Abstract
Most zygotic genes remain transcriptionally silent in Drosophila, Xenopus, and zebrafish embryos through multiple mitotic divisions until the midblastula transition (MBT). Several genes have been identified in each of these organisms that are transcribed before the MBT, but whether precocious expression of specific mRNAs is important for later development has not been examined in detail. Here, we identify a class of protein coding transcripts activated before the MBT by the maternal T-box factor VegT that are components of an established transcriptional regulatory network required for mesendoderm induction in Xenopus laevis, including the Nodal related ligands xnr5, xnr6, and derrière and the transcription factors bix4, and sox17α. Accumulation of phospho-Smad2, a hallmark of active Nodal signaling, at the onset of the MBT requires preMBT transcription and activity of xnr5 and xnr6. Furthermore, preMBT activation of the Nodal pathway is essential for mesendodermal gene expression and patterning of the embryo. Finally, xnr5 and xnr6 can also activate their own expression during cleavage stages, indicating that preMBT transcription contributes to a feed-forward system that allows robust activation of Nodal signaling at the MBT.
Keywords: midblastula transition (MBT), Nodal, VegT, mesoderm, Smad, zygotic gene activation (ZGA)
INTRODUCTION
Following fertilization, early development is primarily under the control of maternal factors until the onset of zygotic gene expression. The timing of the maternal to zygotic transition, which involves degradation of maternal mRNAs and regulated activation of zygotic genes, varies in different organisms (Andeol, 1994; Davidson, 1986; Tadros and Lipshitz, 2009). In Drosophila, Xenopus, and zebrafish, most zygotic genes are silent until large-scale zygotic gene activation (ZGA) begins at the midblastula transition (MBT), 10–14 cleavage divisions after fertilization (Andeol, 1994; Bachvarova et al., 1966; Davidson, 1986; Edgar and Schubiger, 1986; Newport and Kirschner, 1982b; Tadros and Lipshitz, 2009). In contrast, ZGA begins in mouse embryos at the first cell division, but up to 20 hours after fertilization (Clegg and Pikó, 1982; Davidson, 1986).
Although the mechanisms that maintain transcriptional silence before the MBT are not well characterized, early Xenopus and Drosophila embryos contain the components needed for RNA polymerase-II-dependent transcription (Veenstra, 2002), suggesting that zygotic genes are repressed in the preMBT embryo, for example by a factor(s) that is titrated out as the nuclear to cytoplasmic ratio increases with successive nuclear divisions (Dunican et al., 2008; Kimelman et al., 1987; Newport and Kirschner, 1982a, b; Pritchard and Schubiger, 1996). Analysis of chromatin structure in Xenopus tropicalis and zebrafish also reveals that histone H3K4 trimethylation (H3K4Me3), typically a mark of active chromatin, is not detected at protein coding genes until the MBT (Akkers et al., 2009; Vastenhouw et al., 2010), although genes required for induction of the dorsal (Spemann) organizer in Xenopus acquire H3K4Me3 well before their transcription at the MBT (Blythe et al., 2010).
Despite the global transcriptional quiescence of the zygotic genome during cleavage stages, transcription of a small number of protein coding genes before large scale ZGA has been uncovered in many organisms including fly, frog, zebrafish, and mouse (reviewed in (Andeol, 1994; Tadros and Lipshitz, 2009)). In Drosophila, new transcription is detected as early as mitotic cleavage 8 (De Renzis et al., 2007; Edgar and Schubiger, 1986; Porcher et al., 2010; ten Bosch et al., 2006; Tracey et al., 2000). In Xenopus, the Nodal-related genes xnr5 and xnr6 are transcribed up to 6 cell divisions before the MBT (Blythe et al., 2010; Rosa et al., 2009; Takahashi et al., 2006; Yang et al., 2002), and additional high molecular weight polyadenylated RNAs are also transcribed before the MBT (Kimelman et al., 1987; Nakakura et al., 1987; Yang et al., 2002). Similarly, in zebrafish, the dorsal Wnt target gene bozozok is expressed before the MBT (Leung et al., 2003) and several other newly transcribed mRNAs are detected by in situ hybridization prior to the MBT (Mathavan et al., 2005).
The presence of preMBT transcription in diverse organisms suggests that this early transcription may be specifically regulated and serve a developmental function. In Drosophila, the promoters for most preMBT genes contain a heptad repeat that regulates preMBT transcription (De Renzis et al., 2007; ten Bosch et al., 2006). Increasing the number of heptad repeats accelerates expression and removing the heptad repeats delays expression (ten Bosch et al., 2006). This cis-acting element binds the Bicoid Stability Factor (BSF), the zinc finger protein Zelda, and Grainyhead (De Renzis et al., 2007; Harrison et al., 2010; Liang et al., 2008). Furthermore, maternal loss of Zelda globally interferes with preMBT transcription and results in defects in cellularization soon after the MBT (Liang et al., 2008). These observations indicate that a site-specific, activating factor is required (in Drosophila) for preMBT transcription in an otherwise repressive context. Although a similar regulator has not been identified in Xenopus or other vertebrates, we report here that VegT, a maternal T-box transcription factor required for the induction of mesoderm and endoderm, regulates the preMBT expression of multiple genes in Xenopus.
The phenotype observed with mutations in Zelda, a global regulator of preMBT genes in Drosophila, strongly supports that preMBT transcription is critical for development; however, the importance of preMBT transcription for specific mRNAs has not yet been addressed in detail. Activity of the maternal β-catenin/Tcf complex, which mediates the transcriptional output of the canonical Wnt pathway, is required during early cleavage stages for specification of the dorsal-ventral axis in Xenopus. However, β-catenin/Tcf functions in this setting to establish poised chromatin at dorsal genes that are not transcribed until the MBT (Blythe et al., 2010). Wnt signaling also induces preMBT transcription of xnr5 and xnr6 (Yang et al., 2002) but whether their transcription specifically during preMBT stages is developmentally significant has been unclear (Kimelman, 2010).
Transcription of xnr5 and xnr6 after the MBT also requires the maternal T-box transcription factor VegT; we therefore investigated the contribution of VegT to preMBT transcription. We find that multiple VegT target genes, in addition to xnr5 and xnr6, are transcribed well before the MBT in Xenopus. All of these genes were previously identified as components of a gene regulatory network involved in the induction of endoderm and mesoderm (Zorn and Wells, 2007). Maternal loss-of-function and gain-of-function data show that VegT is necessary and sufficient for the preMBT expression of these genes. We also find that transcription of xnr5 and xnr6 before the MBT is essential for activation of the Nodal pathway, induction of mesendodermal genes in the late blastula/early gastrula, and later morphogenesis. Finally, we show that Nodal signaling activates expression of xnr5 and xnr6 before the MBT, indicating that preMBT transcription of Nodal genes contributes to a feed-forward system, ensuring that the pathway is activated by the time the embryo reaches the midblastula transition.
MATERIALS and METHODS
Xenopus Embryo Manipulations
Embryos were obtained by in vitro fertilization, cultured, and injected as described (Sive et al., 2000). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1994). Embryos were maintained at 23°C and the intervals between the first 8 cell divisions were closely monitored for each clutch of eggs. The timing of the 10th through the 12th cleavage divisions was predicted based on the length of the earlier cell cycles (approximately 22 minutes at 23°C) and MBT was defined as the 12th cleavage division based on the work of Newport and Kirschner (Newport and Kirschner, 1982a). Animal cap explants were excised and cultured as described (Sive et al., 2007). Dorsal-ventral dissections were performed by first injecting ventral blastomeres (determined by pigmentation) at the 4-cell stage with fluorescent dextran and then manually dissecting 256-cell stage embryos with a hair knife under epifluoresence. SB505124 (Sigma, St. Louis, MO, USA) was dissolved in DMSO (100mM) and for embryo treatment was added to injection buffer (0.5X Marc’s modified ringers solution (MMR) with 3% Ficoll (Sigma, St. Louis, MO, USA) and 50µg/ml gentamycin) to a final concentration of 200µM in glass culture dishes. To inhibit RNA polymerase II, α-amanitin was injected at 50pg/cell into both cells at the 2-cell stage. α-amanitin did not affect the rate of cell division or morphology at the stages examined (effects were not evident until the gastrula stage, as reported previously (Newport and Kirschner, 1982a; Sible et al., 1997).
mRNA Injection
Synthetic capped mRNAs were produced with the mMessage mMachine kit (Ambion, Austin, TX, USA) using SP6 polymerase. Smad2 and VegT were amplified by PCR and cloned into pCS2-GRZ and pCS2-MT, respectively (Figs 2, 4). The xnr5 open reading frame was PCR-amplified from pNRRX-xnr5 (Takahashi et al., 2000) and cloned into pCS2 to make pCS2-xnr5. pCS2-Smad2-GR, pCS2-VegT, pCS2-xnr1 (Sampath et al., 1997) and pCS2-xnr5 were linearized with Not1 (Figs 2, 3, 4). pCS105-GFP-Smad2 and pCS105-GR-tSmad2 (N-terminal truncation of Smad2 that increases activity; both constructs a kind gift from Chenbei Chang) were linearized with AscI. Where indicated, fixable fluorescent lysine dextran (FLDx, 2.5 ng/cell) was co-injected with mRNAs and anti-fluorescein immunodetection was performed to mark injected cells in fixed embryos. For Fig. 6 mRNAs for xnr1, xnr5 and xnr6 were synthesized as described (Jones et al., 1995; Takahashi et al., 2000).
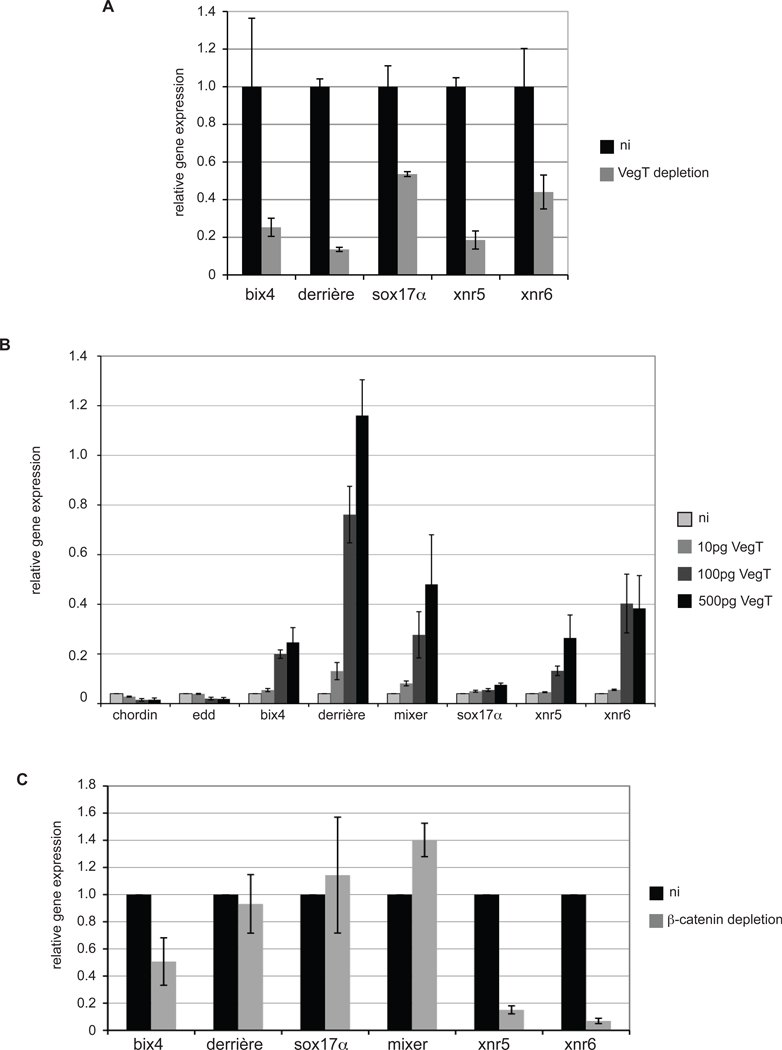
Fig. 2. VegT is required for preMBT transcription.
(A) VegT mRNA was depleted in oocytes with 5ng antisense phosphorthiotate oligos and embryos were recovered using the host-transfer method. Control and VegT depleted embryos were collected at the 1000-cell stage for qRT-PCR. Error bars represent the mean of triplicate measurements in qRT-PCR. (B) VegT mRNA was injected into the animal pole at the 1-cell stage. Animal caps were excised at the 256-cell stage, cultured, and harvested for qRT-PCR at the 1000-cell stage. (C) Antisense morpholino oligonucleotide (MO) targeting β-catenin (40ng/embryo) was injected laterally into two cells at the 2-cell stage. Embryos were harvested for qRT-PCR at the 1000-cell stage. ni, not injected. Error bars represent standard error of the mean for 2 or more experiments. Gene expression was normalized to ODC for each sample.
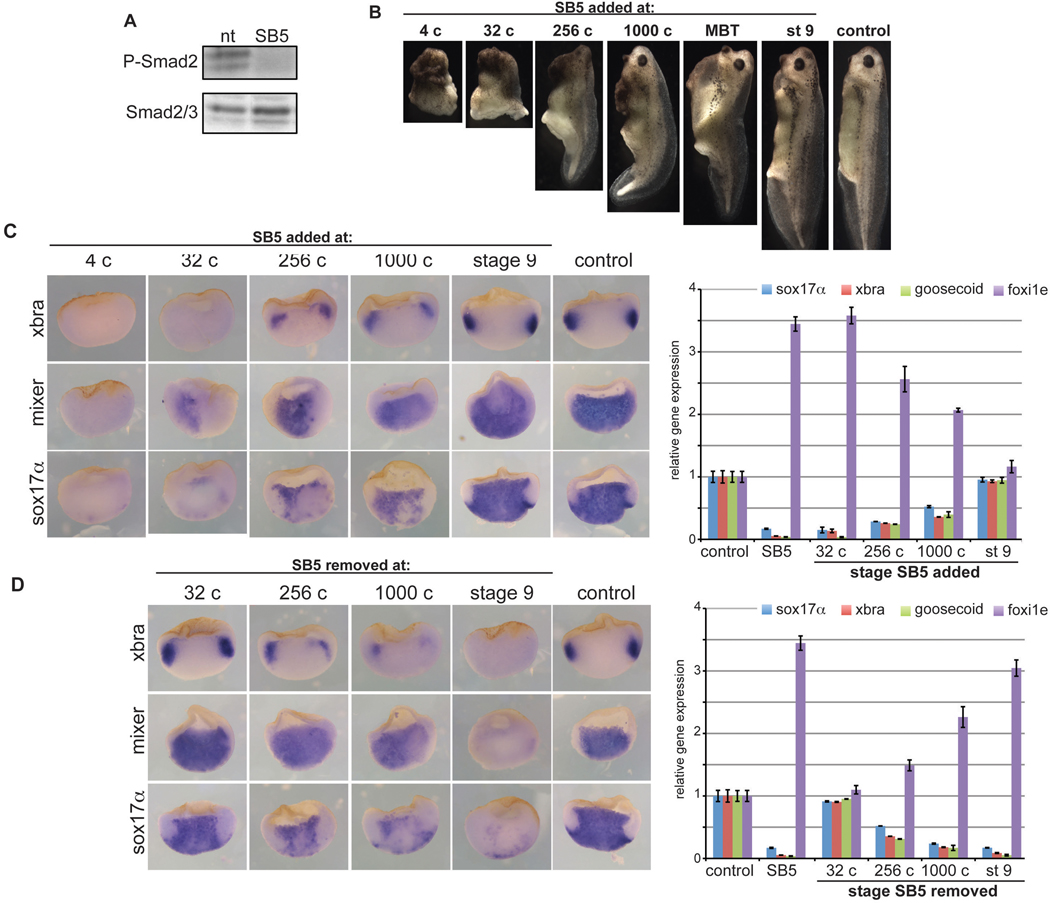
Fig. 4. preMBT Nodal signaling is required for mesendoderm induction.
(A) Embryos were treated with control medium (nt) or SB5 (200µM) from the 4-cell stage to stage 10 and harvested for Western blot for P-Smad2/3 or Smad2/3. (B) Embryos were treated with SB5 from the indicated stages until the onset of gastrulation and cultured until stage 40. Frequency of phenotype is indicated in the lower right corner. (C) Embryos were treated with SB5 beginning at the indicated stages, cultured until stage 10, and analyzed by in situ hybridization (ISH) for mesendodermal gene expression (left). “Control” indicates DMSO (vehicle) only. Embryos were also harvested at early gastrula stage (stage 10) for qRT-PCR for the indicated mesendodermal markers and for the ectodermal marker foxi1e (right panel). All samples were normalized to expression levels in untreated embryos at stage 10 (control). “Control” indicates DMSO; “SB5” indicates treatment from 4-cell until gastrula (stage 10). (D) SB5 was added at the 4-cell stage; embryos were washed at the indicated stages, transferred to control medium, and analyzed by ISH for mesendodermal gene expression at stage 10, as in (C). Histogram on right shows qRT-PCR for mesendodermal and ectodermal markers, as in (C). Note that the “control” and “SB5” (4-cell until stage 10) qRT-PCR data in panel D are identical to those shown in panel C, as qRT-PCR for both groups was performed at the same time.
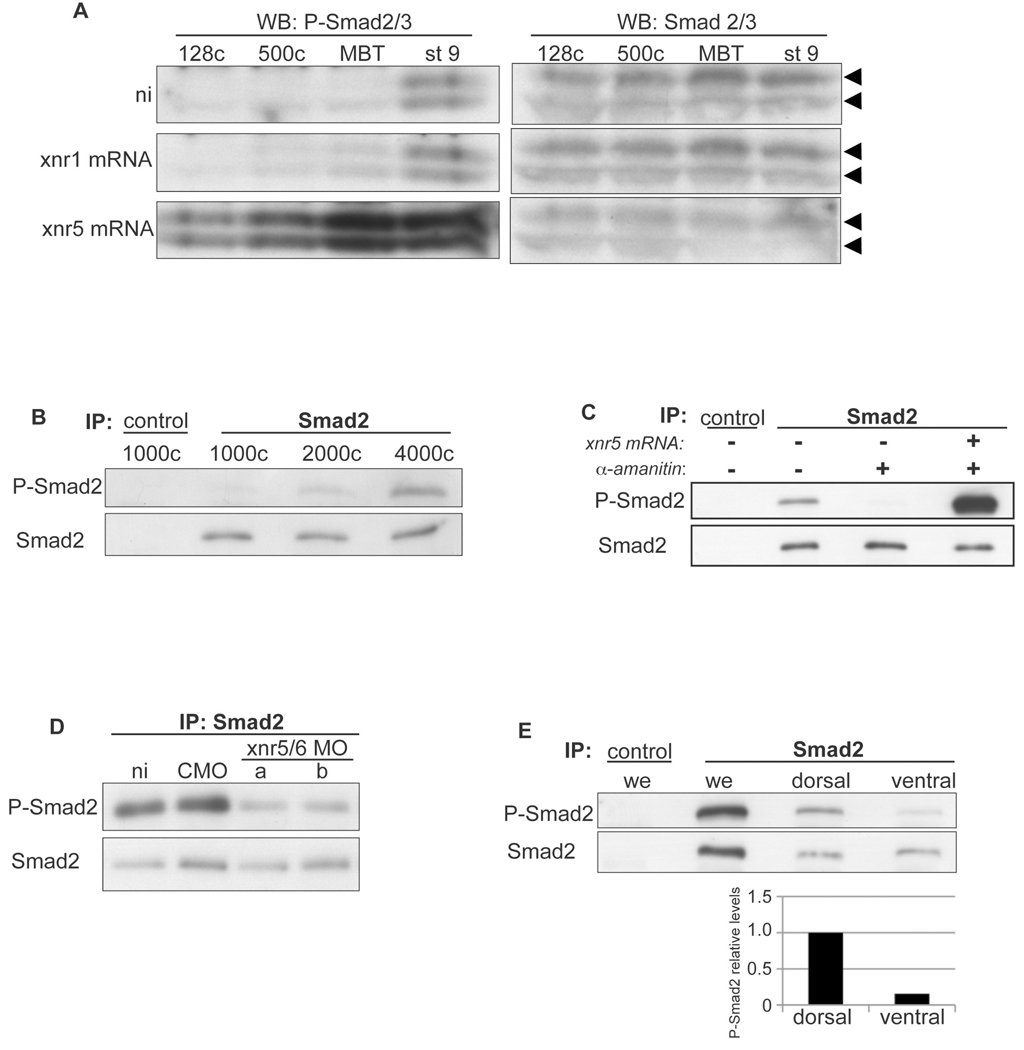
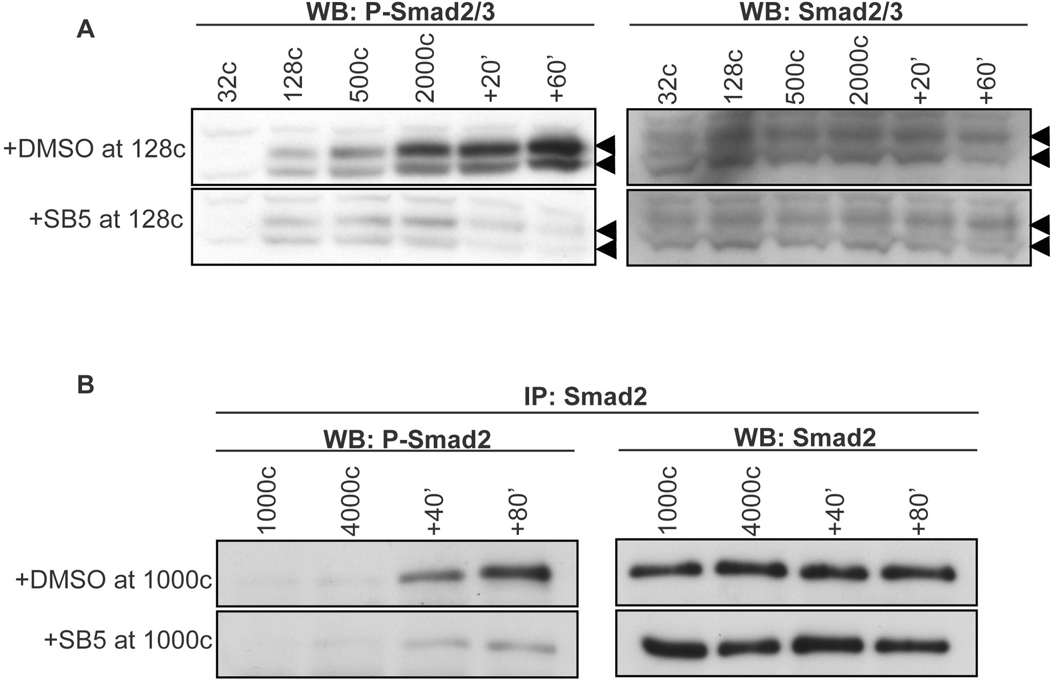
Fig. 3. preMBT Nodal expression activates Smad2/3.
(A) xnr5 mRNA (5pg) or xnr1 mRNA (5pg) was injected vegetally into 1-cell embryos. Embryos were collected at the indicated stages and lysates were analyzed by Western blot for P-Smad2 and Smad2/3. “ni” indicates non-injected controls. Arrowheads indicate P-Smad2 (upper) and P-Smad3 (lower). (B) Non-injected embryos were collected at the indicated stages and subjected to immunoprecipitation with an antibody for Smad2 or for Wnt1 as a negative control (control), then immunoblotted with antibodies for P-Smad2 or Smad2/3. (C) 1-cell embryos were injected laterally with 100pg of the pol-II inhibitor α-amanitin alone or together with 5pg xnr5 mRNA, then collected for IP and immunoblot at the 4000-cell stage (MBT) as in B. (D) xnr5 and xnr6 translation was inhibited by injection in all cells of 4-cell embryos of antisense morpholino oligonucleotides (MO) directed against both xnr5 and xnr6. Xnr5/6MO “a” indicates xnr5 MOa + xnr6 MOa (15 ng each); “b” indicates xnr5 MOb + xnr6 MOb (15 ng each); ni: non-injected; CMO: control MO (30ng). Embryos were collected at the 4000-cell stage for IP and immunoblot as in B. (E) Embryos were injected ventrally at the 4-cell stage with rhodamine dextran, dissected into dorsal and ventral halves at the 256-cell stage, cultured, then harvested at the 4000-cell stage for IP and Western analysis as in B. we, whole embryo. Band intensity was determined by quantifying pixel density from a scanned film using Image J; the level of P-Smad2 was then normalized to total Smad2 and plotted.
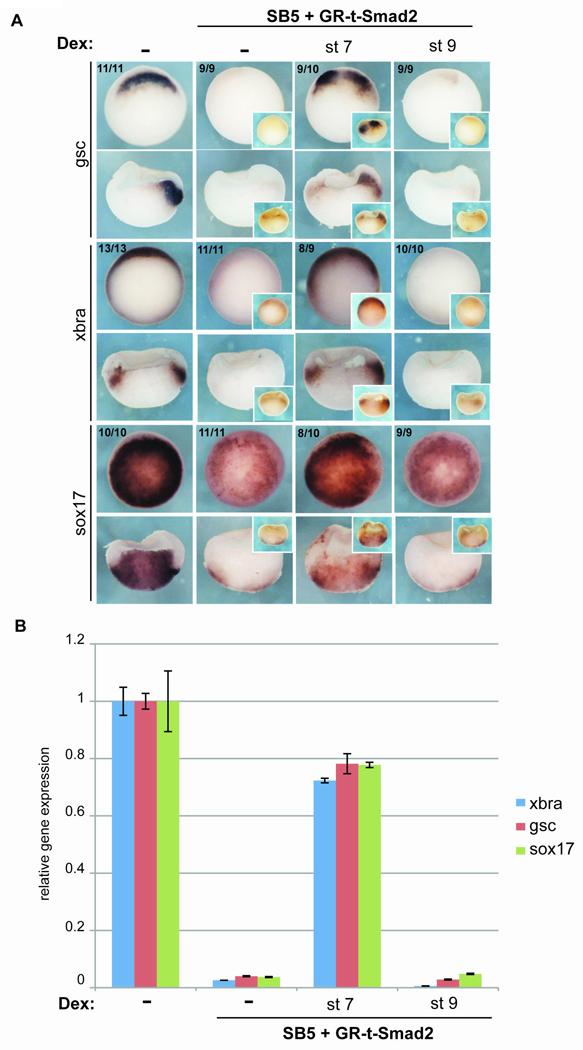
Fig. 6. Smad2 rescues mesendodermal gene expression when activated before but not after the MBT.
Embryos were injected with a hormone inducible, activated form of Smad2 (GR-tSmad2) at the 2-cell stage (along with FLDx as a lineage tracer) and then treated with SB5 beginning at the 4-cell stage. GR-tSmad2 was activated by addition of dexamethasone (dex) before the MBT (stage 7) or after the MBT (stage 9) and embryos were cultured until the onset of gastrulation and then collected for analysis of mesendodermal gene expression by ISH (A) or qRT-PCR (B), as described in Fig. 4. (In panel A, the insets show FLDx immunostaining to reveal the progeny of injected blastoemeres and the numbers in the upper left indicate the frequency of the displayed phenotype/expression pattern). Error bars represent standard deviation for a representative experiment.
Morpholinos and VegT Maternal Depletion
Antisense morpholino oligonucleotides (MOs) targeting xnr5 and xnr6 (Luxardi et al., 2010) and β-catenin (Heasman et al., 2000) were purchased from GeneTools LLC (Philomath, OR, USA). Reduction in biological activity of xnr5 and xnr6 upon MO injection and rescue using recombinant Nodal protein were described previously (Luxardi et al., 2010). Maternal depletion of VegT was carried out as described (Zhang et al., 1998). Briefly, full-grown oocytes were manually defolliculated and cultured in oocyte culture medium. Oocytes were vegetally injected with 5 ng of oligo (C*A*G*CAGCATGTACTT*G*G*C). Oocytes were cultured for 24 hours at 18°C, stimulated with 2 µM progesterone overnight, stained with dyes, and transferred into foster mothers. Two hours after the egg transfer, the host was placed in 1XMMR. After 4 hours, all eggs were collected and fertilized using a sperm suspension. Colored experimental embryos were sorted out and cultured in 0.2XMMR. Depletion of maternal VegT in embryos was confirmed by RT-PCR (data not shown), as described previously (Heasman et al., 2001; Kofron et al., 1999; Kofron et al., 2004; Zhang et al., 1998).
RT-PCR and qRT-PCR
Reverse transcription-polymerase chain reaction (RT-PCR) was performed on staged embryos as described (Blythe et al., 2010), except that non-radioactive nucleotides were used and PCR products were separated via gel electrophoresis on a 2.5% agarose gel and visualized with ethidium bromide. cDNA was synthesized using random primers (Yang et al., 2002).
Quantitative RT-PCR (qRT-PCR) was performed as described (Blythe et al., 2009). For each experiment, individual samples were analyzed in triplicate. Unless otherwise noted, data were analyzed by first normalizing to ODC loading control, and then calculating fold change compared to the indicated control condition using the ΔΔC(t) method (reviewed in (Taneyhill and Adams, 2008)). Embryonic cDNA was synthesized as described above, using random primers. To measure the number of transcripts per embryo, a standard curve of cDNA for each marker was analyzed in parallel with cDNA derived from embryos at different stages. For each primer set, a serial dilution of the plasmids encoding each target gene (104, 103, 102, and 101 copies) was subjected to qPCR in parallel with qRT-PCR for each sample. For the standard curve, C(t) was plotted as a function of copy number, and linear regression was used to determine copy number for a given sample based on the results of the standard curve. The plasmids used in the standard curve were as follows: pBSK-Bix4 (IMAGE clone from Open BioSystems, GenBank: BG022838.1); pCS2-derriere (Sun et al., 1999); pCS2-xnr5 (described above); pBSK-xnr6 (Takahashi et al., 2000); pCS2-chordin (Sasai et al., 1994) pBSK-edd (Sasai et al., 1996).
Immunoprecipitation and Western Blot
For immunoprecipitation, 100–200 embryos per group were lysed and immunoprecipitated with 25 µL polyclonal anti-Smad2 antibody (#sc-6200, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or 25 µL polyclonal anti-Wnt1 antibody (negative control; #sc-6280, Santa Cruz Biotechnology, Santa Cruz, CA, USA) as described (Blythe et al., 2010). Western blots were performed by transferring proteins separated by SDS-PAGE to nitrocellulose, blocking membranes at room temperature for 1 hour in 5% non-fat milk in TBS-T (tris buffered saline with 0.1% tween-20) and immunoblotted with anti-P-Smad2/3 (1:500; #3101, Cell Signaling, Danvers, MA, USA), or anti-Smad2/3 (1:2000; #07–408, Upstate, Billerica, MA, USA) overnight at 4°C in blocking solution. HRP-linked secondary antibody and ECL Plus (GE Healthcare, Bucks, UK) were used to visualize bands. Image J software was used to quantify bands.
In situ hybridization and Antibody Staining
Whole-mount in situ hybridisation (WISH) with digoxygenin labelled probes (Roche, Indianapolis, IN, USA) was carried out as described (Marchal et al., 2009) using the following probes: sox17α (Hudson et al., 1997), Xbra (Smith et al., 1991), goosecoid (Yasuo and Lemaire, 2001), mixer (Henry and Melton, 1998), xnr1 (Jones et al., 1995), xnr5 and xnr6 (Takahashi et al., 2000). To reveal the GFP-Smad2 fusion, we used a rabbit anti-GFP antibody (1:400; Torrey Pines Biolabs, East Orange, NJ, USA,) together with a secondary antibody coupled to Alexa 488 (Molecular Probes, Carlsbad, CA, USA). DAPI (1 µg/ml) was used to stain nuclei.
RESULTS
VegT target genes are transcribed before the midblastula transition
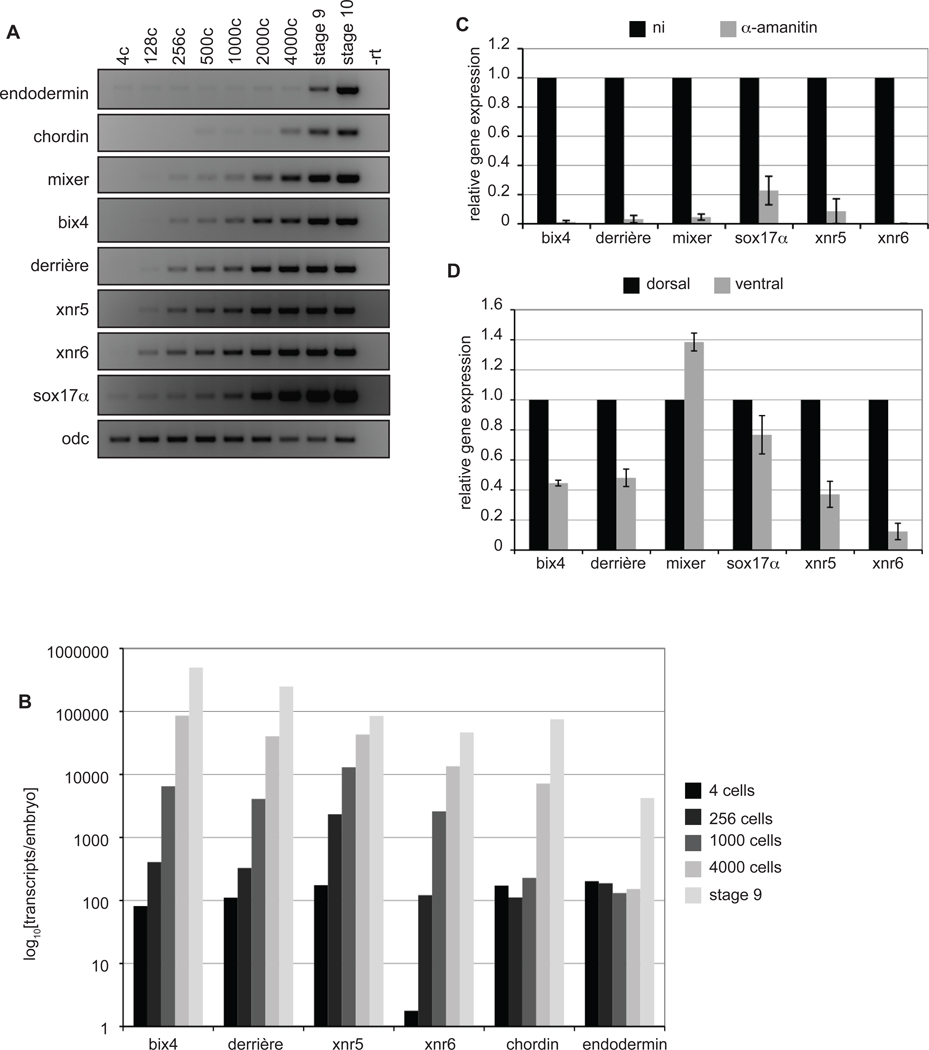
During Xenopus development, the Nodal-related genes xnr5 and xnr6 are expressed before the midblastula transition (MBT) (Blythe et al., 2010; Luxardi et al., 2010; Rosa et al., 2009; Takahashi et al., 2006; Yang et al., 2002). Although the β-catenin-TCF transcription complex, a key modulator of Wnt-dependent transcription, is necessary for this preMBT gene expression (Blythe et al., 2010; Yang et al., 2002), other Wnt target genes, such as siamois, twin, myf5, and engrailed, are not expressed before MBT, suggesting that activity of the β-catenin-TCF complex is not sufficient to drive preMBT transcription. The maternal transcription factor VegT is also required for expression of xnr5 and xnr6; although this has so far only been determined for xnr5 and xnr6 expression after the MBT (Rex et al., 2002; Takahashi et al., 2000; Xanthos et al., 2002), we considered the possibility that VegT could be the maternal factor that determines preMBT expression of these two genes. As VegT directly regulates other mesendodermal genes and can be detected in the nucleus by the 16-cell stage (Stennard et al., 1999), we examined whether other VegT targets are transcribed prior to the MBT in a manner similar to xnr5 and xnr6. We isolated mRNA from multiple pre and post MBT stages and assessed gene expression using RT-PCR. (MBT was defined as the 12th embryonic division/4096-cell stage, as described previously (Newport and Kirschner, 1982a) and was determined experimentally for each clutch of eggs as described in Methods. The VegT targets derrière, bix4, mixer, and sox17α are newly transcribed before the MBT, with the first new transcripts appearing between the 256 and 1000-cell stages (Fig. 1A). In contrast, the direct VegT target genes chordin and endodermin are readily detectable after the 4000-cell stage (MBT).
Fig. 1. Mesendodermal genes transcribed before the MBT.
(A) RT-PCR was performed on embryos collected at the indicated stages. (B) Quantitative RT-PCR was performed on staged embryos and the number of transcripts/embryo was determined as described in Materials and Methods. Log10[transcripts/embryo] was plotted for each gene. (C) α-amanitin (100pg/embryo) was injected into both cells at the 2-cell stage. Injected and non-injected controls were harvested at the 1000-cell stage (preMBT) for qRT-PCR analysis. The results were normalized to expression in the absence of α-amanitin (ni) for each gene. (D) Two ventral cells at the 4-cell stage were injected with rhodamine dextran as a lineage tracer, dissected into dorsal and ventral halves at the 256-cell stage, then harvested at the 1000-cell stage for qRT-PCR. MBT = 4000c; ni, not injected. Error bars represent standard error of the mean for 2 or more experiments.
We also measured the number of mRNA transcripts per embryo for several of these preMBT genes using quantitative reverse transcription-coupled PCR (qRT-PCR) to compare mRNA isolated from staged embryos to a standard curve for each gene (Fig. 1B). We found a baseline of approximately 102 transcripts per embryo at the 4-cell stage for most of the mRNAs, although xnr6 was considerably lower. The number of transcripts for the preMBT genes xnr5, xnr6, derrière, and bix4 increased from 3 to 70-fold from the 4-cell to the 256-cell stage and another 5 to 20-fold from the 256-cell to 1000-cell stage (Fig. 1B). In contrast, chordin and endodermin remained at baseline throughout preMBT stages, with chordin rising sharply at the 4000-cell stage (MBT) and endodermin increasing after the MBT. The number of transcripts/embryo for mixer and sox17α also increased 2 to 5-fold between the 256-cell and 1000-cell stage, although sox17α transcript levels were initially relatively high, even at the 4-cell stage, suggesting a low level maternal contribution to sox17α expression (Howard et al., 2007)(data not shown). To demonstrate that the appearance of these transcripts depends on new transcription, we measured mRNA levels from preMBT embryos with or without α-amanitin, an RNA polymerase II (pol II) inhibitor (Fig. 1C); we found that all preMBT transcripts were greatly reduced in the presence of α-amanitin, although some sox17α remained, presumably representing maternal mRNA. In summary, these data show a rapid, pol II-dependent increase in the number of mRNA transcripts for preMBT genes detectable as early as the 256-cell stage and clearly distinct from typical zygotic genes that are not transcribed until the MBT.
Before the MBT, xnr5 and xnr6 are expressed predominantly in dorsal-vegetal blastomeres (Takahashi et al., 2006; Yang et al., 2002). To test whether dorsal expression is a common feature of other preMBT genes, we dissected 1000-cell embryos into dorsal and ventral halves, isolated RNA, and performed qRT-PCR. Similar to xnr5 and xnr6 expression, bix4 and derrière are preferentially expressed in the dorsal half of the preMBT embryo, whereas new sox17α and mixer transcripts are equally distributed (Fig. 1D). Thus, preMBT transcription is not restricted to dorsal-vegetal cells, consistent with the widespread distribution of maternal VegT protein in the vegetal hemisphere (Horb and Thomsen, 1997).
VegT is required for preMBT gene expression
To determine whether maternal VegT is necessary for preMBT gene expression, we depleted VegT mRNA from oocytes using antisense phosphorothioate oligos and generated VegT-deficient embryos using the host-transfer method (Heasman et al., 2000). Control and VegT-depleted embryos were harvested at the 1000-cell stage (2 cell cycles before the MBT) for qRT-PCR. Expression of bix4, derrière, xnr5, and xnr6 was reduced by 54% – 86% in VegT–depleted embryos (Fig. 2A), demonstrating that maternal VegT is required for the preMBT expression of these genes. sox17α also requires VegT, but was reduced by only 46% in VegT-depleted embryos, reflecting the maternal contribution of sox17α seen in Fig. 1. To test whether VegT is sufficient to induce preMBT gene expression, we expressed VegT in animal pole cells, which normally do not express this vegetally localized RNA. Ectodermal explants were excised at the 1000-cell stage and RNA was isolated for qRT-PCR. As shown in Fig. 2B, VegT induces preMBT transcription of the established VegT target genes bix4, derrière, mixer, sox17α, xnr5, and xnr6 in animal cap explants in a dose-dependent manner. Interestingly, VegT did not induce precocious expression of chordin or endodermin, VegT targets that are normally not expressed until after the MBT (Fig. 2B).
Although expression of xnr5 and xnr6 requires maternal β-catenin as well as VegT, the other preMBT VegT target genes identified here are not dependent on β-catenin. Thus, when β-catenin was depleted with an antisense morpholino oligonucleotide (Heasman et al., 2000), preMBT expression of these VegT target genes was not affected, whereas expression of xnr5 and xnr6 was reduced by 85% and 93%, respectively (Fig. 2C). Together, these data show that VegT is required for preMBT transcription of multiple target genes.
PreMBT expression of xnr5 and xnr6 activates Nodal signaling
Expression of xnr5 and xnr6 in the embryo prior to large-scale ZGA suggests that these Nodal ligands could play an early role in mesendoderm induction. One of the hallmarks of activated Nodal signaling is the presence of phosphorylated Smad2 (Shen, 2007). In previous reports, Smad2 phosphorylation was first detected by immunoblotting in late blastulae (stage 9 to 9.5) (Faure et al., 2000; Lee et al., 2001) and by immunofluoresence at stage 8 (1000-cell stage) (Schohl and Fagotto, 2002). As xnr5 and xnr6 are present prior to the MBT and the Nodal pathway can be activated by ectopic expression of activin prior to the MBT (Lee et al., 2001), we tested whether xnr5 could induce Smad2 phosphorylation prior to the MBT. We injected xnr5 mRNA into embryos and performed Western blot analysis on embryos collected at various time points using an antibody specific for Smad2 phosphorylated at C-terminal serines (P-Smad2). Indeed, expression of exogenous xnr5 mRNA induced P-Smad2 as early as the 128-cell stage (stage 7); in contrast, an equivalent amount of xnr1 mRNA did not induce P-Smad2 until after the MBT (Fig. 3A). Thus the Nodal pathway can function well before MBT and is sensitive to relatively low concentrations of xnr5 mRNA.
As xnr5 and xnr6 are expressed before the MBT, we hypothesized that the endogenous Nodal pathway may also be activated by these ligands before the MBT. To enhance detection of phosphorylated Smad2, we immunoprecipitated Smad2 from Xenopus embryos at different stages of development and then detected P-Smad2 by Western blot. Using this approach, we observed a weak P-Smad2 signal as early as the 1000-cell stage and a stronger signal at the 2000-cell stage (Fig. 3B), indicating that Nodal signaling is initiated at least by the 1000 to 2000-cell stage, consistent with Schohl and Fagotto, who detected P-Smad2 by immunofluoresence at stage 8 (1000-cell stage) (Schohl and Fagotto, 2002). Furthermore, injection of early embryos with the pol-II inhibitor α-amanitin blocked endogenous phosphorylation of Smad2 at the MBT (Fig. 3C), indicating that activation of Nodal signaling requires preMBT transcription. Smad2 phosphorylation can be restored in the presence of α-amanitin by injection of xnr5 mRNA, demonstrating that, aside from the ligand, other essential components of the pathway do not require new transcription before the MBT (Fig. 3C).
The finding that Nodal signaling requires new transcription before the MBT led us to ask if xnr5 and xnr6 are required for this early signaling activity. Targeting xnr5 and xnr6 with antisense morpholino oligonucleotides (previously characterized in (Luxardi et al., 2010)) reduced Smad2 phosphorylation at the MBT by 52% to 74% compared to control MO levels, showing that preMBT expression of xnr5 and xnr6 is required for early activation of Nodal signaling (Fig. 3D). Furthermore, as preMBT xnr5 and xnr6 expression is restricted to dorsal blastomeres, we tested whether P-Smad2 is asymmetrically distributed in early embryos. Embryos were dissected into dorsal and ventral halves, as in Fig. 1D, and P-Smad2 was detected by Western blot. Consistent with the early dorsal distribution of xnr5 and xnr6, the phosphorylation of Smad2 at the MBT is dorsally enriched (Fig. 3E); this observation is also consistent with the immunofluoresence data reported by Schohl and Fagotto (Schohl and Fagotto, 2002), as well as immunostaining and Western blot data in stage 9 embryos reported by others (Faure et al., 2000; Lee et al., 2001). These observations strongly support that Nodal signaling is activated before the MBT and depends on preMBT transcription of Nodal ligands.
Inhibition of Nodal signaling before MBT disrupts mesendoderm induction
Although morpholino oligonucleotides (MOs) offer a powerful approach to knock down gene expression, injection of MOs is only feasible through early cleavage stages. To test the requirement for Nodal signaling at multiple stages from early cleavage through late blastula, we used the small molecule inhibitors SB505124 (SB5) and SB431542 (SB4), which inhibit Nodal signaling by selectively inhibiting the kinase activity of activin receptor-like kinases ALK4, 5, and 7 (DaCosta Byfield et al., 2004; Watabe et al., 2003). These drugs reduce phosphorylation of Smad2 in Xenopus embryos (Luxardi et al., 2010) (Fig. 4A), without affecting Smad1/5/8 (DaCosta Byfield et al., 2004; Inman et al., 2002) (data not shown). When SB5 was applied at the 4-cell stage, embryos failed to gastrulate and by tailbud stage the larvae were severely deformed, lacking axial structures (Fig. 4B) and showing a severe reduction in all mesendodermal markers analyzed (Fig. 4C). Similar morphological defects were observed with SB4 (data not shown). However, in contrast to the marked effects observed when the drug was applied at the 4-cell stage, the severity of the morphological defects declined as the inhibitor was applied at later time points. Although embryos treated at the MBT showed delayed gastrulation and a failure of the blastopore to close completely, consistent with a previous report (Luxardi et al., 2010), the tadpoles nevertheless developed elongated anterior-posterior axes, clear dorsal-ventral polarity, and anterior structures including eyes, brain vesicles, cement gland, dorsal fin, and tail, structures that were not observed when embryos were treated at the 4-cell stage (Fig. 4B). Additionally, the expression of mesendodermal markers was most sensitive to inhibitor added at the 4-cell stage and this effect on mesendodermal gene expression progressively declined as the inhibitor was added at later stages, as analyzed by in situ hybridization and by qRT-PCR (Fig. 4C) on embryos harvested at the onset of gastrulation (stage 10). In contrast, expression of foxI1e, an ectoderm marker that is repressed by Nodal signaling, increased in the presence of the SB5 (see also (Luxardi et al., 2010)), and this increase was similarly more sensitive to the Nodal inhibitor during preMBT stages (Fig. 4C, D). To define further the window of sensitivity to SB5, embryos were placed in medium containing SB5 at the 4-cell stage and then washed and transferred to medium lacking SB5 at the stages indicated in Fig. 4D. Embryos were cultured until the onset of gastrulation and mesendodemal marker expression was analyzed by in situ hybridization and qRT-PCR (Fig. 4D). These data show minimal effect on mesendodermal gene expression when inhibitor is removed during early cleavage stages (32-cell stage) but marked reduction when embryos are removed from SB5 at the preMBT blastula stage (1000-cell).
Important concerns with these experiments are that the SB5 compound may enter cells slowly, requiring prolonged incubation for effective inhibition of Nodal signaling, or that the access of the inhibitor may differ at different developmental stages. To test the “lag time" between addition of SB5 and inhibition of Smad2 phosphorylation, we added SB5 to embryos at the 128-cell (cleavage stage) or the 1000-cell (preMBT blastula) stage and then assessed Smad2/3 phosphorylation at multiple time points after addition of drug (Fig. 5). For the 128-cell group, we expressed xnr5 to enhance detection of Smad2 phosphorylation. When SB5 was added at the 128-cell stage, there was no further increase in Smad2 phosphorylation at the 512-cell or later stages (Fig. 5A). To assess the efficacy of SB5 added in the blastula stage, we examined Smad2 phosphorylation induced by endogenous Nodal signaling, using immunoprecipitation followed by immunoblot, as in Fig. 3B–E. SB5 added to 1000-cell blastulae markedly reduced further Smad2 phosphorylation in post-MBT embryos (Fig. 5B), although a small increase in endogenous phosphorylation was still detectable. These data demonstrate that SB5 readily enters embryonic cells and can inhibit Nodal signaling when added at cleavage or blastula stages.
Fig. 5. Rapid inhibition of Nodal signaling by SB5.
(A) xnr5 mRNA (20 pg) was injected at the 2-cell stage. SB5 (200uM) or vehicle (0.2% DMSO) was added at the 128 cell-stage and embryos were collected for immunoblot at the indicated stages. The left panels are immunoblots with an antibody specific for phospho-Smad2/3 and the right panels show total Smad2/3 as a loading control. Double arrowheads indicate the Smad2 and Smad3 bands. (B) SB5 or vehicle was added to uninjected embryos at the 1000-cell stage and embryos were collected at the indicated times for detection of phospho-Smad2 by immunoprecipitation with anti-Smad2 antibodies followed by immunoblot with either the phospho-Smad2 specific antibody (left panel) or an antibody for total Smad2/3 (right panel). +20, +40, +60 and +80 indicate minutes after the MBT.
These data suggest that Nodal signaling before the MBT plays an important role in mesendodermal development. To test this further, we used a hormone-inducible, activated form of Smad2 fused to the glucocorticoid receptor ligand-binding domain (GR-tSmad2) to rescue the mesendodermal gene expression in the presence of the Nodal inhibitor. Embryos injected with GR-tSmad2 mRNA were treated with SB5 from the 4-cell stage. GR-tSmad2 was then activated by addition of dexamethasone (dex) either before (stage 7) or after the MBT (stage 9). (We have shown previously that dex rapidly enters the embryo, as the lag between dex addition and translation of a detectable amount of protein is 90 minutes in Xenopus embryos (Yang et al., 2002)). Mesendodermal gene expression was then measured at stage 10 by in situ hybridization (Fig. 6A) and qRT-PCR (Fig. 6B). SB5 treatment alone markedly reduced mesendodermal gene expression, as in Fig. 4. Activation of GR-tSmad2 by addition of dex before the MBT (stage 7) restored mesendodermal gene expression to nearly 80% of untreated control embryos. Activation of Smad2 after the MBT (stage 9) was ineffective in restoring mesendodermal gene expression. This shows that preMBT Nodal signaling, stimulated using downstream canonical components, can contribute significantly to the restoration of mesendodermal gene expression.
xnr5 and xnr6 enhance their own expression through a positive-feedback mechanism in the early embryo
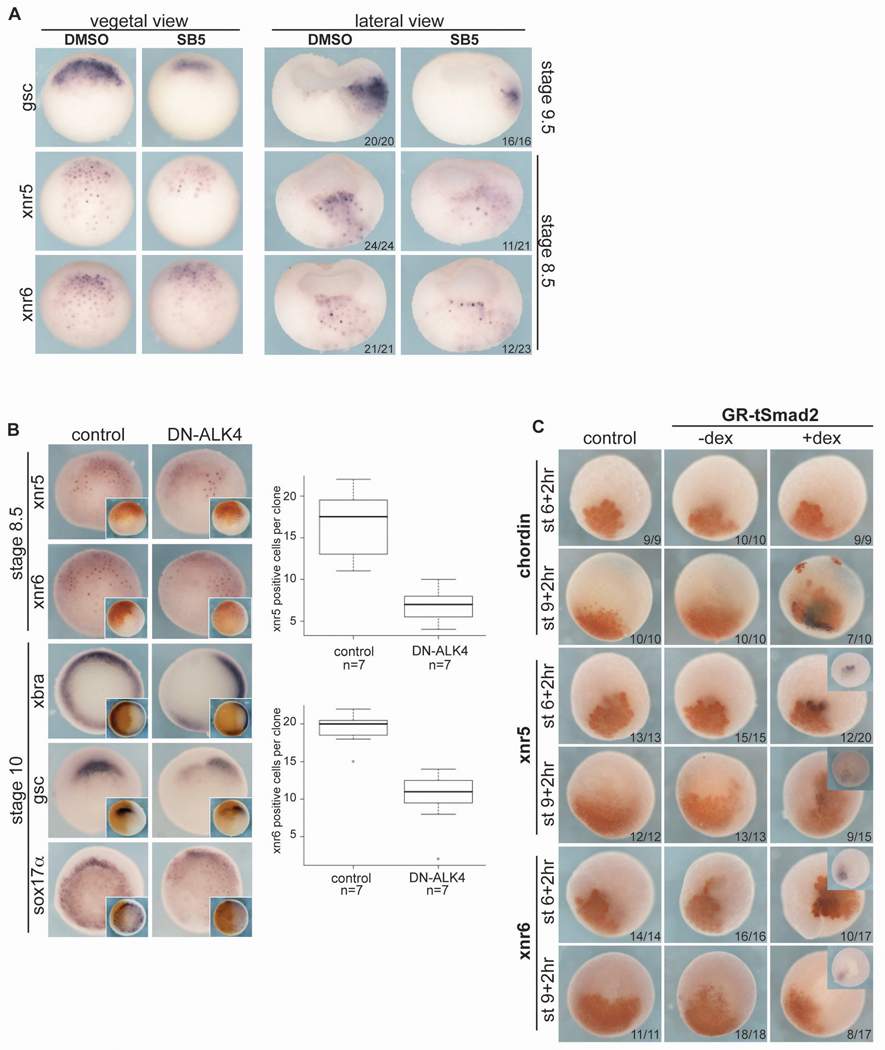
Nodals are known to activate their own expression as part of a positive feedback loop (Osada et al., 2000; Shen, 2007), and we therefore reasoned that preMBT Nodals may utilize a similar amplification system. We first tested the effects of inhibition of Nodal signaling on the expression of xnr5 and xnr6 mRNA. We applied SB5 to embryos at the 4-cell stage and then assayed gene expression at later stages (as indicated in Fig. 7) by in situ hybridization and qPCR. SB5 reduced the number of xnr5 and xnr6 positive cells at stage 8.5 detected by in situ hybridization (Fig. 7A). (SB5 did not affect expression of the preMBT VegT target genes bix4, mixer, sox17a, or derriere; data not shown). We note that SB5 did not completely eliminate xnr5 and xnr6 expression, perhaps because their expression is also dependent on VegT and b-catenin, which are not inhibited by SB5. To confirm Nodal regulation of xnr5 and xnr6 expression, we used the dominant negative Nodal receptor DN-ALK4 to interfere with Nodal signaling and examined gene expression at MBT (stage 8.5) and in the gastrula (stage 10.5). When DN-ALK4 mRNA was injected along with a lineage tracer unilaterally into 4-cell embryos, expression of xnr5 and xnr6 was reduced in the injected half, along with the Nodal target genes xbra, goosecoid, and sox17α (Fig. 7B). Conversely, expression of GR-tSmad2 in animal pole cells induced preMBT expression of xnr5 and xnr6, whereas chordin (chd) was not induced until after the MBT (Fig. 7C). These data taken together imply that Nodal signaling can stimulate expression of xnr5 and xnr6 in the early embryo.
Fig. 7. Nodal signaling contributes to xnr5/6 expression.
(A) SB5 (200 µM) was added at the 4-cell stage and embryos were fixed for ISH at MBT (stage 8.5). Whole-embryos were photographed in vegetal views, dorsal to the top (vegetal view). Embryos bisected along the dorsal/ventral axis prior to ISH are shown with dorsal side to the right (lateral view). The frequency of the expression pattern for each group is indicated in the lower right corner. DMSO was used as a control. (B) To inhibit Nodal signaling, the dominant-negative Nodal receptor (DN-ALK4) was injected (1 ng), along with fluorescent dextran (insets, rust-colored staining), into the 2 right-hand blastomeres of 4-cell embryos. Embryos were fixed at the indicated times and analyzed by ISH. Fluorescent dextran injection alone is shown as control. The number of cells expressing xnr5 or xnr6 in injected clones was counted and significance was determined using the Kruskal-Wallis test. (C) 8-cell embryos were injected in one animal cell with GR-tSmad2 (400 pg) and fluorescent dextran. GR-tSmad2 was activated by addition of dex for a two-hour window beginning at the indicated stages. Embryos were fixed at the end of the window and analyzed by ISH.
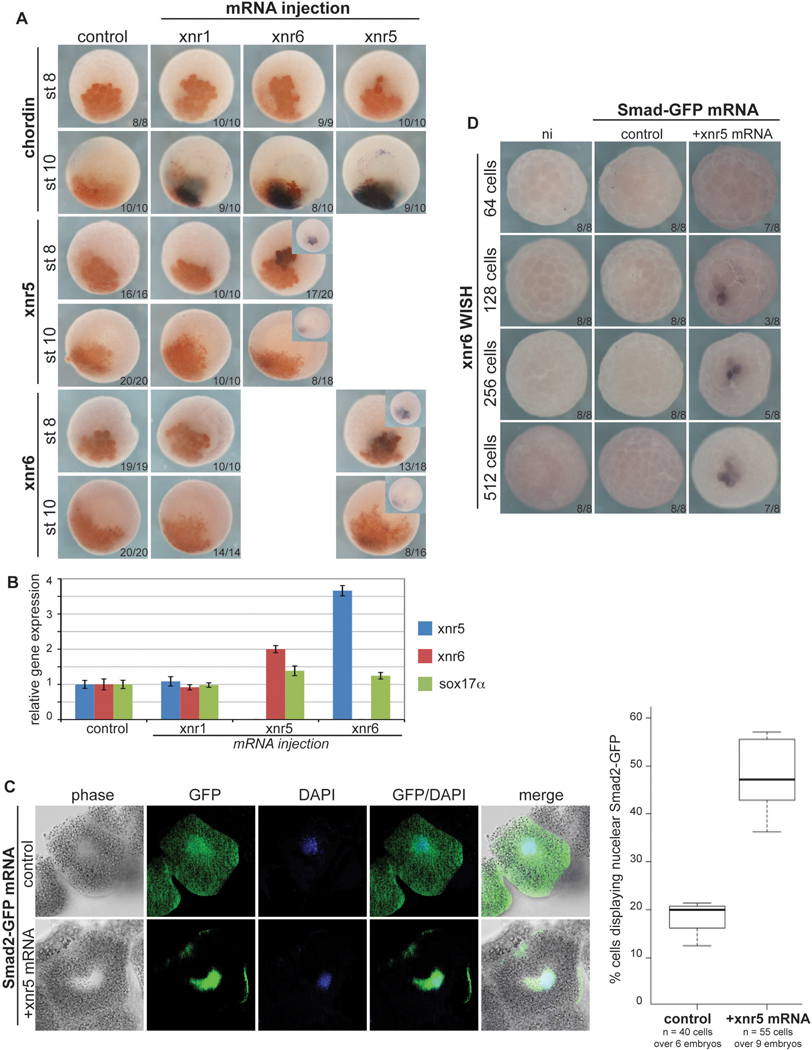
To test whether xnr5 and xnr6 can induce their own expression prior to the MBT, we overexpressed either xnr5 or xnr6 mRNA in the embryo and then assayed for the other by in situ hybridization and qPCR. Overexpression of xnr5 in animal pole cells induced ectopic expression of xnr6 but not chd before the MBT (Fig. 8A). Similarly, xnr6 overexpression induced xnr5 but not chd expression. In contrast, overexpression of xnr1 at a level that was sufficient to induce chd expression did not induce xnr5 and xnr6. When expression levels were measured with qRT-PCR before MBT (stage 8), xnr5 mRNA induced xnr6 expression 2-fold, and xnr6 induced xnr5 3.5-fold, (Fig. 8B). We then tested whether ectopic Nodal activity involved nuclear translocation of Smad2 prior to the MBT. For this, we injected GFP-tagged Smad2 mRNA into animal pole cells with or without xnr5 mRNA. As anticipated, we observed consistent nuclear translocation of Smad2-GFP at preMBT stages in response to xnr5 (Fig. 8C). We used xnr6 ectopic activation as a positive control in this experiment; while Smad2-GFP was insufficient to induce xnr6 expression by itself, we could detect xnr6 activation as early as the 128-cell stage in response to xnr5 (Fig. 8D). These data suggest that a positive feedback loop in the early embryo enhances xnr5 and xnr6 expression through activation of Smad2.
Fig. 8. xnr5 and xnr6 can activate Smad2 and regulate their own expression before the MBT.
(A) 8-cell embryos were injected in one animal cell with fluorescent dextran alone (control) or with xnr1 (50 pg), xnr5 (20pg), or xnr6 (20 pg) mRNAs. Whole mount ISH for xnr5, xnr6, and chd was performed on embryos fixed at the indicated stages. (B) Embryos from A were analyzed by qRT-PCR at stage 8 (preMBT). (C) 8-cell embryos were injected in one animal cell with GFP-Smad2 (200pg) alone or with xnr5 mRNA (50pg). Embryos were fixed at the 512-cell stage, processed for anti-GFP immunostaining, and sectioned for fluorescence imaging. DAPI was used to reveal nuclei. The graph shows the percent of DAPI positive nuclei that also contain GFP-Smad2. (D) Embryos injected as in C were fixed at various stages as indicated and stained for xnr6 expression. ui, uninjected.
DISCUSSION
Essential preMBT functions for zygotic mRNAs
Nodal signaling is required in vertebrates for the establishment and patterning of mesoderm and endoderm. In this work, we find that the maternal T-box transcription factor VegT is required for the preMBT transcription of a group of genes involved in mesendoderm development, including the Nodal genes xnr5 and xnr6; we also show that Nodal signaling in Xenopus embryos is initiated before the MBT and that preMBT transcription of xnr5 and xnr6 is essential for this activity. Furthermore, we show that xnr5 and xnr6 are themselves targets of Nodal signaling and can enhance their own expression, providing robust activation of postMBT Nodal signaling and mesendoderm induction. These findings establish a critical function for zygotic transcription before the MBT in Xenopus embryos.
How preMBT genes bypass early repression is not clear. In Drosophila, most preMBT genes contain a cis-regulatory heptad repeat that binds BSF, Grainyhead, and the zinc finger transcription factor Zelda, which is essential for their preMBT expression (De Renzis et al., 2007; Harrison et al., 2010; Liang et al., 2008). A similar zinc finger protein has not been identified in Xenopus, but the preMBT genes so far identified are regulated by the maternal transcription factor VegT. The presence of maternal transcription factors such as Zelda, Grainyhead and VegT that are required to activate multiple preMBT genes suggests more generally that preMBT gene expression represents a regulated and biologically significant phenomenon. However, whether the early transcription of specific preMBT genes is critical for development has not previously been addressed. Thus, in this work we targeted specific preMBT Nodal transcripts, restored Smad2 signaling conditionally at different developmental stages, and found that Nodal expression and signaling before the MBT is essential for mesendoderm development.
PreMBT transcription is required for early Nodal/Smad2 signaling
In previous work, Smad2 phosphorylation and nuclear localization were first detected in stage 9 blastulae (Faure et al., 2000; Lee et al., 2001; Saka et al., 2007), whereas others detected P-Smad2 by stage 8/1000-cell stage (Schohl and Fagotto, 2002). Similar to Faure et al and Lee et al, we show that activation of Nodal signaling requires zygotic transcription. Furthermore, we show that xnr5 and xnr6 transcribed before the MBT are required for early activation of this pathway, as they are for phosphorylation of Smad2 in the gastrula stage (Luxardi et al., 2010). Detection of phosphorylated Smad2 in the embryo is limited by the sensitivity of the antibody and the detection method; by using a different antibody and immunoprecipitating Smad2, we have enhanced the sensitivity to detect Smad2 phosphorylation as early as the 1000-cell stage, and it is likely that Smad2 is phosphorylated earlier. Schohl and Fagotto speculated that early P-Smad2 is activated by a maternal TGF-β/Nodal signal (Schohl and Fagotto, 2002), but their findings are also consistent with the zygotic expression and activity of xnr5 and xnr6 before the MBT reported here, in addition to a contribution from Vg1 (Birsoy et al., 2006), a maternal TGF-β-related ligand (Weeks and Melton, 1987).
Significance of preMBT transcription for germ layer specification
The importance of preMBT transcription mediated by VegT may reflect the fact that VegT activates a gene regulatory network that is required for establishment of the primary germ layers, one of the earliest cell fate decisions in vertebrate embryos. While the expression of mesoderm and endoderm markers begins, for the most part, after the MBT, earlier Nodal signaling, along with the positive autoregulation through xnr5 and xnr6 reported here, may be required to initiate the postMBT phase of Nodal signaling (xnr1, 2, and 4) and mesendoderm induction. This conclusion is consistent with prior work showing that Nodal/TGF-β signaling is required for the initiation of xnr1, 2, and 4 expression after the MBT (Agius et al., 2000; Luxardi et al., 2010). Agius et al proposed Vg1 as a candidate molecule for this signal, but their data are also consistent with the preMBT activity of xnr5 and xnr6 reported here.
Furthermore, Jones and Woodland reported that mesoderm-inducing signals are present by stage 6 to 6.5, well before the MBT (Jones and Woodland, 1986). Similarly, Ding and colleagues detected a mesoderm-inducing signal between the 16-cell and 128-cell stage (Ding et al., 1998), and Wylie et al reported a weak ventral mesoderm inducing activity present before the MBT, although they also reported that dorsal mesoderm inducing activity was not detectable (in Nieuwkoop recombinants) until after the MBT (Wylie et al., 1996). Jones and Woodland concluded that the mesoderm-inducing signal they detected was most likely encoded by a maternal gene. However, subsequent molecular work showed that VegT-dependent zygotic expression of Nodal-like ligands is required for mesendodermal induction (Agius et al., 2000; Kofron et al., 1999; Zhang et al., 1998). Our data, taken together with evidence for the role of maternal Vg1 in mesoderm induction (Birsoy et al., 2006), could help to reconcile these findings, as we have shown that the zygotic transcription of xnr5 and xnr6 begins by stage 6.5 (64 cells)(Yang et al., 2002) and their preMBT expression is required for Smad2 phosphorylation and mesendoderm induction.
Nodal autoregulation and dorsal-ventral patterning
xnrs 1, 2, 4, 5 and 6 are expressed initially at higher levels in dorsal cells, and Smad2 phosphorylation is also initially enriched in dorsal cells (Lee et al., 2001)(Fig. 3E). Several groups have proposed that dorsal Nodal expression is required for development of the Spemann organizer and expression of organizer specific genes (reviewed in (Whitman, 2001)). Furthermore, Agius et al proposed that VegT and a maternal Nodal-like signal such as Vg1 may synergize with the dorsal specifying activity of the Wnt/β-catenin pathway to generate a zygotic gradient of xnrs in blastula-stage endoderm (Agius et al., 2000). Similarly, Lee et al proposed that the initial dorsal enrichment of Smad2 phosphorylation arises through cooperation between VegT and β-catenin (Lee et al., 2001). Our data strongly support these ideas, as we show that VegT and β-catenin directly activate preMBT expression of xnr5 and xnr6 in a spatially restricted manner in the dorsal vegetal cells known as the Nieuwkoop center and that preMBT transcription is also required for the dorsally enriched phosphorylation of Smad2. These observations are also consistent with a two-step model for mesoderm induction, wherein a low level of Nodal signaling initiates mesoderm induction before the MBT, contributing to a higher level after the MBT (Kimelman and Griffin, 1998; Takahashi et al., 2000; Yasuo and Lemaire, 1999), except that we propose that preMBT zygotic Nodals contribute significantly to the initial mesendoderm-inducing signal. In this framework, the level of Smad2 activation in early embryos, representing contributions from both preMBT and postMBT signaling, would determine the extent of mesendoderm induction.
We found that the Nodal related gene derrière, like xnr5 and xnr6, is also dorsally enriched before the MBT (see also (Eimon and Harland, 2002)), and yet, unlike xnr5 and xnr6, derrière does not appear to be dependent on maternal Wnt/β-catenin signaling; the mechanisms that confer dorsal enrichment of derrière transcripts remain to be determined.
Nodal signaling is known to autoregulate in other contexts, including postMBT mesoderm induction (reviewed in (Shen, 2007). In frog, Nodal signaling activates postMBT expression of xnr1, 2, and 4 (Osada et al., 2000; Rex et al., 2002; Takahashi et al., 2000), but autoregulation had not previously been shown for xnr5 or xnr6. A positive feedback loop involving xnr5 and xnr6 is attractive, as it would contribute to reaching optimal levels of Nodal ligands in cells undergoing mesendodermal induction at the onset of global zygotic gene expression.
Multiple roles for preMBT transcription
PreMBT expression of Nodals and the VegT-induced transcription factors bix4, mixer, and sox17α may serve to segregate distinct functions of Nodal signaling in the early embryo. Previous work showed that early signaling through xnr5 and xnr6 contributes to mesendoderm induction whereas later Nodal signaling through xnr1 and xnr2 activates movement-effector genes involved in gastrulation (Luxardi et al., 2010). Similarly, the Wnt/β-catenin pathway has distinct functions during the blastula stage, and these must be separated temporally in the early embryo (Blythe et al., 2010). Dorsal specifying genes such as siamois and twin are first expressed after the MBT; however, these genes require Wnt/β-catenin signaling during early cleavage stages and become insensitive to the pathway by the MBT (Christian and Moon, 1993; Darken and Wilson, 2001). In this context, dorsal Wnt target genes are primed for expression at the MBT by β-catenin dependent chromatin modifications conferred on promoters during earlier stages of development (Blythe et al., 2010; Kimelman, 2010). Thus maternal Wnt signaling marks dorsal organizer genes before MBT for expression at the MBT. Through this mechanism, organizer genes are protected from postMBT Wnt signaling, which is required for ventral and posterior patterning.
Taken together, these findings demonstrate at least two preMBT functions for maternal transcription factors: Maternal Wnt/β-catenin signaling is required before the MBT to preset organizer genes for later expression at the MBT, whereas VegT directly activates preMBT transcription of genes required for mesendodermal induction. Both mechanisms confer critical temporal control on pathways that set up the earliest cell fate decisions in the embryo, dorsal-ventral specification and primary germ layer induction.
Interestingly, these two mechanisms overlap in the regulation of xnr5 and xnr6, as the initial, early expression of these genes in the Nieuwkoop center depends on both Wnt/β-catenin and VegT function. How VegT targets escape global transcriptional repression in the preMBT embryo remains unclear and is a topic for future study. Whereas Wnt target genes are poised for expression before the MBT, VegT may recruit transcription elongation factors to complete transcription of these target genes. However, VegT by itself is clearly not sufficient to confer preMBT transcription, as at least two direct targets of VegT are not transcribed until after the MBT; perhaps activation by VegT in conjunction with loss of repression by an unidentified factor is required to confer gene expression before the MBT. Future work will investigate what distinguishes pre and postMBT VegT targets and address whether additional maternal transcription factors regulate preMBT gene expression.
Research highlights.
Zygotic gene expression before MBT is required for development in Xenopus laevis
Multiple genes required for mesendoderm induction are activated by VegT before MBT
Zygotic gene expression before MBT is required for early Nodal signaling
preMBT Nodal activation is essential for mesendoderm induction
ACKNOWLEDGEMENTS
We extend special thanks to the late Tom Kadesch for advice and comments. We also thank Daniel Kessler and Shelby Blythe for comments on the manuscript, and Mary Mullins, Jean-Pierre Saint-Jeannet, and members of the Klein lab for helpful discussions. We thank Makoto Asashima, Chenbei Chang, Eddy De Robertis, Dan Kessler, and Hazel Sive for providing plasmids. This work was supported by a grant from the NIH (1R01GM076621) to PSK, the Cell and Molecular Biology Training Grant (T32-GM07229) to JS, and a grant from the French National Research Agency (ANR) to LK. GL was supported by the French Ministry of Research and the Association Against Cancer (ARC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agius E, Oelgeschläger M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–1183. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs K-J, Stunnenberg HG, Veenstra GJC. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andeol Y. Early transcription in different animal species: implication for transition from maternal to zygotic control in development. Roux's Arch Dev Biol. 1994;204:3–10. doi: 10.1007/BF00189062. [DOI] [PubMed] [Google Scholar]
- Bachvarova R, Davidson EH, Allfrey VG, Mirsky AE. Activation of RNA synthesis associated with gastrulation. Proc Natl Acad Sci U S A. 1966;55:358–365. doi: 10.1073/pnas.55.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy B, Kofron M, Schaible K, Wylie C, Heasman J. Vg 1 is an essential signaling molecule in Xenopus development. Development. 2006;133:15–20. doi: 10.1242/dev.02144. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Cha S-W, Tadjuidje E, Heasman J, Klein PS. β-catenin Primes Organizer Gene Expression By Recruiting a Histone H3 Arginine 8 Methyltransferase, Prmt2. Developmental Cell. 2010;19:1–12. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian JL, Moon RT. Interactions between Xwnt-8 and Spemann organizer signaling pathways generate dorsoventral pattern in the embryonic mesoderm of Xenopus. Genes Dev. 1993;7:13–28. doi: 10.1101/gad.7.1.13. [DOI] [PubMed] [Google Scholar]
- Clegg KB, Pikó L. RNA synthesis and cytoplasmic polyadenylation in the one-cell mouse embryo. Nature. 1982;295:343–344. doi: 10.1038/295342a0. [DOI] [PubMed] [Google Scholar]
- DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65:744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- Darken RS, Wilson PA. Axis induction by wnt signaling: Target promoter responsiveness regulates competence. Dev Biol. 2001;234:42–54. doi: 10.1006/dbio.2001.0253. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Gene activity in early development. 3 ed. Academic Press; 1986. [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Hausen P, Steinbeisser H. Pre-MBT patterning of early gene regulation in Xenopus: the role of the cortical rotation and mesoderm induction. Mech Dev. 1998;70:15–24. doi: 10.1016/s0925-4773(97)00163-9. [DOI] [PubMed] [Google Scholar]
- Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135:1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Eimon PM, Harland RM. Effects of heterodimerization and proteolytic processing on Derrière and Nodal activity: implications for mesoderm induction in Xenopus. Development. 2002;129:3089–3103. doi: 10.1242/dev.129.13.3089. [DOI] [PubMed] [Google Scholar]
- Faure S, Lee MA, Keller T, ten Dijke P, Whitman M. Endogenous patterns of TGFbeta superfamily signaling during early Xenopus development. Development. 2000;127:2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Developmental biology. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Heasman J, Wessely O, Langland R, Craig EJ, Kessler DS. Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Dev Biol. 2001;240:377–386. doi: 10.1006/dbio.2001.0495. [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–96. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. A vegetally localized T-box transcription factor in Xenopus eggs specifies mesoderm and endoderm and is essential for embryonic mesoderm formation. Development. 1997;124:1689–1698. doi: 10.1242/dev.124.9.1689. [DOI] [PubMed] [Google Scholar]
- Houston DW, Kofron M, Resnik E, Langland R, Destree O, Wylie C, Heasman J. Repression of organizer genes in dorsal and ventral Xenopus cells mediated by maternal XTcf3. Development. 2002;129:4015–4025. doi: 10.1242/dev.129.17.4015. [DOI] [PubMed] [Google Scholar]
- Howard L, Rex M, Clements D, Woodland HR. Regulation of the Xenopus Xsox17alpha(1) promoter by co-operating VegT and Sox17 sites. Developmental biology. 2007;310:402–415. doi: 10.1016/j.ydbio.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolás FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CV. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development. 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- Jones EA, Woodland HR. Development of the ectoderm in Xenopus: tissue specification and the role of cell association and division. Cell. 1986;44:345–355. doi: 10.1016/0092-8674(86)90769-5. [DOI] [PubMed] [Google Scholar]
- Kimelman D. On the fast track to organizer gene expression. Dev Cell. 2010;19:190–192. doi: 10.1016/j.devcel.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Griffin KJ. Mesoderm induction: a postmodern view. Cell. 1998;94:419–421. doi: 10.1016/s0092-8674(00)81582-2. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Kofron M, Demel T, Xanthos J, Lohr J, Sun B, Sive H, Osada S, Wright C, Wylie C, Heasman J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development. 1999;126:5759–5770. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- Kofron M, Wylie C, Heasman J. The role of Mixer in patterning the early Xenopus embryo. Development. 2004;131:2431–2441. doi: 10.1242/dev.01132. [DOI] [PubMed] [Google Scholar]
- Lee MA, Heasman J, Whitman M. Timing of endogenous activin-like signals and regional specification of the Xenopus embryo. Development. 2001;128:2939–2952. doi: 10.1242/dev.128.15.2939. [DOI] [PubMed] [Google Scholar]
- Leung T, Bischof J, Söll I, Niessing D, Zhang D, Ma J, Jäckle H, Driever W. bozozok directly represses bmp2b transcription and mediates the earliest dorsoventral asymmetry of bmp2b expression in zebrafish. Development. 2003;130:3639–3649. doi: 10.1242/dev.00558. [DOI] [PubMed] [Google Scholar]
- Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxardi G, Marchal L, Thome V, Kodjabachian L. Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway. Development. 2010;137:417–426. doi: 10.1242/dev.039735. [DOI] [PubMed] [Google Scholar]
- Marchal L, Luxardi G, Thome V, Kodjabachian L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc Natl Acad Sci U S A. 2009;106:17437–17442. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S, Lee SGP, Mak A, Miller LD, Murthy KRK, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakura N, Miura T, Yamana K, Ito A, Shiokawa K. Synthesis of heterogeneous mRNA-like RNA and low-molecular-weight RNA before the midblastula transition in embryos of Xenopus laevis. Developmental biology. 1987;123:421–429. doi: 10.1016/0012-1606(87)90400-3. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P, Faber J. Normal Table of Xenopus laevis (Daudin) New York: Garland Publishing Inc.; 1994. [Google Scholar]
- Osada SI, Saijoh Y, Frisch A, Yeo CY, Adachi H, Watanabe M, Whitman M, Hamada H, Wright CV. Activin/nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development. 2000;127:2503–2514. doi: 10.1242/dev.127.11.2503. [DOI] [PubMed] [Google Scholar]
- Porcher A, Abu-Arish A, Huart S, Roelens B, Fradin C, Dostatni N. The time to measure positional information: maternal Hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development. 2010;137:2795–2804. doi: 10.1242/dev.051300. [DOI] [PubMed] [Google Scholar]
- Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes & Development. 1996;10:1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- Rex M, Hilton E, Old R. Multiple interactions between maternally-activated signalling pathways control Xenopus nodal-related genes. Int J Dev Biol. 2002;46:217–226. [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Saka Y, Hagemann AI, Piepenburg O, Smith JC. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007;134:4209–4218. doi: 10.1242/dev.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath K, Cheng AM, Frisch A, Wright CV. Functional differences among Xenopus nodal-related genes in left-right axis determination. Development. 1997;124:3293–3302. doi: 10.1242/dev.124.17.3293. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Piccolo S, De Robertis EM. Endoderm induction by the organizer-secreted factors chordin and noggin in Xenopus animal caps. EMBO J. 1996;15:4547–4555. [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schohl A, Fagotto F. Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development. 2002;129:37–52. doi: 10.1242/dev.129.1.37. [DOI] [PubMed] [Google Scholar]
- Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Sible JC, Anderson JA, Lewellyn AL, Maller JL. Zygotic transcription is required to block a maternal program of apoptosis in Xenopus embryos. Dev Biol. 1997;189:335–346. doi: 10.1006/dbio.1997.8683. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Animal Cap Isolation from Xenopus laevis. Cold Spring Harb Protoc. 2007;2007 pdb.prot4744. [Google Scholar]
- Smith JC, Price BM, Green JB, Weigel D, Herrmann BG. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Stennard F, Zorn AM, Ryan K, Garrett N, Gurdon JB. Differential expression of VegT and Antipodean protein isoforms in Xenopus. Mech Dev. 1999;86:87–98. doi: 10.1016/s0925-4773(99)00119-7. [DOI] [PubMed] [Google Scholar]
- Sun BI, Bush SM, Collins-Racie LA, LaVallie ER, DiBlasio-Smith EA, Wolfman NM, McCoy JM, Sive HL. derrière: a TGF-beta family member required for posterior development in Xenopus. Development. 1999;126:1467–1482. doi: 10.1242/dev.126.7.1467. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Onuma Y, Yokota C, Westmoreland JJ, Asashima M, Wright CVE. Nodal-related gene Xnr5 is amplified in the Xenopus genome. Genesis. 2006;44:309–321. doi: 10.1002/dvg.20217. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto J, Asashima M. Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development. 2000;127:5319–5329. doi: 10.1242/dev.127.24.5319. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Adams MS. Investigating regulatory factors and their DNA binding affinities through real time quantitative PCR (RT-QPCR) and chromatin immunoprecipitation (ChIP) assays. Methods Cell Biol. 2008;87:367–389. doi: 10.1016/S0091-679X(08)00219-7. [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Ning X, Klingler M, Kramer SG, Gergen JP. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ. Early Embryonic Gene Transcription in Xenopus. Adv Dev Biol Biochem. 2002;12:85–105. [Google Scholar]
- Watabe T, Nishihara A, Mishima K, Yamashita J, Shimizu K, Miyazawa K, Nishikawa S-I, Miyazono K. TGF-beta receptor kinase inhibitor enhances growth and integrity of embryonic stem cell-derived endothelial cells. J Cell Biol. 2003;163:1303–1311. doi: 10.1083/jcb.200305147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DL, Melton DA. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell. 1987;51:861–867. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]
- Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Maternal beta-catenin establishes a 'dorsal signal' in early Xenopus embryos. Development. 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Tao Q, Schaible K, Wylie C, Heasman J. The roles of three signaling pathways in the formation and function of the Spemann Organizer. Development. 2002;129:4027–4043. doi: 10.1242/dev.129.17.4027. [DOI] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Wylie C, Heasman J. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development. 2001;128:167–180. doi: 10.1242/dev.128.2.167. [DOI] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Lemaire P. A two-step model for the fate determination of presumptive endodermal blastomeres in Xenopus embryos. Curr Biol. 1999;9:869–879. doi: 10.1016/s0960-9822(99)80391-1. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Lemaire P. Role of Goosecoid, Xnot and Wnt antagonists in the maintenance of the notochord genetic programme in Xenopus gastrulae. Development. 2001;128:3783–3793. doi: 10.1242/dev.128.19.3783. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Molecular basis of vertebrate endoderm development. Int Rev Cytol. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]