Abstract
Human ELAV proteins are implicated in cell growth and differentiation via regulation of mRNA expression in the cytoplasm. In human embryonic teratocarcinoma (hNT2) cells transfected with the human neuronal ELAV-like protein, Hel-N1, neurites formed, yet cells were not terminally differentiated. Cells in which neurite formation was associated with Hel-N1 overexpression, also expressed increased levels of endogenous neurofilament M (NF-M) protein, which distributed along the neurites. However, steady-state levels of NF-M mRNA remained similar whether or not hNT2 cells were transfected with Hel-N1. These findings suggest that turnover of NF-M mRNA was not affected by Hel-N1 expression, despite the fact that Hel-N1 can bind to the 3′ UTR of NF-M mRNA and was found directly associated with NF-M mRNA in transfected cells. Analysis of the association of NF-M mRNA with the translational apparatus in Hel-N1 transfectants showed nearly complete recruitment to heavy polysomes, indicating that Hel-N1 caused an increase in translational initiation. Our results suggest that the stability and/or translation of ARE-containing mRNAs can be regulated independently by the ELAV protein, Hel-N1, depending upon sequence elements in the 3′ UTRs and upon the inherent turnover rates of the mRNAs that are bound to Hel-N1 in vivo.
Keywords: RNA stability, AU-rich elements, ARE, differentiation, translation, hNT2, NF-M
mRNA localization, stability, and translation in the cytoplasm have been recognized as important mechanisms of regulation of gene expression (Dreyfuss et al. 1996; Jacobson and Peltz 1996; Wickens et al. 1997). Many growth-regulatory mRNAs contain AU-rich sequence elements (AREs) in their 3′-untranslated regions (UTRs) that have been implicated in regulating and influencing mRNA decay and translation. Recently, several reports have demonstrated that human ELAV proteins are trans-acting factors affecting ARE-mediated mRNA stability both in vivo and in vitro (Jain et al. 1997; Fan and Steitz 1998a; Keene 1999; Levy et al. 1998; Peng et al. 1998; Ford et al. 1999).
elav was discovered as a gene essential for the development and maintenance of the nervous system of Drosophila (Campos et al. 1985; Robinow and White 1988). Four homologs of the ELAV protein have been identified in vertebrates and the cDNAs cloned: HuA (HuR)—expressed in all tissues; HuB (Hel-N1)—expressed in ovaries, testes, and neurons; and HuC and HuD—expressed only in neurons (Good 1995; Antic and Keene 1997). All elav genes encode proteins containing three RNA recognition motifs (RRMs). Mammalian ELAV proteins are tumor-specific antigens in paraneoplastic neurologic disorders (Darnell 1996), and were found to bind with high affinity to mRNAs containing AREs in their 3′ UTRs including c-myc, c-fos, and GM-CSF (Levine et al. 1993; Antic and Keene 1997; Keene 1999). These RNA sequences resemble the AREs in the 3′ UTRs of many growth-regulatory and cytokine mRNAs (Caput et al. 1986) that have been shown to mediate mRNA decay (Shaw and Kamen 1986). Subsequent studies confirmed interactions of ELAV proteins with these sequences in numerous mRNAs (Gao et al. 1994; King et al. 1994; Liu et al. 1995; Abe et al. 1996; Chagnovich and Cohn 1996; Chung et al. 1996; Chung et al. 1997; Myer et al. 1997)
Transfection studies have implicated ELAV proteins more directly in post-transcriptional regulation of mRNA expression (Jain et al. 1997; Fan and Steitz 1998a; Levy et al. 1998; Peng et al. 1998). For example, 3T3 L1 cells, which are capable of being induced chemically to differentiate into adipocytes, showed an enhanced differentiation phenotype following ectopic expression of Hel-N1 (Jain et al. 1997). This was caused, in part, by increased expression of the glucose transporter 1 (GLUT1) protein following addition of a differentiation inducer to the transfected cells. These experiments indicated that endogenous mRNA encoding GLUT1 protein was both stabilized and translated more efficiently in Hel-N1-transfected cells. Although GLUT1 is considered to be an early-response gene (ERG) (Cornelius et al. 1991), the more classical growth-regulatory gene, c-myc, was also found to be stabilized following Hel-N1 expression (L.G. Andrews, R.G. Jain, N. Lu, P.H. Pekala, J.D. Keene, unpubl.). However, in both of these experiments, interactions between the protein and the endogenous mRNAs were only demonstrated in vitro. Thus, the observed change in the metabolism of GLUT1 and c-myc mRNAs and the phenotype of 3T3 L1 cells was correlated with Hel-N1 expression. Similarly, transient cotransfection of mouse L929 cells with HuA (HuR) and β-globin-reporter cDNA constructs containing c-fos or GM-CSF AREs resulted in their coincident stabilization presumably by interactions with HuR (Fan and Steitz 1998a; Peng et al. 1998). However, direct association of HuR with the reporter mRNAs in these transfected cells was not demonstrated. Likewise, HuR binds to the instability element in the 3′ UTR of vascular endothelial growth factor (VEGF) mRNA in vitro and overexpression of HuR in 293T cells was correlated with VEGF mRNA stabilization under hypoxic conditions. Although VEGF mRNA stabilization was observed by adding recombinant HuR to cell extracts, direct association of HuR with VEGF mRNA was not demonstrated in transfected cells or in extracts of transfected cells (Levy et al. 1998). Another study addressing a role of Hu proteins in neurogenesis demonstrated that misexpression of chicken HuD (cHuD) in cultured neural crest cells correlated with the development of neuronal morphology, expression of neuronal markers, and dependence on neurotrophin suggesting involvement of cHuD in regulation of neuronal differentiation. However, neither RNA interactions nor the underlying mechanisms of this phenomenon were addressed (Wakamatsu and Weston 1997).
To investigate the role of human neuronal ELAV proteins in regulating cell growth and differentiation we have stably transfected the human ELAV cDNA encoding Hel-N1 into human embryonic teratocarcinoma (hNT2) cells, a line that can be induced to differentiate into neurons by retinoic acid (RA) treatment. We report that Hel-N1-transfected hNT2 cells developed neurite-like processes even in the absence of RA induction, and showed an enhanced differentiation phenotype upon subsequent stimulation with RA. Furthermore, hNT2 cells failed to develop neurites, and did not express detectable amounts of either ELAV or neurofilament M (NF-M) proteins, following transfection with antisense Hel-N1 cDNA and induction with RA, suggesting an essential function of the HuB gene class in neuronal differentiation. Analysis of the expression of neuronal markers in Hel-N1-transfected hNT2 cell lines showed elevated expression of NF-M protein, which was caused by increased translation of NF-M mRNA. We demonstrate direct interaction of Hel-N1 protein with the NF-M mRNA in vivo in conjuction with nearly complete recruitment of the NF-M mRNA to heavy polysomes. Although Hel-N1 was bound to NF-M mRNA in vivo, the steady-state levels of this mRNA did not appear to be affected. We conclude that the ELAV protein, Hel-N1, can affect the translation of an inherently stable target mRNA that encodes a differentiation marker, NF-M, by direct interaction with its mRNA in transfected human teratocarcinoma cells. We propose that the fate of certain ARE-containing mRNAs can be controlled by cis sequences that interact with ELAV proteins and engage them in translation without necessarily affecting mRNA turnover.
Results
ELAV proteins and neuronal differentiation
To investigate the functions of human ELAV proteins in regulating gene expression during neuronal differentiation we used hNT2 cells as a model system because they can be induced to differentiate into neurons (hNTN) by RA treatment (Andrews 1984; Pleasure and Lee 1993). Such treatment of hNT2 cells results in the formation of cultures containing ∼5% terminally differentiated neurons that coexist with a population of unidentified large, flat, dividing cells and a residual number of undifferentiated stem cells (Andrews 1984; Pleasure et al. 1992). In contrast to these dividing cells, terminally differentiated neurons in these cultures are resistant to treatment with mitotic inhibitors. Approximately two weeks after differentiation the hNTN cells migrate to form large cellular aggregates that are connected by numerous neuronal processes (Andrews 1984; Pleasure et al. 1992).
hNT2 cells have been characterized also for the expression of neuron-specific markers both before and during the induction of differentiation (Pleasure and Lee 1993). Following RA induction of hNT2 cells, we confirmed the previously reported (Pleasure and Lee 1993) appearance of neuronal markers such as NF-M, NFM(P+++), and MAP2 (Fig. 1). The up-regulation of f-tau and down-regulation of cytokeratin expression were similar to these previous results (data not shown). Whereas Pleasure and Lee (1993) detected small amounts of NF-M expression in uninduced hNT2 cells, our cells did not express detectable levels of NF-M unless they were induced with RA. We examined the expression of ELAV proteins and other known neuronal markers by double immunostaining of cells with antibodies. As expected, neuronal ELAV proteins were not expressed in uninduced hNT2 cells (Fig. 1A) but ELAV became apparent by day 5 after RA induction (Fig. 1B). Although some differentiating cells were stained only for ELAV proteins at this stage (Fig. 1B, red), a few cells were coexpressing ELAV and another early neuronal marker, NF-M (Fig. 1B, yellow). Expression of ELAV proteins remained high throughout differentiation and in mature neurons (Fig. 1, red), as shown in other cell systems (Gao and Keene 1996; Antic and Keene 1997; Antic and Keene 1998). These results are consistent with a role of ELAV proteins at an early step in neuronal differentiation.
Figure 1.
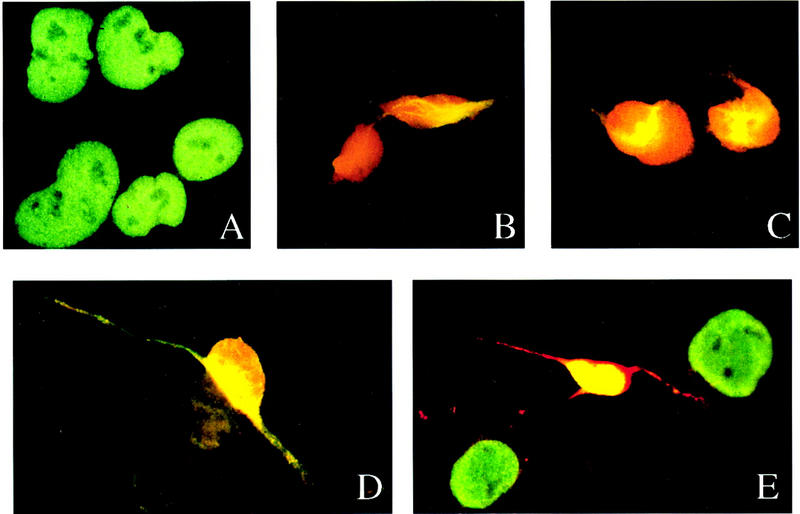
ELAV proteins are up-regulated during neuronal differentiation of human embryonal teratocarcinoma (hNT) cells. (A) Uninduced hNT2 cells double stained with anti-Hel-N1 antibody (red) and anti-hnRNP C antibody (green); (B) hNT cells at day 5 of induction stained with anti-Hel-N1 antibody (red) and anti-NF-M antibody (green, and appears yellow because of overlap with red); (C) hNT cells at day 15 of induction stained with anti-Hel-N1 antibody (red) and anti-NF-M(P+++) (phosphorylated NF-M) antibody (green, and appears as yellow because of overlap with red). (D,E) hNTN cells at day 32 (4 weeks) of induction stained with (D) anti-Hel-N1 antibody (red) and anti-MAP2 antibody (green, and appears as yellow because of overlap with red) and (E) anti-Hel-N1 antibody (red) and anti-hnRNP C antibody (green, and yellow because of overlap with red). (D,E) Only mature neurons and not the supporting nonneuronal cells (stained green in E) are expressing ELAV proteins.
To determine whether ELAV proteins are themselves sufficient to drive differentiation, we carried out both stable and transient transfections. We used the HuB (Hel-N1) cDNA because it is the earliest ELAV protein expressed during neurogenesis (Okano and Darnell 1997; Wakamatsu and Weston 1997) and it is expressed early in embryonic development (Good 1995; Antic and Keene 1997). Uninduced hNT2 cells were transfected with the Hel-N1 cDNA in the sense antisense orientation fused to the GFP gene. As a control, cells were transfected with vector containing only the GFP gene [Fig. 2A; marked as ‘vector’ (V) throughout the text]. All stable cell lines were compared ultimately to untransfected control cells. As demonstrated in Figure 2, vector-transfected or antisense-transfected cells displayed phenotypes identical to untransfected hNT2 cells (Fig. 2B,D,F). Surprisingly, Hel-N1-transfected cells exhibited a neuron-like morphology, that is, they developed neurite-like processes (Fig. 2H) and resembled the RA-differentiated hNTN cells (Fig. 2C). To determine if Hel-N1 transfection caused terminal neuronal differentiation, we treated transfected cells with a cocktail of mitotic inhibitors that resulted in death of all cells in the culture (not shown). We conclude that the Hel-N1-transfected cells were not terminally differentiated.
Figure 2.
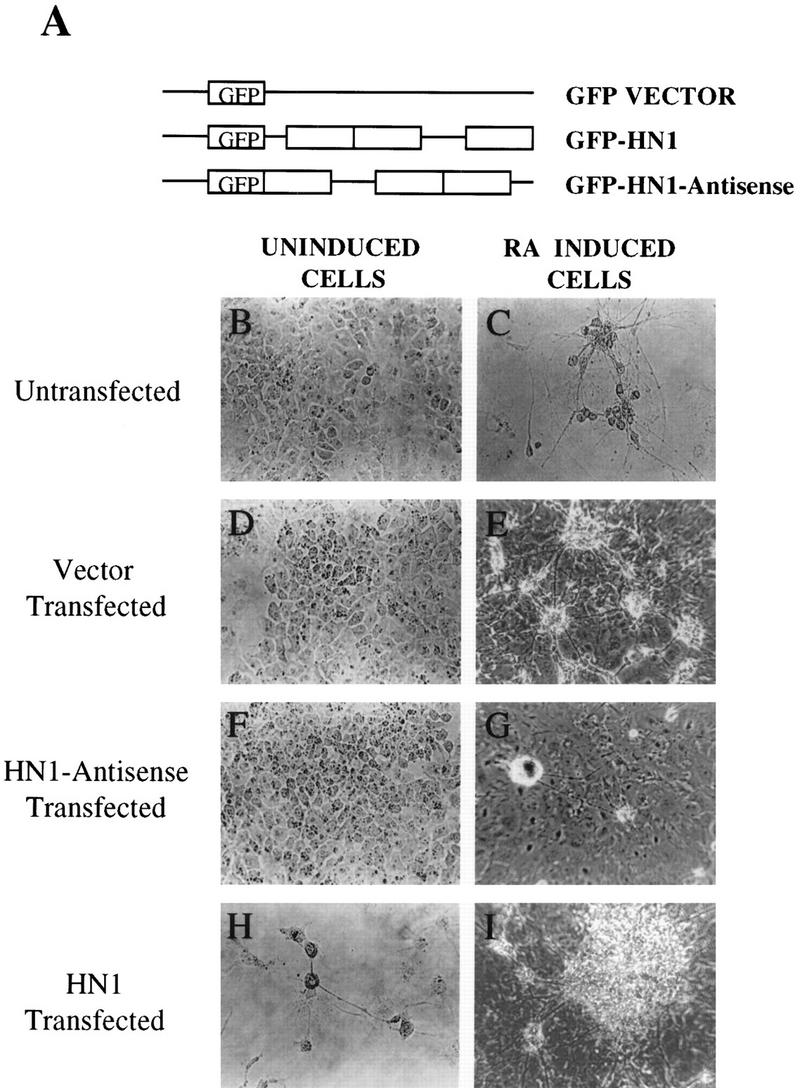
Morphology of stably transfected hNT2 cell lines. (A) Constructs used for transfections with ELAV protein, Hel-N1, were subcloned either into pHβAPr-1–neo or into pGFPC1 plasmids (see Materials and Methods). Untransfected (B,C), vector transfected (D,E), Hel-N1–antisense transfected (F,G), and Hel-N1-transfected (H,I) cells before (B,D,F,H) or after (C,E,G,I) the RA treatment. (C,H) Photographed with a 32× lens and are focused on the area containing isolated neurons (C) or neuron-like cells (H). Other panels were photographed with a 10× lens using a Zeiss inverted microscope and show clumps containing large numbers of neurons. Untransfected, RA-induced cells were morphologically indistinguishable at all magnifications from the vector-transfected, RA-induced cells shown in E.
In exploring the capacity of transfected cells to differentiate into neurons when induced with RA, we first demonstrated that vector-transfected cells were able to differentiate just like untransfected control cells (Fig. 2, cf. E and C; note different magnifications). However, cells transfected with Hel-N1 showed what appeared to be enhanced differentiation, following RA induction, as measured by the 1.5–2 times increase in the number of terminally differentiated neurons (see Materials and Methods), compared to vector transfected cells (Fig. 2, cf. panels I and E) . Moreover, Hel-N1-transfected cells invariably formed much larger neuronal clumps (Fig. 2, cf. I and E; both with lower magnification) that were distributed evenly throughout the culture. Interestingly, antisense Hel-N1-transfected cells appeared to be significantly impaired in their ability to differentiate into neurons following RA treatment (Fig. 2, cf. G and E). Typically, only a few clusters of neurons were observed per plate (Fig. 2G) suggesting an important role of the elav gene(s) for neuronal differentiation. Furthermore, transfection of another related RRM protein, Hel-N2, into hNT2 cells did not cause development of processes (not shown) demonstrating specificity of the Hel-N1 effect.
Expression of introduced cDNAs in transfected cells was examined and compared to the expression of endogenous ELAV proteins by Western blotting (not shown). Induction of differentiation of vector and Hel-N1-transfected cells (which were both capable of differentiating into neurons) caused dramatic up-regulation of endogenous ELAV protein expression. On the contrary, endogenous ELAV proteins were not up-regulated in antisense-Hel-N1-transfected cells following RA treatment. Although all of the antisense-Hel-N1-transfected lines were impaired greatly in their ability to produce neurons after the RA treatment, we observed small differences in the number of neuronal clumps obtained after differentiation. Examination of these lines by Western blotting revealed that the levels of endogenous ELAV proteins varied from very low to undetectable (not shown). The observed reduction in levels of all ELAV proteins by transfected antisense-Hel-N1 cDNA could be because of interaction of the full-length antisense-Hel-N1 cDNA with mRNAs encoding other members of the ELAV family. Alternatively, expression of HuB, the earliest elav gene expressed in neurogenesis (Pleasure et al. 1992; Good 1995; Okano and Darnell 1997; Wakamatsu and Weston 1997; Atasoy et al. 1998) may be required for the expression of all subsequently expressed neuronal ELAV proteins. We believe that the second possibility is most likely because we could not detect expression of other neuronal markers, such as NF-M, NF-M(P+++), or MAP2, in our RA-induced antisense-Hel-N1 transfected cells.
The results presented above implicate the HuB gene in an important pathway regulating the expression of proteins involved in the development of neuronal processes. Therefore, the expression of neuron-specific markers was examined in transfected hNT2 cell lines.
Hel-N1-transfected hNT2 cells up-regulate NF-M protein without RA induction
The development of the neuron-like morphology by Hel-N1-transfected cells without the addition of RA to induce differentiation (Fig. 2H) was unexpected. Because it is likely that these cells express some neuron-specific markers, we performed immunofluorescence with a panel of antibodies against neuronal proteins (NF-M, NF-M(P+++), MAP2, and f-tau) which are characteristically up-regulated during differentiation, as well as antibodies against cytokeratins that are down-regulated following RA induction of differentiation (Pleasure and Lee 1993). As expected, we found that the expression of cytokeratins was not altered in Hel-N1-transfected cells (not shown) because transfectants did not differentiate (see above) but only developed a partial neuronal morphology. When examined by immunofluorescence we found that NF-M was up-regulated in Hel-N1-transfected cells (Fig. 3C,D), but not in vector (Fig. 3A) or antisense-Hel-N1 transfected cells (Fig. 3 B). Surprisingly, only ∼50% of the cells, in each Hel-N1-transfected stable cell line, were stained for NF-M. It is possible that the levels of NF-M protein were too low to be detected by immunofluorescence in all cells; alternatively, only a subpopulation of cells have the capacity to become neurons and the potential to express NF-M. Not all of the Hel-N1-transfected cells developed neuron-like morphology (Fig. 2H). Even under optimal conditions, after RA induction of hNT2 cells only ∼5% of cells in the population represent terminally differentiated neurons (Pleasure et al. 1992), consistent with precursor hNT2 cells being heterogeneous in their ability to differentiate into mature neurons. We observed that after ∼30 passages in culture, Hel-N1-transfected cells reverted to the non-neuronal phenotype that was associated with the loss of the detectable levels of the ectopically expressed GFP–Hel-N1 protein. Whereas these results further correlated the expression of the NF-M protein with expression of Hel-N1, they clearly indicated a limitation in the ability to continuously propagate and analyze stably transfected hNT2 cell lines.
Figure 3.
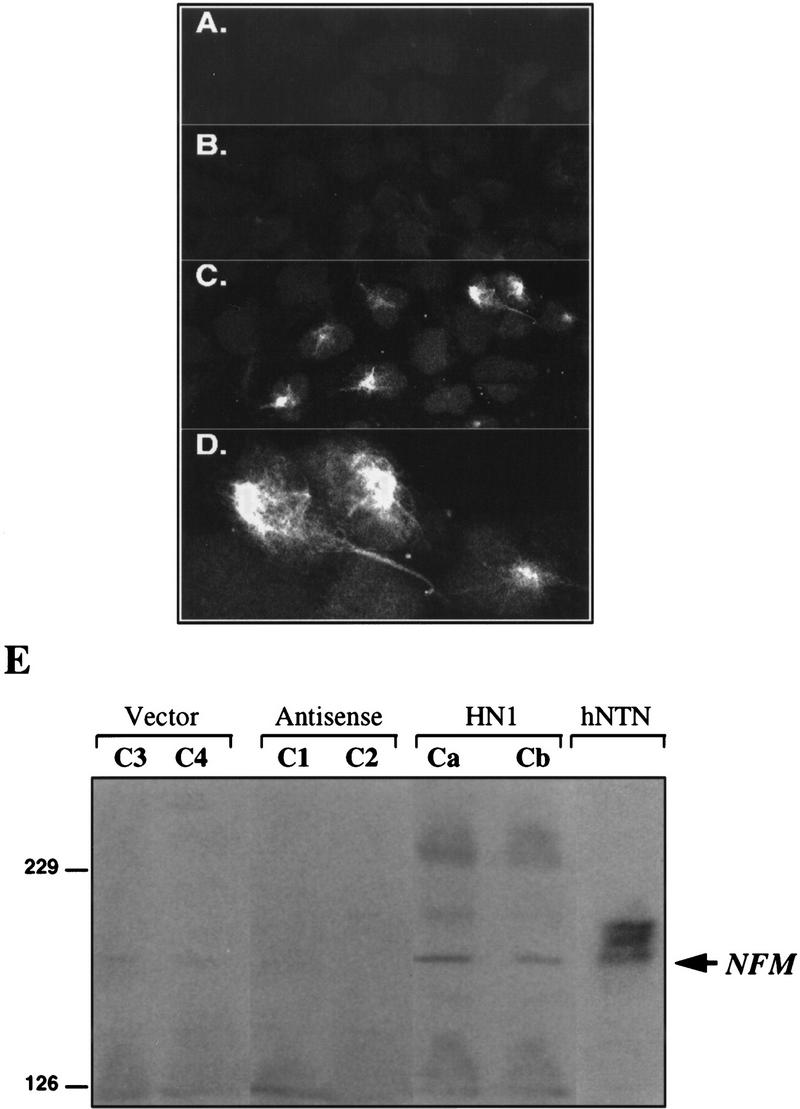
Expression of NF-M protein in cells stably transfected with Hel-N1. Confocal microscopy images of vector-transfected cells (A), antisense-Hel-N1-transfected cells (B), and Hel-N1-transfected cells immunostained with anti-NF-M antibody(C,D). (D) Magnified top right corner of C. (E) Immunoblot stained with anti-NF-M antibody. Total cell extracts (150 μg) from two vector-transfected clones (C3 and C4), two antisense-Hel-N1-transfected clones (C1 and C2), two Hel-N1-transfected clones (Ca and Cb), and untransfected differentiated neurons (hNTN) were loaded onto 6% acrylamide gels and analyzed for NF-M expression by ECL immunoblotting.
Expression of NF-M protein, in the Hel-N1-transfected cell lines, was confirmed by Western blotting of cell extracts (Fig. 3E) prepared from two vector clones (C3 and C4), two antisense Hel-N1 clones (C1 and C2), and two Hel-N1-expressing clones (Ca and Cb) of hNT2 cells. We used extracts prepared from untransfected mature neurons (hNTN) as a positive control. The three bands observed in hNTN neurons correspond to NF-M (lower of the three bands indicated by arrow) and to two phosphorylated forms of NF-M (two upper bands) which we also detected using an antibody directed against phosphorylated NF-M peptides, NF-M(P+++) (Black and Lee 1988; Pleasure and Lee 1993; and data not shown).
We conclude that Hel-N1-transfected hNT2 cells express NF-M protein, which forms filamentous structures (Fig. 3D), thus leading to the appearance of neurite-like processes.
Regulation of NF-M expression
The observed expression of NF-M protein in uninduced Hel-N1-transfected cells could involve regulation at any of several levels of gene expression including transcription, post-transcriptional RNA processing, and/or translation. To address these possibilities, we investigated the presence of NF-M mRNA in untransfected and transfected hNT2 cells. We also used HepG2 epithelial cells as a negative control for the neuronal hNT2 precursors. We found that NF-M mRNA was expressed both in untransfected and in all transfected cell lines (not shown).
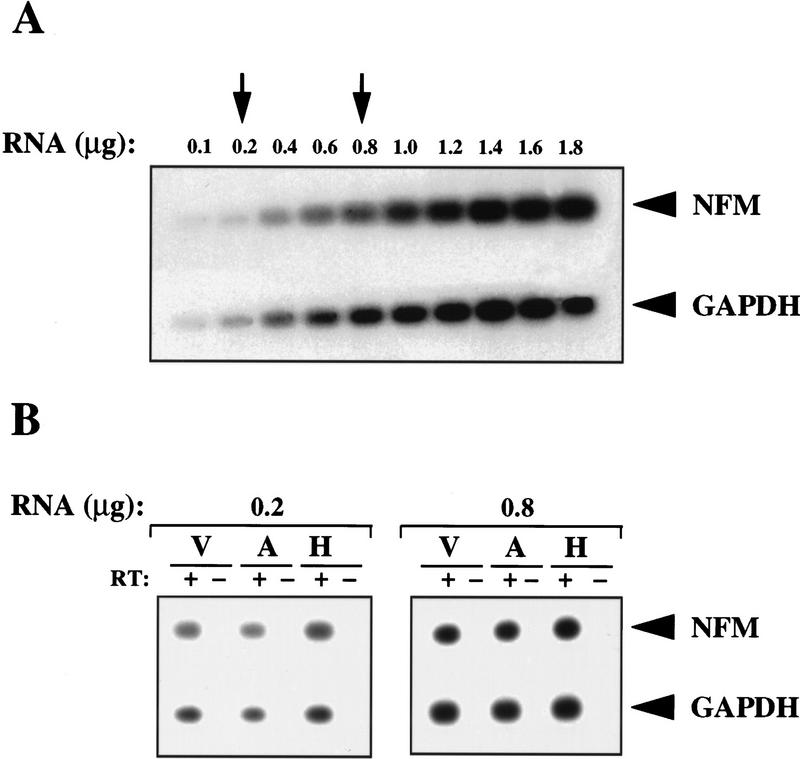
To compare mRNA levels in different transfectants we carried out a quantitative RT–PCR analysis. This approach was necessary as levels of NF-M mRNA in both untransfected or transfected cells were too low to be detected by Northern blotting or by S1 nuclease protection assays (not shown). Quantitation was performed over a range of input total RNA concentrations (Fig. 4A) and the number of PCR cycles was determined empirically so that concentration-dependent curves could be obtained (not shown). To standardize the assay, the RT reaction was carried out using random hexanucleotides, and subsequent PCR reactions were performed using specific primers. Two concentrations of total RNA (indicated by arrows in Fig. 4A) were chosen for the assay. The concentration of 0.8 μg/ml represents the middle range of the dose-response curve and the concentration of 0.2 μg/ml was chosen as an amount of total RNA that would be sufficiently small to avoid any possible saturation in the PCR reactions. Because GAPDH mRNA was not bound by Hel-N1 protein in vivo (Fig. 6, below) and was present in approximately the same amounts in all transfected cells as determined by Northern blotting (not shown), we have normalized NF-M mRNA to the levels of GAPDH mRNA (Fig. 4B). Using either of the two total RNA concentrations noted above, (0.8 or 0.2 μg/ml), we obtained similar results indicating that NF-M mRNA levels did not differ significantly between the vector (V), antisense-Hel-N1 (A), and Hel-N1 (H) transfected cell lines. These findings are consistent with the conclusion that the turnover of this mRNA was not affected significantly by overexpression of Hel-N1. Interestingly, as reported previously (Schwartz et al. 1992) and confirmed by us (not shown), NF-M is a stable mRNA whose half-life is longer than 4 hr. It is likely that the atypical AREs in the 3′ UTR of NF-M mRNA differ from canonical AREs found in unstable mRNAs encoding c-fos, c-myc, GM-CSF, or GLUT1 in that they do not confer properties of instability, yet they still bind to Hel-N1 in vivo (Jain et al. 1997; Fan and Steitz 1998a; Peng et al. 1998) Therefore, it is likely that increased expression of NF-M protein in Hel-N1-transfected cells was caused by increased translation of this mRNA.
Figure 4.
Comparative levels of NF-M mRNA in stably transfected cell lines as determined by quantitative RT–PCR. (A) Southern blot—titration of RNA concentrations using total RNA from Hel-N1 transfected cells: dose-response for the NF-M (20 PCR cycles) and GAPDH mRNAs (15 PCR cycles). (B) Southern blots—quantitative analysis of NF-M mRNA in two pooled vector transfected clones (V), two pooled antisense-Hel-N1 transfected clones (A), or in two pooled Hel-N1 transfected clones (H) using 0.2 or 0.8 μg of total RNA. The RT step was performed either with (+) or without (−) reverse transcriptase. Values obtained for the NFM/GAPDH ratios were 0.5, 0.4, and 0.7 using 0.2 μg; and 0.7, 0.8, and 0.9 using 0.8 μg of V, A, and H RNA, respectively. These values were within experimental error based upon numerous repeats.
Figure 6.
Immunoprecipitation of NF-M mRNA GFP–Hel-N1 complexes from stably transfected cells demonstrates their direct association in vivo. Cell extracts of two pooled vector transfected clones (V), two pooled antisense Hel-N1 transfected clones (A), or two pooled Hel-N1-transfected clones (H) were used for immunoprecipitation with the following monoclonal antibodies (MAb): anti-gene 10 (g10), anti-GFP (GFP), or anti-U1–70K (70K). RNA was extracted from immunoprecipitated pellets and used for RT–PCR either with (+ RT) or without (−RT) reverse transcriptase. Total RNA lane (t) was used as a positive control. Primers specific for NF-M mRNA, GAPDH mRNA, or U1 RNA were employed both for the RT and for the PCR steps. (C,D) Southern blots of agarose gels shown in A and B probed with an oligonucleotide to NF-M (NFM probe) and GAPDH (GAPDH probe), respectively. (E) Southern blot probed simultaneously with an oligonucleotide specific for the U1 RNA (U1 probe) and with an oligonucleotide specific for the NF-M mRNA (NFM-probe).
Binding of Hel-N1 to the 3′ UTR of NF-M mRNA
Direct post-transcriptional or translational regulation of NF-M mRNA expression by Hel-N1 protein would require physical interaction between Hel-N1 and NF-M mRNA. To investigate a direct RNA–protein interaction between these molecules we began with binding studies in vitro. The 3′ UTR of NF-M mRNA contains several U-rich motifs, as noted by asterisks (Fig. 5A). These motifs do not represent typical AREs containing clustered AUUUA pentamers, like those found in many proto-oncogene and cytokine mRNAs (Caput et al. 1986; Shaw and Kamen 1986), but they are similar to the non-AUUUA type, class III ARE (Peng et al. 1996). As presented in Figure 5, Hel-N1 binding to the 3′ UTR of NF-M mRNA was demonstrated by both mobility shift (Fig. 5B) and immunoprecipitation (Fig. 5C) of bound complexes. Interestingly, binding was observed only when most of the 3′ UTR (DraI and TaqI fragments) was used in the reactions. Also, the DraI fragment (Fig. 5A) contains only one additional U-rich motif compared to the Afl II fragment (Fig. 5A), suggesting that the conformation of the larger fragment was necessary for Hel-N1 binding. However, binding observed with the DraI fragment was four times less efficient than binding with the TaqI fragment, as determined by immunoprecipitated radiolabeled RNA molecules (Fig. 5C, cf. lanes 4 and 5), suggesting that the last three U-rich motifs are necessary for the optimal interaction between NF-M mRNA and Hel-N1 protein. To determine whether the last four U-rich motifs are sufficient for interaction with Hel-N1 protein, we tested binding of Hel-N1 to the small fragment (Fig. 5A, fragment 6) containing these four U-rich motifs. We found that Hel-N1 protein was only able to partially shift this RNA in the mobility shift assay (Fig. 5B, lanes 6). Binding of this small fragment was confirmed by immunoprecipitation experiments (not shown). However, the amount of RNA bound in this assay was ∼10 times less than that of the TaqI fragment, further confirming earlier data suggesting that the most efficient binding was obtained when all U-rich elements were present in the nearly full-length 3′ UTR of NF-M mRNA. It is important to note that for all RNA-binding experiments, RNA concentrations were adjusted to equimolar amounts.
Figure 5.
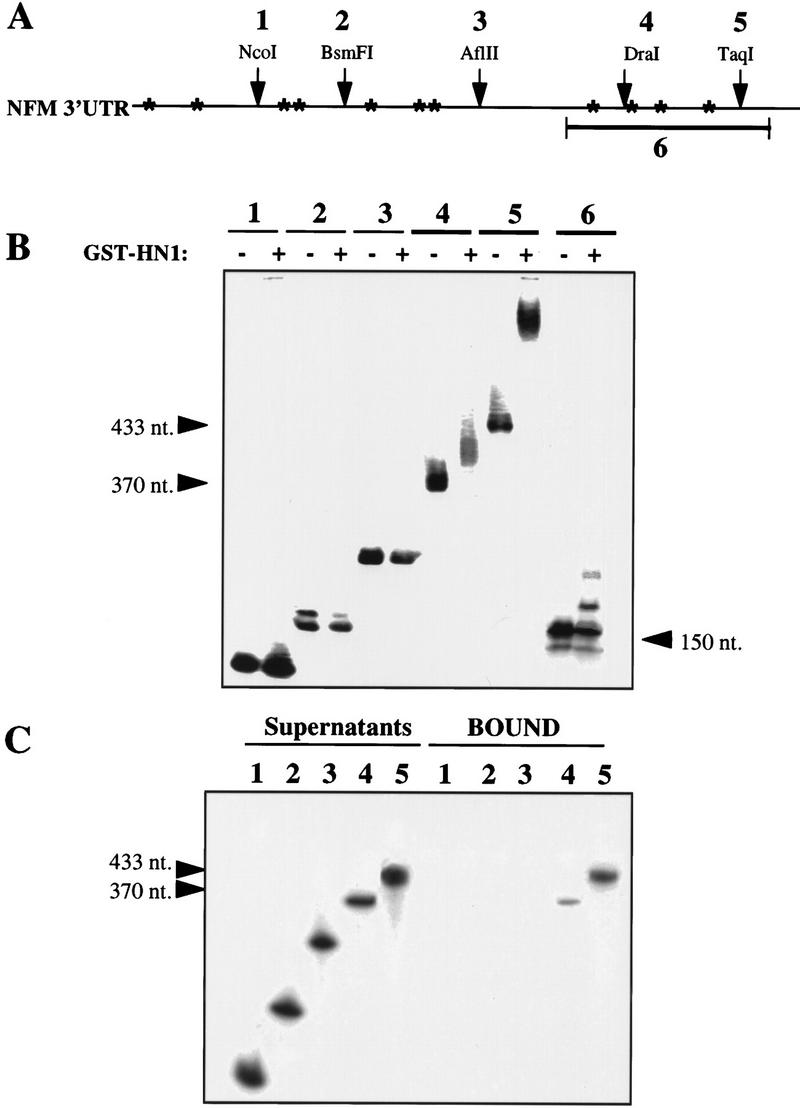
RNA binding by mobility shift or immunoprecipitation of NF-M 3′ UTR using recombinant GST–Hel–N1 fusion protein. (A) Partial restriction map and position of U-rich sequences (marked by *) in the 3′ UTR of NF-M mRNA. Numbers 1–6 indicate fragments of the 3′ UTR used for RNA-binding experiments. (B) Mobility shift of in vitro transcribed RNAs representing each fragment of the NF-M 3′ UTR (as in A). RNAs that were bound by recombinant GST–Hel-N1 fusion protein are marked by arrowheads and their size is labeled. RNAs were analyzed on nondenaturing polyacrylamide gels (C) Immunoprecipitation of in vitro-transcribed RNAs using recombinant GST–Hel-N1 fusion protein: RNAs representing fragments 1–5 were bound to GST–Hel-N1 protein and immunoprecipitated with anti-Hel-N1 antibody and protein A beads. Bound RNAs were extracted from the beads (bound) and remaining unbound RNAs were extracted from the supernatants. RNAs were analyzed on denaturing urea-polyacrylamide gels. Bound fragments 4 and 5 are marked by arrowheads.
These data demonstrate interactions between the Hel-N1 protein and the 3′ UTR of NF-M mRNA in vitro. However, this protein–RNA interaction would be relevant to the biological effects observed only if it occurs in vivo.
Association of Hel-N1 protein with NF-M mRNA in vivo
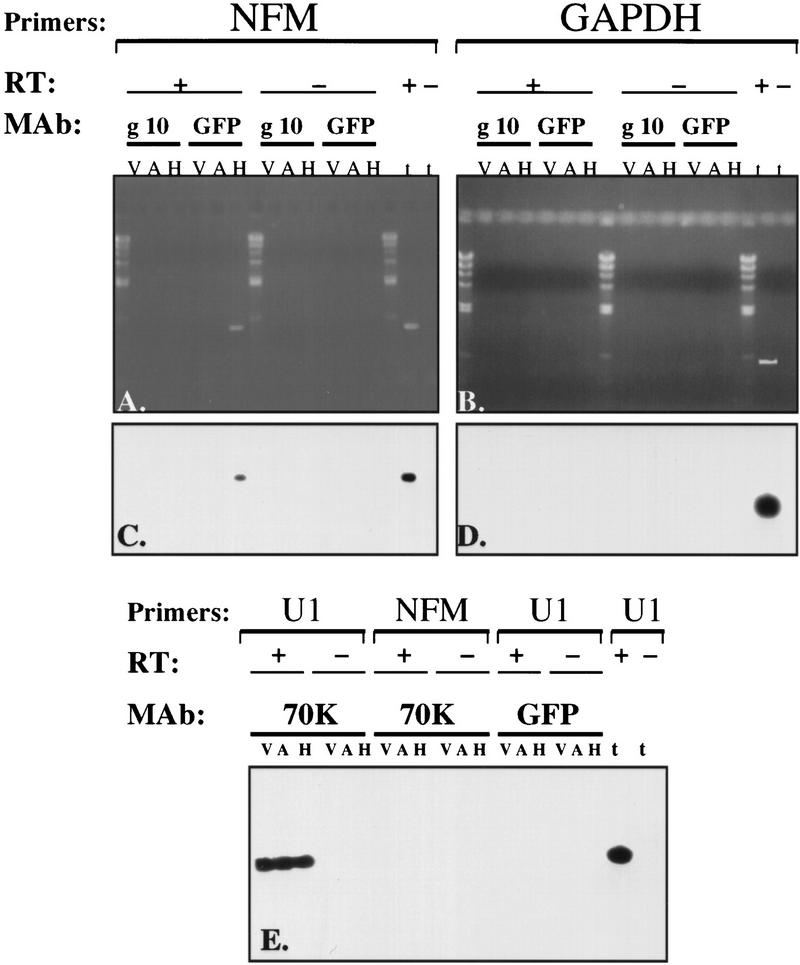
Experimental support for direct association of ELAV proteins with specific mRNAs has not been possible, in part, because they are in RNP complexes and not accessible to antibodies specific for ELAV (Levine et al. 1993; Gao et al. 1994; D. Antic and J.D. Keene, unpubl.). To investigate whether this would be possible using an epitope tag, we carried out mRNP immunoprecipitation of cell extracts prepared from vector-transfected, antisense Hel-N1-transfected, and Hel-N1-transfected cells using a monoclonal anti-GFP antibody (Fig. 6A–D). We also used anti-U1-70K monoclonal antibody as a control for RNP precipitation (Fig. 6E). Nonspecific background immunoprecipitation was evaluated using the anti-gene10 monoclonal antibody (Fig. 6A–D) described by Tsai et al. 1992 as control. The efficiency of immunoprecipitation was determined by examination of protein and RNA in the pellets (Fig. 6).
After demonstrating quantitative removal of the GFP-tagged Hel-N1 from the extract after immunoprecipitation by Western blotting (not shown) we analyzed immunoprecipitated Hel-N1 complexes for the presence of specific RNAs by RT–PCR. Using specific primers in the RT reaction and a nested set of primers for the PCR reaction we searched for the presence of NF-M, GAPDH, and U1 RNAs. We chose GAPDH as an internal control RNA because it is expressed in all the cell lines used (not shown) and its coding or noncoding regions do not contain AU-rich sequences. The specificity of RT–PCR fragments derived from each sample was confirmed by Southern blotting using specific internal oligonucleotides as probes (Fig. 6C,D,E).
As shown in Fig. 6A, C, and E, NF-M mRNA was immunoprecipitated only from the GFP–Hel-N1 transfected cell extracts when the anti-GFP antibody was used [Fig. 6A and C; compare vector (V), antisense (A), and Hel-N1 (H) transfected cell lysates, GFP lanes]. Furthermore, the RT–PCR product was obtained only in the presence of reverse transcriptase demonstrating that the material detected indeed represented RNA and was free of genomic DNA contaminants (Fig. 6A and C, + RT samples and − RT samples). When the control antibodies, anti-gene10 (Fig. 6A and C, g10 lanes) or anti-U1-70K (Fig. 6E., 70K and NFM lanes) were used, NF-M mRNA was not detected in any of the immunoprecipitated samples. Moreover, RNA obtained from anti-GFP precipitated samples did not contain U1 RNA (Fig. 6E, GFP and U1 lanes) and it also did not contain GAPDH (Fig. 6, B and D, GFP lanes). As expected, anti-U1–70K antibody precipitated U1 RNA from all cell lines (Fig. 6E, 70K and U1 lanes) and anti-gene 10 antibody did not immunoprecipitate GAPDH mRNA (Fig. 6B and D, g10 lanes) or U1 RNA (not shown). The reactions were again positive only when reverse transcriptase was included (Fig. 6E, +RT and −RT lanes). Likewise, another positive control for RT–PCR with each set of primers was performed using total RNA in the presence or the absence of reverse transcriptase (Fig. 6, t lanes, + or −RT) and reactions were positive only when the RT was included.
In total, these data demonstrate a direct interaction in vivo between ectopically expressed Hel-N1 protein and the neuron-specific NF-M mRNA.
NF-M mRNA is recruited efficiently to heavy polysomes in Hel-N1-transfected cells
To investigate how NF-M mRNA translation was affected in Hel-N1 transfectants, we examined the distribution of GAPDH and NF-M mRNAs on polysomal gradients. This experimental approach is used conventionally to investigate translational regulation of gene expression in vivo (Hershey 1991; Savant-Bhonsale and Cleveland 1992; Rousseau et al. 1996).
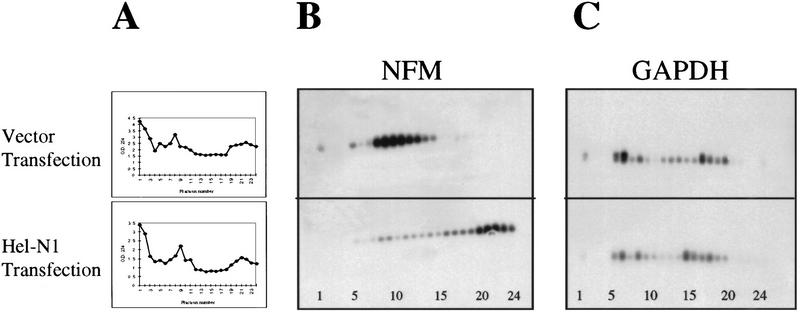
hNT2 cells were transiently transfected with vectors expressing gene10–Hel-N1 fusion protein or with a control GFP protein. This assay allowed us to determine the immediate effect of Hel-N1 protein, which was expressed at very high levels (not shown), on the distribution of NF-M mRNA in polysomes. Also, the use of GFP as a control permitted estimation of transfection efficiency for each experiment by direct immunofluorescence. Once we determined that each transfection occurred at a high efficiency (between 60% and 70% of the cells were transfected), cytoplasmic extracts were prepared from both the GFP- and Hel-N1-transfected cells and loaded onto 15%–45% sucrose gradients. Each gradient was fractionated after centrifugation, and the optical density at 254 nm of each fraction was determined (Fig. 7A). The remaining material was used for RNA extraction. GAPDH and NF-M mRNAs were detected by RT–PCR across the gradients (Fig. 7B,C). Furthermore, the distribution of Hel-N1 protein in polysomal gradients was determined by Western blotting (not shown).
Figure 7.
NF-M mRNA is recruited to heavy polysomes in cells transiently transfected with Hel-N1. (A) Polysome profiles of cytoplasmic cell extracts from vector transfected or Hel-N1-transfected cells. (B) Distribution of NF-M mRNA and (C) GAPDH mRNA in polysome gradients that were loaded with vector or Hel-N1-transfected cytoplasmic cell extracts. Gradient fraction numbers (1–24) are shown at the bottom of the panels.
Whereas the OD254 profiles (Fig. 7A) and distribution of GAPDH mRNA (Fig. 7C) were similar between the two gradients representing vector and Hel-N1-transfected cells , the distribution of NF-M mRNA differed greatly (Fig. 7B). NF-M mRNA in vector-transfected cell lysates was mainly distributed in the lighter fractions of the polysomal gradient, consistent with it not being translated efficiently. However, in Hel-N1-transfected cells, ∼80% of the NF-M mRNA migrated with heavy polysomes, demonstrating that it was actively translated in these cells. Interestingly, when we examined the distribution of Hel-N1 protein, we found that it migrated throughout the gradient, but was more concentrated in the bottom few heaviest fractions (data not shown), similarly to that shown previously for endogenous Hel-N1 (Antic and Keene 1998).
Consistent with reports in literature (Hershey 1991; Savant-Bhonsale and Cleveland 1992; Rousseau et al. 1996), this finding suggests that Hel-N1 expression resulted in increased re-initiation of translation on NF-M mRNA, thus forming heavy polysomes and accumulating higher levels of NF-M protein (Fig. 3). Because the polysome association of the control GAPDH mRNA was not affected in Hel-N1-transfected cells, we conclude that the recruitment to polysomes was specific to NF-M mRNA. These results demonstrate the effect of Hel-N1 on translation, rather than turnover of NF-M mRNA.
Discussion
In this study we explored the role of the human neuronal ELAV protein, Hel-N1, in neurogenesis using human embryonic teratocarcinoma cells (hNT2) (Pleasure et al. 1992; Pleasure and Lee 1993). Our results implicate the Hel-N1 (HuB) gene in neuronal differentiation by up-regulating translation of at least one important neuron-specific mRNA, NF-M. In exploring the mechanism underlying this upregulation, we found that overexpressed Hel-N1 did not affect the steady-state levels of NF-M mRNA in uninduced hNT2 cells, but rather, it caused efficient recruitment of NF-M mRNA into heavy polysomes. We showed that this was a direct and specific effect of Hel-N1 protein on NF-M mRNA by immunoprecipitating mRNP complexes containing NF-M mRNA and Hel-N1 protein.
NF-M ARE directs translation regulation via interactions with Hel-N1 protein
Analysis of the 3′ UTR of NF-M mRNA revealed that it has a non-AUUUA type ARE similar to the class III AREs (Chen and Shyu 1995). Our data indicate that binding of Hel-N1 to this putative class III ARE in the NF-M 3′ UTR did not affect the steady-state level of the mRNA, but rather it recruited the mRNA to the translational apparatus and increased its translation in vivo. Thus, we conclude that ELAV proteins, in either proliferating or differentiating cells, are able to regulate expression at the levels of mRNA stability and/or translatability, which may be linked in some cases. Previous studies implicating HuR in ARE-mediated mRNA stabilization (Fan and Steitz 1998a; Levy et al. 1998; Peng et al. 1998) did not address the translation of the target mRNAs.
It is assumed that different AREs recruit different sets of trans-acting factors, including ELAV proteins, which then determine the outcome of protein expression. Our previous work demonstrated that neuronal ELAV proteins bind to a subset of poly(A)+ mRNA and in association with other proteins form cytoplasmic mRNPs (α complexes) that attach to microtubules in cells of neuronal origin (Antic and Keene 1998). This association with the cytoskeleton was found to be important for presentation of neuronal ELAV mRNPs to the components of the translational apparatus and for the assembly of the translational machinery (β complexes) (Antic and Keene 1998). Consistent with our previous findings, the up-regulation of NF-M translation in Hel-N1-transfected cells can be explained by direct interaction between NF-M mRNA and Hel-N1 protein resulting in the assembly of mRNPs, possibly α complexes. These ELAV mRNPs can then interact with components of the translational apparatus resulting in active translation of the engaged NF-M mRNA. This interpretation is supported by our data demonstrating mobilization of NF-M mRNA into heavy polysomes (Fig. 7). Numerous reports have suggested that mobilization of mRNA into large polysomes is a result of an increase in the translation re-initiation rate (Hershey 1991; Savant-Bhonsale and Cleveland 1992; Rousseau et al. 1996). Further studies will be needed to address the role of translation initiation factors and other proteins in regulating stability and/or translatability of growth-regulatory mRNAs through interactions with ELAV proteins.
Overexpression of Hel-N1 protein in hNT cells leads to up-regulation of NF-M expression and appearance of neurites
Neurofilaments L, M, and H are the three protein subunits that form 8–10 nm intermediate filaments in mature neurons. Transfection of cells with either of the three subunits demonstrated that they can coassemble with vimentin to form a filamentous network (Chin and Liem 1989; Monteiro and Cleveland 1989). Furthermore, NF-L can coassemble into a filamentous network either with NF-M or with NF-H; however NF-M and NF-H cannot coassemble into filaments with each other (Chin and Liem 1990; Ching and Liem 1993; Lee et al. 1993; Leung and Liem 1996).
Previous studies of neurofilaments in hNT2 cells demonstrated that NF-L and NF-M were detectable at low levels in uninduced precursor cells and that they were upregulated during induction with RA (Pleasure and Lee 1993). However, we could not detect expression of NF-M protein in our uninduced hNT2 cells by immunofluorescence or by Western blotting (Fig. 3). It is not clear why our cells differed in this respect when all other morphological and differentiation characteristics were identical. However, we did observe expression of unphosphorylated NF-M protein in uninduced hNT2 cells stably transfected with Hel-N1 cDNA (Fig. 3E). The NF-M protein formed filamentous networks (Fig. 3) in the cytoplasm of Hel-N1-transfected cells that also extended into processes. This is consistent with the report of Pleasure et al. (1992) demonstrating that the hypophosphorylated form of NF-M was localized in the cell body and to short, tapering processes (i.e., in somatodendritic domains of terminally differentiated hNTN neurons); and that highly phosphorylated NF-M was found in long thin processes corresponding to axons (Pleasure et al. 1992). It is possible that the NF-M filaments observed in our Hel-N1-transfected cells consisted of both NF-M and NF-L proteins or that NF-M protein formed filamentous networks by coassembly with the endogenous vimentin network (Chin and Liem 1989; Monteiro and Cleveland 1989).
Although our study focused on the interaction of the overexpressed Hel-N1 protein with the neuron-specific NF-M mRNA, we anticipate similar interactions with other differentiation-specific mRNAs (Gao et al. 1994; Antic and Keene 1998). Hence, if interactions of Hel-N1 with target mRNAs upregulate levels of corresponding neuron-specific proteins, it is conceivable that they would affect cell morphology and neuronal differentiation as shown above (Fig. 2). Thus, Hel-N1 appears to be an early regulator of post-transcriptional expression events associated with neuronal differentiation. Consistent with this observation, expression of antisense Hel-N1 cDNA diminished endogenous ELAV protein levels and blocked neuronal differentiation of the host cells following RA treatment. Whereas the HuB class of ELAV proteins is localized to a subset of neurons in the brain (King et al. 1994) every neuron expresses at least one ELAV protein class in addition to the HuA (HuR) class (Okano and Darnell 1997; Wakamatsu and Weston 1997). Thus, it is possible that HuB, HuC, and HuD regulate subsets of neuronal mRNAs in different regions of the central nervous system.
In recent years, many cis-acting sequence elements located in the coding or noncoding regions of various growth-regulatory mRNAs have been characterized and shown to be important for mRNA turnover, translation and/or localization. However, no trans-acting factors have been shown to interact with these mRNAs in vivo and to directly affect their fate. Results presented in this report together with previously published data (Jain et al. 1997; Fan and Steitz 1998a; Peng et al. 1998; Ford et al. 1999) suggest that transport and targeting of cellular mRNAs towards the decay and/or translation pathways most likely depends on the configuration of the ARE cis-elements, their direct interaction with ELAV proteins, and presumably, other trans-acting factors (Fan and Steitz 1998b; Keene 1999). Additional studies will be needed to understand the precise interplay between mRNAs and the trans-acting factors which influence cell proliferation and differentiation.
Materials and methods
hNT2 cells and transfections
Human embryonic teratocarcinoma (hNT2) cells were obtained from Stratagene and were propagated as described previously (Pleasure et al. 1992) and as recommended by the manufacturer. All untransfected and transfected cell lines were propagated at the identical, high plating density (>5 × 106 cells/75-cm2 flask) and for the induction of differentiation 2 × 106 cells were plated in a 75-cm2 flask and were induced with 1 × 10−5 m all-trans retinoic acid (Sigma Chemicals) dissolved in DMSO. Terminally differentiated neurons were purified as described previously (Pleasure et al. 1992) and their number was determined after replate 3 by hemocytometer cell counting in the presence of trypan blue. Induction of differentiation of all transfected clones was repeated at least three times and similar results were obtained each time.
Stable transfections were performed using lipofectin (GIBCO-BRL) and transfectants were selected with a medium containing 0.3 mg/ml G418 (Sigma Chemicals) for 3 days following by further selection with 0.4 mg/ml G418. Individual colonies were picked (∼2 weeks after transfection) and propagated separately. Cell lines were constantly propagated in DMEM-HG containing 0.3 mg/ml G418.
Transient transfections were performed using Superfect (Qiagen) reagent and the protocol was optimized for hNT2 cells. Cells were plated into T75 flasks and were grown to 80% confluency. Plasmid DNA (8 μg) was mixed with 0.4 ml of PBS and 70 μl of Superfect, incubated at room temperature for 10 min and mixed with 4 ml of serum containing DMEM with high glucose (DMEM-HG). Cells were washed twice with PBS, overlaid with the plasmid mix, placed in the 37°C incubator supplemented with 5% CO2, and incubated for 4 hr. Transfection mix was removed, cells were washed twice with PBS and fed with fresh serum-containing medium, which was replaced again after 16 hr of incubation. Cells were collected and cell extracts were prepared 27 hr after transfection when maximum expression of introduced proteins was observed.
The morphology of hNT2 cells was documented on a Zeiss inverted IM35 microscope.
Plasmids used for transfections and preparation of cell extracts
For stable transfections (as shown in Fig. 2A) Hel-N1 constructs were cloned into two plasmids, pHbAPr-1–neo (Gunning et al. 1987) and pGFPC1 (Clontech). Stable cell lines, containing either set of plasmid constructs were obtained and analyzed. The two plasmids were used to obtain different levels of protein expression because high levels of ectopically expressed ELAV proteins resulted in death of certain cell lines (unpublished data). Therefore, pHbAPr-1–neo plasmid in which introduced genes are driven by the actin promoter provided a medium level of expression in stably transfected embryonal carcinoma (EC) cells, whereas pGFPC1 plasmid in which the genes of interest are placed under the control of CMV promoter provided a low level of expression in stable EC cell lines. For simplicity, GFP vector (Fig. 2A) control was marked as vector (V) throughout the text.
Transient transfections were performed with pAlphaGFP cycle 3 plasmid (Crameri et al. 1996; Affymax, Palo Alto, CA) or with the pAlpha plasmid in which GFP was replaced by gene10-tagged-Hel-N1. Both cDNAs were driven by an SV40 early promoter which was fused to the R segment and part of the U5 sequence (R-U5) from the HTLV-1 LTR (Takebe et al. 1988) providing very high expression levels of introduced genes in transiently transfected hNT2 cells.
Cell extracts were made as described previously (Antic and Keene 1998). Protein concentration was determined by Bio-Rad protein assay.
Immunofluorescence
hNT2 cells were grown in chamber slides (Nunc), washed with PBS prior to fixation with 3% paraformaldehyde in PBS and stained as described previously (Gao and Keene 1996). To observe GFP fluorescence, transfected cells were resuspended in PBS, inoculated onto a glass slide, covered with a coverslip, and monitored without fixation. Immunofluorescence of samples was documented as described previously (Antic and Keene 1998).
Sources of antibodies
Rabbit polyclonal anti-Hel-N1 antibody was produced as described (Gao et al. 1994). Anti-GFP monoclonal antibody was purchased from Clontech. Antibodies against NF-M (RMO254) and NF-M(P+++) (HO14) were kindly provided by V.M.-Y. Lee, University of Pennsylvania (Pleasure and Lee 1993). Anti-MAP2 antibody was purchased from Boehringer Mannheim (clone AP20); anti-tau (T14) was acquired from Zymed Laboratories, So. San Francisco, CA; or from Boehringer Mannheim; anti-human cytokeratin antibody (CAM 5.2) was obtained from Becton-Dickinson. Anti U1-70K was obtained from S. Hoch (Agauron Institute, LaJolla, CA); anti-gene 10 monoclonal antibody was obtained from Novagen. Anti-hnRNP C antibody was kindly provided by G. Dreyfuss (University of Pennsylvania, Philadelphia). All secondary antibodies used for indirect immunofluorescence (Jackson ImmunoResearch Laboratories, Inc., PA) were F(ab)2 fragments with minimal species crossreactivity and secondary antibodies used for immunostaining of Western blots were conjugated to horseradish peroxidase (Amersham Life Sciences).
Protein gels and Western blotting
Protein samples were separated either on 6% acrylamide–0.08% bis-acrylamide or on 15% acrylamide–0.2% bis-acrylamide denaturing gels and were transferred electrophoretically to Hybond-ECL nitrocellulose membranes (Amersham Life Sciences). Immunostaining was visualized by ECL (Amersham Life Sciences).
RNA isolation
Total cellular RNA was extracted using Trizol reagent (GIBCO-BRL) as described by the manufacturer. To avoid DNA contamination, RNA samples were treated with RQ1 DNase (Promega). RNA isolation from immunoprecipitated samples or from fractions of polysome gradients was performed with proteinase K (0.4 mg/reaction) digestion (Sambrook et al. 1989) followed by phenol/chloroform/isoamylalcohol (PCI) extraction and ethanol precipitation.
Immunoprecipitation of mRNP complexes from cell lysates
Immunoprecipitation of Hel-N1 mRNPs was obtained only when EDTA was added to our cell lysates (10mm). Under these conditions, polysomes and ribosomes were dissociated into subunits and ELAV proteins still remained associated with mRNA (Antic and Keene 1998). Also, whereas anti-Hel-N1 antibody was not able to immunoprecipitate RNP complexes under any experimental conditions, anti-gene10 or anti-GFP monoclonal antibodies precipitated corresponding Hel-N1 fusion proteins efficiently from transfected cell lysates in the presence of 10 mm EDTA.
Immunoprecipitation using anti-GFP monoclonal antibody and cell lysates from stably transfected cell lines, or anti-gene10 monoclonal antibody and cell lysates of transiently transfected cells were both performed as follows: cell lysates were incubated with 2.5 mg of protein A beads (Sigma Chemicals) for 2 hr at 4°C with constant rotation. Beads were pelleted and supernatants were transferred to new tubes. Monoclonal antibody (2–3 μl) was added to the precleared extracts and reactions were incubated for 1.5 hr at 4°C, with constant rotation. Protein A beads (2.5 mg) were added to reactions and incubation was continued for another 30 min at 4°C with constant rotation. Beads and immunoprecipitated complexes were pelleted and supernatants were transferred to new tubes. While supernatants were analyzed for immunodepletion of specific proteins, beads were washed five times with ice-cold B4 buffer [50 mm Tris (pH 7.6); 150 mm KCl; 3 mm MgCl2] containing 1% NP-40. Finally, beads were resuspended in 150 μl of STE buffer (10 mm Tris at pH 8.0; 100 mm NaCl; 1 mm EDTA) and were treated with 5 μl of RQ1 DNase (Promega) at 37°C for 15 min. The reaction was stopped by the addition of 150 μl of 2× NENS buffer (100 mm sodium acetate at pH 5.1; 200 mm NaCl; 20 mm EDTA; 1% SDS) and 20 μl of proteinase K (20 mg/ml stock). Digestion was allowed to proceed at 37°C for 30 min with periodic mixing of each sample. After proteinase K digestion, RNA was extracted with PCI and ethanol precipitated prior to RT–PCR.
RT–PCR
Qualitative analysis of RNA was performed as a one-tube RT–PCR reaction as described by GIBCO-BRL with minor modifications: RNA samples were heated to 80°C for 5 min, cooled to 42°C, and Superscript II, diluted in buffer (GIBCO-BRL), was added. After the RT reaction, RNA was digested with 1 μl of RNase One (Promega); and PCR was carried out with Taq polymerase (GIBCO-BRL). All steps of the RT–PCR reaction were done in a Perkin Elmer Gene Amp PCR System 2400.
The RT–PCR experiment described in Figure 6 was performed as follows: RNA obtained after immunoprecipitations was resuspended in 20 μl H2O and 2 μl was used for each RT reaction. For the RT step, we used 100 ng of specific oligonucleotides: NFM–RT (nucleotides 5442–5413), GAPDH–RT (nucleotides 1040–1020), or with the U1–RT (nucleotides 133–119). RT reactions were amplified through 25 PCR cycles (PCR I) using specific oligonucleotides, as described above [NFM–RT and NFM1 (nucleotides 5024–5050); GAPDH–RT and GAPDH1 (nucleotides 366–385); or U1–RT and U1–3′ (nucleotides 3–17). Whereas amplification of U1 RNA was carried out only through one round of PCR (PCR I), NF-M and GAPDH mRNAs were amplified through two rounds of PCR using nested oligonucleotides: NFM2 (nucleotides 5411–5397) and NFM3 (nucleotides 5100–5114); GAPDH2 (nucleotides 1009–992) and GAPDH3 (nucleotides 517–534). Amplification was analyzed by agarose–gel electrophoresis and Southern blotting using internal specific oligonucleotides as probes: NFM probe (nucleotides 5301–5321), GAPDH probe (nucleotides 827–847), and U1 probe (nucleotides 61–81).
Quantitative RT–PCR (Fig. 4) was carried out with total RNA isolated either from the vector or from Hel-N1-transfected cells using the method described previously (Gause and Adamovicz 1995) with minor modifications. RT reactions used a great excess of dNTPs (20 mm of each dNTP) and random hexamers (300 ng) as primers. RNA samples were heated at 80°C for 5 min, cooled to 25°C for 10 min, and the reaction was allowed to proceed at 42°C when the enzyme, diluted in buffer, was added. An aliquot of the cDNA ( ) was amplified through one round of PCR, with a great excess of specific primers (400 ng of NFM–RT and 400 ng of NFM-1; or 400 ng of GAPDH–RT and 400 ng of GAPDH1) and dNTPs (20 mm each). To obtain dose-response curves, RNA samples were titrated from 0.1 to 1.8 μg of total RNA and number of PCR cycles was varied from 15 to 25. Once the optimal RNA concentration (<1 μg) and the number of cycles (15 or 20) was determined, RNA samples isolated from different transfected cell lines were subjected to these optimized RT–PCR conditions (as described in Results).
) was amplified through one round of PCR, with a great excess of specific primers (400 ng of NFM–RT and 400 ng of NFM-1; or 400 ng of GAPDH–RT and 400 ng of GAPDH1) and dNTPs (20 mm each). To obtain dose-response curves, RNA samples were titrated from 0.1 to 1.8 μg of total RNA and number of PCR cycles was varied from 15 to 25. Once the optimal RNA concentration (<1 μg) and the number of cycles (15 or 20) was determined, RNA samples isolated from different transfected cell lines were subjected to these optimized RT–PCR conditions (as described in Results).
Semiquantitative RT–PCR (Fig. 7) was similar to the quantitative RT–PCR in that RNA from each gradient fraction was resuspended in 10 μl of H2O and the entire amount was used for the RT reaction by priming with 300 ng of random hexamers and an excess of dNTPs (20 mm each). One-fifth of each RT reaction was amplified through 25 PCR cycles with 200 ng of each specific primer (NFM–RT and NFM1; or GAPDH–RT and GAPDH1) and 20 mm of each dNTP. An aliquot (1/5th) of the PCR reaction was analyzed by agarose-gel electrophoresis and Southern blotting, as described previously (Sambrook et al. 1989), using specific oligonucleotides (NFM probe and GAPDH probe) as probes.
Quantitative analysis of Southern blots was performed using PhosphorImager and ImageQuant program (Molecular Dynamics).
Purification of recombinant GST–Hel-N1 fusion protein
The Hel-N1-coding region (King 1994) was subcloned into pGEX vector (Pharmacia). Induction of the GST–Hel-N1 protein and purification of the recombinant fusion protein over glutathione beads was carried out as described by the manufacturer.
Subcloning of the NF-M 3′ UTR, in vitro transcription, mobility shift, and immunoprecipitation of in vitro assembled RNP complexes
Plasmid containing human genomic NF-M sequence was provided by R.A. Lazzarini, Mt. Sinai Medical School (Myers et al. 1987). The 3′ UTR of NF-M was PCR amplified using plasmid DNA as a template and specific primers NFM5 and NFM6. A SacI restriction site was added to primer NFM6 (nucleotides 5552–5569) and a site for EcoRI was added to primer NFM5 (nucleotides 6216–6236) so that the entire 3′ UTR was subcloned as a SacI–EcoRI fragment into pBluescript SK+ (Stratagene). In vitro transcripts of the subcloned 3′ UTR were obtained with T3 polymerase (Promega), as described by the manufacturer and they contained only 10 nucleotides of the vector sequence. Fragment 6 was PCR amplified from the 3′ UTR clone using specific primers. The downstream primer contained the T7 promoter sequence allowing in vitro transcription of fragment 6. RNA-binding reactions using in vitro-transcribed RNAs and the recombinant GST–Hel-N1 protein were performed as described previously (Jain et al. 1997; Levine et al. 1993).
Polysome gradients
Cytoplasmic cell extracts were loaded over linear, 11-ml, sucrose gradients (15%–45%) which were preformed with a polystaltic pump and gradient maker (Buchler Instruments, A Labconco Company, Fort Lee, NJ). The gradient medium was dissolved in B4 buffer as described (Antic and Keene 1998). Samples were centrifuged for 2 hr at 35,000 rpm in a Sorval SW41 rotor. After centrifugation, 24 fractions were collected from the top using an Auto Densi-Flow IIC collector (Buchler Instruments, A Labconco Company, Fort Lee, NJ). An aliquot of each fraction (50 μl) was used for OD254 readings and the remaining material was used for RNA extraction.
Acknowledgments
We thank Virginia Lee for antibodies to NF-M and Robert A. Lazzarini for a cDNA clone encoding NF-M. This work was supported by research grant R01 CA60083 from the National Cancer Institute to J.D.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL antic@abacus.mc.duke.edu; FAX (919) 684-8735.
References
- Abe R, Yamamoto K, Sakamoto H. Target specificity of neuronal RNA-binding protein, Mel-N1: Direct binding to the 3′ untranslated region of its own mRNA. Nucleic Acids Res. 1996;24:2011–2016. doi: 10.1093/nar/24.11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D, Keene JD. Messenger ribonucleoprotein complexes containing human ELAV proteins: Interactions with cytoskeleton and translational apparatus. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- Atasoy U, Watson J, Keene JD. Ubiquitously expressed ELAV RNA-binding protein, HuA, localizes to both the nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Black MM, Lee VM. Phosphorylation of neurofilament proteins in intact neurons: Demonstration of phosphorylation in cell bodies and axons. J Neurosci. 1988;8:3296–3305. doi: 10.1523/JNEUROSCI.08-09-03296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AR, Grossman D, White K. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J Neurogenet. 1985;2:197–218. doi: 10.3109/01677068509100150. [DOI] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnovich D, Cohn SL. Binding of a 40-kDa protein to the N-myc 3′-untranslated region correlates with enhanced N-myc expression in human neuroblastoma. J Biol Chem. 1996;271:33580–33586. doi: 10.1074/jbc.271.52.33580. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin SS, Liem RK. Expression of rat neurofilament proteins NF-L and NF-M in transfected non-neuronal cells. Eur J Cell Biol. 1989;50:475–490. [PubMed] [Google Scholar]
- Chin SS, Liem RK. Transfected rat high-molecular-weight neurofilament (NF-H) coassembles with vimentin in a predominantly nonphosphorylated form. J Neurosci. 1990;10:3714–3726. doi: 10.1523/JNEUROSCI.10-11-03714.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching GY, Liem RK. Assembly of type IV neuronal intermediate filaments in nonneuronal cells in the absence of preexisting cytoplasmic intermediate filaments. J Cell Biol. 1993;122:1323–1335. doi: 10.1083/jcb.122.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- Chung S, Eckrich M, Perrone-Bizzozero N, Kohn DT, Furneaux H. The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. J Biol Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- Cornelius P, Marlowe M, Call K, Pekala PH. Regulation of glucose transport as well as glucose transporter and immediate early gene expression in 3T3-L1 preadipocytes by 8-bromo-cAMP. J Cell Physiol. 1991;146:298–308. doi: 10.1002/jcp.1041460215. [DOI] [PubMed] [Google Scholar]
- Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- Darnell RB. Onconeural antigens and the paraneoplastic neurologic disorders: At the intersection of cancer, immunity, and the brain. Proc Natl Acad Sci. 1996;93:4529–4536. doi: 10.1073/pnas.93.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Hentze M, Lamond AI. From transcript to protein. Cell. 1996;85:963–972. doi: 10.1016/s0092-8674(00)81298-2. [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998a;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci. 1998b;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stablize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes & Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- Gause WC, Adamovicz J. RNA purification. In: Dieffenbach CW, Dveksler GS, editors. PCR primer: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Labortory Press; 1995. pp. 113–132. [Google Scholar]
- Good PJ. A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, Leavitt J, Muscat G, Ng SY, Kedes L. A human β-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King PH. Hel-N2: A novel isoform of Hel-N1 which is conserved in rat neural tissue and produced in early embryogenesis. Gene. 1994;151:261–265. doi: 10.1016/0378-1119(94)90668-8. [DOI] [PubMed] [Google Scholar]
- King PH, Levine TD, Fremeau RT, Jr, Keene JD. Mammalian homologs of Drosophila ELAV localized to a neuronal subset can bind in vitro to the 3′ UTR of mRNA encoding the Id transcriptional repressor. J Neurosci. 1994;14:1943–1952. doi: 10.1523/JNEUROSCI.14-04-01943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Xu Z, Wong PC, Cleveland DW. Neurofilaments are obligate heteropolymers in vivo. J Cell Biol. 1993;122:1337–1350. doi: 10.1083/jcb.122.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CL, Liem RKH. Characterization of interactions between the neurofilament triplet proteins by the yeast two-hybrid system. J Biol Chem. 1996;271:14041–14044. [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- Liu J, Dalmau J, Szabo A, Rosenfeld M, Huber J, Furneaux H. Paraneoplastic encephalomyelitis antigens bind to the AU-rich elements of mRNA. Neurology. 1995;45:544–550. doi: 10.1212/wnl.45.3.544. [DOI] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Monteiro MJ, Cleveland DW. Expression of NF-L and NF-M in fibroblasts reveals coassembly of neurofilament and vimentin subunits. J Cell Biol. 1989;108:579–593. doi: 10.1083/jcb.108.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MW, Lazzarini RA, Lee VM, Schlaepfer WW, Nelson DL. The human mid-size neurofilament subunit: A repeated protein sequence and the relationship of its gene to the intermediate filament gene family. EMBO J. 1987;6:1617–1626. doi: 10.1002/j.1460-2075.1987.tb02409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: Evidence for a novel class of AU-rich elements. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SSY, Chen CYA, Xu NH, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Lee VM. NTera 2 cells: A human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J Neurosci Res. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Page C, Lee VM. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J Neurosci. 1992;12:1802–1815. doi: 10.1523/JNEUROSCI.12-05-01802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savant-Bhonsale S, Cleveland DW. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes & Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- Schwartz ML, Shneidman PS, Bruce J, Schlaepfer WW. Actinomycin prevents the destabilization of neurofilament mRNA in primary sensory neurons. J Biol Chem. 1992;267:24596–24600. [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SR alpha promoter: An efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai DE, Kenan DJ, Keene JD. In vitro selection of an RNA epitope immunologically cross-reactive with a peptide. Proc Natl Acad Sci. 1992;89:8864–8868. doi: 10.1073/pnas.89.19.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: Messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: Key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]