Abstract
Poly(ADP-ribose) polymerase1 (PARP1) is a global regulator of different cellular mechanisms, ranging from DNA damage repair to control of gene expression. Since PARP1 protein and pADPr have been shown to persist in chromatin through cell cycle, they may both act as epigenetic markers. However, it is not known how many loci are occupied by PARP1 protein during mitosis genome-wide. To reveal the genome-wide PARP1 binding sites, we used the ChIP-seq approach, an emerging technique to study genome-wide PARP1 protein interaction with chromatin. Here, we describe how to perform ChIP-seq in the context of PARP1 binding sites identification in chromatin, using human embryonic kidney cell lines.
Keywords: PARP1, pADPr, genomics, human embryonic cells, mitotic arrest, ChIP, ChIP-seq
1. Introduction
PARP1 is an abundant and ubiquitous nuclear enzyme that catalyzes the transfer of poly(ADP-ribose) (pADPr) moiety from NAD to either a protein acceptor or an existing pADPr chain (1). PARP1 regulates many cellular functions, including stress-induced apoptosis, DNA damage detection and repair (2–4), transcriptional regulation and chromatin remodeling (5–7), and the control of gene expression by the induction of chromatin loosening at targeted genetic loci (8). The distribution of PARP1 in chromatin is nonrandom and globally regulates transcription (9). The dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin is mediated by nucleosomal core histones (10). For example, PARP1 and histone H1 exhibit a reciprocal pattern of chromatin binding at many RNA polymerase II-transcribed promoters (11). Since PARP1 is involved in the regulation of so many cellular mechanisms, we were inspired to study its genome-wide locations in the human genome in interphase and mitotic cells. Results of this work reveal the true loci of PARP1 in mitotic chromatin, allowing us to further understand the molecular mechanisms of PARP1-dependent processes.
In order to identify PARP1 protein binding sites in the human genome, we applied chromatin immunoprecipitation followed by sequencing (ChIP–seq). In ChIP–seq experiments, the precipitated ChIP-DNA fragments of interest are sequenced directly. In comparison to microarray, ChIP–seq has higher resolution, generates fewer artifacts, and provides greater coverage and a larger dynamic range. ChIP-seq studies have been used to characterize transcription factor binding (12–14), genome-wide nucleosome positioning (15), and to determine epigenetic changes (16). ChIP-seq technology does not require very long sequencing reads. Large numbers of short reads (35 bp) are sufficient for mapping binding sites in most organisms. Therefore, Illumina/Solexa and ABI/SOLiD have been favored over Roche/454 because they both generate millions of very short reads (about 35 bases/read), whereas Roche/454 generates fewer reads, but longer length (200–300 bases/read). These three main sequencing technologies are utilized on the basis of their applications. As a control, input DNA, consisting of nonimmunoprecipitated, sonicated and cross-linked DNA, has great importance in ChIP-Seq studies, as ChIP DNA samples are normally scored against the input DNA for transcription factor binding site (TFBS) identification (17). Even after successfully extracting ChIP-seq raw data, determination of binding sites from the data remains a formidable challenge. Therefore, many research groups published different algorithms that allow determining binding sites (18 – 24).
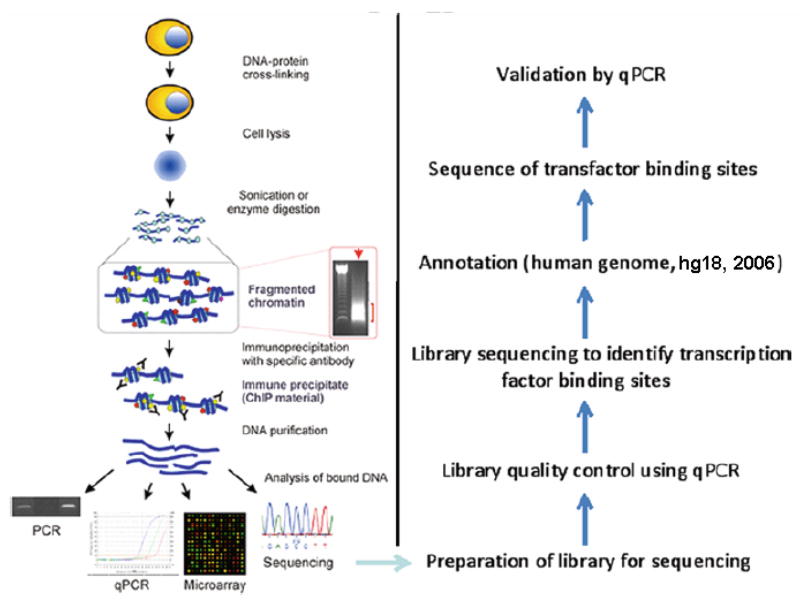
ChIP-seq can be divided in to the following steps (Figure 1): 1) ChIP; 2) Library preparation (end repair; addition of an ‘A’ base to the 3′-end of DNA fragments; ligation of adapters to DNA fragments; amplification of adapter-modified DNA fragments and gel purification; pre-sequencing control assays (enrichment check using positive/negative control primers)); and 3) library sequencing (annotation, sequence of DNA and validation by quantitative PCR (qPCR)).
Fig. 1.
ChIP-seq flow chart. All the steps are same as ChIP up to DNA precipitation; afterward, ChIP-seq steps are followed, adapted from Collas and Dahl (Frontiers in Bioscience) (2008).
2. Materials
2.1. Cell Culture and Lysis
Dulbecco’s modified eagle’s medium (DMEM) (Gibco/BRL) supplemented with 10% fetal bovine serum (FBS) (HyClone).
Solution of 0.25% trypsin and 1 mM ethylenediaminetetraacetic acid (EDTA) (Gibco/BRL).
Teflon cell scrapers (Fisher), Pipettes, Flasks (T75).
Human embryonic kidney 293 cell lines (HEK293).
10X Phosphate-buffered saline (10X PBS) (Gibco/BRL).
Nocodazole (Sigma).
2.2. Western blotting to check mitotic arrest of synchronized cells
PAGE Gel (4–12%) and transfer of gel setup apparatus (Invitrogen).
10X stock Tris-buffered saline with Tween (10X TBS-T): 1.37 M NaCl, 27 mM KCl, 250 mM Tris-HCl, pH 7.4, 1% Tween-20 and SDS lysis buffer (2X).
Blocking buffer: 5% (w/v) nonfat dry milk in 1X TBS-T.
Primary antibody dilution buffer: 1X TBS-T supplemented with 2% (w/v) fraction bovine serum albumen (BSA).
Primary antibodies: rabbit anti-histone H3 phosphor-serine10 (Millipore), anti-histone H3 phospho-threonine 3 (Millipore), anti-PARP1 (Abcam) and anti-tubulin antibody (Sigma) (see Note 1).
Secondary antibody: goat anti-rabbit and anti-mouse IgG conjugated to horse radish peroxidase (Sigma).
Enhanced chemiluminescent (ECL) reagents (GE) and Bio-Max ML film (Kodak).
2.3. Chromatin Immunoprecipitation (ChIP) Assay
37% formaldehyde, molecular biology grade (Sigma).
2 M glycine (Sigma).
Miracloth tissue.
Vacuum chamber.
Liquid nitrogen.
Double distilled autoclaved water.
Vortex.
Nutator.
Falcon tubes for 50 and 15 mL.
Refrigerated centrifuge (Eppendorf).
Sonicator (Bioruptor).
Protein A Agarose mix (Invitrogen).
1 M Tris-HCl, pH 6.5.
5 M NaCl (Sigma).
0.5 M EDTA (Sigma).
Heating block at 65°C.
10 mg/ml proteinase K (Invitrogen).
Novagen pellet paint (CN Biosciences).
10 mg/ml RNase A (Qiagen).
Chloroform (Sigma).
Ethanol.
Protease inhibitor tablets (Roche). To prepare 25X protease inhibitor cocktail (25X PIC), dissolve 1 tablet in 2 mL of water.
Antibodies: Anti-rabbit polyclonal antibodies against PARP1 (Abcam) and anti-IgG (Abcam) are used for the ChIP assay.
Extraction buffer 1 (for 100 mL): 0.4 M sucrose (20 mL of 2 M stock); 10 mM Tris, pH 8.0 (1 mL of 1 M stock); 5 mM β-mercaptoethanol, molecular biology grade (β-ME) (35 μL of 14.3 M stock); 0.1 mM PMSF (50 μL of 0.2 M stock); 1X PIC (should be added immediately before use).
Extraction buffer 2 (for 10 mL): 0.25 M sucrose (1.25 mL of 2 M stock); 10 mM Tris, pH 8.0 (100 μL of 1 M stock); 10 mM MgCl2 (100 μL of 1 M stock); 1% Triton X-100 (0.5 mL of 20% stock); 5 mM β-ME (3.5 μL of 14.4 M stock); 0.1 mM PMSF (5 μL of 0.2 M stock); 1X PIC as in buffer 1.
Extraction buffer 3 (for 10 mL): 1.7 M sucrose (8.2 mL of 2 M stock); 10 mM Tris, pH 8.0 (100 μL of 1 M stock); 0.15% Triton X-100 (75 μL of 20% stock); 2 mM MgCl2 (20 μL of 1 M stock); 5 mM β-ME (3.5 μL of 14.3 M stock); 0.1 mM PMSF (5 μL of 0.2 M stock); 1X PIC as in buffer 1.
Nuclei lysis buffer: 50 mM Tris, pH 8.0; 10 mM EDTA; 1% SDS. Preheat at 40°C. 1X PIC as in buffer 1.
ChIP dilution buffer: 1.1% Triton X-100; 1.2 mM EDTA; 16.7 mM Tris, pH 8.0; 167 mM NaCl. Preheat at 40°C. 1X PIC is added at the time of use.
Elution buffer: 1% SDS and 0.1 M NaHCO3. Prepare fresh each time.
Low salt buffer: 150 mM NaCl; 0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris, pH 8.0.
High salt buffer: 500 mM NaCl; 0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris, pH 8.0. Store at 4°C.
Lithium chloride wash buffer: 0.25 M LiCl; 1% NP-40; 1% sodium deoxycholate; 1 mM EDTA; 10 mM Tris, pH 8.0. Store at 4°C.
TE buffer: 10 mM Tris, pH 8.0; 1 mM EDTA. Store at 4°C.
QIAquick PCR purification kit (Qiagen).
2.4. ChIP PCR Control
SYBR Green and Taq Polymerase (Applied Biosystems).
-
The following oligonucleotides were used for positive (PVALB, GDF15, KDM2A) and negative (GAPDH, actin) PCR control experiments, and to validate the ChIP-seq data in subheadings 3.2. and 3.6. (see Note 2):
PVALB Fwd 5′ GCT CCC CTA TCT GCA CAC TC 3′
PVALB Rev 5′ CAA AGG CTG TTT GGA AGC TC 3′
GDF15 Fwd 5′ CTC AGA TGC TCC TGG TGT TG 3′
GDF15 Rev 5′ CTC GGA ATC TGG AGT CTT CG 3′
GAPDH Fwd 5′ CGA CCA CTT TGT CAA GCT CA 3′
GAPDH Rev 5′ AGG GGT CTA CAT GGC AAC TG 3′
Actin Fwd 5′ GCT GTT CCA GGC TCT GTT CC 3′
Actin Rev 5′ ATG CTC ACA CGC CAC AAC ATG C 3′
KDM2A Fwd 5′ ATT GCT AAT GAA GTT TCG GGG 3′
KDM2A Rev 5′ CTG CTC TCA AAC CAT GTC T 3′
2.5. Sequencing Library
End-It DNA END Repair Kit (Epicentre).
Klenow fragment polymerase (3′- to 5′-exo-minus), 5,000 U/mL (NEB).
100 mM dATP (Invitrogen).
LigaFast DNA ligase, 3 U/μL (Promega).
Oligo-only Kit for single-end read sequencing containing 100 μM genomic adapter oligo mix; 25 μM genomic PCR primers (Illumina).
Phusion DNA polymerase (NEB).
QIAquick PCR purification kit (Qiagen).
QIAquick gel extraction kit (Qiagen).
NanoDrop 1000 spectrophotometer is used to determine the concentration of double-stranded DNA samples.
2.6. Pre-sequencing Control
Positive and negative control primers for qPCR (subheading 2.4.).
SYBR Green (Applied Biosystems).
3. Methods
ChIP combines immunoprecipitation of chromatin fragments and qPCR of precipitated DNA to map the binding sites of protein-DNA interaction in vivo. ChIP procedures fall into two main categories: those that use native chromatin prepared by nuclease digestion (designated N-ChIP) and those that use chromatin in which DNA and proteins are cross-linked, either chemically or with UV light (designated X-ChIP). Each procedure has its own advantages and drawbacks. Here, we outline the methods currently in use in our laboratory to isolate and immunoprecipitate cross-linked chromatin from cultured cells and to isolate and analyze immunoprecipitated protein and DNA.
3.1. Preparation of samples for Western blot and ChIP
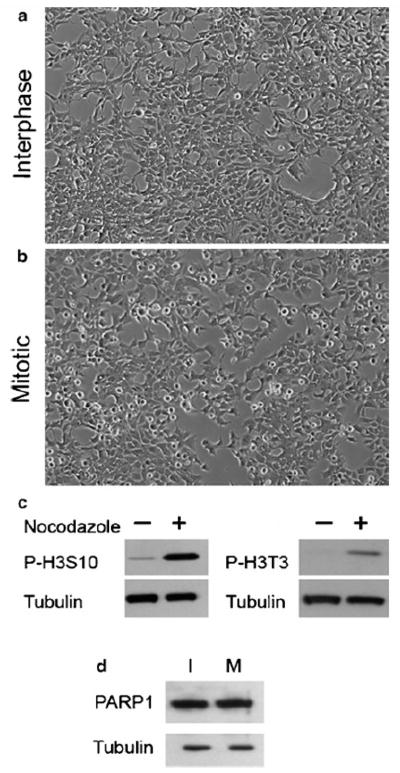
Human embryonic kidney 293 (HEK293) cells were cultured in two plates of 10 cm (for the Western blot analysis) and four plates of 15 cm for ChIP. At confluence of 70%, half of the plates (for Western blot and ChIP) were treated with 60ng/mL nocodazole for 18hrs to synchronize the cells in prometaphase stage. The morphology of treated and untreated cells was the same (Figure 2A, B).
Fig. 2.
Human embryonic kidney 293 cells (HEK293) untreated (interphase) (a) and synchronized cells in prometaphase treated with nocodazole (b). (c) Histone H3 phosphorylation on serine 10 (H3S10) and on threonine 3 (H3T3) increased in nocodazole-treated HEK293 cells, (d) Specificity of PARP1 antibody checked by Western blot before ChIP. In this figure, PARP1 (113.0 kDa) specificity is shown in interphase (I) and synchronized HEK293 cells (M) with tubulin as a loading control (55 kDa).
3.1.1. Western blotting to check synchronization of cells after nocodazole and specificity of PARP1
Scrape the cells into two separate 50 mL 1X PBS aliquots containing PIC diluted 1/100 (add 20 μL of 25X stock solution).
Centrifuge cells for 5 minutes at 5000g at 4°C. Preheat the SDS lysis buffer to room temperature (RT) to dissolve precipitated SDS after lysis.
Remove the supernatant and dissolve the pellet in SDS lysis buffer; then sonicate the cells for 30 sec to reduce viscosity.
The two samples are then separated by SDS-Page gel (4–12%, Invitrogen). After completion of gel run, separated proteins are transferred to nitrocellulose membranes by iBlot gel transfer (Invitrogen), according to manufacturer’s protocol.
Once the transfer is complete, membrane is taken out and incubated in 10 mL blocking buffer for 1 h at room temperature on a rocking platform.
The blocking buffer is discarded, and the membrane is quickly rinsed prior to addition of 1:2000 dilution of anti-phospho-histone H3 serine10 (H3S10), anti-phospho-histone H3 threonine3 (H3T3) antibody (1:7500), anti-PARP1 (1:5000) and 1μgm/ml dilution of anti-tubulin in blocking buffer for 1 h at room temperature on a rocking platform.
The primary antibody is then removed and the membrane is washed three times for 5 min each with 10 mL TBS-T.
The secondary antibody is freshly prepared at 1:2500-fold dilution in blocking buffer and added to the membrane for 30 min at room temperature on a rocking platform.
The secondary antibody is discarded and the membrane is washed five times for 10 min each with TBS-T.
During the final wash, 2 mL aliquots of each portion of the ECL reagent are heated separately to room temperature, and the remaining steps are done in a dark room under safe light conditions. Once the final wash is removed from the blot, the ECL reagents are mixed together and then immediately added to the blot, which is then rotated by hand for 1 min to ensure even coverage.
The blot is removed from the ECL reagents, soaked with buffer with Kim-Wipes, and put into an X-ray film cassette.
The film is exposed for a few minutes. The results are shown in Figure 2C, D.
3.1.2. Chromatin Immunoprecipitation (ChIP)
Four plates of 15 cm provide cell material for 4 immunoprecipitation reactions (approximately 107 cells per IP).
Two of them are treated with nocodazole for 18h and followed by formaldehyde treatment to cross-link the cells.
Cross-linking: 37% formaldehyde is added directly to the culture medium of untreated and treated cells so that its final concentration is 1%, then mixed and incubated for 10 minutes at room temperature.
Wash: Medium is aspirated thoroughly; cells are washed twice with ice cold 1X PBS. Keep cells on ice.
The cells are scraped into two separate 50 mL 1X PBS aliquots containing PIC diluted 1/100 (add 20 μL of 25X stock solution).
Centrifuge cells for 5 minutes at 5000g at 4°C. Preheat the nuclei lysis buffer to room temperature to dissolve precipitated SDS.
Resuspend each cell pellet in 300 μL of nuclei lysis buffer containing 1X PIC (add 12 μL), transfer them into two separate 1.5 mL Eppendorf tubes, and incubate for 10 minutes on ice.
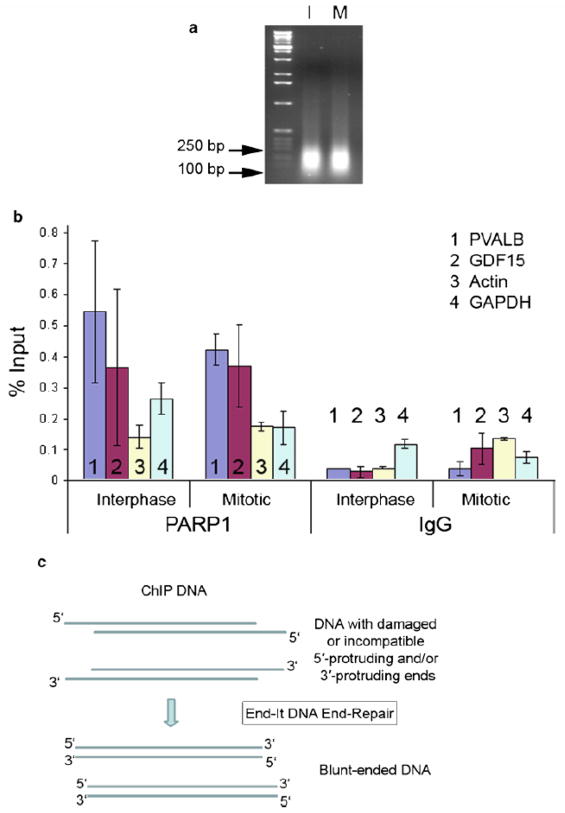
Sonicate the lysate to shear DNA fragments ranging in size from 150 to 250 bp (Figure 3A) and keep the samples on ice. We used Bioruptor sonicator for four cycles of 10 minutes, with 30 seconds on/off (see Note 3, 4 and 5).
Centrifuge the lysate for 10 minutes at 20,000g at 4°C and make two aliquots of 100μl from each of the samples in 1.5 mL Eppendorf tubes, i.e., 4 samples (two untreated and two treated with nocodazole). At this stage, we froze the 50 μL aliquot from each at −80°C as input control aliquots.
Dilute the sonicated extract 10-fold in ChIP dilution buffer with 1X PIC by adding 800 μL ChIP dilution buffer and 40 μL of 25X PIC to the 100 μL sonicated extracts for a final volume of 1 mL.
To reduce nonspecific background, preclear the 1 mL diluted cell extract with 60 μL of agarose beads (Invitrogen) (see Note 6) for 1 hr at 4°C with rotation.
Centrifuge agarose beads for 1 min at 3000g and collect the supernatant fractions in new tubes.
Add anti-PARP1 (specificity previously confirmed by Western blot) and IgG antibody (Abcam, host rabbit for both) (10 μg each) (see Note 7) to 4 samples (two of each, untreated and treated, respectively) of 1mL supernatant fractions and incubate overnight at 4°C with rotation on nutator.
Add 60μL of agarose beads (Invitrogen) to each sample and incubate 1hr at 4°C with rotation on nutator.
Collect beads with antibody/protein/DNA complexes by gentle centrifugation (1400g at 4°C for 1 min). Carefully remove the supernatant that contains unbound, nonspecific DNA. Wash the beads/antibody/protein/DNA complexes for 15 minutes on a rotating platform with 1mL of each of the buffers listed in the order given here: 1X low salt immune complex wash buffer; 1X high salt immune complex wash buffer; 1X LiCl immune complex wash buffer; 2X TE.
Elute the protein/DNA complexes from the beads/antibody by adding 250μL elution buffer (fresh) to the pelleted beads/antibody/protein complex from step 15 (above). Vortex briefly to mix and then incubate at room temperature for 15 minutes with rotation on nutator. Spin the beads at 1400g for 1 minute and carefully collect the supernatant fraction to 1.5 ml tubes. Repeat elution again using 250μL of elution buffer.
Take 50 μL of the frozen input (from step 9) and dilute it with 10X ChIP dilution buffer (add 450 μL).
Add 20 μL 5M NaCl to the combined eluates (500 μL) and input and reverse protein-DNA cross-links by heating at 65°C for 6 hrs.
Add 10 μL of 0.5 M EDTA; 20 μL of 1 M Tris-HCl, pH 6.5; and 2 μL of 10 mg/mL proteinase K to the combined eluates and incubate for 1 hr at 45°C.
Recover DNA in 100 μL of nuclease-free water by using Qiagen PCR purification kit followed (see Note 8) by RNA digestion with 100 μg/mL RNAse and quantitate the DNA using the sensitive Quant-iT™ PicoGreen assay (Invitrogen) (see Note 9).
Fig. 3.
(a) Sonicated fragments of chromatin are in the range of 150–250 bp for ChIP-seq in interphase (I) and synchronized cells (M). (b) qPCR with PVALB, GDF15, actin, and GAPDH to check the quality of ChIP-DNA. (c) DNA with damaged or incompatible 5′-protruding and/or 3′-protruding ends repaired by End-It DNA End-Repair Reaction kit.
3.2. qPCR to check the specific binding of PARP1
Before moving forward to ChIP-seq, precipitated ChIP DNA was checked by qPCR (StepOnePlus™, Applied Biosystems) using known PARP1 binding genes (positive control) (see Note 10).
Normalize the primer (subheading 2.4.) concentrations and mix gene-specific forward and reverse primer pair. Each primers’ (forward or reverse) concentration in the mixture is 10 pM/μL.
Set up the experiment and the following PCR program on StepOnePlus™ (Applied Biosystems): Step 1 (1 cycle): 95°C 10 min; Step 2 (40 cycles): 95°C 15 sec, 60°C 30 sec, 72°C 30 s; Step 3 (1 cycle): 72°C 10 min.
The real-time PCR reaction mixture total volume is 20 μL. Prepare the following mixture in each optical tube to detect PVALB and GDF15 (positive controls), actin and GAPDH (negative controls) genes: 2x SYBR Green Mix (10 μL); ChIP DNA (2 μl); 10 pM/μL Primer F (1 μL); 10 pM/μL Primer R (1 μL); H2O (6 μL).
Proceed with real-time PCR reactions and analyze the results with the StepOnePlus™ software (Applied Biosystems) (Figure 3B).
3.3. Preparation of the Sequencing Library
Here, we describe the library preparation protocol for Illumina sequencing platforms, an important step for ChIP-seq. This protocol is based on the Illumina Sample Preparation Kit for Genomic DNA with minor modifications. Currently, most ChIP-Seq studies have been performed on Illumina. During Illumina library generation, oligonucleotide adapters are introduced at the ends of the small ChIP DNA fragments. Protocols differ depending on the sequencing platform used; 454/Roche, Solexa/Illumina, SOLiD/ABI, and Helicos each use different strategies to create a library representing the population of short DNA fragments selected by ChIP.
3.3.1. End Repair
End repair is performed using the “End-It DNA End-Repair Kit” (Epicentre Biotechnologies). The End-It™ DNA End-Repair Kit was used to convert DNA with damaged or incompatible 5′-protruding and/or 3′-protruding ends to 5′-phosphorylated, blunt-end DNA for next-generation DNA sequencing adapters (Figure 3C).
ChIP DNA from subheading 3.1.2. step 20 (100μL) for end repair reaction can be concentrated to 34 μL using a lyophilizer.
Prepare the following mixture (for a total reaction volume of 50 μL): Lyophilized ChIP DNA (34 μL); 10X end repair buffer (5 μL); 10 mM ATP (5 μL); 2.5 mM dNTP Mix (5 μL); end repair enzyme mix (1 μL).
Incubate at room temperature for 45 min.
Stop the reaction by heating at 70°C for 10 minutes.
Purify DNA using a PCR purification kit (Qiagen). Elute in 34 μL of the kit’s elution buffer.
3.3.2. Addition of an ‘A’ Base to the 3′-End of DNA Fragments
Prepare the following mixture (for a total reaction volume of 50 μL): DNA from subheading 3.3.1. (34 μL); 10X Klenow buffer (5 μL); 1 mM dATP (10 μL); Klenow fragment (3′- to 5′-exo-minus) (1μL) (5 U).
Incubate for exactly 30 min at 37°C. Purify DNA using a PCR purification kit (QIAquick PCR purification kit).
Elute by adding 12 μL of the kit’s elution buffer.
3.3.3. Ligation of Adapters to DNA Fragments
Prepare the following mixture (for a total reaction volume of 30 μL): DNA from subheading 3.3.2. (12 μL); 2X DNA ligase buffer (15 μL); genomic adapter oligo mix (1 μL); LigaFast DNA ligase (2 μL). The genomic adapter oligo mix from Illumina should be diluted 1:10 in water before use.
Incubate for 15 min at room temperature.
Purify DNA using a PCR purification kit (Qiagen).
Elute with 25 μL of the kit’s elution buffer.
3.3.4. Amplification of Adapter-Modified DNA Fragments and Gel Purification
Prepare the following mixture (for a total reaction volume of 50 μL): DNA from subheading 3.3.3. (24 μL); 2X Phusion DNA polymerase (25 μL); Illumina Genomic PCR primer 1.1 (0.5 μL); Illumina Genomic PCR primer 2.1 (0.5 μL).
Follow the PCR protocol used to amplify ChIP-DNA: Step 1: 30 s at 98°C; Step 2 (15 cycles): 10 s at 98°C, 30 s at 65°C, 30 s at 72°C; Step 3: 5 min at 72°C.
Purify and concentrate DNA using a PCR purification kit (Qiagen).
Elute with 10 μL of the kit’s elution buffer.
Run purified DNA product on a 2% agarose gel next to 50 bp or 100 bp ladders.
Excise DNA between 150 and 500 bp. This step ensures the emoval of unused primers and adapters and selection of proper fragment size for sequencing.
Purify DNA using a gel extraction kit.
Elute DNA in 30 μL of the kit’s elution buffer and measure DNA concentration by NanoDrop.
Follow the instructions of the sequencing facility for sample submission and further steps for sequencing.
3.4. Presequencing Control Assays: Enrichment Check Using Positive/Negative Control Primers Library
To verify that a library has maintained a specific enrichment of ChIP target sites, an input library derived from the 10% input sample serves as a control to normalize the qPCR data to, in turn, determine the relative enrichment of a given target.
3.4.1. Prepare Input Library
Using 50–200 ng of input chromatin extract, construct an input library as described above. Using qPCR analysis, check the ChIP-seq library as well as input library (see Note 11).
Prepare the following mixture (for a total reaction volume of 20 μL): 2 ng library from (subheadings 3.3.4 and 3.4.1) (2 μL); nuclease-free H2O (6 μL); 2X SYBR Green mix (Applied Biosystems) (10 μL); 10pm/μL target primer mix (1 μL from each).
Perform qPCR reactions using the following protocol: Step 1: 3 min at 95°C; Step 2 (40 cycles): 30 s at 95°C, 30 s at 60°C; Step 3: Include a 70–95°C melting curve at the end of the qPCR program.
3.4.2. Determination of Enrichment
Analyze the qPCR results by manually determining the cycle threshold for each reaction across the plate within the linear range of the amplification curve. Divide the relative DNA amount of the ChIP-seq library by the relative DNA amount of the input library for a given primer set. The resultant quotient corresponds to the enrichment value of a target in the library over the input library. The enrichment value for a target primer set should be at least 20-fold greater than the enrichment of a negative control primer set.
3. 5. Library Sequencing
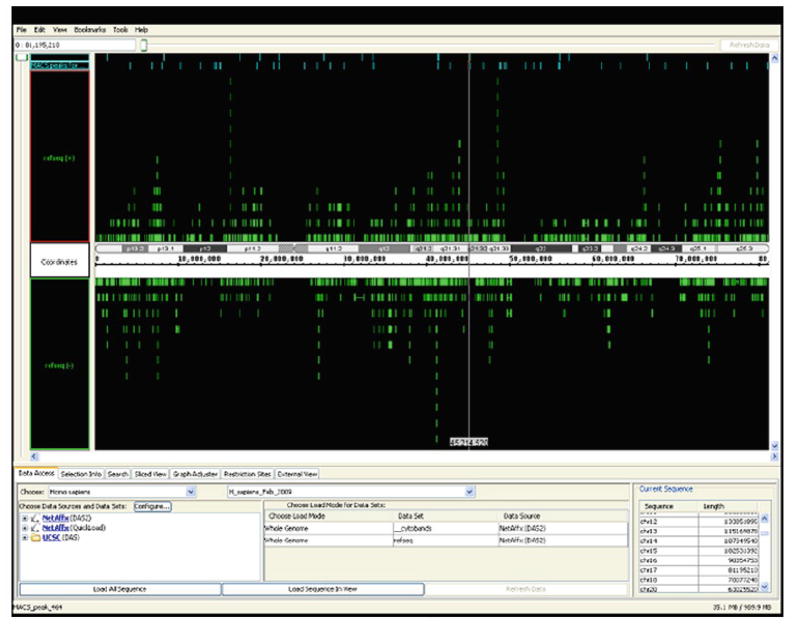
On average, 10–20 million uniquely mapped tags are sufficient to identify all binding sites for a site-specific binding transcriptional factor. Ideally, these reads should come from two independent ChIP samples, with the binding sites identified in each replicate having at least a 60% overlap. After the sequencing is performed, the short tags (approx. 25–50 nt) are mapped to the human genome (hg18, 2006). The tags that map uniquely to only one location in the genome are selected, and then the unique tags are extended to the average size of the library fragments (approx. 200 nt) and mapped into consecutive maps running the length of each chromosome. The collected data can be visualized using the UCSC browser (http://www.genome.ucsc.edu) or the Affymetrix Integrated Genome Browser (http://www.affymetrix.com/partners_programs/-programs/developer/tools/download_igb.affx) (Figure 4). Target sites can be identified using a variety of peak calling methods (12–14, 19).
Fig. 4.
Affymetrix-Integrated Genome Browser. Transcription factor (PARP1) binding sites are located on chromosome of reference genome.
3.6. Validation of ChIP-seq by qPCR using primers within KDM2A gene locus
The ChIP-seq experiment can be validated by qPCR using primers designed to amplify a mapped sequence to the reference genome of a transcription factor. ChIP samples of specific, nonspecific and input are used as template. These input samples should be diluted 1:10.
Prepare the following mixture (for a total reaction volume of 20 μL): SYBR Green (10 μL); 10 pM/μL KDM2A forward primer (1 μL); 10 pM/μL KDM2A reverse primer (1 μL); template ChIP-DNA (2 μL) and H2O (6 μL).
Perform real-time PCR reactions and analyze the result with StepOnePlus™ software (Applied Biosystems). The results indicating the specific and nonspecific regions of the KDM2A gene are shown in Figure 5A.
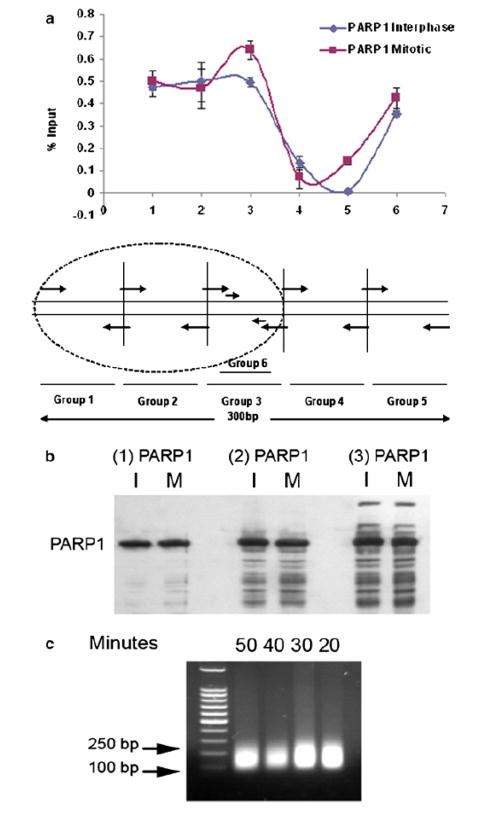
Fig. 5.
(a) Occupancy of PARP1 on lysine (K)-specific demethylase 2A (KDM2A) locus (300 bp). (b) Western blot showing the specificity of three different PARP1 antibodies. (c) Sonicated chromatin was checked on agarose gel after different time intervals of sonication.
ChIP-seq is an unbiased technique for genome-wide mapping of transcription factor binding sites, nucleosome mapping, and determining histone modifications at a high resolution. For successful ChIP-seq, both antibody specificity and sonication size of chromatin play very important roles. Before experimentation begins, specificity of antibody should be checked by Western blotting. The size of DNA fragments after sonication should be in range of 150–250 bp. Analysis of the ChIP-seq data explores unknown new binding sites and the location of binding sites relative to nearby potential new genes that may affect the functions of transcription factors.
Acknowledgments
We thank D. Martin and K. Pechenkina for comments on the manuscript. We also thank Greg Donahue (University of Pennsylvania Medical School) and Yan Zhou (Fox Chase Cancer Center) for advice on ChIP-seq data analysis. The research was supported by grants from the National Institutes of Health (R01 DK082623) to A.V.T.
Footnotes
The choice of an antibody is very important for successful ChIP-seq. Therefore, it is essential to keep detailed records on all antibodies that include their catalog and lot numbers. We checked antibody by Western blot before the ChIP experiment. The major band on the blot should be the correct size for the protein of interest. Antibodies that work in a test immunoprecipitation will most likely work in a ChIP assay (Figure 5B).
We designed the 100–150 bp oligos for amplicon. If possible, design replicate oligos for positive and negative controls, as well as for the test.
Micrococcal nuclease or sonication can be used to shear the chromatin. Both techniques work fine. The protocol discussed here is based on shearing by sonication.
To sonicate the chromatin, we used BioRuptor on a high setting for 5 cycles of 10 min with 30 sec on/off. Sonication depends on cell number, time, solution volume, and extent of cross-linking. To optimize sonication time, we collected 20 μL aliquots of chromatin after each 10 min cycle and analyzed size of DNA fragment on 1.5% agarose gel (Figure 5C).
The size of sonicated chromatin should be in the range of 150–250 bp.
We did not use agarose beads blocked with salmon sperm DNA as this could cause sequencing of the salmon sperm DNA. This would result in low-quality ChIP-seq data. Here, we used agarose beads from Invitrogen.
If possible, use nonspecific and specific antibody from the same batch.
After proteinase K (Invitrogen) and RNAse (Qiagen) digestion, purify the DNA by Qiagen PCR purification kit. This step prevents the DNA loss and saves time.
The precipitated ChIP DNA should be quantitated by Quant-iT™ PicoGreen assay (Invitrogen) before proceeding to library preparation.
Before library preparation, oligo pairs should be checked by SYBR Green-based q-PCR. To do this, use input DNA. For the dissociation curve, there should be a single peak of desired amplicon.
Input DNA should be processed with specific and nonspecific antibody for library preparation. Input library is critical for determining a baseline genome for identification of binding sites.
References
- 1.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–68. [PMC free article] [PubMed] [Google Scholar]
- 2.Nosseri C, Coppola S, Ghibelli L. Possible involvement of poly(ADP-ribosyl) polymerase in triggering stress-induced apoptosis. Exp Cell Res. 1994;212:367–373. doi: 10.1006/excr.1994.1156. [DOI] [PubMed] [Google Scholar]
- 3.Dantzer F, Amé JC, Schreiber V, Nakamura J, Ménissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 4.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 6.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- 9.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 11.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 12.Cuddapah S, Jothi R, Schones DE, Roh T-Y, Cui K, Zhao K. Global analysis of the insulator CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 14.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:1–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 15.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marks H, Chow JC, Denissov S, Françoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;8:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, nyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, Euskirchen G, Krzywinski M, Birol I, Snyder M, Hoodless PA, Hirst M, Marra MA, Jones SJ. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Wei CL, Lin F, Sung WK. An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics. 2008:2344–2349. doi: 10.1093/bioinformatics/btn402. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]