Abstract
Vertebrate Hox-clusters contain protein-coding genes that regulate body axis development and microRNA (miRNA) genes whose functions are not yet well understood. We over-expressed the Hox-cluster microRNA miR-196 in zebrafish embryos and found four specific, viable phenotypes: failure of pectoral fin bud initiation, deletion of the 6th pharyngeal arch, homeotic aberration and loss of rostral vertebrae, and reduced number of ribs and somites. Reciprocally, miR-196 knockdown evoked an extra pharyngeal arch, extra ribs, and extra somites, confirming endogenous roles of miR-196. miR-196 injection altered expression of hox genes and the signaling of retinoic acid through the retinoic acid receptor gene rarab. Knocking down rarab mimicked the pectoral fin phenotype of miR-196 over-expression, and reporter constructs tested in tissue culture and in embryos showed that the rarab 3’UTR is a miR-196 target for pectoral fin bud initiation. These results show that a Hox-cluster microRNA modulates development of axial patterning similar to nearby protein-coding Hox genes, and acts on appendicular patterning at least in part by modulating retinoic acid signaling.
Keywords: microRNA (miRNA), Mir196 (miR-196), axial skeletal patterning, pectoral fin, pharyngeal arch, retinoic acid
INTRODUCTION
Hox-cluster genes control animal body patterning in radiata and in bilateria, including both protosotomes and deuterostomes (Finnerty et al., 2004; Postlethwait and Schneiderman, 1969; Wellik, 2009). In vertebrate deuterostomes, Hox-cluster genes control the anterior-posterior body axis, including the identity of vertebrae and pharyngeal arches and the axes of body appendages (Krumlauf, 1994), and they are important for the development of mesodermal organ systems (Di-Poi et al., 2010). Hox clusters evolved by tandem gene duplication followed by whole genome duplication events in vertebrates that provided tetrapods with four Hox clusters and most teleost fish with seven or eight (Amores et al., 1998; Amores et al., 2004; Chambers et al., 2009; Gehring et al., 2009; Graham et al., 1989; Woltering and Durston, 2006).
Hox genes are expressed in a collinear fashion along the anterior-posterior body axis during early development, with genes located 3’ in the cluster controlling anterior development and those located 5’ regulating more posterior organ development (Duboule and Morata, 1994; Graham et al., 1989); as a result, Hox gene mutations can delete vertebrae or transform vertebral identity and remove or reduce limb skeletal elements (Chen and Capecchi, 1997; Davis et al., 1995). Hox genes act by controlling downstream transcription factors that regulate signaling events controlling body segmentation and organ initiation. Some Hox genes are themselves directly regulated by the extracellular signaler retinoic acid (RA), which controls axis and pectoral appendage development (Grandel et al., 2002; Hoffman et al., 2002; Nolte et al., 2003).
Bilaterian Hox clusters contain protein-coding genes and genes encoding microRNAs (miRNAs), small non-coding RNAs that generally bind to 3’ untranslated regions (UTRs) of messenger RNAs and regulate their stability or translation (Fjose and Zhao, 2010; Vella et al., 2004). The human genome has three Hox-cluster miRNA genes, MIR10, MIR196, and MIR615. The MIR10 gene is broadly distributed among bilaterians; MIR196 is conserved among vertebrates; and MIR615 is restricted to mammalian genomes (see miRBase collection at http://www.miRBase.org (Griffiths-Jones et al., 2008; Yekta et al., 2008)). In zebrafish, the hoxdb cluster lost all of its protein-coding genes (Amores et al., 1998), but surprisingly, retained mir10 (Woltering and Durston, 2006). The similarity of Hox-cluster miRNA expression patterns to those of nearby hox genes suggested that Hox-cluster miRNAs and Hox-cluster genes share regulatory mechanisms (Wienholds et al., 2005). Furthermore, the discovery that the 3’ UTRs of several Hox-cluster genes contain predicted binding sites for either miR-196 or miR-10 suggested that some Hox genes might be regulated by Mir10 and/or Mir196 (He et al., 2009; Hornstein et al., 2005; Kawasaki and Taira, 2004; Woltering and Durston, 2008; Yekta et al., 2004; Yekta et al., 2008). For example, mir10 is involved in the regulation of metastasis by controlling Hoxd10 in cell culture and hoxb1a and hoxb3a in vivo (Lund, 2009; Ma et al., 2007; Woltering and Durston, 2008). miR-196 binds to Hoxb8 mRNA, thereby accelerating its cleavage, and this interaction has been hypothesized to be important for the outgrowth of hindlimb buds (Hornstein et al., 2005; Kawasaki and Taira, 2004; Yekta et al., 2004). In the CNS, miR-196 restricts motor neuron differentiation by regulating Hoxb8 (Asli and Kessel, 2010). miR-196 is also involved in cancer progression by interaction with other Hox8 paralogs (Li et al., 2010)(Chen et al., 2010). miRNA-196 can also repress BACH1 expression in human liver cells (Hou et al., 2009) and is important for tail regeneration in the axolotl (Sehm et al., 2009). Knockdown of miR-196 in chick embryos leads to a homeotic transformation of a cervical vertebra to thoracic identity (McGlinn et al., 2009). Because no phenotype has yet been described for the over-expression of mir196 in embryos and no phenotype has been described in other tissues where it is expressed, we do not yet fully understand its roles in development or the mechanisms by which it acts.
Here we show that precise levels of mir196 are required to initiate development of the pectoral appendage, to develop the correct number of pharyngeal arches, and to specify the number and identity of rostral vertebrae and ribs. We show that miR-196 can alter hox gene expression patterns and that miR-196 acts on pectoral appendage development by altering retinoic acid signaling via fine-tuning the expression of the retinoic acid receptor Rarab.
RESULTS
mir196 genomics
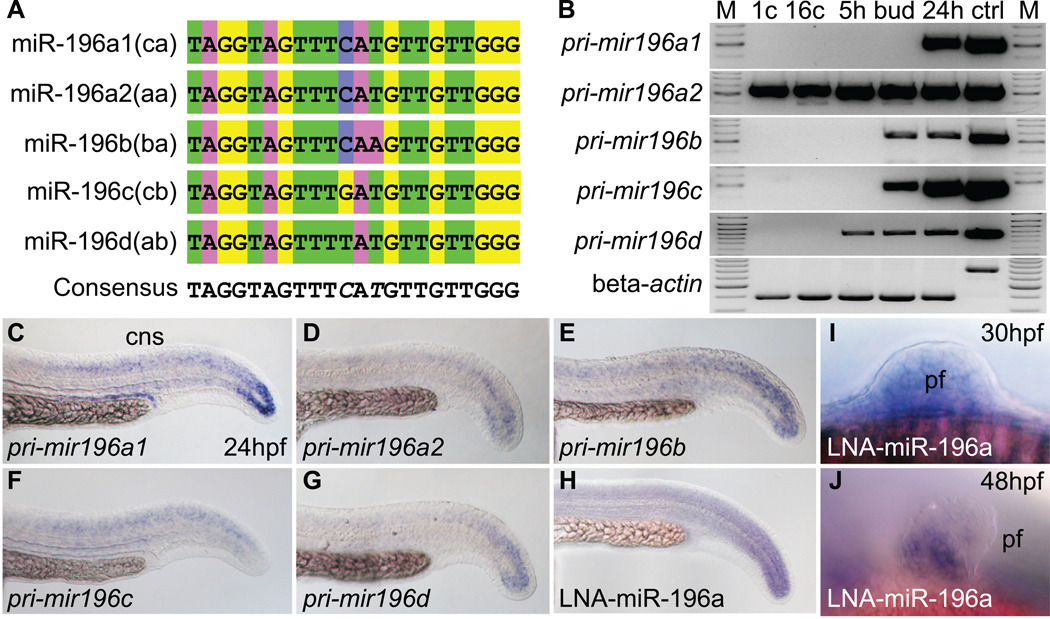
The human genome has three copies of MIR196 located between paralogy groups 9 and 10 (Yekta et al., 2008), but due to the teleost genome duplication (Amores et al., 1998; Postlethwait et al., 1998; Taylor et al., 2003), zebrafish has five mir196 genes (Supplementary Fig. S1A). The teleost whole genome duplication would have initially produced six mir196 genes, but one of the two hoxbb mir196 duplicates was lost and duplicates of only the hoxa and hoxc cluster genes were maintained. The five zebrafish mir196 paralogs encode four different mature miR-196 sequences with a central nucleotide trio containing (C/G/T) A (A/T). The duplicate hoxa and hoxc clusters have mir196 paralogs that differ by one nucleotide [(C/T) AT and (C/G)AT], respectively (Fig. 1A). Because miRNAs often bind their targets with some mismatch (He and Hannon, 2004; Yekta et al., 2004), all four miR-196 sequences probably regulate the same targets.
Figure 1.
Sequence and expression of mir196 genes. (A) Alignment of mature miR-196 encoded by five mir196 genes. (B) Expression of mir196 paralogs studied by RT-PCR. Beta-actin (bactin1) was used as control for contaminating genomic DNA (ctrl lane). M, size marker; 1c, 1 cell stage; 16c, 16 cell stage; 5h, 5 hpf (hours post-fertilization); bud, bud stage, about 10 hpf; 24h, 24 hpf; ctrl, genomic DNA control. (C–H) Whole mount in situ hybridization for mir196 primary transcripts showed expression in the tail bud and neural tube. (H) Linked nucleic acid (LNA) probe for miR-196a showed an expression pattern similar to the primary transcript. (I, J) LNA probes for miR-196a in the pectoral fin bud at 30 and 48 hpf. cns, central nerve system; pf, pectoral fin.
mir196 expression patterns
mir196 genes and nearby hox genes share spatial expression patterns in the central nervous system (CNS) and pectoral fin bud (Wienholds et al., 2005; Woltering and Durston, 2006; Yekta et al., 2008) (compare Fig. 1C–J and Supplementary Fig. S1B–M). This result suggests that hox-cluster miRNAs may share regulatory mechanisms with neighboring hox genes. To investigate temporal aspects of mir196 expression, we used gene-specific primers for mir196 primary transcripts and RT-PCR to discover that mir196a1(hoxca) transcript had begun to accumulate at 24 hours post fertilization (hpf), but mir196a2(hoxaa) transcript, which encodes the same mature miRNA sequence as mir196a1(hoxca), was maternally expressed (Fig. 1B). Transcripts from mir196b(hoxba) and mir196c(hoxcb) genes first appeared at bud stage, and transcript from mir196d(hoxab) first accumulated at 5 hpf (Fig. 1B) when gastrulation initiates. This gene-specific timing suggests that different mir196 genes experience different regulation and may play different roles in development. In addition, whole mount in situ hybridization experiments showed that mir196 genes are expressed in a pattern similar to but different from each other at 24 hpf (Fig. 1C–J).
mir196 and hox targets
Sequence comparisons revealed that the 3’ UTRs of several zebrafish hox-cluster genes surrounding mir196 contain predicted miR-196 targets ((Yekta et al., 2008) and Supplementary Fig. S1A, Supplementary Table S1). The mir196 genes lie between posterior hox paralogy groups 9 and 10, and ten hox-cluster genes ranging from paralogy group 5 to 13 contain predicted miR-196 binding sites. Conversely, mir10 genes lie between anterior paralogy groups 4 and 5 and predicted targets are in anterior hox paralogy groups 1 to 4 and in hoxd10a (Supplementary Fig. S1A and Supplementary Table S1). This conserved non-random organization of hox-cluster miRNA genes and their predicted targets suggests a conserved functional role between hox-cluster genes and their neighboring miRNA genes (Yekta et al., 2008).
To test whether miR-196 regulates transcript levels of predicted hox-cluster targets, we over-expressed miR-196 duplex or knocked down mir196 expression with morpholino antisense oligonucleotide (Mo) designed to inhibit miR-196 maturation or control morpholino and then examined transcript levels by in situ hybridization in 24 hpf embryos using the hindbrain marker egr2a as internal control. Results showed that after over-expression of mir196, hox genes that have mismatch target sites and are expressed early in the region of the pectoral fin bud, including hoxb5a, hoxb5b, hoxb6b and hoxc6a, mostly retained their native expression level, but with a weakened anterior boundary (Supplementary Fig. S2A–L). Quantifying miR-196 by qPCR confirmed that over-expression elevated miR-196 levels 10–20 times normal and that knockdown resulted in 20% or less of normal miR-196 amounts, but that mis-matched miR-196 over-expression and control morpholino had no effect (Fig. S3A). We conclude that miR-196 can fine-tune the anterior expression border of hoxb5a, hoxb5b, hoxb6b, and hoxc6a either as direct targets or because they are downstream of a miR-196 target.
Zebrafish hoxb8a mRNA has a perfect target site for miR-196a like its ortholog HOXB8 in human and mouse (Yekta et al., 2008). After over-expression of mir196, the level of hoxb8a transcript was diminished (Supplementary Fig. S2M–P). This result can be explained if hoxb8a transcript is degraded after miRNA binding like other transcripts with perfect matches between miRNAs and their target sites (Hornstein et al., 2005; Kawasaki and Taira, 2004; McGlinn et al., 2009; Yekta et al., 2004). To confirm the miR-196-related inhibition of hoxb8a, we made a reporter gene construct by attaching the 3’UTR of hoxb8a to the coding region of GFP (Supplementary Fig. S2Q). Results showed that co-injection of mRNA for the reporter with either miR-196a or miR-196b duplex inhibited fluorescence signal, and reciprocally, fluorescence increased after co-injection of the reporter mRNA and mir196-morpholino (Supplementary Fig. S2R–U). These results showed that both endogenous and injected miR-196 inhibit hoxb8a expression by degrading hoxb8a mRNA in zebrafish as in other species (Hornstein et al., 2005; Yekta et al., 2004).
miR-196 can block induction of zebrafish pectoral fin initiation
To learn the roles of miR-196 in embryonic development, we over-expressed miR-196 duplex by injection into early cleavage embryos and inhibited miR-196 processing and binding with morpholinos. Resulting animals survived to adulthood but showed highly specific phenotypes in the pectoral appendage, pharyngeal arches, and rostral vertebrae and ribs.
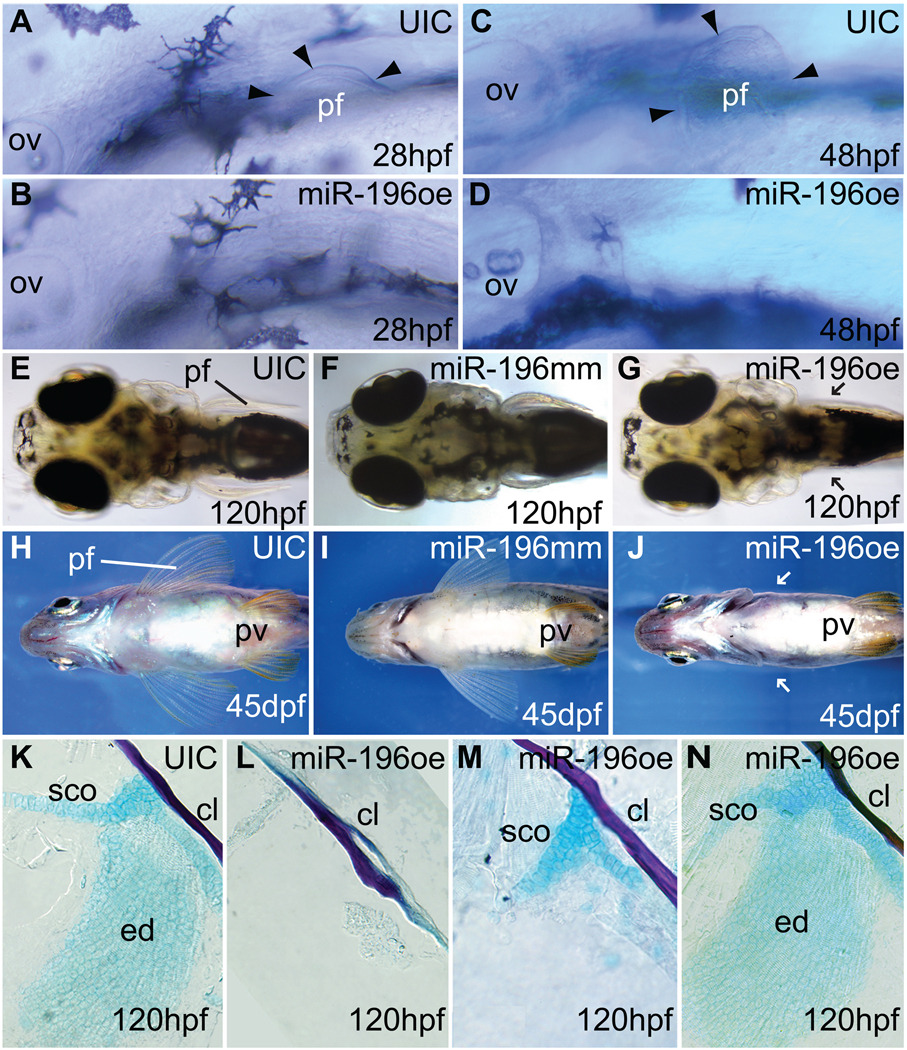
After miR-196 duplex injection, at least one pectoral fin was absent in 161 of 183 injected animals (Fig. 2A–G) and fin loss persisted into adulthood (Fig. 2H–J). At 5 dpf (days post fertilization), the pectoral apparatus from 234 animals over-expressing miR-196 either lacked the endochondral disc and scapulocoracoid (42.3%, Fig. 2L), lacked the endochondral disc only (6.0%, Fig. 2M), or had normal fin buds (51.7%, Fig. 2N). The cleithrum, a dermal bone that does not form in fin mesenchyme (Mercader, 2007), was always present (Fig. 2K–N). We conclude that miR-196 blocks an early stage in pectoral fin development.
Figure 2.
miR-196 over-expression inhibits pectoral fin initiation. (A, C) Pectoral fin buds are readily detectable at 28 hpf and 48 hpf in normally developing uninjected control animals, but embryos over-expressing miR-196 (B, D) showed no evidence of a pectoral fin bud. Arrowheads mark the edges of pectoral fin buds. (E, F, H, I) Uninjected controls or miR-196 mismatch injected controls showed normal pectoral fins by 5 and 45 days post fertilization (dpf), but larvae over-expressing miR-196 had not recovered pectoral fins by 5 dpf and became paraplegic adults (G, J, arrows point to missing pectoral fins). Pelvic fins were normal in fish with pectoral fin defects (J). (K–N) Dissected pectoral fins of 5 dpf larvae stained with Alcian blue for cartilage and Alizarin red for bone showed skeletal defects ranging from absence of the endochondral disc and scapulocoracoid (L, 42.3%, 234 total fins) to missing the endochondral disc but having part of scapulocoracoid (M, just 6%) to normal (N, 51.7%,). Abbreviations: cl, cleithrum; ed, endochondral disc; ov, otic vesicle; pf, pectoral fin or bud; pv, pelvic fin; sco, scapulocoracoid.
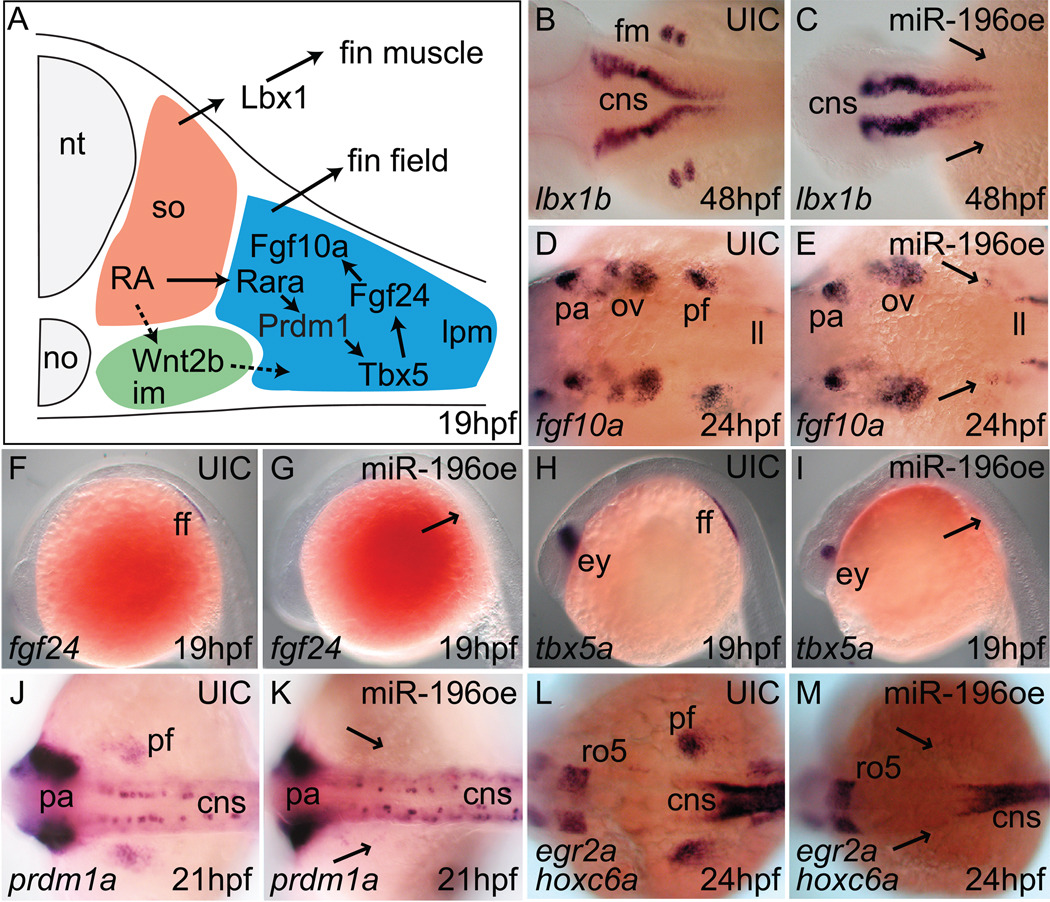
To learn how miR-196 acts to block pectoral fin formation, we interrogated steps in appendage development. In pectoral fin development (Fig. 3A), somite-derived retinoic acid (RA) acts on intermediate mesoderm to induce wnt2ba, which, along with RA acting via the retinoic acid receptor Rarab and prdm1a (Linville et al., 2009; Mercader et al., 2006), causes lateral plate mesoderm (LPM) to express tbx5a (Garrity et al., 2002), which turns on fgf24 leading to expression of fgf10a, which activates the apical epidermal fold (AEF), thereby promoting fin bud outgrowth followed by the development of lbx1b-expressing fin muscle (Mercader, 2007; Wotton et al., 2008). To discover which step is sensitive to miR-196, we examined pectoral fin gene expression after mir196 manipulation. miR-196-injected embryos lost expression of lbx1b, showing that miR-196 acts before fin muscle induction (Fig. 3B, C). Working backward through development, miR-196 injection blocked expression of fgf10a, fgf24, and tbx5a, the earliest expressed pectoral-fin specific gene (Fig. 3D–I). Because wnt2ba is weakly expressed even in wild types (Koudijs et al., 2008; Mercader et al., 2006), it was difficult to detect whether its expression changed in the fin field region after miR-196 over-expression (data not shown). Expression of prdm1a is downstream of RA signaling in the pectoral fin bud (Mercader et al., 2006), and we found that miR-196 injection inhibited the expression of prdm1a in the pectoral fin field without affecting its expression in the CNS or pharyngeal arches (Fig. 3J, K), implying that miR-196 acts upstream of prdm1a in pectoral fin. RA induces Hoxc6 expression in the mesenchyme of chick wing bud (Oliver et al., 1990) and we found that miR-196 injection inhibited the expression of hoxc6a in the pectoral fin field (Fig. 3L, M). This result is consistent either with the direct action of miR-196 on hoxc6a activity or an indirect action via retinoic acid signaling. The knockdown of miR-196 disrupted neither the expression of fgf24 and tbx5a in the pectoral fin field (data not shown) nor the development of pectoral fins.
Figure 3.
miR-196 duplex injection inhibits expression of zebrafish fin development genes. (A) Model of the pectoral fin developmental pathway (after (Mercader, 2007)). In situ hybridization for lbx1b (B, C), fgf10a (D, E), fgf24 (F, G), tbx5a (H, I), prdm1a (J, K), hoxc6a (L, M), on controls (B, D, F, H, J, L) and miR-196 over-expression (oe) animals (C, E, G, I, K, M) at ages indicated in the lower right of each panel. Arrowheads show the presumptive pectoral fin region in miR-196 duplex-injected animals. Results showed that miR-196 acts at or before the earliest stages of fin bud initiation. Abbreviations: cns, central neural system; ff, fin field; fm, pectoral fin muscle; im, intermediate mesoderm; ll, lateral line primordium; no, notochord; nt, neural tube; ov, otic vesicle; pa, pharyngeal arch; pf, pectoral fin bud; ro5, rhombomere 5.
miR-196, fin buds, and retinoic acid signaling
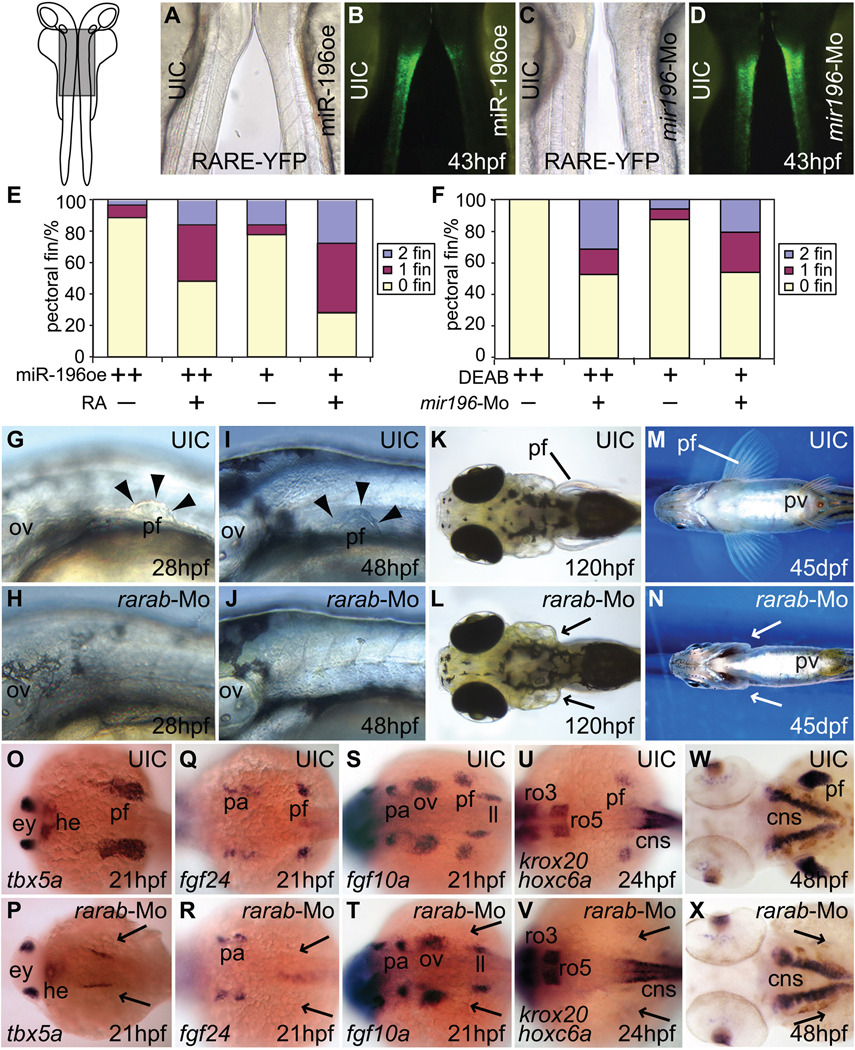
Our gene expression analyses showed that the mechanism of action of miR-196 lies upstream of prdm1a, and hence pointed to a problem in RA signaling. To learn if mir196 over-expression disrupts RA signaling, we utilized transgenic animals in which RA signaling activates a retinoic acid response element leading to expression of Yellow Fluorescent Protein (YFP) (Perz-Edwards et al., 2001) in the CNS by 43 hpf (Fig. 4A–D, left embryo). These reporter animals were injected with miR-196 duplex, or with mis-matched miR-196 duplex as control, or with miR-196 morpholino. Embryos over-expressing miR-196 displayed less fluorescence than controls, signifying reduced RA signaling (Fig. 4A, B, right embryo, Supplementary Fig. S4). Conversely, mir196 knockdown gave elevated fluorescence compared to controls, and hence enhanced RA signaling (Fig. 4C, D, right embryo, Supplementary Fig. S4). These results demonstrate that miR-196 can inhibit RA signaling in the CNS, and, coupled with the fact that mutation of aldh1a2, which encodes an RA-synthesizing enzyme can delete the fin bud (Begemann et al., 2001), suggest the hypothesis that the pectoral fin phenotype of miR-196 over-expression results from decreased RA signaling.
Figure 4.
miR-196 activity modulates RA signaling. (A) Brightfield microscopy of a 43 hpf uninjected control transgenic RA-signaling reporter individual and a miR-196 injected 43 hpf RA-reporter embryo oriented back-to-back as in the insert at left of part (A), with the boxed region blown up in A–D. (B) The animals in (A) viewed in fluorescence microscopy. The intensity of green fluorescence is proportional to the level of RA signaling, and was greatly reduced after miR-196 over-expression. (C) Brightfield and (D) fluorescence views of a control (left) and a miR-196-morpholino injected 43 hpf embryo (right). The elevated expression of YFP found after miR-196 knockdown is opposite to the phenotype found after miR-196 over-expression. (E, F) Rescue experiments. (E) Although injecting cleavage embryos with 5nL of miR-196 duplex and treating them with DMSO as controls (++) caused a greater fraction of animals to develop without pectoral fins (89%, n=63) than injections with 1nL (+) (78%, n=76), both can be rescued by RA treatment (+) (48%, n=81 and 28% n=61 without fin respectively). This experiment was repeated four times. (F) Treating embryos with 1×10−5M (++) or 5×10−6 M (+) DEAB to decrease RA levels resulted in all (100%, n=31) or most of the animals (87.5%, n=16) lacking pectoral fins respectively, but first injecting cleavage embryos with 1nL of 3mM mir196-Mo (+) to diminish the inhibition of RA signaling before treating embryos with DEAB, led to partial rescue of the fin phenotype (just 53%, n=51 or 54%, n=48, respectively). Compared to controls (G, I, K, M), rarab-Mo injection (H, J, L, N) inhibited pectoral fin outgrowth already by 28 hpf (G, H) and this phenotype was maintained through 48 hpf (I, J), 120 hpf (K, L), and into adulthood (M, N) verifying the specificity of the effect. Arrowheads mark the edge of the pectoral fin and arrows direct attention to missing pectoral fins and buds. (O–V) rarab-Mo inhibited expression of the pectoral fin genes tbx5a (O, P), fgf24 (Q, R), fgf10a (S, T), and hoxc6a (U, V), as well as the fin muscle marker gene lbx1b (W, X) specifically in the fin field region while leaving other expression domains intact, the same result as from miR-196 injection. Arrows: missing pectoral fin buds. cns, central neural system; ey, eye; he, heart; im, intermediate mesoderm; ll, lateral line primordium; ov, otic vesicle; pa, pharyngeal arch; pf, pectoral fin; pv, pelvic fin; ro3, -5, rhombomere3, -5.
The hypothesis that miR-196 negatively regulates RA signaling in fin bud initiation predicts that the inhibition of RA signaling by other methods should lead to the same phenotype. To test this prediction, we used DEAB, a reversible inhibitor of RA-synthesizing enzymes (Perz-Edwards et al., 2001). RA inhibition, like miR-196 over-expression (Fig. 2), caused a loss of pectoral fin outgrowth (Supplementary Fig. S5A–F) that persisted to adulthood (Supplementary Fig. S5G, H) and inhibited expression of fgf24, tbx5a, fgf10a, and lbx1b (Supplementary Fig. S5I–R). These results are as expected if miR-196 blocks fin bud development by inhibiting RA signaling.
If miR-196 inhibits RA signaling, then excess RA should partially reverse this inhibition and rescue the fin phenotype of miR-196 over-expression; conversely, knockdown of endogenous miR-196 should partially relieve the inhibition of RA signaling and thus rescue a situation with diminished RA signaling. We increased RA signaling either by exposing 12 hpf embryos to synthetic RA (10−7 M) for two hours or by reducing RA degradation using a mutation in cyp26a1, which encodes the enzyme that destroys RA (Emoto et al., 2005). At 120 hpf, both treatments resulted in short or absent pectoral fins (Grandel et al., 2002)(Supplementary Fig. S6A–F). In homozygous cyp26a1rw716 mutants, the endochondral disc was more sensitive than the scapulocoracoid (Supplementary Fig. S6G–J). While 89% (n=63) of animals over-expressing miR-196 (5 nL) had no pectoral fin (Fig. 4E), only 48% of animals co-injected with miR-196 and treated with 10−7 M RA lacked pectoral fins (n=81). Reciprocally, the knockdown of endogenous miR-196 partially rescued defective fins resulting from DEAB-inhibited RA signaling (Fig. 4F). We interpret this rescue to mean that with less endogenous miR-196, its target becomes more active, which partially overcomes diminished RA signaling caused by DEAB. These results show that not only injected miR-196, but also endogenous miR-196 inhibits pectoral fin bud initiation by interfering with RA signaling.
A target for miR-196 in fin bud development
To identify a molecular target related to RA signaling, we looked for potential miR-196 binding sites in the 3’UTRs of genes that are related to RA signaling, as well as genes known to be involved in pectoral fin initiation. A check of binding site prediction software (miRbase and MicroInspector (Rusinov et al., 2005)) showed that transcripts encoding Cyp26a1 and the retinoic acid receptor Rarab have predicted miR-196 target sites (Supplementary Fig. S6K and Fig. 6F, respectively), but other RA-pathway genes, including aldh1a2, all other rar and rxr genes, prdm1a, fibin, tbx5a, wnt2b and fgf ligands and receptor genes lacked such sites.
Figure 6.
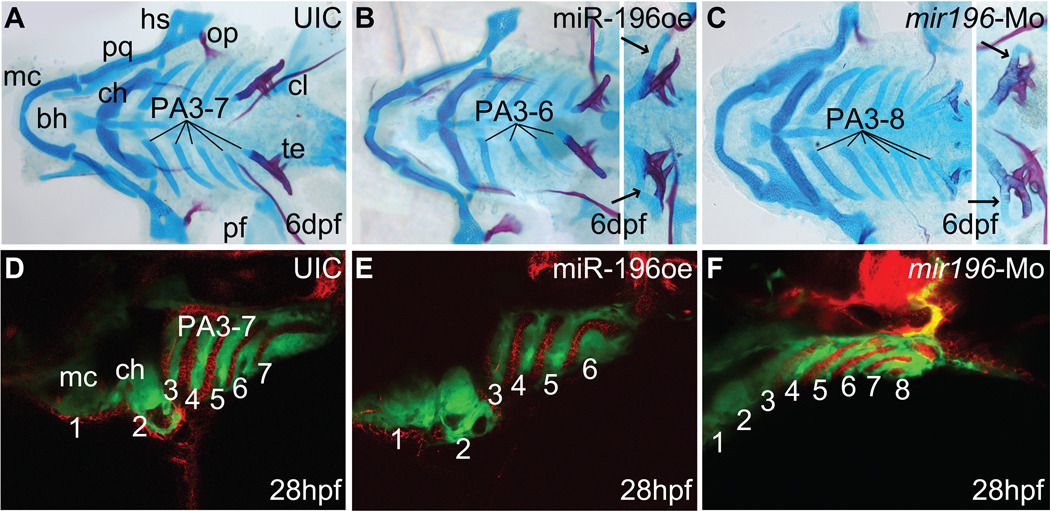
miR-196 regulates pharyngeal arch segmentation via pharyngeal endoderm. (A–C) Zebrafish embryos injected with miR-196 duplex or mir196-Mo were raised to 6 dpf and stained with Alcian blue for cartilage and Alizarin red for bone. (A) Control larvae had seven pharyngeal arches, including Meckels and ceratohyal cartilages (in PA1 and PA2) and five branchial arches (in PA3-7). (B) Animals over-expressing miR-196 lacked one branchial arch. Some animals with four branchial arches (insert) possessed a skeletal element attached to the last arch (arrows), suggesting that the missing arch was the one anterior to the tooth-bearing pharyngeal arch-7. (C) Injection of mir196-Mo resulted in animals with extra branchial arches. Some animals with seven pharyngeal arches (insert) displayed extra skeletal elements attached to pharyngeal arch-7 (arrows), suggesting that the extra arch was the one just anterior to PA7. (D–F) Transgenic animals expressing GFP in the neural crest (green) that were either over-expressing (E) or knocked down (F) for miR-196 showed defective segmentation of endodermal pouches stained with zn-8 antibody (red) and pharyngeal arches (green) consistent with observed alterations in arch numbers. Abbreviations: bh, basihyal; ch, ceratohyal; cl, cleithrum; hs, hyosymplectic; mc, Meckels cartilage; op, opercle; pa, pharyngeal arch; pf, pectoral fin; pq, palatoquadrate; te, teeth.
The target of miR-196 should possess at lease three properties: knockdown of the target should (a) mimic the loss of fin bud initiation caused by miR-196 over-expression, (b) delete the pectoral fin bud expression domain of tbx5a as does miR-196 over-expression, and (c) lead to decreased RA signaling as we observed after miR-196 over-expression. A comparison of cyp26a1 and rarab in these regards should suggest which is a better candidate as a miR-196 target. In cyp26a1 mutants, the pectoral fin bud in zebrafish initiates and the animals form the scapulocoracoid (Supplementary Fig. S6G-J and (Emoto et al., 2005)), but after rarab knockdown and miR-196 over-expression, animals fail to initiate fin bud development and become adults lacking pectoral fins (Fig. 4G–N and (Linville et al., 2009)). After rarab knockdown, pectoral fins either lacked the endochondral disc and scapulocoracoid or had normal fins (70% and 30%, respectively, n=101). The expression domain of tbx5a is shifted anteriorly in cyp26a1 mutants (Emoto et al., 2005) but in rarab knockdown and in miR-196 over-expression, we found that the tbx5a expression domain was deleted (Fig. 3H, I; Fig. 4O, P). Finally, inhibition of Cyp26a1, an RA degrading enzyme, augments RA signaling rather than reduces RA signaling as after rarab knockdown and mir196 over-expression. Furthermore, over-expressing rarab by injecting rarab mRNA did not give a pectoral fin phenotype (data not shown), mimicking results for mir196 knockdown. To further test whether Cyp26a1 is a target for miR-196 in pectoral fin initiation, we made a luciferase reporter construct by ligating the cyp26a1 3’UTR to the firefly luciferase coding region (Supplementary Fig. S6L). Co-transfection of tissue culture cells with this construct along with miR-196 showed that cyp26a1 3’UTR is insensitive to miR-196 (Supplementary Fig. S6M). These data make it unlikely that cyp26a1 is the major miR-196 target relevant for the fin bud phenotype but are all consistent with rarab being the target.
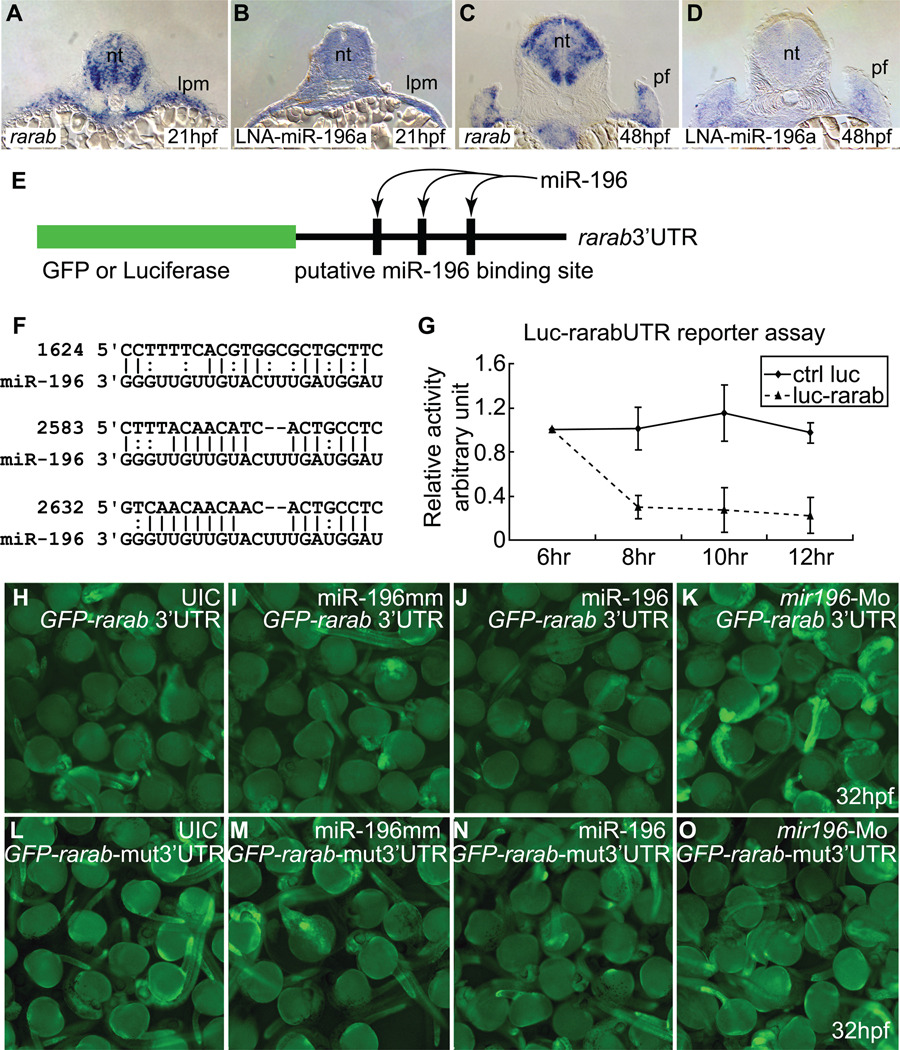
If miR-196 attenuates rarab, then both mir196 and rarab transcripts should be expressed in the same cells. Analysis showed that the expression of rarab (Hale et al., 2006; Linville et al., 2009) is similar to that of miR-196 in fin bud initiation (Fig. 5A–D, and shown in two-color double in situ hybridizations in Supplementary Fig. S7, along with double in situs of rarab and tbx5a). To see if miR-196 could target the rarab 3’UTR, we used a firefly luciferase assay in cultured cells using Renilla luciferase as an internal standard. We attached the rarab 3’UTR with its three predicted miR-196 binding sites to luciferase coding sequence (Fig. 5E, F) and co-transfected this construct and miR-196 into human 293T cells. miR-196 led to reduced luciferase fluorescence from the rarab 3’UTR construct compared to the control (Fig. 5G), showing that miR-196 can act on the rarab 3’UTR to inhibit message stability and/or translation. To test this interaction in living embryos, we co-injected a construct containing the GFP coding region followed by the rarab 3’UTR (Fig. 5E) and then over-expressed or knocked down miR-196. Embryos injected with miR-196 had less GFP fluorescence than uninjected controls or controls injected with miR-196 mismatch duplex, but embryos experiencing miR-196 knockdown had more fluorescence than controls (Fig. 5H–K, Supplementary Fig. S8). Deletion of the three predicted binding sites for miR-196 in the rarab 3’UTR led to no significant difference in GFP fluorescence compared to controls (Fig. 5L–O). This reporter assay confirmed direct interaction between miR-196 and the rarab 3’UTR in living embryos. We conclude that rarab is the miR-196 target responsible for the pectoral fin initiation phenotype. Because miR-196 over-expression or knockdown did not change the amount of rarab transcript (Supplementary Fig. S9), miR-196 is likely to act more strongly on rarab translation than on message stability. To determine whether miR-196 is itself a target of RA signaling, we treated animals with RA or with DEAB, an inhibitor of RA synthesis, and measured levels of miR-196 by qPCR. Results showed that, relative to untreated controls and DMSO carrier treated controls, RA treatments slightly decreased miR-196 levels and RA knockdown increased miR-196 levels to a small degree (Fig. S3B). We conclude that, if there is an effect of RA on miR-196 levels, it is in the direction of RA down-regulating miR-196 expression, but the effect is not strong.
Figure 5.
rarab is a target of miR-196. (A–D) In situ hybridization on tissue sections for rarab (A, C) and miR-196 (using LNA probe, B, D) at 21 hpf (A, B) and 48 hpf (C, D) shows that rarab and miR-196 are co-expressed in the lateral plate mesoderm at 21 hpf (A, B) and medially in the pectoral fin bud at 48 hpf (C, D). (E–O) Reporter assays. (E) Reporter assay constructs used luciferase in (G) and GFP in (H–K). (F) The 3’UTR of rarab has three predicted binding sites for miR-196 (nucleotide position according to NM_131399, binding sites were retrieved or predicted from miRBase.com and MicroInspector). (G) miR-196 can interfere with expression of a luciferase reporter construct bearing the 3’UTR of rarab compared to control. This experiment was repeated three times. (H–K) GFP-rarab 3’UTR mRNA was injected into early cleavage stage embryos either by itself (H) or co-injected with miR-196 mismatch (I), with miR-196 duplex (J), or with mir196-Mo (K). miR-196 depressed fluorescence but mir196 knockdown enhanced fluorescence, as predicted by the hypothesis that miR-196 acts directly on the 3’UTR of rarab. This experiment was repeated four times. (L–O) The three predicted miR-196 binding sites were removed from the GFP-rarab 3’UTR construct to make a GFP-rarab-mut3’UTR. GFP-rarab mut3’UTR mRNA was injected into early cleavage stage embryos either by itself (L) or co-injected with miR-196 mismatch (M), with miR-196 duplex (N), or with mir196-Mo (O). miR-196 did not depress fluorescence (N) and mir196 knockdown did not enhance fluorescence (O) from this mutant construct. This experiment was repeated three times. lpm; lateral plate mesoderm; pf, pectoral fin bud.
mir196 inhibits branchial arch segmentation
Animals over-expressing mir196 lacked not only pectoral fins, but also lacked one, and only one, pharyngeal arch (PA). Zebrafish have seven PAs, including mandibular (PA1), hyoid (PA2), and five branchial (gill) arches (PA3-7). After miR-196 over-expression, PA1 and PA2, as well as the tooth-bearing PA7, were normal. In 84% (n=254) of miR-196 duplex injected animals, however, one or both sides of the animal had four rather than five branchial arches (Fig. 6A, B). PA3 to PA5 were always normal in morphology, but occasionally a short PA6 was fused to PA7 after miR-196 over-expression (Fig. 6B, insert). This malformation suggests that the missing arch was PA6. This result shows that over-expression or ectopic expression of mir196 is sufficient to reduce the number of branchial arch segments. We infer that miR-196 inhibits a process necessary to add branchial arches, probably PA6.
To distinguish between the hypotheses that the loss of PA6 arises from over-expression or ectopic expression of miR-196, we knocked down miR-196 with a morpholino designed to inhibit miRNA processing. Results showed that 33% of injected animals (n=140) had six rather than the normal five branchial arches (Fig. 6C). Some animals with five branchial arches had an additional skeletal element fused to PA7, suggesting that endogenous miR-196 normally inhibits the arch just anterior to PA7. This result shows that endogenous miR-196 is necessary to prevent the addition of supernumerary branchial arches. Because the miR-196 knockdown phenotype is opposite to that of miR-196 over-expression, we conclude that precisely adjusted levels of miR-196 are required for proper branchial arch segmentation.
Cartilage-forming cells in pharyngeal arches arise from post-migratory neural crest that is divided into several streams by endodermal pouches (Fig. 6D and ref. (Crump et al., 2004)). To determine whether miR-196 acts primarily on the endodermal pouches or only on the neural crest, we used transgenic fli-GFP fish in which the skeletogenic crest fluoresces green (Lawson and Weinstein, 2002) and then stained animals with the antibody zn-8 as a marker for pharyngeal endoderm (Trevarrow et al., 1990). In contrast to normal 36 hpf embryos with seven pharyngeal arches (Fig. 6D), mir196 over-expression embryos lacked one endodermal pouch (Fig. 6E). Conversely, mir196 knockdown embryos had an extra endodermal pouch that resulted in an extra pharyngeal arch precursor (Fig. 6F). These results in embryos correspond to the cartilage phenotype of miR-196 manipulated larvae and support the hypothesis that miR-196 derived defects in pharyngeal arch patterning arise from initial effects on the patterning of pharyngeal pouch endoderm. Together, these results lead to the conclusion that mir196 acts in pharyngeal arch endoderm to suppress the formation of a posterior pharyngeal pouch.
Normal miR-196 levels are essential to pattern the axial skeleton
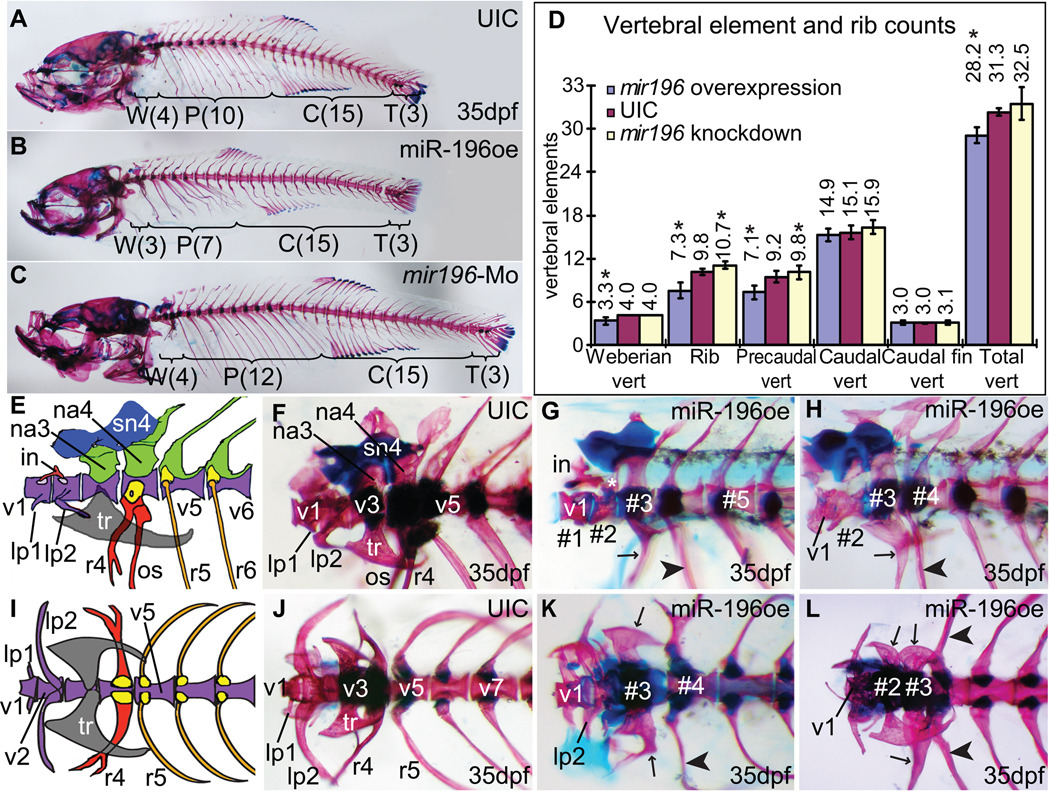
Controlled levels of miR-196 are important for proper segmentation not only of branchial arches but also of the rostral axial skeleton. The anterior four vertebrae of zebrafish and other Otophysi form the Weberian apparatus, bones that transmit sound from the swim bladder to the inner ear (Grande and Young, 2005). Caudal to the Weberian apparatus, normal animals have about ten rib-bearing precaudal vertebrae (Fig. 7A, D and ref. (Bird and Mabee, 2003)). Adult fish developing from embryos injected with miR-196 duplex showed disrupted rostral axial skeletons, including axial shifts in the patterning of the Weberian apparatus and fewer precaudal vertebrae and ribs (Fig. 7B, D, E–L). Adult fish developing from embryos injected with miR-196 Mo did not show defects in the Weberian apparatus but they had more ribs and rib bearing precaudal vertebrae than normal (Fig.7C, D). Statistically, miR-196 injected animals had on average 0.7 fewer segments in the Weberian apparatus than wild types (73.2% of 56 injected animals were affected), and 2.5 fewer ribs (81.8% affected), and 2.1 fewer rib-bearing precaudal vertebrae than normal (36.2% affected) (Fig. 8D).
Figure 7.
Over- and under-expression of miR-196 disrupts patterning of the axial skeleton. (A) Normal axial skeleton stained for cartilage (Alcian blue) and bone (Alizarin red) showing 4, 10, 15 and 3 vertebrae in the Weberian apparatus (region W), rib-bearing precaudal vertebra (region P), caudal vertebra (region C) and caudal fin vertebra (region T), respectively. (B, D) Over-expression of mir196 showing deletion of one Weberian vertebra and three ribs with their precaudal vertebrae. (C, D) Morpholino knockdown of miR-196 resulted in extra ribs and extra rib-bearing precaudal vertebrae. (D) Skeletal element counts after miR-196 manipulation showing reciprocal effects of over-expression and knockdown in the rostral axial skeleton. Asterisks indicate statistical significance compared to UIC (uninjected control) (Student T-test, p < 0.001). UIC, n=79; miR-196oe (over-expression), n=56; mir196-Mo (miR-196 knockdown), n=122. (E, F, I, J) Weberian apparatus sketch (after (Grande and Young, 2005)) and skeleton staining, lateral and dorsal views, respectively. (G, H, K, L) Over-expression of miR-196, lateral (G, H) and ventral (K, L) views of the Weberian apparatus from four different individuals. In wild-type fish, the Weberian apparatus has four highly modified vertebrae, each with bone and cartilage morphologies that specify their identity (E, F, I, J). Typically, the first vertebra (v1) has a pair of anterior-laterally projecting bones called the lateral process-1 (lp1) and dorsally, a small bone called intercalarium (in) that contacts with vertebra-2 (v2). V2 normally has a pair of laterally projecting bones (the lateral process-2, lp2) on each side. Lp2 is larger than lp1 (E, I). Vertebra-3 (v3) has a pair of lateral tripus (tr) bones, a triangular shaped bone, and neural arch-3 (na3) dorsally. Vertebra-4 (v4) has a pair of highly modified ribs (r4) protruding ventral-laterally that bears os suspensorium bones ventrally and dorsally, a neural arch-4, on top of which, supranueral-4 (sn4) usually fuses anteriorly with supranueral-3 (sn3) on v3. Precaudal vertebrae are defined by their parapophyses, which support ribs, and their lack of hemal arches and hemal spines, which are found on v15–v28 (Bird and Mabee, 2003). Treated animals are discussed in the text. Abbreviations: #1, #2, #3, vertebrae numbers from anterior without regard to identity; in, intercalarium; lp1, lp2, lateral processes; na3, na4, neural arches; os, os suspensorium; r4, r5, ribs; tr, tripus; v1, v2, v3, v4, v5, v7 vertebrae with identities.
In wild-type fish, the Weberian apparatus has four highly modified vertebrae, each with bone and cartilage morphologies that specify their identity (Fig. 7E, F, I, J). The lateral process-1 (lp1), -2 (lp2), the intercalarium (in), the tripus bones (tr), the neural arch-3 (na3), neural arch-4 (na4), the highly modified rib-4 (r4) ,os suspensorium (os), supranueral-3 (sn3), -4 (sn4) are attached to one of the four vertebra in the Weberian apparatus. Analysis of these skeletal structures showed that the over-expression of miR-196 provoked fate transformations in the Weberian apparatus. For example, in the animal in Figure 8G, the second segment (segment #2) had the joint for the intercalarium, which is appropriate for a normal vertebra-2, but this segment possessed a dorsal projection that appeared similar to, but not as broad as, neural arch-3 (asterisk in Fig. 7G). The morphology of this animal’s segment #3 was similar to the normal v4 with a neural arch-4 and rib-4 like rib (arrow in Fig. 7G). Segment #4 had a rib appropriate for v5 (arrowhead in Fig. 7G). We conclude that in this fish, the identity of segments #2–4 were partially transformed into more posterior fates, an apparent homeotic transformation. Alternatively, v2 or v3 was deleted and segment #2 assumed an identity intermediate between v2 and v3. The animal in Fig. 7H had a normal v1 with a normal lateral process-1, but segment #2 had an abnormally short lateral process-2; segment #3 had a ventral projection intermediate in character between the tripus (Fig. 7E, F) and anteriorly projecting, forked rib-4 (r4) which is more appropriate for the next segment more posterior (arrow in Fig. 7H), and a neural arch more similar to neural arch-4 than neural arch-3; in addition, segment #4 had a rib with the morphology of rib-5 rather than rib-4 (arrowhead in Fig. 7H). Figure 7K is a ventral view of the Weberian apparatus of an animal over-expressing miR-196 with relatively normal v1 and v2, but v3 showed partial posterior transformation on the right side with a forked tripus anteriorly oriented like rib-4, although the left side was relatively normal; segment #4 had a rib on the left side that was appropriate for rib-4, but on the right side, the rib pointed backwards and was not forked, which are characteristics of the more posterior rib-5. The Weberian apparatus of the animal in Figure 7L had a segment #2 with a normal tripus on the right side (rather than on the normal v3) and a malformed tripus on the left side; segment #3 of this animal had an rib-4-like structure on the right side and a nearly normal tripus on the left side (arrows in Fig. 7L); segment #4 on both sides had ribs appropriate for rib-5 (arrowheads in Fig. 7L). This phenotype occurred repeatedly in affected animals (See supplementary Table S2). In summary, analysis of miR-196 over-expression animals suggests that the identities of rostral vertebrae are transformed into structures appropriate for more posterior elements.
In contrast to the over-expression results, in which the Weberian apparatus showed posterior phenotypes and fewer than normal ribs, knockdown of miR-196 resulted in fish that had a morphologically normal Weberian apparatus but tended to have extra ribs and extra rib-bearing precaudal vertebrae (Fig. 7C, D). This result shows that native miR-196 expression levels are important for axial segment morphology. With regard to rib and precaudal vertebrae number, the results from miR-196 knockdown (segment gain) were opposite that of miR-196 over-expression (segment loss). In summary, the manipulation of miR-196 levels resulted in altered numbers and homeotic-like fate transformations and segmentation abnormalities along the axial skeletal system.
Axial skeletal segmentation depends on early somitogenesis (Bagnall et al., 1988)(Morin-Kensicki et al., 2002)(Sparrow et al., 2007). To understand how miR-196 regulates axial skeleton segmentation, we scored larval somite numbers after miR-196 over-expression or knockdown (Supplementary Fig. S10). By 4 dpf, wild type zebrafish embryos had developed about 30 or 31 somites (78.4%, n=74). We found that over-expression of miR196 reduced total somite number (81.1% of 53 animals had fewer than 30 somites), while knockdown of mir196 induced more somites than control fish (62.1% of 66 animals had more than 31 somites). This result is consistent with our counts of ribs and vertebrae in adults, and shows that miR-196 regulates somitogenesis,
DISCUSSION
Over-expressing miR-196 caused four highly specific phenotypes – failure of pectoral appendage initiation, deletion of one pharyngeal arch, transformation of vertebral identity and number, and change in somite number.
The interaction of hox-cluster protein-coding genes and mir196 genes
The similar expression patterns of miR-196 genes and their neighboring hox-cluster protein-coding genes would be expected if they share regulatory mechanisms (Yekta et al., 2008). Hox-cluster protein-coding genes are rich in miR-196 targets in zebrafish as they are in tetrapods (Yekta et al., 2008). Like its tetrapod ortholog, zebrafish hoxb8a is has a site with perfect complementarity to miR-196 and our results showed that in zebrafish, as in tetrapods, miR-196 acts on hoxb8a expression by decreasing messenger stability. Other predicted hox-cluster target genes for miR-196 have only partially complementary matches with miR-196, suggesting that miR-196 does not regulate these genes by message degradation. We observed changes in transcript distribution for hoxb5a, hoxb5b, hoxb6b and hoxc6a at their anterior expression borders, suggesting that miR-196 may regulate a factor upstream of these hox-cluster protein-coding genes. Mutations in genes that encode retinoic acid synthesizing or degrading enzymes (raldh1a2 or cyp26a1, respectively) (Begemann et al., 2001; Emoto et al., 2005) shift the anterior border of hox-cluster protein-coding genes, with up-regulation of RA signaling causing rostral expansion, implying that the phenotype we saw may be a combination of direct control (miR-196 binds and inhibits translation of hox target genes) and indirect regulation (miR-196 regulates RA signaling, which secondarily regulates hox gene expression); alternatively, miR-196 may regulate these hox-cluster protein coding genes by an unknown regulatory mechanism.
miR-196 inhibits pectoral fin induction by inhibiting rarab expression
Animals injected with miR-196 duplex lacked pectoral fins and showed diminished expression of even the earliest fin bud markers. We conclude that miR-196 over-expression inhibits the initiation, rather than the patterning or elongation of pectoral fin buds. No conclusions can be drawn with respect to the effect of miR-196 on the pelvic fin bud because it develops more than two weeks after fertilization (Grandel et al., 2002), which is long after the injected miRNA would be effective. Other miRNAs are involved in zebrafish fin regeneration (Thatcher et al., 2008). Retinoic acid signaling acts upstream of pectoral appendage initiation (Gibert et al., 2006; Grandel et al., 2002; Linville et al., 2009; Mercader et al., 2006; Waxman et al., 2008). Because we found that miR-196 knockdown partially rescued diminished RA-signaling, we conclude that endogenous miR-196 plays a role in normally developing zebrafish. Transcript from rarab appears in both the CNS and lateral plate mesoderm (Hale et al., 2006; Linville et al., 2009) and contains three predicted partially complementary binding sites for miR-196, suggesting the hypothesis that miR-196 attenuates RA signaling by lowering levels of Rarab protein production. Furthermore, rarab knockdown blocks pectoral appendage initiation (Linville et al., 2009), which would be expected if miR-196 inhibits pectoral fin initiation by limiting rarab function. Likewise, miR-196 over-expression diminished RA signaling in the CNS as detected in the RARE-YFP transgenic line, and the same result was obtained in the same line after direct rarab knockdown (Linville et al., 2009). Finally, a luciferase assay in tissue culture cells and a GFP assay in living embryos both showed that miR-196 functions via the 3’UTR of rarab. Because miR-196 over-expression does not inhibit the accumulation of significant quantities of rarab transcript and similar experiments showed a substantial decrease of transcript levels for hoxb8a, which serves as a positive control, we conclude that miR-196 acts more on translation than on message stability to modulate expression of rarab. These results are as predicted by the hypothesis that miR-196 over-expression blocks fin bud initiation, at least partially, by binding the rarab 3’UTR in lateral plate mesoderm to inhibit its translation, thereby diminishing the ability of lateral plate cells to detect somite-derived RA, which leads to lack of tbx5a induction, without which the fin bud can not initiate. Although miR-196 morpholino knockdown did not give a pectoral fin bud phenotype in otherwise normal development, evidence that endogenous miR-196 is involved in pectoral fin bud initiation comes from the demonstration that miR-196 knockdown rescued diminished RA signaling. Thus, we conclude that rarab is a target of miR-196 for pectoral fin bud initiation in zebrafish.
Tetrapods have three Rar genes (Rara, Rarb, Rarg), and zebrafish has duplicate copies of Rara and Rarg but no Rarb gene (Hale et al., 2006). The functions of the three tetrapod Rar genes appear to have partitioned among the four zebrafish raraa, rarab, rarga and rargb genes (Linville et al., 2009). As in zebrafish, pectoral appendage initiation in mouse requires RA signaling (Niederreither et al., 1999), and double knockout of Rara and Rarg in mouse causes hypoplastic pectoral limb buds (Wendling et al., 2001). Thus, the mechanism of pectoral appendage initiation is generally conserved between zebrafish and tetrapods, but because none of the Rar or Rxr genes in mouse, human, or chicken have predicted miR-196 targets, the role of miR-196 may differ in zebrafish and tetrapods.
Although several hox genes that are expressed in pectoral fins have predicted miR-196 binding sites, they are unlikely to be responsible for the fin bud phenotype because most are expressed downstream of RA signaling in the fin bud and their temporal-spatial expression patterns exclude them from functioning during pectoral fin bud induction. For example, although hoxb5b is an RA-responsive gene expressed in the forelimb field, its function is dispensable for forelimb formation (Waxman et al., 2008). Furthermore, although miR-196 causes a decrease in hoxb8a message levels in zebrafish as in tetrapods (Kawasaki and Taira, 2004; McGlinn et al., 2009; Yekta et al., 2004), hoxb8a is unlikely to be the gene responsible for miR-196 regulation of pectoral fin bud initiation because (1) it is expressed in zebrafish pectoral fin buds well after initiation, (2) because our experiments showed that hoxb8a knockdown in zebrafish does not affect pectoral fin development (data not shown), and (3) because hoxb8a mutants in medaka initiate fin bud development normally but are defective in the maintenance of pectoral fin bud outgrowth (Sakaguchi et al., 2006). In addition, we used morpholinos to knockdown individually the expression of hoxb5a, hoxb5b, hoxb6b, hoxc6a and none of them showed pectoral fin defects (data not shown). Thus none of the five hox genes we checked are required for pectoral fin initiation. We conclude that miR-196 regulates pectoral fin bud initiation primarily by inhibiting rarab expression.
miR-196 and pharyngeal arch patterning
Over-expression experiments showed that miR-196 inhibits the formation of a posterior arch, probably PA6, and knockdown experiments showed that this mechanism applies to endogenous miR-196 as well. In principle, miR-196 could perform this role by modulating either hox gene expression, RA signaling, or FGF signaling. In mouse, Hox2 paralogs help control PA2 and PA1 identity (Minoux et al., 2009) and Hox3 paralogs help specify PA3 and PA4 (Minoux et al., 2009), but the mechanisms that control the identity of PA5 to PA7 are as yet unclear. In zebrafish, hoxb5a is expressed strongly and hoxb5b weakly in PA3-7 (Bruce et al., 2001; Jarinova et al., 2008), suggesting that Hox5 paralogs could help pattern posterior arches. Although hoxb5a and hoxb5b are both predicted miR-196 targets, are both expressed in the posterior PAs, and our experiments show that both are sensitive to miR-196, the knockdown of hoxb5a and hoxb5b in zebrafish and Hoxb5 in mouse does is not reported to give an arch phenotype (McIntyre et al., 2007; Waxman et al., 2008)(and our unpublished experiments). These considerations make it unlikely that hoxb5 genes are the miR-196 targets responsible for the arch phenotype.
Because retinoic acid is a posteriorizing factor in pharyngeal endoderm (Bayha et al., 2009) and suppression of RA signaling deletes branchial arches (Begemann et al., 2001; Birkholz et al., 2009), it is possible that miR-196 acts on pharyngeal endoderm to inhibit RA signaling. The rather broad and general phenotype arches show after the manipulation of RA signaling, however, contrasts with the highly specific effect of miR-196 over-expression. Inhibiting RA signaling progressively between 16 and 30 hpf results in fewer deleted PAs (Kopinke et al., 2006), suggesting that, if miR-196 acts through RA signaling to suppress PA6, it must act at or after 30 hpf. Double knockout of Rara and Rarb alters development of posterior PAs in mouse (Dupe et al., 1999) in ways that mimic miR-196 over-expression in zebrafish. These considerations suggest that pharyngeal phenotypes of miR-196 manipulated zebrafish might, like the fin phenotype, be mediated by rarab. Although rarab is expressed in PAs (Hale et al., 2006; Linville et al., 2009), rarab morpholino knockdown had no effect on the branchial arch phenotype ((Linville et al., 2009) and our unpublished data). In contrast, knockdown of rarga, which does not have predicted miR-196 binding sites, did alter gill arch formation (Linville et al., 2009). Thus, either rarab inhibition does not explain the gill arch phenotype of miR-196 manipulation, or the knockdown of rarab activity by miR-196 over-expression is more profound than rarab knockdown by morpholino. The endoderm of the pharyngeal arches expresses cyp26a1 and the cyp26a1 3’UTR has miR-196 binding sites, but PA3-7 are normal in cyp26a1 mutants (Emoto et al., 2005), thus ruling out cyp26a1 as the miR-196 target responsible for the arch phenotype. Thus, in pharyngeal arches, miR-196 appears likely to act either on a component of the pharyngeal segmentation mechanism or on an as yet unknown gene essential for the specification of PA6.
Mesoderm- and CNS-derived Fgf3 and Fgf8 help direct the segmentation of pharyngeal pouches (Crump et al., 2004), suggesting that miR-196 may alter Fgf signaling. Our finding that manipulating miR-196 levels alters RA signaling in the hindbrain could lead to alterations in Fgf signaling that promote the loss or gain of PA6. Again, however, the great specificity of the miR-196 phenotype contrasts to the broader perturbation of PA3-7 development caused by Fgf manipulations, and suggests that, if miR-196 acts on Fgf signaling, it must be through a tissue-specific downstream target of Fgf because none of the zebrafish fgf, fgfr and other Fgf pathway genes have predicted miR-196 binding sites.
These considerations lead us to propose that in normal development, miR-196 attenuates action of a gene essential to direct the formation of pharyngeal pouches between PA5 and PA7. The identity of this target is as yet unknown.
miR-196 and axial skeleton patterning
Manipulating miR-196 levels provoked patterning anomalies specifically in regions of the axial skeleton that express hox genes with predicted miR-196 targets. The predicted miR-196 target hoxb5a is expressed in somites-2 and -3 but the target hoxb5b is not expressed in the somites (Bruce et al., 2001; Jarinova et al., 2008). Somites-1 and -2 and the anterior of somite-3 do not contribute to vertebrae in zebrafish or tetrapods, but in tetrapods at least, they contribute to the caudal part of the skull (Huang et al., 2000; Morin-Kensicki et al., 2002), which was morphologically normal in zebrafish over- or under-expressing miR-196. Furthermore, altered patterning of the zebrafish axial skeleton is not seen after knockdown of hoxb5a or hoxb5b ((Waxman et al., 2008), confirmed in our unpublished experiments). In mouse, Hoxb5 mutants show an anteriorizing homeotic transformation of the caudal cervical and first thoracic (rib bearing) vertebrae (Rancourt et al., 1995); while, conversely, we observed a posteriorizing effect in the homologous vertebrae after miR-196 over-expression. This result argues against the hypothesis that miR-196 acts on hoxb5a or hoxb5b to regulate anterior axial skeleton patterning.
Five predicted miR-196 targets (hoxb6b, hoxc6a, hoxb8a, hoxb8b and hoxc8a) are expressed with anterior borders in somites-3 to -7, which form the Weberian apparatus (Bruce et al., 2001; Morin-Kensicki et al., 2002; Prince et al., 1998). Morpholino knockdown of these genes, however, did not result in changes in the Weberian apparatus (our data, not shown). Correspondingly, we found that miR-196 injection led to posteriorizing homeotic transformations or vertebral segment deletion in Weberian vertebrae, which are homologous to vertebrae surrounding the cervical-to-thoracic transition in tetrapods (Burke et al., 1995; Morin-Kensicki et al., 2002). If miR-196 inhibits expression of these hox genes, then miR-196-injected animals should show hox loss-of-function phenotypes and miR-196 knockdown animals should show hox gain-of-function phenotypes, as observed in chick after miR-196 knockdown (McGlinn et al., 2009). In contrast, loss of function mutations for mouse orthologs of hoxb6b, hoxc6a, hoxb8a and hoxb8b give rise to anteriorizing, not posteriorizing, homeotic transformations at the cervical/thoracic transition (Garcia-Gasca and Spyropoulos, 2000; Rancourt et al., 1995; van den Akker et al., 2001), and Hoxc8 mutations cause anteriorization of the caudal thoracic vertebrae (van den Akker et al., 2001). How can we understand this discrepancy?
In contrast to most Hox genes, Hoxa5 and Hoxa6 mutants show a posterior homeotic transformation in the rostral mouse vertebral column (Jeannotte et al., 1993; Kostic and Capecchi, 1994). Mouse Hoxa5 is expressed with an anterior border in the third cervical vertebra and continuing expression into thoracic vertebrae (Jeannotte et al., 1993); this region is homologous to the zebrafish Weberian vertebrae. Zebrafish has no hoxa6 gene and has a single hoxa5 gene (Amores et al., 1998), which is not a predicted miR-196 target and is not expressed in somites (Thisse, 2005); the final zebrafish hox5 paralogy group gene, hoxc5a, is also not expressed in somites (Ericson et al., 1993). In addition, genes of the hoxb cluster (hoxb1a and hoxb1b) have newly assumed, or have maintained ancestral, functions equivalent to the same paralogy group but to a different cluster in mouse (Hoxa1), a process called ‘function shuffling’ (McClintock et al., 2002). The hox1 findings suggest the analogous hypothesis that function shuffling could have occurred between the zebrafish hoxb5 duplicates and the mouse Hoxa5 gene. According to this interpretation, the posterior transformations at the cervical/thoracic transition found after Hoxa5 knockout in mouse, which are similar to the posterior transformations of the homologous region in zebrafish after miR-196 action, are caused by the miR-196-induced inhibition of zebrafish hoxb5a, which is expressed in a pattern homologous to that of mouse paralog Hoxa5 and which we showed to respond to miR-196, at least in the CNS, by changed transcript patterns.
An alternative explanation for miR-196 induced re-patterning is that a Weberian v ertebra is missing rather than showing homeotic transformations. Loss of Hoxa3 and Hoxd3 function deletes a cervical vertebra in mouse (Horan et al., 1995), and deletion of a zebrafish homolog of a cervical vertebra could mimic the observed posteriorization, causing, for example, the third segment to have the morphology of the fourth vertebra.
miR-196 alters the number of elements in the axial skeleton
Besides pattern changes among Weberian vertebrae, increased and decreased miR-196 levels produced animals with fewer and more ribs than normal, respectively. Morpholino knockdown of hoxb5a, hoxb5b, hoxb6b and hoxc6a did not cause changes in rib and vertebral number, while knockdown of hoxb8a resulted in one extra rib in 54.5% (n=33) of the animals checked, contradicting the prediction that knockdown of Hox gene targets for mir196 should result in missing ribs and vertebrae. The predicted miR-196 target hoxa10b is expressed in somites that give rise to rib-bearing (pre-caudal) vertebrae with the same anterior border as the non-targets hoxb10a and hoxd10a (Morin-Kensicki et al., 2002). In mouse, knockdown of Hoxa10, a predicted target of miR-196, gives rise to posterior transformations of caudal thoracic vertebrae (Rijli et al., 1995). Over-expression of miR-196 should knockdown hoxa10b function, and an accompanying posterior transformation as in mouse could change pre-caudal, rib-bearing vertebrae into caudal, non-rib-bearing vertebrae, thereby decreasing rib number. Conversely, knockdown of miR-196 could cause over-expression or ectopic expression of hoxa10b, which could transform caudal vertebrae to pre-caudal, rib-bearing vertebrae, thereby increasing the number of vertebrae, as we observed.
A simple one-to-one fate transformation model caudal to the Weberian apparatus does not provide a full explanation of our results because the total number of vertebrae decreased after miR-196 over-expression (up to seven fewer somites than controls) and, reciprocally, increased after miR-196 knockdown (up to four more somites). In normal zebrafish, length variation arises mostly from variation in caudal vertebrae (Morin-Kensicki et al., 2002), whereas length variation in miR-196-manipulated fish involved mostly Weberian and precaudal vertebrae, indicating a difference between the mechanism of miR-196 action and the origin of naturally occurring variation. Somite number variation suggests that miR-196 may interfere with the segmentation clock, the mechanism that dictates the rhythm of somitogenesis from pre-segmental mesoderm (Lewis et al., 2009). Direct players in the zebrafish segmentation clock (her1, her4, her7, notch1a, notch1b, notch5, notch6, deltaC, deltaD, Mespa/b, ephA4, ephrinAl1 and ephrin-B2) (Lewis et al., 2009) do not have predicted miR-196 binding sites, suggesting that miR-196 does not act on somite number directly by inhibiting expression of these genes. The expression of miR-196 and several caudal hox genes with predicted miR-196 binding sites are co-expressed in the tailbud, and one or more of these are potential targets to explain the somite number effect of miR-196. Our experiments suggest that miR-196 plays a role in somite number, but further work is required to identify the mechanism by which it controls the length of time the segmentation clock continues to run.
Conclusions
These experiments revealed four exquisitely specific viable phenotypes caused by up- or down-regulation of miR-196 levels in zebrafish embryos. Analysis showed that the miR-196-induced failure of fin bud initiation arises from the suppression of retinoic acid signaling in lateral plate mesoderm by fine-tuning expression of the retinoic acid receptor rarab, which had previously been shown to be essential for fin bud outgrowth. The inhibition of pharyngeal arch 6 by endogenous or exogenous miR-196 does not arise from the inhibition of any single predicted hox target, but from the inhibition of pharyngeal pouch segmentation, about which we currently know little. The posteriorizing effect of miR-196 on vertebrae at a level that corresponds to the cervical-to-thoracic transition are best understood by the differential sorting out of ancestral functions common to Hoxa and Hoxb genes in zebrafish and tetrapod lineages. Finally, the inhibitory effect of exogenous and endogenous miR-196 on somite number is due to its inhibition of an unknown target, perhaps one or more hox genes. These experiments show remarkable parallels between the patterning functions of the protein-coding genes of the Hox clusters and a microRNA gene embedded between them.
METHODS
Animals
Wild-type fish were ABC/TU hybrids and the Tg(fli1:EGFP)y1 line (alias fli-GFP) (Lawson and Weinstein, 2002) provided animals with labeled cranial crest. The Tg(RARE-gata2:NTD-eYFP)ld1 line (Perz-Edwards et al., 2001) provided RA signaling reporters. The cyp26a1rw716 mutant fish was kindly provided by Lei Feng (C. Moens laboratory). Skeleton preparations were as described (Walker and Kimmel, 2007). Experiments involving animals used protocols approved by the University of Oregon IACUC.
Injections
Morpholino oligonucleotides (MO, Gene Tools) sequences were: mir196a-MO: AATCCCAACAACATGAAACTACCTAA, mir196b mutiple blocking (MB)-MO: ACGTCCAGCCCAACAACTTGAAACTACCTAA. Experiments utilized the GeneTools ‘control’ morpholino CCTCTTACCTCAGTTACAATTTATA. We injected one-cell stage zebrafish embryos with approximately 3 nL of these two MOs at a final concentration of 1.5 mM mir196aMO and 0.5 mM mir196bMB-MO. The rarab-MO was as reported (Linville et al., 2009). RNA oligonucleotide (Integrated DNA Technology) sequences were: miR-196a: UAGGUAGUUUCAUGUUGUUGGG; miR-196a*: CGACAACAAGAAACUGCCUUGA; miR-196b UAGGUAGUUUCAAGUUGUUGGG; miR-196b*: CAGGAACCUGAAACUGCCUGAA; miR-196bmm (mismatch control): UUCCGUCAAUCAAGUUGUUGGG. Because 3 nL of 12.5 µM stock of miR-196a or miR-196b duplexes yielded identical phenotypes, we used miR-196b duplex for most experiments.
Reporter constructs
From genomic DNA, we amplified a 1349 nt fragment of the 3’UTR of rarab (primers: rarab+314: GTAGACTTTGACCCGGACTGAACA and rarab-1639: AGAAGGCTTTTGGGTGAACTATCC) containing all three predicted miR-196 binding sites and inserted it into pCR4-TOPO (Invitrogen). To fuse GFP with the rarab 3’UTR, we used Notl and Spel to liberate the rarab 3’UTR from pCR4-TOPO and used NotI and XhoI to extract GFP from pEGFP-N3 (Clontech). To make the GFP-rarab3’UTR construct, we ligated fragments into PCRII-TOPO (Invitrogen) between SpeI and XhoI sites. To make the luciferase reporter, ptkLuc+ vector was digested with NgoMIV and KpnI and the fragment was ligated to the rarab 3’UTR that was cloned from genomic DNA by the above primers containing NgoMIV and KpnI sites (primers: rarab+314NgoMIV: gggccggcGTAGACTTTGACCCGGACTGAACA and rarab-1639KpnI: ggggtaccAGAAGGCTTTTGGGTGAACTATCC; small letters represent linkers added for cloning). To make GFP-rarab3’UTR mRNA, the construct PCRII-GFP-rarab3’UTR was linearized with SpeI and transcribed in vitro using mMESSAGE mMACHINE T7 kit (Ambion). GFP-rarab3’UTR mRNA was purified with an RNA clean-up kit (Zymo Research) and diluted to 15 µL with nuclease-free water to store in −80° C. For co-injections, we injected first 200 ng/µL of the synthetic GFP-rarab 3’UTR mRNA and then mir196 morpholino mix or 12.5 µM miR-196 duplex or miR-196bmm control.
To make the GFP-rarab-mut3’UTR construct, the following primers were used to skip the predicted binding sites for miR-196 in the 3’UTR of rarab: rarab+314NotI gggcggccgcTAGACTTTGACCCGGACTGAACA; rarabUTR-430XhoI ggctcgagGCTCTTGTAGTCGCTGAATC; rarabUTR+468XhoI ggctcgagCTTCACAGAGATGACAGAACA; rarabUTR−1393sacII ttggccgcggTAAAGTACAGAAGAAGAGGAA; rarabUTR+1479sacII ttccgcggTGTGACAATCACTTCAAGTAA; and rarabUTR−1639SpeI ggactagtAGAAGGCTTTTGGGTGAACTATCC. PCR products were digested by restriction enzymes identified in the primer names and were sequentially ligated for insertion into GFP downstream of the same vector for GFP-rarab3’UTR. mRNA was synthesized and injected and scored as for GFP-rarab3’UTR.
Similarly, The hoxb8a 3’UTR was cloned into pCR4 vector by the primer pairs: hoxb8a+1081 CCGGCGAAGACTGCGACAA; hoxb8a-1731 ACCCCAAGAAAGGAAGACAACAAA and replaced rarab 3’UTR in pCRII-GFP for GFP reporter assay. The 3’UTR of cyp26a1 was cloned using the primer pair: cyp26a1+NgoMIV ggcgccggcGGACCCCCGACAATGAAAAC; cyp26a1-KpnI: ggcggtacCGAACAGTCTGGGTATGTTAAAT (small letters represent linkers added for the cloning) and replaced rarab 3’UTR of ptkLuc-rarab3’UTR for the cyp26a1 3’UTR reporter assay.
Reporter assay
One cell embryos were injected with 3µL of a 200 ng/µL solution of GFP-rarab3’UTR or GFP-hoxb8a3’UTR mRNA and then were co-injected with 1µL of a solution of 12.5 µM miR-196 duplex or 2 mM mir196-MO. Embryos raised at 28.5 °C to 28 hpf were imaged in 3 % methylcellulose. To quantify GFP intensity, we used Photoshop, selected fuzziness to 40 and used the “select” and “color range” function to set a threshold, then used the histogram function to calculate the numbers and standard deviation of the green pixels. For the luciferase assay in 293T cells, we used the original ptkLuc+ vector for which luciferase expression is driven by a thymidine kinase promoter with luciferase flanked by either rarab 3’UTR or SV40 polyadenylation signal as a control. For the assay, either 50 ng of ptkLuc+ control plasmid or ptKLuc-rarab 3’UTR with 10 ng of CMV-Renilla plasmid and 200 ng miR-196 duplex were co-transfected. Cells were harvested at 2-hour intervals from 4 to 12 hours after transfection. Firefly and Renilla (sea pansy) luciferase activities were both directly quantified within each sample directly after cell lysis (Dual-Luciferase Reporter Assay System, Promega) and the firefly luciferase activity was calculated relative to the Renilla luciferase control. In addition, the relative luciferase activity of ptkLuc-rarab 3’UTR construct was normalized to the ptkLuc+ control.
For the ptkLuc-cyp26a13’UTR assay, 50 ng of ptkLuc-cyp26a13’UTR construct or ptkLuc+ control vector together with 10 ng of renilla luciferase construct, 40 ng of miR196 duplex or miR196mm duplex per well were mixed and transfected with 0.3 ul FuGENE HD transfection reagent (http://www.roche.com) into 293T cells in a 96-well plate. 24 hours after transfection, firefly and renilla luciferase activity was assayed by using the Promega Dual-Glo Luciferase Assay System (http://www.promega.com). This experiment was repeated 6 times. Standard deviation was plotted as error bars.
RA treatment
A stock solution of 10−3 M all-trans retinoic acid (RA) (Sigma cat# R2625) was prepared in DMSO and stored at − 80 °C. Zebrafish embryos were dechorionated in embryo medium and the stock DEAB solution was added to 5 mL embryo medium at a final concentration of 10−5 M or 5×10−6 M. 12 hpf embryos were treated for 2 hours to elevate the level of RA during pectoral fin induction. Treatment was carried out in a dark environment. After treatment, embryos were washed in fresh medium twice and then cultured in fresh medium until fixed. 0.5ul DMSO was added to the control embryo medium as a negative control.
DEAB treatment
A stock solution of 10−2 M 4-Diethylaminobenzaldehyde (DEAB) (Sigma cat# R86256) was prepared in DMSO and stored at − 80 °C. Zebrafish embryos were dechorionated in embryo medium before treatment. DEAB at a final concentration of 10µM was added into fresh embryo medium from 100% epiboly to 24 hpf and then were fixed at various times later. Treatment was carried out in a dark environment. After treatment, embryos were washed twice in embryo medium and cultured in fresh medium until fixed. DMSO with the same concentration was used as a negative control.
Cloning and in situ hybridization
To generate PCR products containing partial mir196 primary transcript, we used 1 dpf zebrafish whole embryo cDNA reverse transcribed with oligo-dT primer. Cloning of mir196 primary transcripts used the primers: mir196a1+523 ATTAAATGAACGCTAGCGGCTGTATGATG, mir196a1−1014 TTTTGCTAGCGCTTTGTCTTTGTAACCA; mir196a2+1349 GCAGACAGGAGAGCGGCAAGAA, mir196a2−1891 AGCAGGCAAGGCAAGATTATGGTA; mir196b+756 GTATCTCTTTGCCCCGCTGTGG, mir196b−1292 TGGAAAAACGATGGGAAAGTATTG; mir196c+1016 ATTGCTTTAGATTATGCGCGGGTATTT, mir196c−1339 CAAGCTATGTCAAGGCGTGTCTGTCT; mir196d+467 TATGCTACCTGGTGCCGTGAAG, mir196d−1325 CCGCTGATAATGGAAGACAACC. Gene sequences were submitted to NCBI GenBank with the following nucleotide sequence accession numbers: dre-mirn196a-1, GU188984; dre-mirn196a-2, GU188985; dre-mirn196b, GU188986; dre-mirn196c, GU188987; and dre-mirn196d, GU188988. Other probes were cloned with primers: beta actin+184 TGGTTGGCATGGGACAGAAAGA, bactin-556 ATGGCATGGGGAAGAGCGTAAC; tbx5+305 TCAACAGGGAATGGAGGGAATCAAA, tbx5−1213 AGAGTAGCTTAGGGGCCGGTAGTAGTGGT; fgf10a+11 ATGCCCCTCGTCGCCTCTTATTCTG, fgf10a−1458 TTCCCTGGTGCCAATAACTTAAACAA; wnt2b+344 GGTGGTACATTGGTGCGTTAGGAG, wnt2b−1304 GCCAGTCGGGTTTCTTGTGTAGTT; prdm1+2208 GAGGGCATGGTGGAGAAGCAGATA, prdm1−3391 AAAGGCCGAGGTGACGTGAAGAGT; lbx1b was from Dr. Haruki Ochi (nt 67 to 810 of NM_001025532) and fgf24 probe was as described (Fischer et al., 2003). PCR products were cloned into pCR4-TOPO (Invitrogen) vector and in situ hybridization was as described (Hale et al., 2006). Antisense LNA (locked nucleic acid) probe for miR-196a with the sequence, 5’Dig/CCCAACAACATGAAACTACCTA/3’Dig, was ordered from Exiqon (http://Exiqon.com) and in situ hybridization was according to the manufacturer.
RT-PCR and qPCR Analysis
From each experimental and control group, total RNA was isolated from 50 zebrafish embryos at 27 hpf using the RoboZol RNA extraction reagent for total RNA including microRNA (Amresco Cat:N580-100ML). Two micrograms total RNA from each sample was reverse transcribed into cDNA for qPCR using Mir-X™ miRNA First-Strand Synthesis and SYBR® qRT-PCR (Clontech, cat:PT4445-1). A 1/70th aliquot of each microRNA cDNA reaction was used in a 20 µL qPCR amplification reaction and qPCR assayed in StepOnePlus (Applied Biosystem). Primer sequences were miR-196a TAGGTAGTTTCATGTTGTTGGG and miR-196b TAGGTAGTTTCAAGTTGTTGGG. The relative expression of miR-196a and miR196b were normalized to the U6 spliceosomal RNA provided in the Mir-X™ miRNA First-Strand Synthesis and SYBR® qRT-PCR kit (Clontech). The comparative CT experimental method was used to calculate the normalized relative expression level of the target gene from triplicate measurements. Averaged plots for triplicate qPCR reactions for each relative quantitation are shown in Supplemental Figure 10. Cycling conditions for KAPA SYBR® FAST ABI Prism® 2X qPCR Master Mix (KAPA Biosystems, Cat: KK4603) consisted of 95°C, 10 min followed by 40 cycles of 95°C, 60°C 20s and then dissociate at 95°C, 1 min, 55°C, 30s and 95°C, 30s.
Highlights Zebrafish miR-196.
> Injection of the Hox-cluster microRNA-196 in zebrafish embryos causes four phenotypes. > Phenotypes: loss of pectoral fin, arch-6, some ribs and somites, and vertebral homeosis. > miR-196 knockdown caused extra pharyngeal arch, ribs, and somites.> miR-196 altered hox expression and retinoic acid signaling through the rarab receptor. > Reporters showed that rarab 3’UTR is a miR-196 target for pectoral fin bud initiation.
Supplementary Material
Zebrafish mir10 and mir196 genes lie in hox clusters and nearby hox genes that contain predicted binding sites for these miRNAs. (A) Arrows represent hox genes and their transcription direction. Grey and black rectangles represent the five mir196 and five mir10 genes. Rows represent hox clusters and columns represent paralogy groups. Grey arrows indicate predicted hox targets for mir196 and black marks predicted hox targets of mir10 from miRBase and Microinspector (Griffiths-Jones et al., 2008; Rusinov et al., 2005). (B–L) In situ hybridization for hox gene neighbors of mir196; whole-mounted zebrafish embryos at 24 hours post-fertilization (hpf) (B–G) and pectoral fins at about 48 hpf (H–I). The hoxcb cluster has no hox9 or hox10 member. In situ hybridization for mir196d primary transcript on fin bud (M).
miR-196 inhibits somite segmentation. The number of somites was scored after either miR-196 overexpression or knockdown. Of 74 untreated control fish, 78.4% developed 30 to 31 somites at 4 dpf. After miR-196 over-expression, only 13.2% of 53 fish developed 30 or 31 somites but most fish (81.1%) developed somites with fewer than 30 somites. After miR-196 knockdown by injection of mir196-Mo, 33.3% of 74 fish developed 30 or 31 somites while the remaining 62.1% developed more than 31 somites.
Manipulation of mir196 expression alters expression of hox-cluster genes. Animals were either untreated (A, D, G, J, M), or were injected in early cleavage with miR-196 duplex (miR-196oe, B, E, H, K, N) or mir196 morpholino (mir196-Mo, C, F, I, L, O) and then at 24 hpf, were prepared for in situ hybridization with the predicted miR-196 targets hoxb5a (A–C), hoxb5b (D–F), hoxb6b (G–I), hoxc6a (J–L), or hoxb8a (M–O) along with egr2a (krox20a) to mark rhombomere-3 and rhombomere-5 in the hindbrain. The anterior border and overall level of hoxb5a, hoxb5b, hoxb6b and hoxc6a expression in the neutral tube became weaker after miR-196 over-expression (A, B, D, E, G, H, J, K, M, N) but was not significantly altered by miR-196 morpholino knockdown (C, F, I, L), asterisk in B indicates expanded posterior CNS expression of hoxb5a. Expression of hoxb8a was strongly inhibited after miR-196 duplex injection (M, N), but was not greatly affected after miR-196 knockdown (O). Like Hoxb8 in tetrapods, the zebrafish hoxb8a 3’UTR has a perfect match to miR-196, but with wobble at two nucleotides (T). To test whether the miR-196 inhibition of hoxb8a expression is mediated by its 3’UTR, we ligated the GFP coding region to the hoxb8a 3’UTR (U) and injected the mRNA into animals either maintained as controls (P), or injected with miR-196a duplex (Q) or miR-196b duplex (R), or with a mixture of miR-196a and miR-196b morpholino (S). Results showed that miR-196 duplex suppressed and miR-196 morpholino augmented GFP expression from this construct. Abbreviations: asterisk in panel B, increased posterior CNS expression of hoxb5a; cns, central nervous system; ro3 and ro5, rhombomere-3 and -5.
Quantifying miR-196. A. To deterimine the effect of morpholino knockdown and miR-196 over-expression on the quantity of miR-196 in the animals, we injected MO or miR-196 duplex into early cleavage stage embryos and evaluated the levels of miR-196 at 27 hpf by qPCR after normalizing values to the U6 RNA control and setting uninjected controls to 1.0. Results showed that mis-matched morpholino (MMO) and mis-matched miR-196 duplex (MMC) had little effect on miR-196 levels, but that maturation-blocking morpholino reduced miR-196 levels at least 20% and that miR-196 injection increased miR-196 levels more than 10 fold for both miR-196a and miR-196b. The graph is broken between relative expression values 2 and 10 and the scale is changed to accommodate the data. B. To learn whether miR-196 is a target of retinoic acid signaling, we treated animals with the diluent DMSO, with RA, or with DEAB, an inhibitor of RA synthesis, and measured levels of miR-196 by qPCR normalized to uninjected controls shown in Fig. s3A.
Retinoic acid signal is regulated by miR-196. YFP signal from individual fish was analyzed by counting the number of YFP pixels. The average number of pixels above threshold was plotted in horizontal bars for each category. The Student t-test showed that miR-196 over-expression significantly inhibited YFP signal (p < 0.001) and miR-196 knockdown significantly increased YFP signal (p = 0.03).
Inhibition of retinoic acid synthesis by DEAB blocks pectoral appendage outgrowth and the expression of pectoral appendage genes. (A, C, E, G, I, K, M, O, Q) DMSO treated controls. (B, D, F, H, J, L, N, P, R) Animals treated with 5×10−7 M DEAB in fish water from 100% epiboly to 24 hpf. At (A, B) 28 hpf, (C, D) 48 hpf, (E, F) 120 hpf, (G, H) 45 dpf, DEAB delivered in this protocol specifically blocks pectoral fin initiation without substantial effects on other developmental systems (Gibert et al., 2006; Wakahara et al., 2007). In situ hybridization analysis of gene expression for (I, J) fgf24 at 19 hpf; (K, L) fgf24 at 24 hpf; (M, N) tbx5a at 24 hpf; (O, P) fgf10a at 24 hpf; (Q, R) lbx1b at 48 hpf. Inhibiting RA synthesis specifically inhibits the expression of genes in the program of pectoral fin development and leaves other expression domains intact. cns, central nervous system; ey, eye; he, heart; ov, otic vesicle; pa, pharyngeal arch; pf, pectoral fin.
An excess of RA perturbs pectoral fin outgrowth. (A) DMSO-treated controls at 120 hpf have normal pectoral fins. (B, C) 120 hpf animals that had been treated with 10−7 M RA from 12 to 14 hpf have aberrant pectoral fins and other abnormalities. (D) Untreated wild-type control at 120 hpf. (E, F) Homozygous cyp26a1rw716 mutants at 120 hpf show phenotypes characteristic of excess RA. (G) Wild-type pectoral fin bud. (H–J) Fin buds of cyp26a1rw716 mutants. (K) The predicted binding site in the 3’UTR of cyp26a1 was aligned with miR-196 showing seed region pairing. (L) Schematic drawing of Luciferase reporter construct for cyp26a1 3’UTR. (M) Firefly luciferase reporter with cyp26a1 3’UTR was interrogated with miR196. Relative firefly luciferase intensity was normalized with control renilla luciferase. Result shows that miR-196 manipulation did not affect the luciferase signal. Standard deviations for 6 independent repeats were plotted as error bars.These results show that an excess of RA causes variable reductions in fin bud development due to problems with the endochondral disc, although the shoulder girdle is largely normal. Abbreviations: cl, cleithrum; ed, endochondral disc; pf, pectoral fin; sco, scapulocoracoid.
Two-color double label expression analyses for rarab, tbx5, and mir196. (A–F) Expression of rarab (A, D, green) showing expression in the central nervous system (cns) and lateral plate mesoderm (lpm) and expression of tbx5a (B, E, red) showing expression in the lateral plate mesoderm and pectoral fin bud (pf) and the merged images (C, F) at 24 hpf (A–C) and 48 hpf (D–F). (G–L) Expression of rarab (G, J, green) showing expression in the central nervous system (cns) and lateral plate mesoderm (lpm) and expression of mir196 (H, K, red) showing broad expression including the central nervous system, lateral plate mesoderm and pectoral fin bud and the merged images (I, L) at 24 hpf (G–I) and 48 hpf (J–L).
The 3’UTR of rarab responds to miR-196. Levels of GFP expression above background (yolk cell) for the experiment in Fig. 5H–K were quantified by setting a threshold using Adobe Photoshop to calculate the average number of green pixels per fish for each treatment. Standard deviations for 4 independent repeats were plotted as error bars.
miR-196 has no effect on rarab transcript accumulation. (A–C) raraa expression. (D–F) rarab expression. (A, D) Uninjected controls. (B, E) miR-196 injected embryos. (C, F) mir196-morpholino treated embryos. miR-196 levels do not effect quantities of raraa or rarab transcript. These results suggest that miR-196 does not act to strongly decrease the level of rarab transcript quantity, so it likely exerts its demonstrated effect on rarab-mediated retinoic acid signaling at the level of translation.
ACKNOWLEDGMENTS
We thank Charles Kimmel, Judith Eisen, and Cristian Cañestro for helpful discussions, Haruki Ochi for the gift of a plasmid, Lei Feng for cyp26a1 mutant fish, Ruth BreMiller for help with histology, Amanda Rapp for animal care, J .T. Neal help for QPCR, Catherine Wilson for help with statistics, and Jesse Buss and Jiin Park for undergraduate research help. The work was supported by DAAD through the Graduate School of Life Sciences, University of Würzburg (XH), the Deutsche Forschungsgemeinschaft (grant GRK 1048 (MS)), Alexander von Humboldt Foundation (JHP), and National Institutes of Health (grants 5 P01 HD22486, 1 R01 RR020833, and 1 U01 DE020076 (JHP)). Support agencies played no role in study design, in the collection, analysis or interpretation of data, in the writing of the report, or in the decision to submit the paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Xinjun He, Email: xhe@uoneuro.uoregon.edu.
Yi-Lin Yan, Email: yan@uoneuro.uoregon.edu.
Johann K. Eberhart, Email: eberhart@mail.utexas.edu.
Amaury Herpin, Email: amaury.herpin@biozentrum.uni-wuerzburg.de.
Toni U. Wagner, Email: toni.wagner@biozentrum.uni-wuerzburg.de.
Manfred Schartl, Email: phch1@biozentrum.uni-wuerzburg.de.
John H. Postlethwait, Email: jpostle@uoneuro.uoregon.edu.
REFERENCES
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Amores A, Suzuki T, Yan YL, Pomeroy J, Singer A, Amemiya C, Postlethwait JH. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asli NS, Kessel M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev Biol. 2010;344:857–868. doi: 10.1016/j.ydbio.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Bagnall KM, Higgins SJ, Sanders EJ. The contribution made by a single somite to the vertebral column: experimental evidence in support of resegmentation using the chick-quail chimaera model. Development. 1988;103:69–85. doi: 10.1242/dev.103.1.69. [DOI] [PubMed] [Google Scholar]
- Bayha E, Jorgensen MC, Serup P, Grapin-Botton A. Retinoic acid signaling organizes endodermal organ specification along the entire antero-posterior axis. PLoS One. 2009;4:e5845. doi: 10.1371/journal.pone.0005845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Bird NC, Mabee PM. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae) Dev Dyn. 2003;228:337–357. doi: 10.1002/dvdy.10387. [DOI] [PubMed] [Google Scholar]
- Birkholz DA, Olesnicky Killian EC, George KM, Artinger KB. Prdm1a is necessary for posterior pharyngeal arch development in zebrafish. Dev Dyn. 2009;238:2575–2587. doi: 10.1002/dvdy.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AE, Oates AC, Prince VE, Ho RK. Additional hox clusters in the zebrafish: divergent expression patterns belie equivalent activities of duplicate hoxB5 genes. Evol Dev. 2001;3:127–144. doi: 10.1046/j.1525-142x.2001.003003127.x. [DOI] [PubMed] [Google Scholar]
- Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development. 1995;121:333–346. doi: 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- Chambers KE, McDaniell R, Raincrow JD, Deshmukh M, Stadler PF, Chiu CH. Hox cluster duplication in the basal teleost Hiodon alosoides (Osteoglossomorpha) Theory Biosci. 2009;128:109–120. doi: 10.1007/s12064-009-0056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Capecchi MR. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev Biol. 1997;181:186–196. doi: 10.1006/dbio.1996.8440. [DOI] [PubMed] [Google Scholar]
- Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development. 2004;131:5703–5716. doi: 10.1242/dev.01444. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Di-Poi N, Koch U, Radtke F, Duboule D. Additive and global functions of HoxA cluster genes in mesoderm derivatives. Dev Biol. 2010;341:488–498. doi: 10.1016/j.ydbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Dupe V, Ghyselinck NB, Wendling O, Cham bon P, Mark M. Key roles of retinoic acid receptors alpha and beta in the patterning of the caudal hindbrain, pharyngeal arches and otocyst in the mouse. Development. 1999;126:5051–5059. doi: 10.1242/dev.126.22.5051. [DOI] [PubMed] [Google Scholar]
- Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol. 2005;278:415–427. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Ericson JU, Krauss S, Fjose A. Genomic sequence and embryonic expression of the zebrafish homeobox gene hox-3.4. Int J Dev Biol. 1993;37:263–272. [PubMed] [Google Scholar]
- Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science. 2004;304:1335–1337. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- Fischer S, Draper BW, Neumann CJ. The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development. 2003;130:3515–3524. doi: 10.1242/dev.00537. [DOI] [PubMed] [Google Scholar]
- Fjose A, Zhao XF. Exploring microRNA functions in zebrafish. N Biotechnol. 2010 doi: 10.1016/j.nbt.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Garcia-Gasca A, Spyropoulos DD. Differential mammary morphogenesis along the anteroposterior axis in Hoxc6 gene targeted mice. Dev Dyn. 2000;219:261–276. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1048>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129:4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Kloter U, Suga H. Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol. 2009;88:35–61. doi: 10.1016/S0070-2153(09)88002-2. [DOI] [PubMed] [Google Scholar]
- Gibert Y, Gajewski A, Meyer A, Begemann G. Induction and prepatterning of the zebrafish pectoral fin bud requires axial retinoic acid signaling. Development. 2006;133:2649–2659. doi: 10.1242/dev.02438. [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Grande T, Young B. The ontogeny and homology of the Weberian apparatus in the zebrafish Danio rerio (Ostariophysi: Cypriniformes) Zoological Journal of the Linnean Society. 2005;104:241–254. [Google Scholar]
- Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Kuchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale LA, Tallafuss A, Yan YL, Dudley L, Eisen JS, Postlethwait JH. Characterization of the retinoic acid receptor genes raraa, rarab and rarg during zebrafish development. Gene Expr Patterns. 2006;6:546–555. doi: 10.1016/j.modgep.2005.10.007. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He X, Eberhart JK, Postlethwait JH. MicroRNAs and micromanaging the skeleton in disease, development, and evolution. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L, Miles J, Avaron F, Laforest L, Akimenko MA. Exogenous retinoic acid induces a stage-specific, transient and progressive extension of Sonic hedgehog expression across the pectoral fin bud of zebrafish. Int J Dev Biol. 2002;46:949–956. [PubMed] [Google Scholar]
- Horan GS, Kovacs EN, Behringer RR, Featherstone MS. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Hou W, Tian Q, Zheng J, Bonkovsky HL. Zinc Mesoporphyrin Induces Rapid Proteasomal Degradation of Hepatitis C Nonstructural 5A Protein in Human Hepatoma Cells. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhi Q, Patel K, Wilting J, Christ B. Contribution of single somites to the skeleton and muscles of the occipital and cervical regions in avian embryos. Anat Embryol (Berl) 2000;202:375–383. doi: 10.1007/s004290000131. [DOI] [PubMed] [Google Scholar]
- Jarinova O, Hatch G, Poitras L, Prudhomme C, Grzyb M, Aubin J, Berube-Simard FA, Jeannotte L, Ekker M. Functional resolution of duplicated hoxb5 genes in teleosts. Development. 2008;135:3543–3553. doi: 10.1242/dev.025817. [DOI] [PubMed] [Google Scholar]
- Jeannotte L, Lemieux M, Charron J, Poirier F, Robertson EJ. Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Dev. 1993;7:2085–2096. doi: 10.1101/gad.7.11.2085. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symp Ser (Oxf) 2004:211–212. doi: 10.1093/nass/48.1.211. [DOI] [PubMed] [Google Scholar]
- Kopinke D, Sasine J, Swift J, Stephens WZ, Piotrowski T. Retinoic acid is required for endodermal pouch morphogenesis and not for pharyngeal endoderm specification. Dev Dyn. 2006;235:2695–2709. doi: 10.1002/dvdy.20905. [DOI] [PubMed] [Google Scholar]
- Kostic D, Capecchi MR. Targeted disruptions of the murine Hoxa-4 and Hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech Dev. 1994;46:231–247. doi: 10.1016/0925-4773(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Koudijs MJ, den Broeder MJ, Groot E, van Eeden FJ. Genetic analysis of the two zebrafish patched homologues identifies novel roles for the hedgehog signaling pathway. BMC Dev Biol. 2008;8:15. doi: 10.1186/1471-213X-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lewis J, Hanisch A, Holder M. Notch signaling, the segmentation clock, and the patterning of vertebrate somites. J Biol. 2009;8:44. doi: 10.1186/jbiol145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang M, Chen H, Dong Z, Ganapathy V, Thangaraju M, Huang S. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;70:7894–7904. doi: 10.1158/0008-5472.CAN-10-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linville A, Radtke K, Waxman JS, Yelon D, Schilling TF. Combinatorial roles for zebrafish retinoic acid receptors in the hindbrain, limbs and pharyngeal arches. Dev Biol. 2009;325:60–70. doi: 10.1016/j.ydbio.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AH. miR-10 in development and cancer. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- McGlinn E, Yekta S, Mansfield JH, Soutschek J, Bartel DP, Tabin CJ. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre DC, Rakshit S, Yallowitz AR, Loken L, Jeannotte L, Capecchi MR, Wellik DM. Hox patterning of the vertebrate rib cage. Development. 2007;134:2981–2989. doi: 10.1242/dev.007567. [DOI] [PubMed] [Google Scholar]
- Mercader N. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev Growth Differ. 2007;49:421–437. doi: 10.1111/j.1440-169X.2007.00942.x. [DOI] [PubMed] [Google Scholar]
- Mercader N, Fischer S, Neumann CJ. Prdm1 acts downstream of a sequential RA, Wnt and Fgf signaling cascade during zebrafish forelimb induction. Development. 2006;133:2805–2815. doi: 10.1242/dev.02455. [DOI] [PubMed] [Google Scholar]
- Minoux M, Antonarakis GS, Kmita M, Duboule D, Rijli FM. Rostral and caudal pharyngeal arches share a common neural crest ground pattern. Development. 2009;136:637–645. doi: 10.1242/dev.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Melancon E, Eisen JS. Segmental relationship between somites and vertebral column in zebrafish. Development. 2002;129:3851–3860. doi: 10.1242/dev.129.16.3851. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Nolte C, Amores A, Nagy Kovacs E, Postlethwait J, Featherstone M. The role of a retinoic acid response element in establishing the anterior neural expression border of Hoxd4 transgenes. Mech Dev. 2003;120:325–335. doi: 10.1016/s0925-4773(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Oliver G, De Robertis EM, Wolpert L, Tickle C. Expression of a homeobox gene in the chick wing bud following application of retinoic acid and grafts of polarizing region tissue. Embo J. 1990;9:3093–3099. doi: 10.1002/j.1460-2075.1990.tb07506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz-Edwards A, Hardison NL, Linney E. Retinoic a cid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229:89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Schneiderman HA. A clonal analysis of determination in Antennapedia a homeotic mutant of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1969;64:176–183. doi: 10.1073/pnas.64.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Rancourt DE, Tsuzuki T, Capecchi MR. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 1995;9:108–122. doi: 10.1101/gad.9.1.108. [DOI] [PubMed] [Google Scholar]
- Rijli FM, Matyas R, Pellegrini M, Dierich A, Gruss P, Dolle P, Chambon P. Cryptorchidism and homeotic transformations of spinal nerves and vertebrae in Hoxa-10 mutant mice. Proc Natl Acad Sci U S A. 1995;92:8185–8189. doi: 10.1073/pnas.92.18.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Nakatani Y, Takamatsu N, Hori H, Kawakami A, Inohaya K, Kudo A. Medaka unextended-fin mutants suggest a role for Hoxb8a in cell migration and osteoblast differentiation during appendage formation. Dev Biol. 2006;293:426–438. doi: 10.1016/j.ydbio.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Sehm T, Sachse C, Frenzel C, Echeverri K. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev Biol. 2009;334:468–480. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Turnpenny PD, Dunwoodie SL. Disruption of the somitic molecular clock causes abnormal vertebral segmentation. Birth Defects Res C Embryo Today. 2007;81:93–110. doi: 10.1002/bdrc.20093. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- van den Akker E, Fromental-Ramain C, de Graaff W, Le Mouellic H, Brulet P, Chambon P, Deschamps J. Axial skeletal patterning in mice lacking all paralogous group 8 Hox genes. Development. 2001;128:1911–1921. doi: 10.1242/dev.128.10.1911. [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakahara T, Kusu N, Yamauchi H, Kimura I, Konishi M, Miyake A, Itoh N. Fibin, a novel secreted lateral plate mesoderm signal, is essential for pectoral fin bud initiation in zebrafish. Dev Biol. 2007;303:527–535. doi: 10.1016/j.ydbio.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM. Hox genes and vertebrate axial pattern. Curr Top Dev Biol. 2009;88:257–278. doi: 10.1016/S0070-2153(09)88009-5. [DOI] [PubMed] [Google Scholar]
- Wendling O, Ghyselinck NB, Chambon P, Mark M. Roles of retinoic acid receptors in early embryonic morphogenesis and hindbrain patterning. Development. 2001;128:2031–2038. doi: 10.1242/dev.128.11.2031. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Durston AJ. The zebrafish hoxDb cluster has been reduced to a single microRNA. Nat Genet. 2006;38:601–602. doi: 10.1038/ng0606-601. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton KR, Weierud FK, Dietrich S, Lewis KE. Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates. BMC Evol Biol. 2008;8:171. doi: 10.1186/1471-2148-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Zebrafish mir10 and mir196 genes lie in hox clusters and nearby hox genes that contain predicted binding sites for these miRNAs. (A) Arrows represent hox genes and their transcription direction. Grey and black rectangles represent the five mir196 and five mir10 genes. Rows represent hox clusters and columns represent paralogy groups. Grey arrows indicate predicted hox targets for mir196 and black marks predicted hox targets of mir10 from miRBase and Microinspector (Griffiths-Jones et al., 2008; Rusinov et al., 2005). (B–L) In situ hybridization for hox gene neighbors of mir196; whole-mounted zebrafish embryos at 24 hours post-fertilization (hpf) (B–G) and pectoral fins at about 48 hpf (H–I). The hoxcb cluster has no hox9 or hox10 member. In situ hybridization for mir196d primary transcript on fin bud (M).
miR-196 inhibits somite segmentation. The number of somites was scored after either miR-196 overexpression or knockdown. Of 74 untreated control fish, 78.4% developed 30 to 31 somites at 4 dpf. After miR-196 over-expression, only 13.2% of 53 fish developed 30 or 31 somites but most fish (81.1%) developed somites with fewer than 30 somites. After miR-196 knockdown by injection of mir196-Mo, 33.3% of 74 fish developed 30 or 31 somites while the remaining 62.1% developed more than 31 somites.
Manipulation of mir196 expression alters expression of hox-cluster genes. Animals were either untreated (A, D, G, J, M), or were injected in early cleavage with miR-196 duplex (miR-196oe, B, E, H, K, N) or mir196 morpholino (mir196-Mo, C, F, I, L, O) and then at 24 hpf, were prepared for in situ hybridization with the predicted miR-196 targets hoxb5a (A–C), hoxb5b (D–F), hoxb6b (G–I), hoxc6a (J–L), or hoxb8a (M–O) along with egr2a (krox20a) to mark rhombomere-3 and rhombomere-5 in the hindbrain. The anterior border and overall level of hoxb5a, hoxb5b, hoxb6b and hoxc6a expression in the neutral tube became weaker after miR-196 over-expression (A, B, D, E, G, H, J, K, M, N) but was not significantly altered by miR-196 morpholino knockdown (C, F, I, L), asterisk in B indicates expanded posterior CNS expression of hoxb5a. Expression of hoxb8a was strongly inhibited after miR-196 duplex injection (M, N), but was not greatly affected after miR-196 knockdown (O). Like Hoxb8 in tetrapods, the zebrafish hoxb8a 3’UTR has a perfect match to miR-196, but with wobble at two nucleotides (T). To test whether the miR-196 inhibition of hoxb8a expression is mediated by its 3’UTR, we ligated the GFP coding region to the hoxb8a 3’UTR (U) and injected the mRNA into animals either maintained as controls (P), or injected with miR-196a duplex (Q) or miR-196b duplex (R), or with a mixture of miR-196a and miR-196b morpholino (S). Results showed that miR-196 duplex suppressed and miR-196 morpholino augmented GFP expression from this construct. Abbreviations: asterisk in panel B, increased posterior CNS expression of hoxb5a; cns, central nervous system; ro3 and ro5, rhombomere-3 and -5.
Quantifying miR-196. A. To deterimine the effect of morpholino knockdown and miR-196 over-expression on the quantity of miR-196 in the animals, we injected MO or miR-196 duplex into early cleavage stage embryos and evaluated the levels of miR-196 at 27 hpf by qPCR after normalizing values to the U6 RNA control and setting uninjected controls to 1.0. Results showed that mis-matched morpholino (MMO) and mis-matched miR-196 duplex (MMC) had little effect on miR-196 levels, but that maturation-blocking morpholino reduced miR-196 levels at least 20% and that miR-196 injection increased miR-196 levels more than 10 fold for both miR-196a and miR-196b. The graph is broken between relative expression values 2 and 10 and the scale is changed to accommodate the data. B. To learn whether miR-196 is a target of retinoic acid signaling, we treated animals with the diluent DMSO, with RA, or with DEAB, an inhibitor of RA synthesis, and measured levels of miR-196 by qPCR normalized to uninjected controls shown in Fig. s3A.
Retinoic acid signal is regulated by miR-196. YFP signal from individual fish was analyzed by counting the number of YFP pixels. The average number of pixels above threshold was plotted in horizontal bars for each category. The Student t-test showed that miR-196 over-expression significantly inhibited YFP signal (p < 0.001) and miR-196 knockdown significantly increased YFP signal (p = 0.03).
Inhibition of retinoic acid synthesis by DEAB blocks pectoral appendage outgrowth and the expression of pectoral appendage genes. (A, C, E, G, I, K, M, O, Q) DMSO treated controls. (B, D, F, H, J, L, N, P, R) Animals treated with 5×10−7 M DEAB in fish water from 100% epiboly to 24 hpf. At (A, B) 28 hpf, (C, D) 48 hpf, (E, F) 120 hpf, (G, H) 45 dpf, DEAB delivered in this protocol specifically blocks pectoral fin initiation without substantial effects on other developmental systems (Gibert et al., 2006; Wakahara et al., 2007). In situ hybridization analysis of gene expression for (I, J) fgf24 at 19 hpf; (K, L) fgf24 at 24 hpf; (M, N) tbx5a at 24 hpf; (O, P) fgf10a at 24 hpf; (Q, R) lbx1b at 48 hpf. Inhibiting RA synthesis specifically inhibits the expression of genes in the program of pectoral fin development and leaves other expression domains intact. cns, central nervous system; ey, eye; he, heart; ov, otic vesicle; pa, pharyngeal arch; pf, pectoral fin.
An excess of RA perturbs pectoral fin outgrowth. (A) DMSO-treated controls at 120 hpf have normal pectoral fins. (B, C) 120 hpf animals that had been treated with 10−7 M RA from 12 to 14 hpf have aberrant pectoral fins and other abnormalities. (D) Untreated wild-type control at 120 hpf. (E, F) Homozygous cyp26a1rw716 mutants at 120 hpf show phenotypes characteristic of excess RA. (G) Wild-type pectoral fin bud. (H–J) Fin buds of cyp26a1rw716 mutants. (K) The predicted binding site in the 3’UTR of cyp26a1 was aligned with miR-196 showing seed region pairing. (L) Schematic drawing of Luciferase reporter construct for cyp26a1 3’UTR. (M) Firefly luciferase reporter with cyp26a1 3’UTR was interrogated with miR196. Relative firefly luciferase intensity was normalized with control renilla luciferase. Result shows that miR-196 manipulation did not affect the luciferase signal. Standard deviations for 6 independent repeats were plotted as error bars.These results show that an excess of RA causes variable reductions in fin bud development due to problems with the endochondral disc, although the shoulder girdle is largely normal. Abbreviations: cl, cleithrum; ed, endochondral disc; pf, pectoral fin; sco, scapulocoracoid.
Two-color double label expression analyses for rarab, tbx5, and mir196. (A–F) Expression of rarab (A, D, green) showing expression in the central nervous system (cns) and lateral plate mesoderm (lpm) and expression of tbx5a (B, E, red) showing expression in the lateral plate mesoderm and pectoral fin bud (pf) and the merged images (C, F) at 24 hpf (A–C) and 48 hpf (D–F). (G–L) Expression of rarab (G, J, green) showing expression in the central nervous system (cns) and lateral plate mesoderm (lpm) and expression of mir196 (H, K, red) showing broad expression including the central nervous system, lateral plate mesoderm and pectoral fin bud and the merged images (I, L) at 24 hpf (G–I) and 48 hpf (J–L).
The 3’UTR of rarab responds to miR-196. Levels of GFP expression above background (yolk cell) for the experiment in Fig. 5H–K were quantified by setting a threshold using Adobe Photoshop to calculate the average number of green pixels per fish for each treatment. Standard deviations for 4 independent repeats were plotted as error bars.
miR-196 has no effect on rarab transcript accumulation. (A–C) raraa expression. (D–F) rarab expression. (A, D) Uninjected controls. (B, E) miR-196 injected embryos. (C, F) mir196-morpholino treated embryos. miR-196 levels do not effect quantities of raraa or rarab transcript. These results suggest that miR-196 does not act to strongly decrease the level of rarab transcript quantity, so it likely exerts its demonstrated effect on rarab-mediated retinoic acid signaling at the level of translation.