Al-Lazikani B, Hopkins AL. Opinion - How many drug targets are there? Nature Reviews Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 7.Hanson MA, Stevens RC. Discovery of New GPCR Biology: One Receptor Structure at a Time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossfield A. Recent progress in the study of G protein-coupled receptors with molecular dynamics computer simulations. Biochimica et Biophysica Acta. 2011 doi: 10.1016/j.bbamem.2011.03.010. in press. [DOI] [PubMed] [Google Scholar]
- 9.Polli D, Altoe P, Weingart O, Spillane KM, Manzoni C, Brida D, Tomasello G, Orlandi G, Kukura P, Mathies RA, et al. Conical intersection dynamics of the primary photoisomerization event in vision. Nature. 2010;467:440–443. doi: 10.1038/nature09346. [DOI] [PubMed] [Google Scholar]
- 10.Gascon JA, Batista VS. QM/MM study of energy storage and molecular rearrangements due to the primary event in vision. Biophys J. 2004;87:2931–2941. doi: 10.1529/biophysj.104.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gascon JA, Sproviero EM, Batista VS. Computational studies of the primary phototransduction event in visual rhodopsin. Acc Chem Res. 2006;39:184–193. doi: 10.1021/ar050027t. [DOI] [PubMed] [Google Scholar]
- 12.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 angstrom crystal structure. Journal of Molecular Biology. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- **13.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi HJ, DeVree BT, Sunahara RK, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–U129. doi: 10.1038/nature09665. The longest single atomistic molecular dynamics simulation of a membrane protein published to date.
- 14.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, et al. Anton, a special-purpose machine for molecular dynamics simulation. Communications of the Acm. 2008;51:91–97. [Google Scholar]
- 15.Allen F, Almasi G, Andreoni W, Beece D, Berne BJ, Bright A, Brunheroto J, Cascaval C, Castanos J, Coteus P, et al. Blue Gene: A vision for protein science using a petaflop supercomputer. Ibm Systems Journal. 2001;40:310–327. [Google Scholar]
- 16.Almasi G, Asaad S, Bellofatto RE, Bickford HR, Blumrich MA, Brezzo B, Bright AA, Brunheroto JR, Castanos JG, Chen D, et al. Overview of the IBM Blue Gene/P project. Ibm Journal of Research and Development. 2008;52:199–220. [Google Scholar]
- 17.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters; Proceedings of the ACM/IEEE Conference on Supercomputing (SC06); Tampa, Florida. November 11-17.2006. [Google Scholar]
- 18.Fitch BG, Germain RS, Mendell M, Pitera J, Pitman M, Rayshubskiy A, Sham Y, Suits F, Swope W, Ward TJC, et al. Blue Matter, an application framework for molecular simulation on Blue Gene. Journal of Parallel and Distributed Computing. 2003;63:759–773. [Google Scholar]
- *19.Grossfield A, Zuckerman DM. Quantifying uncertainty and sampling quality in biomolecular simulations. Annual Reports in Computational Chemistry. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. A recent overview of statistical assessments of simulation studies performed on large biomolecular systems at long timescales.
- 20.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 21.Marrink SJ, Fuhrmans M, Risselada HJ, Periole X. The MARTINI forcefield. In: Voth G, editor. Coarse graining of condensed phase and biomolecular systems. CRC Press; 2008. [Google Scholar]
- 22.Christen M, van Gunsteren W. On searching in, sampling of and dynamically moving through conformational space of biomolecular systems: a review. Journal of Computational Chemistry. 2008;29:157–166. doi: 10.1002/jcc.20725. [DOI] [PubMed] [Google Scholar]
- 23.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation - Umbrella sampling. Journal of Computational Physics. 1977;23:187–199. [Google Scholar]
- 24.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules .1. The Method. Journal of Computational Chemistry. 1992;13:1011–1021. [Google Scholar]
- 25.Barducci A, Bussi G, Parrinello M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Physical Review Letters. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 26.Ludemann SK, Lounnas V, Wade RC. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. Journal of Molecular Biology. 2000;303:797–811. doi: 10.1006/jmbi.2000.4154. [DOI] [PubMed] [Google Scholar]
- 27.Ambrosio M, Zurn A, Lohse MJ. Sensing G protein-coupled receptor activation. Neuropharmacology. 2011;60:45–51. doi: 10.1016/j.neuropharm.2010.08.006. [DOI] [PubMed] [Google Scholar]
- **28.Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JET, Lazareno S, Molloy JE, Birdsall NJM. Formation and dissociation of M-1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. The first single-molecule study of G protein-coupled receptor oligomerization.
- 29.Fonseca JM, Lambert NA. Instability of a Class A G Protein-Coupled Receptor Oligomer Interface. Molecular Pharmacology. 2009;75:1296–1299. doi: 10.1124/mol.108.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorsch S, Klotz KN, Engelhardt S, Lohse MJ, Bunemann M. Analysis of receptor oligomerization by FRAP microscopy. Nature Methods. 2009;6:225–230. doi: 10.1038/nmeth.1304. [DOI] [PubMed] [Google Scholar]
- 31.Kasai RS, Suzuki KG, Prossnitz ER, Koyama-Honda I, Nakada C, Fujiwara TK, Kusumi A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Hurst DP, Grossfield A, Lynch DL, Feller S, Romo TD, Gawrisch K, Pitman MC, Reggio PH. A Lipid Pathway for Ligand Binding Is Necessary for a Cannabinoid G Protein-coupled Receptor. Journal of Biological Chemistry. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. Ligand entry pathway from lipid bilayer elucidated by standard molecular dynamics simulations.
- 33.Provasi D, Bortolato A, Filizola M. Exploring Molecular Mechanisms of Ligand Recognition by Opioid Receptors with Metadynamics. Biochemistry. 2009;48:10020–10029. doi: 10.1021/bi901494n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Wang T, Duan Y. Ligand Entry and Exit Pathways in the beta(2)-Adrenergic Receptor. Journal of Molecular Biology. 2009;392:1102–1115. doi: 10.1016/j.jmb.2009.07.093. Exhaustive ligand binding pathway exploration for a G protein-coupled receptor.
- 35.Khelashvili G, Grossfield A, Feller SE, Pitman MC, Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with rhodopsin identified from microsecond length molecular dynamics simulations. Proteins-Structure Function and Bioinformatics. 2009;76:403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dror RO, Arlow DH, Borhani DW, Jensen MO, Piana S, Shaw DE. Identification of two distinct inactive conformations of the beta(2)-adrenergic receptor reconciles structural and biochemical observations. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romo TD, Grossfield A, Pitman MC. Concerted Interconversion between Ionic Lock Substates of the beta(2) Adrenergic Receptor Revealed by Microsecond Timescale Molecular Dynamics. Biophysical Journal. 2010;98:76–84. doi: 10.1016/j.bpj.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ** 38.Sgourakis NG, Garcia AE. The Membrane Complex between Transducin and Dark-State Rhodopsin Exhibits Large-Amplitude Interface Dynamics on the Sub-Microsecond Timescale: Insights from All-Atom MD Simulations. Journal of Molecular Biology. 2010;398:161–173. doi: 10.1016/j.jmb.2010.02.032. Exceptionally large atomistic simulation of a G protein coupled receptor-G protein complex.
- 39.Niv MY, Skrabanek L, Filizola M, Weinstein H. Modeling activated states of GPCRs: the rhodopsin template. Journal of Computer-Aided Molecular Design. 2006;20:437–448. doi: 10.1007/s10822-006-9061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tikhonova IG, Best RB, Engel S, Gershengorn MC, Hummer G, Costanzi S. Atomistic insights into rhodopsin activation from a dynamic model. Journal of the American Chemical Society. 2008;130:10141–10149. doi: 10.1021/ja0765520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angewandte Chemie, International Edition. 2006;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 42.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 43.Provasi D, Filizola M. Putative Active States of a Prototypic G-Protein-Coupled Receptor from Biased Molecular Dynamics. Biophysical Journal. 2010;98:2347–2355. doi: 10.1016/j.bpj.2010.01.047. A general enhanced sampling method to identify metastable states of G protein-coupled receptors along putative activation pathways.
- 44.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 45.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Sequence of late molecular events in the activation of rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauß N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011 doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 47.Neri M, Vanni S, Tavernelli I, Rothlisberger U. Role of Aggregation in Rhodopsin Signal Transduction. Biochemistry. 2010;49:4827–4832. doi: 10.1021/bi100478j. [DOI] [PubMed] [Google Scholar]
- 48.Periole X, Huber T, Marrink SJ, Sakmar TP. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. Journal of the American Chemical Society. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- * 49.Provasi D, Johnston JM, Filizola M. Lessons from Free Energy Simulations of delta-Opioid Receptor Homodimers Involving the Fourth Transmembrane Helix. Biochemistry. 2010;49:6771–6776. doi: 10.1021/bi100686t. The first computational attempt to provide estimates of lifetimes of G protein-coupled receptor dimers in a lipid bilayer.
- 50.Johnston JM, Aburi M, Provasi D, Bortolato A, Urizar E, Lambert NA, Javitch JA, Filizola M. Making Structural Sense of Dimerization Interfaces of Delta Opioid Receptor Homodimers. Biochemistry. 2011;50(10):1682–1690. doi: 10.1021/bi101474v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone JE, Hardy DJ, Ufimtsev IS, Schulten K. GPU-accelerated molecular modeling coming of age. Journal of Molecular Graphics & Modelling. 2010;29:116–125. doi: 10.1016/j.jmgm.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang W, Hardy DJ, Phillips JC, MacKerell AD, Schulten K, Roux B. High-Performance Scalable Molecular Dynamics Simulations of a Polarizable Force Field Based on Classical Drude Oscillators in NAMD. Journal of Physical Chemistry Letters. 2011;2:87–92. doi: 10.1021/jz101461d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. Journal of Molecular Biology. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]