Abstract
We report the identification of a transcription elongation factor from HeLa cell nuclear extracts that causes pausing of RNA polymerase II (Pol II) in conjunction with the transcription inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB). This factor, termed DRB sensitivity-inducing factor (DSIF), is also required for transcription inhibition by H8. DSIF has been purified and is composed of 160-kD (p160) and 14-kD (p14) subunits. Isolation of a cDNA encoding DSIF p160 shows it to be a homolog of the Saccharomyces cerevisiae transcription factor Spt5. Recombinant Supt4h protein, the human homolog of yeast Spt4, is functionally equivalent to DSIF p14, indicating that DSIF is composed of the human homologs of Spt4 and Spt5. In addition to its negative role in elongation, DSIF is able to stimulate the rate of elongation by RNA Pol II in a reaction containing limiting concentrations of ribonucleoside triphosphates. A role for DSIF in transcription elongation is further supported by the fact that p160 has a region homologous to the bacterial elongation factor NusG. The combination of biochemical studies on DSIF and genetic analysis of Spt4 and Spt5 in yeast, also in this issue, indicates that DSIF associates with RNA Pol II and regulates its processivity in vitro and in vivo.
Keywords: DRB, DSIF, transcription elongation, transcription elongation factor, transcription inhibitor, protein phosphorylation, NusG
The regulation of gene expression is most commonly achieved by controlling transcription. Potentially, transcription can be controlled at many steps, including initiation, elongation, and termination (Roeder 1996 and references therein). The regulation of transcription by RNA polymerase II (Pol II) is an area of intense interest, and many studies have shown that regulation can occur at the level of transcription initiation. Whereas initiation has been extensively characterized, much less is understood about transcriptional elongation and the factors that control it, particularly in eukaryotes (Reines et al. 1996).
One promising approach to the study of transcriptional elongation in eukaryotes has been to study the effects of the transcription inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB). DRB was originally discovered as an inhibitor of the synthesis of heterogeneous nuclear RNA in human, murine, avian, and insect cells (Egyhazi 1974; Granick 1975; Sehgal et al. 1976a; Tamm et al. 1976). Many of these studies showed that DRB inhibits the synthesis of full-length RNA transcripts by enhancing the pausing or premature termination of transcription by RNA Pol II. Although DRB was originally proposed to inhibit transcription initiation (Egyhazi 1974, 1975, 1976; Sehgal et al. 1976b; Zandomeni et al. 1983), it has now clearly been shown to inhibit at the level of elongation (Chodosh et al. 1989).
DRB-mediated transcription inhibition appears to result from the inhibition of one or more protein kinases necessary for transcription. DRB has been shown to block the activity of casein kinase II (Zandomeni et al. 1986; Meggio et al. 1990), and of the TFIIH-associated protein kinase (Yankulov et al. 1995, 1996). Recent studies have suggested that by inhibiting the cdc2-related protein kinase called PITALRE, responsible for the positive transcription elongation factor b (P-TEFb), DRB inhibits the phosphorylation of the carboxy-terminal domain (CTD) of the RNA Pol II largest subunit, thereby blocking the transition from an initiated complex to an elongation complex (Mancebo et al. 1997; Zhu et al. 1997).
To identify factors that are directly involved in DRB-mediated inhibition and, therefore, presumably involved in transcription elongation, we have studied DRB-sensitive transcription in vitro. Because the inhibitory effect of DRB is not observed when transcription is reconstituted by a partially purified transcription system (Chodosh et al. 1989), we have used this system to assay for, and to purify, a factor required for DRB sensitivity. The purified DRB sensitivity-inducing factor (DSIF) from HeLa cell nuclear extracts is composed of two polypeptides of 14 kD (p14) and 160 kD (p160), and DSIF works as a negative elongation factor in the absence of DRB. DSIF is also required for the inhibitory effect of H8 on transcription elongation. Sequence analysis shows that p160 is a human homolog of the Saccharomyces cerevisiae transcription factor Spt5 (Swanson et al. 1991). Furthermore, recombinant Supt4h protein, the human homolog of yeast Spt4 (Malone et al. 1993; Hartzog et al. 1996), is functionally equivalent to DSIF p14, strongly suggesting that DSIF is composed of the human homologs of Spt4 and Spt5. In support of a role of DSIF in elongation, we provide several lines of evidence indicating that DSIF associates with RNA Pol II. Furthermore, sequence comparisons reveal that the central region of p160 has significant similarity to the bacterial transcription elongation factor NusG, which interacts with RNA polymerase and regulates its activity (Li et al. 1992; Sullivan and Gottesman 1992). In addition to its negative effect on elongation, DSIF stimulates the rate of transcription elongation under limiting nucleotide conditions in a DRB-sensitive fashion. Thus, DSIF is a novel transcription factor that regulates RNA Pol II elongation and that may be functionally conserved between prokaryotes and eukaryotes. Hartzog et al. (1998) show that the yeast Spt4 and Spt5 complex works as an elongation regulator in vivo, suggesting that DSIF plays a key role in transcription in vivo in human cells.
Results
DRB inhibits transcription elongation
To analyze the mechanism of inhibition by DRB, an in vitro transcription reaction was divided into two steps: a preincubation step with hexokinase-treated HeLa cell nuclear extracts and a DNA template for 45 min, followed by an initiation/elongation step for 10 min in the presence of A/C/UTP, that was sufficient to produce full-length G-free transcripts (data not shown). We used plasmid pTF3-6C2AT-100 or pTF3-6C2AT as a template DNA because transcription from these templates was efficiently inhibited by DRB (Wada et al. 1991; data not shown). The former produces full-length 100-nucleotide G-free transcripts, and the latter produces full-length 380-nucleotide G-free transcripts (Wada et al. 1991).
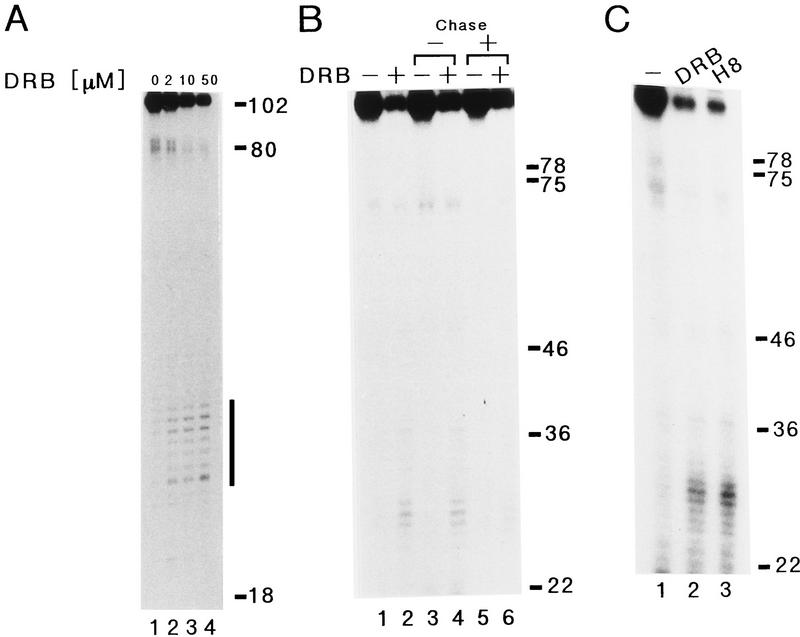
Chodosh et al. (1989) demonstrated previously that DRB inhibits transcription at the level of elongation. To test this conclusion under our conditions, a kinetically synchronized transcription reaction was carried out and transcripts synthesized from the pTF3-6C2AT-100 template were analyzed on a 20% sequencing gel. The synthesis of the 100-nucleotide transcript was strongly inhibited by increasing concentrations of DRB. Short-length transcripts of 25–35 nucleotides, however, accumulated with increasing concentrations of DRB (Fig. 1A, horizontal bar). At 50 μm DRB, the 100-nucleotide transcript was reduced to 20% of the level synthesized in the absence of DRB, and the decrease was roughly equal to the amount of the short-length transcripts accumulated in the reaction (Fig. 1B, lanes 1,2). The accumulated shorter RNA transcripts were eliminated by further incubation of the reaction in the presence of cold ATP, CTP, and UTP to 1 mm each (Fig. 1B, lanes 4,6). The chase in lane 6 reduced the level of short-length transcripts and increased the level of the full-length 100-nucleotide transcripts (cf. lane 2 and 6), suggesting that DRB treatment caused transcription elongation complexes to pause near the initiation site. We also observed the same accumulation of 25- to 35-nucleotide transcripts by the addition of H8 to 50 μm to a transcription system (Fig. 1C, lane 3). These results suggest that the two drugs, DRB and H8, inhibit transcription by inducing the arrest of elongating RNA Pol II.
Figure 1.
Effect of DRB or H8 on transcription elongation. (A) Kinetically synchronized reactions proceeded as described in Materials and Methods except reaction products from the pTF3-6C2AT-100 template were analyzed by a 20% sequencing gel. Numbers at the right indicate the positions of markers (nucleotides) and a solid bar corresponds to positions of RNA transcripts accumulated by addition of DRB. DRB was added to the reaction after the 45-min preincubation to various concentrations indicated above the gel. (B) Reactions proceeded as described in A except reactions were further incubated for 30 min after the 10-min initiation/elongation step in the absence [lanes 3,4, chase (−)] or presence [lanes 5,6, chase (+)] of cold ATP, CTP, and UTP (final concentration, 1 mm each). Reaction products from the pTF3-6C2AT-100 template in the absence (lanes 1,3,5) or presence (lanes 2,4,6) of 50 μm DRB, which was added to the reaction after the 45-min preincubation, were analyzed by a 10% sequencing gel. (C) Reactions proceeded as described in A. Reaction products from the pTF3-6C2AT-100 template in the presence of 50 μm DRB (lane 2) or 50 μm H8 (lane 3), which was added to the reaction after the 45-min preincubation, were analyzed by a 10% sequencing gel. Numbers at the right side indicate the positions of markers (nucleotides).
Purification of a DRB sensitivity-inducing factor
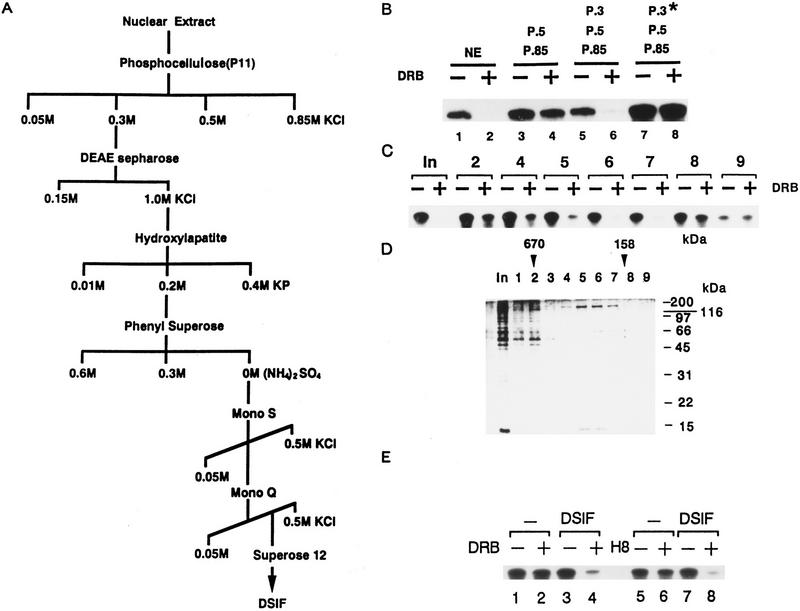
It has been reported that DRB does not inhibit a partially purified reconstituted transcription system but that DRB sensitivity can be conferred on the system by adding back a fraction that does not contain any of the essential transcription factors (Chodosh et al. 1989). To investigate the mechanism of transcription inhibition by DRB in detail, we sought to purify the factor(s) that could confer DRB sensitivity on a reconstituted in vitro transcription system. Accordingly, HeLa cell nuclear extracts were chromatographed on phosphocellulose (Samuels et al. 1982), and fractions were tested for their ability to confer DRB sensitivity on transcription. The 0.5 m (P.5) and 0.85 m (P.85) KCl fractions from the phosphocellulose column are sufficient for the reconstitution of transcription in vitro (Samuels et al. 1982), and we found that these fractions were sufficient for synthesizing 380-nucleotide-long transcripts from the G-free template (Fig. 2B, lane 3). Inhibition by DRB, however, was not observed by use of these fractions (Fig. 2B, lane 4). Addition of the 0.3 m (P.3) KCl fraction, which is not required for accurate transcription initiation in vitro, conferred DRB sensitivity on transcription (Fig. 2B, lanes 5,6). We also found that the activity conferring DRB sensitivity was heat labile (Fig. 2B, lanes 7,8), suggesting that it consists of a protein factor(s). These data agree with results reported previously (Chodosh et al. 1989). We refer to the protein factor that confers DRB sensitivity as DSIF. DSIF was purified from the P.3 fraction by use of conventional chromatography (Fig. 2A). At the final step of purification, gel filtration, DSIF activity was found between fractions 4 and 7 (Fig. 2C), which on silver-stained gels revealed two major proteins p160 and p14 (Fig. 2D, cf. lanes 4 and 7). On the basis of gel filtration, purified DSIF forms a native complex of >300 kD, suggesting the possibility of multimer formation by p160 and p14 (Fig. 2D). We also found that purified DSIF confers H8 sensitivity on a partially reconstituted transcription system (Fig. 2E), suggesting that both DRB and H8 exert their inhibitory effects on transcription elongation through a common factor DSIF.
Figure 2.
Purification of DSIF. (A) The purification scheme for DSIF is illustrated and is described in Materials and Methods. (B) Kinetically-synchronized transcription reaction was carried out with 2 μl of the concentrated P.5 and P.85 fractions from the phosphocellulose column (except lanes 1 and 2, which contained 4 μl of HeLa cell nuclear extracts). Even numbered lanes contained DRB to 50 μm. P.3 (6 μl) was added to a transcription reaction (lanes 5,6). P.3 (6 μl) was incubated at 68°C for 10 min prior to its addition to the reaction (lanes 7,8). (C) Fractions (1 μl) containing DSIF activity from a Superose 12 column were assayed for their ability to confer DRB sensitivity on a partially purified transcription system. Reactions were as described in Materials and Methods. Reaction products were resolved by 8% urea–PAGE. (In) Column input fraction (0.5 μl). (D) Aliquots (1 μl) of the Superose 12 column fractions were analyzed in a 12.5% SDS–polyacrylamide gel and the proteins were visualized by silver. Numbers to the right of gel indicate the position of protein molecular size standards. (E) Purified DSIF was assayed for its ability to confer DRB or H8 sensitivity on a partially purified transcription system. Reactions proceeded as described in B. DRB to 50 μm (lanes 2,4) or H8 to 50 μm (lanes 6,8) was added to the reaction after the 45-min preincubation. Lanes 3, 4, 7, and 8 contained 10 ng of purified DSIF.
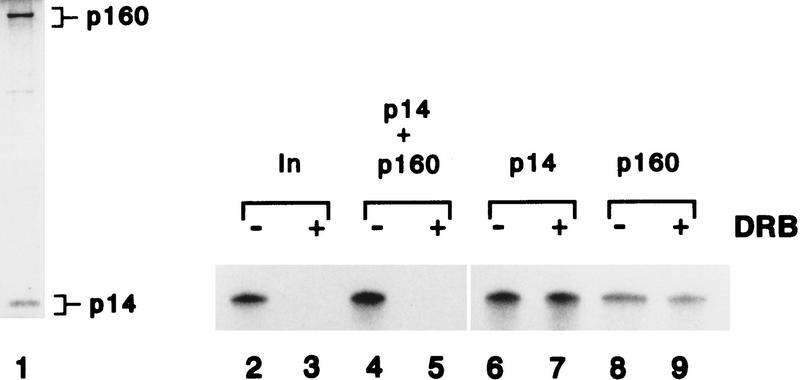
To further examine whether p160 and p14 can confer DRB sensitivity on transcription, fraction 5 was subjected to SDS-PAGE and proteins were recovered from the gel and renatured as described previously (Wada et al. 1991). When p160 and p14 were renatured together, DSIF activity was reconstituted (Fig. 3, lanes 4,5). We also observed the inhibition by DRB when each protein was renatured independently and then added together to a transcription reaction (data not shown). Neither p160 nor p14 alone, however, possessed DSIF activity (Fig. 3, lanes 6–9). These results show that p160 and p14 are necessary and sufficient for DSIF activity.
Figure 3.
Renaturation of DSIF activity. Purified DSIF (fraction 5 from Superose 12, 40 μl) was separated by a 12.5% SDS–polyacrylamide gel. Bands corresponding to 160- and 14-kD polypeptides were cut out of gel. The renaturation method was described in Materials and Methods. Aliquots (1 μl) of the fraction 5 were subjected to 12.5% SDS-PAGE and proteins were visualized by silver staining (lane 1). Renatured proteins were tested for their ability to confer DRB sensitivity on a partially purified reconstituted transcription system (lanes 2–9), and reaction products were resolved by 8% urea–PAGE. DRB was added at the final concentration of 50 μm to lanes 3, 5, 7, and 9. Transcription reactions were performed with purified DSIF (lanes 2,3, 10 ng). p160 and p14 were renatured together and added lanes 4 and 5 (7.5 ng of each protein), and renatured p14 (15 ng) alone was added to lanes 6 and 7, and renatured p160 (15 ng) alone was added to lanes 8 and 9.
Activity of DSIF on transcription
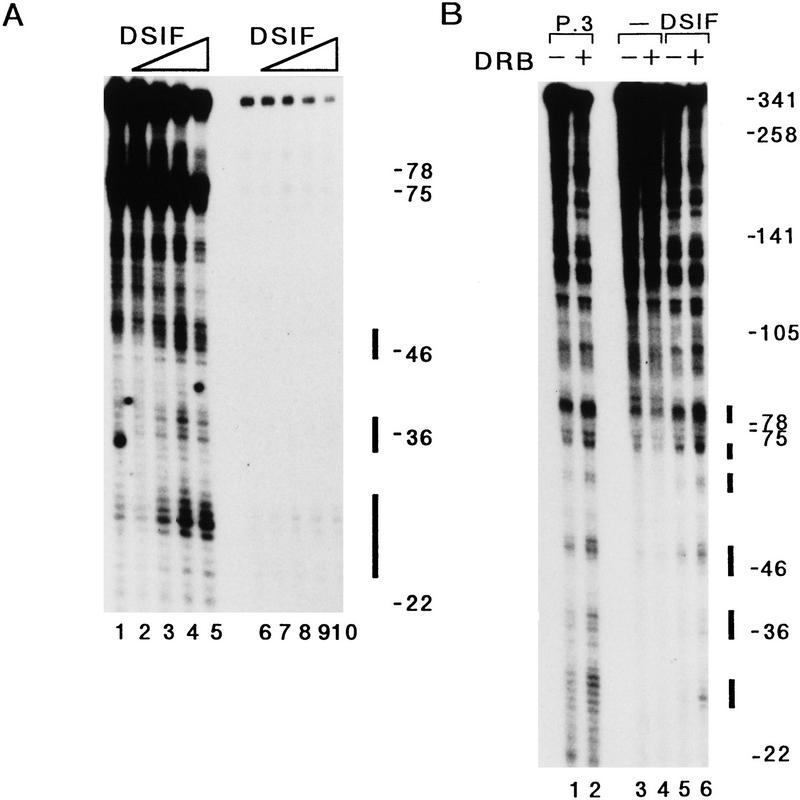
The effect of DRB is at the level of transcription elongation, suggesting that DSIF could act as a negative elongation factor in the absence of DRB. To test this, we added purified DSIF to a partially reconstituted transcription system and a minimal transcription system that was reconstituted by recombinant TBP, TFIIB, TFIIF, and highly purified RNA Pol II (Usuda et al. 1991). TAFII250 (Dikstein et al. 1996), TFIIH, P-TEFb, and other kinases are presumably absent, barring association with the highly purified RNA Pol II complex. The synthesis of the 100-nucleotide transcript was inhibited by increasing amounts of DSIF in both systems. RNA transcripts of ∼28, 38, and 48 nucleotides, and RNA transcripts of ∼28 nucleotides accumulated with increasing amounts of purified DSIF in a partially reconstituted transcription system and in a minimally reconstituted transcription system, respectively (Fig. 4A). These results show that DSIF induces the arrest of elongating RNA Pol II in the absence of DRB, indicating that DSIF works as a negative elongation factor. We further examined the effects of DSIF and DRB on transcription elongation with a 10% sequencing gel. In the absence of DSIF, short RNA transcripts of <75 nucleotides were hardly produced in a partially reconstituted transcription system, regardless of the presence or absence of DRB (Fig. 4B, lanes 3,4). Addition of either the P.3 fraction or purified DSIF, however, increased RNA transcripts of <85 nucleotides (Fig. 4B, lanes 1,5, solid, horizontal bars), and the level of these shorter transcripts was further increased by the addition of DRB to the system (lanes 2,6). This result indicates that one of DSIF’s normal functions (in the absence of DRB) is as a negative elongation factor that regulates RNA Pol II processivity. This, in turn, suggests that DRB inhibits transcription elongation through an effect on DSIF activity. Therefore, DSIF is a negative elongation factor that may be normally inhibited by a DRB-sensitive protein kinase. We also carried out a DRB-sensitive transcription assay by use of a minimally reconstituted transcription system with or without TFIIE and TFIIH. In this system DRB had no effect on transcription in the presence of DSIF (T. Takagi and H. Handa, unpubl.), suggesting that the DRB-sensitive protein kinase may exist in the P.5 and P.85 fractions.
Figure 4.
Characterization of DSIF normal function in vitro. (A) Purified DSIF was added to a partially reconstituted transcription system (lanes 1–5) or a minimal transcription system (lanes 6–10) that was reconstituted by 10 ng of purified recombinant TBP, 30 ng of purified recombinant TFIIB, 16 ng of purified recombinant TFIIF, and 0.5 μl of highly purified RNA Pol II (Usuda et al. 1991). Kinetically synchronized reactions proceeded as described in Materials and Methods except reactions contained the pTF3-6C2AT-100 template. Reaction products were analyzed by a 10% sequencing gel. Numbers to the right of gel indicate the position of markers (nucleotides) and solid bars correspond to positions of RNA transcripts accumulated by the addition of DSIF. (Lanes 2,7) 8 ng of DSIF; (lanes 3,8) 16 ng of DSIF; (lanes 4,9) 32 ng of DSIF; (lanes 5,10) 64 ng of DSIF. (B) Effect of DRB on transcription was tested in the absence or presence of DSIF. Reactions proceeded as described in A. Even numbered lanes contained DRB to 50 μm, which was added to the reaction after the 45-min preincubation. P.3 (6 μl) was added to a partially reconstituted transcription system (lanes 1,2) and DSIF (24 ng) was added to the system (lanes 5,6). Numbers to the right of gel indicate the position of markers (nucleotides) and solid bars correspond to positions of RNA transcripts induced by the addition of DSIF.
DSIF p160 is a homolog of the yeast transcription factor Spt5
The identity of DSIF polypeptide p160 was determined by microsequencing. To do this, 7.5 μg of p160 was subjected to digestion by lysyl endopeptidase, the resulting polypeptides were separated by reverse-phase, high-pressure liquid chromatography, and four peptide sequences were obtained by microsequence analysis. On the basis of the peptide sequences, we cloned a cDNA encoding the p160 subunit of DSIF as described in Materials and Methods. On the basis of several criteria, this cDNA was judged to encode full-length p160. First, all four peptide sequences obtained from microsequencing were found in the predicted coding region (data not shown). Second, a rabbit reticulocyte lysate programmed with RNA transcribed from this cDNA produced a protein with a mobility indistinguishable from that of p160 (Y. Yamaguchi and H. Handa, unpubl.). Finally, Northern analysis of polyadenylated RNA isolated from several different types of human cells detected a single 3.6-kb species, a length consistent with that of the cDNA isolated and adequate to encode a polypeptide of 1087 amino acid protein in the open reading frame of 3261 bp (Y. Yamaguchi and H. Handa, unpubl.).
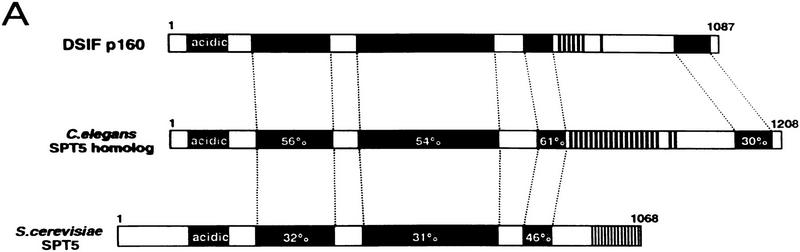
Sequence analysis of the cDNA for the p160 subunit of DSIF revealed that p160 is a homolog of S. cerevisiae Spt5 and of a Caenorhabditis elegans Spt5 homolog of unknown function (Fig. 5A; Chiang et al. 1996a; Stachora et al. 1997). DSIF p160 has two striking features: a very acidic amino terminus and a carboxy-terminal amino acid repeat. S. cerevisiae Spt5 contains 15 copies of the 6-amino-acid repeat SA/TWGGA/Q, but in the C. elegans (20 copies) and the human (7 copies) Spt5 homologs, the 6-amino-acid repeat is K/RTPA/MYG/D/E. The conserved acidic domain of p160 (residues 1–118) was 47% glutamic and aspartic acids and also included a number of potential sites for phosphorylation by casein kinase II. Thus, DSIF p160 is encoded by a human homolog of SPT5, a yeast gene that is essential for growth and normal transcription (Swanson et al. 1991), suggesting that DSIF is critical for normal transcription in mammalian cells.
Figure 5.
DSIF is composed of human Spt4 and Spt5 homologs. (A) Schematic comparison of DSIF p160 and yeast Spt5 and C. elegans Spt5 homologs. The DSIF p160 is compared with the C. elegans Spt5 homolog and yeast Spt5 proteins. The positions of the acidic domain at the amino terminus, highly conserved regions, and the carboxy-terminal repeats are marked by closed boxes. DSIF p160, C. elegans Spt5 homolog, and yeast Spt5 are highly related over their entire lengths. Numbers in boxes indicate percentages of identical amino acid within the region. (B) Recombinant Supt4h protein was tested for its ability to confer DRB sensitivity on a partially purified reconstituted transcription system. The DSIF p160 and rSupt4h were renatured independently as described in Materials and Methods. DRB was added to a final concentration of 50 μm lanes 2, 4, 6, and 8. Transcription reactions were performed with 10 ng of purified DSIF (lanes 1,2), 12.5 ng of p160 (lanes 3–6), 12.5 ng of rSupt4h (lanes 5–8). Reaction products were resolved by 8% urea–PAGE. (In) DSIF input fraction for SDS-PAGE before separation of p160 and p14 on the gel.
DSIF p14 is a homolog of yeast Spt4
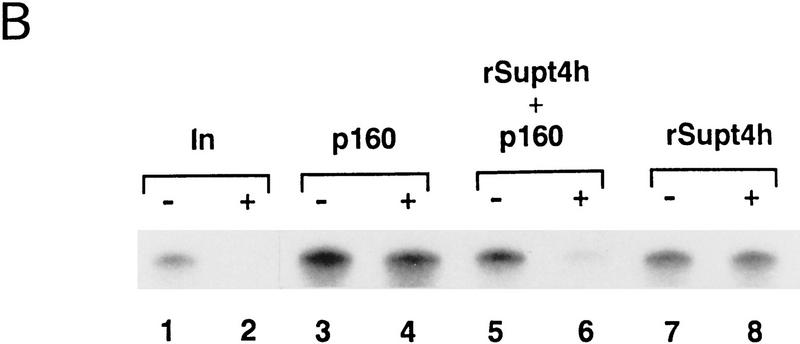
Both biochemical and genetic evidence indicate that Spt4 and Spt5 function in a complex (Swanson and Winston 1992; Hartzog et al. 1998), suggesting that the DSIF p14 may be encoded by SUPT4H, a functional human homolog of the yeast SPT4 gene (Chiang et al. 1996b; Hartzog et al. 1996). To test this hypothesis, the PCR-amplified SUPT4H cDNA was inserted into the vector pET14b and a histidine-tagged fusion Supt4h protein was expressed in bacteria and purified by affinity chromatography (data not shown). Following SDS-PAGE and renaturation, the recombinant Supt4h protein (rSupt4h) was tested for its ability to confer DRB sensitivity on transcription in the presence or absence of p160. When p160 and rSupt4h were mixed and added to a transcription reaction, DSIF activity was reconstituted. Alone, neither protein had any DSIF activity (Fig. 5B). This result shows that Supt4h is functionally equivalent to DSIF p14. Thus, DSIF is composed of human Spt4 and Spt5 homologs.
DSIF confers DRB sensitivity on transcription through its interaction with RNA Pol II
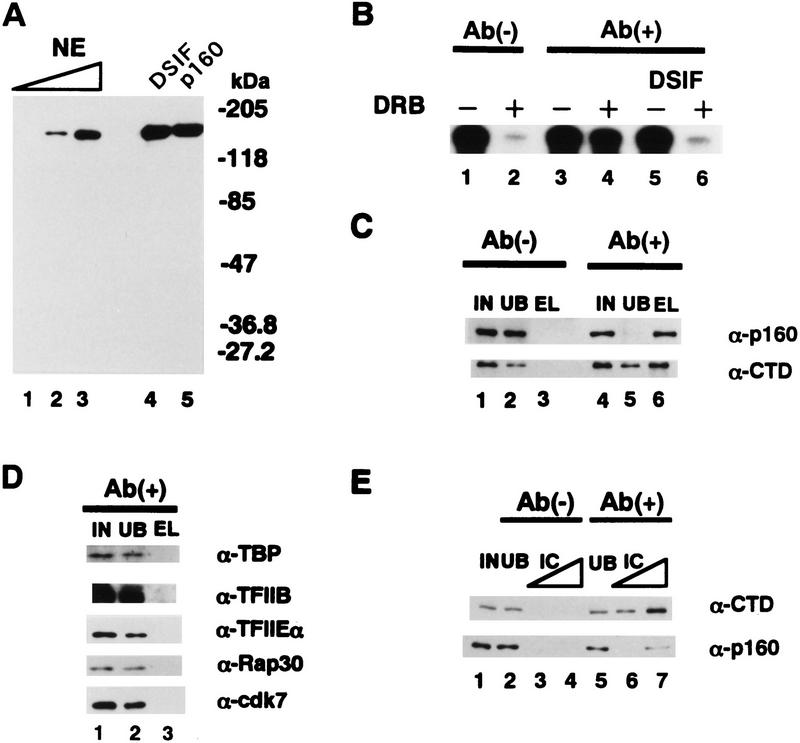
To elucidate how DRB modifies the effect of DSIF on transcription, immunodepletion of DSIF from a crude HeLa cell extract was performed with the α-p160 affinity resin as described in Materials and Methods. To do this, a monoclonal antibody (α-p160) was raised against the carboxy-terminal 250-amino-acid residues of p160. The antibody reacted with p160 in HeLa cell nuclear extracts, p160 in purified DSIF, and recombinant DSIF p160 (Fig. 6A). We estimate that ∼60,000 molecules of p160 exist in a HeLa cell nucleus. In the immunodepletion experiments, a control extract treated by the protein G resin shows DRB-sensitive transcription (Fig. 6B, lanes 1,2). In contrast, immunodepletion of DSIF prevented DRB from working, although it had no effect on transcription in the absence of DRB (Fig. 6B, lanes 1–4). Add-back of purified DSIF to the depleted extract conferred DRB sensitivity on transcription (Fig. 6B, lanes 5,6). These results indicate that depletion of DSIF p160 abolishes DRB-sensitive transcription, suggesting that DRB inhibition is DSIF dependent. Western blotting showed that p160 was removed by the α-p160 affinity resin to <10% compared with that before immunodepletion but not by a control resin and it eluted only from the affinity resin (Fig. 6C).
Figure 6.
DSIF confers DRB sensitivity on transcription through its interaction with RNA Pol II. (A) Western blotting with α-p160. HeLa cell nuclear extracts, purified DSIF and DSIF p160 were analyzed by Western blotting with α-p160. (Lanes 1–3) 0.0625, 0.25, and 1 μl of nuclear extracts, respectively; (lane 4) 4 ng of p160 in DSIF; (lane 5) 4 ng of recombinant p160. (B) Immunodepletion of DSIF. Nuclear extracts were treated twice by the protein G resin [Ab(-) and the α-p160 affinity resin Ab(+)] as described in Materials and Methods. A kinetically synchronized transcription reaction with the pTF3-6C2AT template was carried out with 4 μl of nuclear extracts treated by the resin. DRB was added to 50 μm (lanes 2,4,6). Purified DSIF (10 ng) was added in lanes 5 and 6. Reaction products were resolved by 8% urea–PAGE. (C,D) Western blotting of materials in immunodepletion experiments with α-p160, α-CTD, and specific antibodies against general transcription factors indicated in D. (IN) Nuclear extracts (1 μl); (UB) second unbound fraction (1 μl); (EL) eluate from the resin (2.5 μl). Ab(−) and Ab(+) were described in B. (E) Immunoprecipitation with α-CTD and Western blotting with α-p160 and α-CTD. Reactions were performed as described in Materials and Methods. (IC) Immunoadsorbed complexes (lanes 3,6) 0.3 μl, (lanes 4,7) 1 μl.
Because DSIF p160 has some sequence homology with Escherichia coli transcription elongation factor NusG that binds to E. coli RNA polymerase (see below; Hartzog et al. 1998), we tested whether p160 interacts with RNA Pol II. To do this, we performed Western blotting with α-CTD (carboxy-terminal domain) antibodies and observed that the largest subunit of RNA Pol II was also removed by the α-p160 affinity resin to a level of ∼65% compared with that before immunodepletion of p160. The largest RNA Pol II subunit was also found in the eluate from the affinity resin but not in that from the control resin (Fig. 6C). Silver staining showed that all the subunits of RNA Pol II were found in the eluate from the α-p160 affinity resin (data not shown). As seen in Figure 6D, general transcription factors tested were not found in the eluate of the α-p160 affinity resin, suggesting the specific association of DSIF with RNA Pol II. We also performed immunoprecipitation of RNA Pol II with α-CTD and detected the largest subunit of RNA Pol II and p160 by Western blotting. Immunoadsorbed complexes of α-CTD contained both the largest subunit of RNA Pol II and p160 (Fig. 6E). These results indicate that at least DSIF p160 and RNA Pol II immunoprecipitate together, suggesting that DSIF associates with RNA Pol II.
DSIF can stimulate transcription elongation under certain conditions
An extensive homology search revealed that the central region of p160 contains four tandem KOW motifs that have been found in both the bacterial transcription elongation factor NusG and ribosomal proteins (Fig. 7A; Kyrpides et al. 1996). All four motifs conserved in Spt5 genes were similar in length and the strongest sequence homology was with the KOW motifs of NusG and ribosomal protein RL24 (Fig. 7A). In addition, an invariant glycine residue in the motif was found in all proteins except yeast Spt5. Thus, p160 is related to NusG, which is a member of a class of essential transcription elongation factors that have been implicated in a variety of cellular and viral termination and antitermination processes (Li et al. 1992, 1993; Sullivan and Gottesman 1992; Burns and Richardson 1995; Mogridge et al. 1995). A recent study has shown that E. coli NusG stimulates the rate of transcription elongation both in vivo and in vitro and this stimulation appears to result from the suppression of RNA polymerase pausing (Burova et al. 1995).
Figure 7.
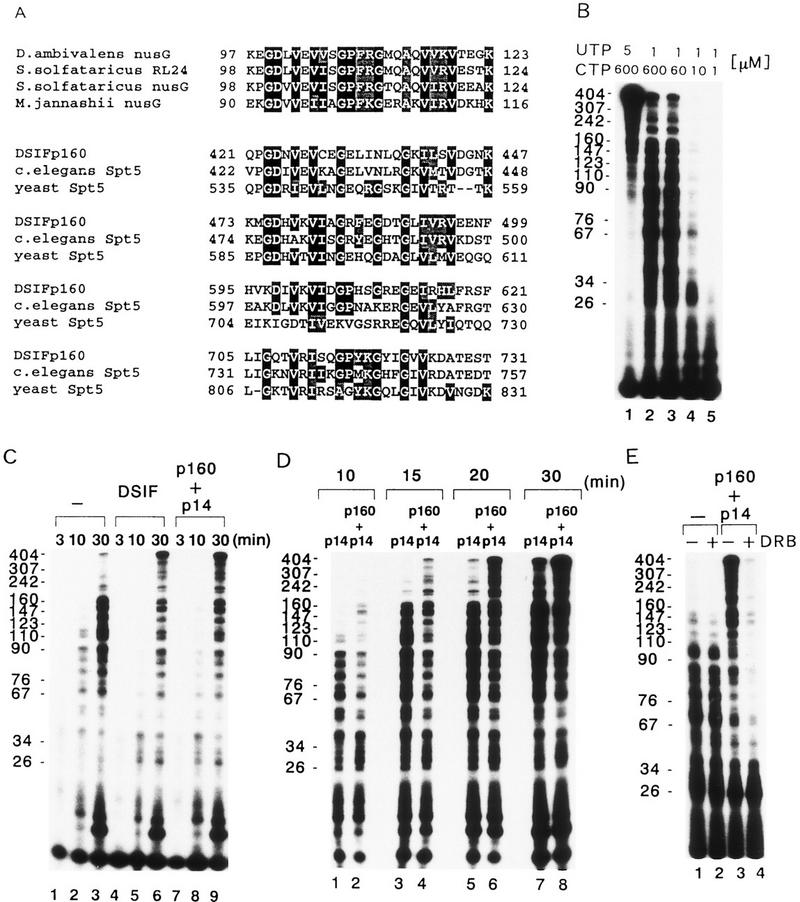
Characterization of the transcription elongation activity of DSIF in vitro. (A) Multiple sequence alignment of the KOW motif as found in bacterial NusG, ribosomal protein RL24, human DSIF p160, C. elegans Spt5 homolog and yeast Spt5. The amino acids of each protein are numbered next to the sequences. A BLAST search of the GenBank database with the amino-terminal 800 amino acids residues of p160 revealed that the central region of p160 shares significant sequence similarity with the KOW motif in Drosophila ambivalens NusG (P = 0.022), Sulfolobus solfataricus RL24 (P = 0.022), S. solfataricus NusG (P = 0.0012), and Methanococcus jannaschii NusG (P = 0.00019). The alignment and shading of multiple sequences was performed with Multiple sequence alignment with hierarchical clustering (Multalin, Corpet 1988) and BOXSHADE 3.21 software under default conditions. (B) Kinetically synchronized transcription reaction containing various concentrations of ribonucleoside triphosphates. Reactions were carried out with 2 μl of the concentrated P.5 and P.85 fractions from phosphocellulose column fractionation as described in Materials and Methods except that a 20 min incubation proceeded after addition of 60 μm ATP, indicated concentrations of CTP and UTP containing 5 μCi of [α-32P]UTP (800 Ci/mmole). Numbers on the left indicate the position of markers (nucleotides). (C) Transcriptional activity of native and recombinant DSIF. A kineticallysynchronized transcription reaction was carried out as described inB except that an incubation after addition of 60 μm ATP, 10 μm CTP, and 1 μm UTP containing 5 μCi of [α-32P]UTP (800 Ci/mmole) proceeded for the indicated times (top, in minutes) in the absence (lanes 1–3) or presence of 16 ng of purified DSIF (lanes 4–6), or 2 ng of p14 and 16 ng of p160 (lanes 7–9). The pTF3-6C2AT template was used. Numbers at left indicate the position of markers (nucleotides). (D) Comparison of specific activities of p14 and a mixture of p14 and p160. Reactions were carried out as described in C except an incubation after addition of ribonucleoside triphosphates proceeded for the indicated times (top, in minutes) in the presence of 2 ng of p14 and 16 ng of p160 (lanes 2,4,6,8). Numbers at left indicate the position of markers (nucleotides). (E) DRB-sensitive transcription under limiting concentrations of ribonucleoside triphosphates. Reactions were carried out as described in D except a 20 min incubation proceeded after addition of ribonucleoside triphosphates in the presence of 2 ng of p14 and 16 ng of p160 (lanes 3,4). Lanes 2 and 4 contained 50 μm of DRB. Numbers on the left indicate the position of markers (nucleotides).
Motivated by these findings, we examined the ability of DSIF to stimulate the rate of accumulation of full-length G-free transcripts. In these experiments we employed the pTF3-6C2AT template, which produces a longer RNA transcript than the pTF3-6C2AT-100 template. The longer RNA transcripts allowed the examination of the effect of DSIF on the rate of transcription elongation. Preinitiation complexes were assembled at the pTF3-6C2AT template, and transcription was initiated by addition of various concentrations of ribonucleoside triphosphates (Fig. 7B). Under decreased nucleotide levels, the rate of RNA chain elongation was very slow, and short-length RNA transcripts accumulated by pausing or arrest of elongating RNA Pol II. To examine the effect of DSIF on the rate of transcription elongation, we used 1 μm UTP and 10 μm CTP in the subsequent experiments. In the absence of DSIF, most of the transcripts that accumulated 30 min after addition of ribonucleoside triphosphates were <180 nucleotides in length (Fig. 7C, lane 3). The amount of full-length RNA transcripts increased on addition of native DSIF or a mixture of the recombinant p14 and p160 subunits (Fig. 7C) but not on addition of the p160 alone (data not shown), suggesting that DSIF stimulates the rate of accumulation of full-length transcripts.
To test whether the rate of transcription elongation can be accelerated and whether such acceleration would be dependent on the p160 subunit, we performed transcription reactions in the presence of p14 alone or with a mixture of the p14 and p160 subunits with different incubation times after addition of ribonucleoside triphosphates. In the presence of p160, full-length RNA transcripts were first detected ∼15 min after addition of ribonucleoside triphosphates and increased in amount with time (Fig. 7D). In contrast, in the absence of p160, full-length transcripts were first detected ∼20 min after addition of ribonucleoside triphosphates. Notably, when the incubation time after addition of ribonucleoside triphosphates was >15 min, the amount of full-length RNA transcripts that accumulated in reactions containing both p14 and p160 was greater than in those containing p14 alone. Moreover, the accumulation of transcripts between 30 and 180 nucleotides long decreased in the presence of both p14 and p160. These results suggest that DSIF increases the rate of transcription elongation by RNA Pol II in a reaction that is strongly dependent on the p160 subunit. To examine the effect of DRB on transcription elongation under decreased nucleotide levels, DRB was added to the reaction at a final concentration of 50 μm (Fig. 7E). DRB had little effect on transcription in the reaction without DSIF. In contrast, production of full-length RNA transcripts and transcripts longer than 50 nucleotides were sensitive to DRB in a reaction containing recombinant DSIF (Fig. 7E). This result supports the idea that DRB inhibits transcription dependent on DSIF. Thus, DRB itself has no effect on in vitro transcription elongation by RNA Pol II in the absence of DSIF; however, the drug enhances the negative effect of DSIF on transcription elongation when an in vitro reaction contains DSIF.
Discussion
Studies of the mechanism by which DRB inhibits transcription elongation in vitro have identified a new cellular factor, DSIF. Our results show that DSIF can play a negative role in transcription elongation; therefore, it is different from eukaryotic elongation factors described previously (for review, see Reines et al. 1996). The two subunits of DSIF, p160 and p14, are human homologs of S. cerevisiae Spt5 and Spt4, respectively, strongly suggesting that DSIF is conserved between humans and yeast. The S. cerevisiae SPT5 and SPT4 genes are members of a large set of SPT genes originally identified by mutations that suppress the transcriptional defects caused by insertions of the retrotransposon Ty or its long terminal repeat, δ, in promoter regions (Winston 1992). This convergence of biochemical and genetic studies on DSIF and Spt4–Spt5 suggests that these functions are critical for normal transcription in vivo throughout eukaryotes. We also found that DSIF p160 has a region homologous to the bacterial transcription elongation factor NusG and that under the limiting conditions, DSIF stimulates the rate of transcription elongation by RNA Pol II in a manner similar to NusG. Results in Hartzog et al. (1998) show that the Spt4–Spt5 complex plays a role in transcription elongation in vivo. Therefore, DSIF function may be evolutionarily conserved in the control of transcription elongation between prokaryotes and eukaryotes.
Transcription inhibition by DSIF and either DRB or H8
Current evidence suggests that both DRB and H8 inhibit transcription elongation by blocking the activity of a protein kinase (Yankulov et al. 1995, 1996; Marshall et al. 1996). The discovery that DSIF is required for both DRB- and H8-mediated transcription inhibition suggests three general models for the role of DSIF in transcription elongation: (1) DSIF and DRB are both required to block the phosphorylation of a component of the preinitiation complex (PIC) and that block subsequently impairs elongation; (2) DSIF is a potent negative elongation factor that is normally inactivated by phosphorylation; DRB inhibits the phosphorylation of DSIF; and (3) DRB inhibits the phosphorylation of a PIC component and that makes RNA Pol II sensitive to pausing or termination by DSIF. All three models are possible on the basis of the current data; however, model 1 is unlikely, because cells are not normally exposed to DRB. Models 2 and 3 both postulate a negative role for DSIF in transcription elongation. We showed here that DSIF functions as a negative elongation factor in the absence of DRB. Model 2, however, is unlikely, because DSIF p160 is phosphorylated in a DRB-insensitive manner in crude HeLa cell nuclear extracts under our transcription conditions (T. Takagi and H. Handa, unpubl.). Therefore, we prefer model 3. We have no evidence indicating that DSIF itself is a protein kinase, ruling it out as a direct target of DRB (unpubl.).
Importantly, we showed here that DSIF-mediated transcription inhibition by H8 (50 μm), a well-characterized protein kinase inhibitor, and transcription inhibitor, as well as DRB, in our reconstituted transcription system (Fig. 1C). The simplest interpretation of the results is that both inhibitors target the same protein kinase, which alleviates the negative effect of DSIF on transcription elongation. H8 inhibits transcription in some crude transcription systems (Yankulov et al. 1995; Marshall et al. 1996) but does not in a highly purified system (Serizawa et al. 1993). The difference between them may be attributed to the presence of DSIF. In contrast, H8 (>200 μm) is capable of inhibiting transcription from the murine dehydrofolate reductase (DHFR) promoter in a defined transcription system devoid of DSIF (Akoulitchev et al. 1995). Thus, transcription inhibition by a high concentration of H8 may not be dependent on DSIF, or there may exist multiple kinases with different sensitivities to H8. At least in our system, DSIF is an essential component of both DRB- and H8-mediated transcription inhibition.
Understanding the mechanism by which DRB inhibits elongation will help to elucidate the role of DSIF. Previous studies have shown that DRB can block the activity of particular protein kinases, including casein kinase II (Zandomeni et al. 1986; Meggio et al. 1990), TFIIH-associated protein kinase (Yankulov et al. 1995), and P-TEFb (Marshall and Price 1995; Marshall et al. 1996). On the other hand, the mechanistic basis for the transcription inhibition by H8 remains elusive. Studies on the DHFR promoter have suggested that H8 (>200 μm) inhibits the formation of the first phosphodiester bond (Buermeyer et al. 1992; Akoulitchev et al. 1995), but other studies have suggested that H8 (<200 μm) has an inhibitory effect on transcription elongation (Yankulov et al. 1995; Marshall et al. 1996). H8 is known to be a protein kinase inhibitor that inhibits a broad spectrum of protein kinases including cAMP-dependent protein kinase (Hidaka et al. 1984). Among these kinases, TFIIH-associated kinase and P-TEFb have been suggested to be the target for transcription inhibition by H8 (Akoulitchev et al. 1995; Yankulov et al. 1995; Marshall et al. 1996). It is interesting that these two protein kinases are also sensitive to DRB and can phosphorylate the Pol II CTD. P-TEFb, identified previously in Drosophila in an in vitro complementation assay as an elongation factor that enhances productive elongation in a DRB-sensitive manner, has been shown recently to be a CTD kinase (Marshall and Price 1995; Marshall et al. 1996). In addition, most recent studies provide the strongest evidence that the protein kinase responsible for P-TEFb activity is a previously known cdc2-related kinase called PITALRE (Mancebo et al. 1997; Zhu et al. 1997). The carboxy-terminal domain of the Pol II largest subunit is phosphorylated extensively during the transition from initiation to elongation phase of the transcription cycle (Dahmus 1996 and references therein), and its phosphorylation may play a regulatory role in transcription elongation (Yankulov et al. 1995, 1996; Jones 1997). Therefore, phosphorylation of the Pol II CTD by either TFIIH-associated kinase, p-TEFb, or both, may be the target of DRB, H8, or both, that alleviates the negative effect of DSIF. It has been reported that the presence of the CTD, or its phosphorylation is not required for transcription from some promoters (Buermeyer et al. 1992; Serizawa et al. 1993; Akoulitchev et al. 1995; Makela et al. 1995). In contrast, DSIF is potentially a general negative regulator of transcription elongation (see below). Thus, the elucidation of an interaction between the CTD and DSIF awaits future studies.
DSIF contains homologs of S. cerevisiae Spt4 and Spt5
The identification of DSIF subunits as mammalian homologs of the S. cerevisiae Spt4 and Spt5 proteins provides strong support for the idea that DSIF plays a critical role in transcription in vivo. Genetic studies in yeast have shown that Spt5 is essential for growth and is important for transcription (Swanson et al. 1991). In addition, between studies of human DSIF and yeast Spt4–Spt5 there are some clear similarities. First, as for DSIF, genetic analysis suggests that Spt4 and Spt5 are negative regulators of transcription. This hypothesis is based on the observation that spt4/spt5 mutations suppress mutations that impair promoter function. For example, both spt4 and spt5 mutations suppress loss of a UAS, suggesting that in the absence of UAS function, Spt4 and Spt5 negatively control transcription (Swanson and Winston 1992). Second, in both mammalian and yeast cells, Spt4 and Spt5 are found tightly associated (this work; Hartzog et al. 1998). Third, the human SPT4 gene, SUPT4H, functions very well in yeast (Hartzog et al. 1996). In addition, DSIF p14, Supt4h, has been shown to be a nuclear protein in HeLa cells (Hartzog et al. 1996). These results support the hypothesis that DSIF and Spt4–Spt5 carry out similar functions in vivo.
DSIF as a general regulator of transcription elongation
What is the physiological function of DSIF? We showed here that DSIF represses transcription elongation in the absence of DRB (Fig. 4A). We also showed that DSIF stimulates the rate of elongation by RNA Pol II under nucleotide-limiting conditions (Fig. 7). In addition, immunological analysis indicates the association of DSIF with RNA Pol II (Fig. 6). These data indicate that DSIF associates with RNA Pol II and regulates its processivity. Several lines of evidence suggest that DSIF acts as a general regulator of transcription elongation. First, DRB affects all the class II genes tested to date. Second, DSIF inhibits transcription from the adenovirus E4 promoter containing only TATA box. Third, DSIF works as an elongation inhibitor in the minimally reconstituted transcription system employing recombinant TBP, TFIIB, TFIIF, and purified-RNA Pol II. Fourth, DSIF associates with RNA Pol II. In agreement with this finding, Hartzog et al. (1998) report genetic and biochemical evidence for an interaction between yeast Spt4–Spt5 and RNA Pol II.
Initial studies of Spt4–Spt5 in yeast have suggested a role in chromatin-mediated transcription initiation. This hypothesis is based on the ability of spt4 and spt5 mutations to suppress defects in Snf/Swi, a nucleosome-remodeling complex, and on the very strong phenotypic similarities between spt4, spt5, and particular histone mutants (Swanson and Winston 1992; Winston and Carlson 1992). In addition, spt4 and spt5 mutations show many genetic interactions with spt6 mutations. Spt6 has been shown recently to interact directly with histones and spt6 mutations alter chromatin structure in vivo (Bortvin and Winston 1996). Results in Hartzog et al. (1998), however, strongly suggest that Spt4–Spt5 also plays a role in elongation. In addition, the Snf–Swi complex has been shown to play a role in transcription elongation in vitro (Brown et al. 1996). Therefore, the suppression of snf/swi mutations by spt4 and spt5 mutations could also be at the level of elongation. In turn, Spt4–Spt5 may be able to interact with both components of the chromatin template and the transcription machinery, including RNA Pol II. In addition, structural and functional similarities between DSIF p160 and other Spt5 proteins with E. coli NusG provide strong evidence that DSIF associates with RNA Pol II and regulates its processivity at the elongation level (our results; Hartzog et al. 1998).
DSIF stimulates the rate of elongation in an in vitro transcription reaction under decreased nucleotide levels
Immunodepletion of DSIF p160 abolished DRB-sensitive transcription but had no effect on the production of full-length transcripts when reactions contained the optimal concentration of ribonucleoside triphosphates, suggesting that under these conditions DSIF activity has no effect on transcription elongation by RNA Pol II (Fig. 6). As described above, we prefer a model in which DRB inhibits the phosphorylation of a preinitiation complex component (presumably by P-TEFb) and that makes RNA Pol II sensitive to pausing or termination by DSIF. Therefore, we propose that the phosphorylation is required to abrogate a negative effect of DSIF on transcription, but not for the production of full-length transcripts.
We were also able to examine the effect of DSIF on RNA Pol II processivity under conditions that promote pausing by limiting the concentration of ribonucleoside triphosphates in transcription reactions. There is no evidence that a G-free cassette has natural transcriptional pause or arrest sites; however, low nucleotide concentration did cause pausing and arrest in vitro (Fig. 7). The processivity of RNA Pol II, allowing the synthesis of DRB-sensitive long transcripts, requires DSIF activity under these nucleotide-limiting conditions. This suggests that DSIF stimulates the rate of transcription elongation in vitro by the suppression of pausing or arrest by RNA Pol II, similar to E. coli NusG (Burova et al. 1995). This is consistent with the finding that many spt4 and spt5 mutants cause sensitivity to 6-azauracil (6-AU), which reduces UTP and GTP levels in vivo (Hartzog et al. 1998). Thus, both in vivo and in vitro, DSIF appears to be an essential transcription elongation factor of when nucleotides are limiting, strongly suggesting that DSIF has an important role in transcription elongation in vivo.
Antitermination mechanisms in eukaryotes
Antitermination mechanisms may be divided into two categories; processive and nonprocessive mechanisms (Greenblatt et al. 1993). Processive mechanisms allow the RNA polymerase to transcribe through multiple transcription terminators by modifying the RNA polymerase. Antitermination by the λ N protein involves the host proteins, NusA, NusB, NusG, ribosomal protein S10 and RNA elements at the 5′ end of nascent viral transcripts. Whereas processive mechanisms found in phage λ have been extensively characterized (Greenblatt et al. 1993 and references therein), the mechanisms in eukaryotes are much less understood, except for the Drosophila hsp70 gene (Rougvie and Lis 1988; O’Brien and Lis 1991), the c-myc gene (Krumm et al. 1992; Strobl and Eick 1992), and factor TFIIF (Price et al. 1989; Bengal et al. 1991), TFIIS (Sekimizu et al. 1976), SIII (Elongin) (Bradsher et al. 1993a,b; Aso et al. 1995), P-TEFb (Marshall and Price 1995; Marshall et al. 1996), and HIV-1 Tat (Marciniak and Sharp 1991; Kato et al. 1992; Zhou and Sharp 1995; Mancebo et al. 1997; Zhu et al. 1997). Tat activation of HIV-1 transcription is interesting because Tat enhances the processivity of transcription complexes in a manner reminiscent of λ N (Greenblatt et al. 1993), and Tat activation is sensitive to DRB (Marciniak and Sharp 1991; Zhou and Sharp 1995; Mancebo et al. 1997; Zhu et al. 1997). In the present study, we showed that DSIF-mediated stimulation of RNA Pol II processivity is sensitive to DRB and the DSIF p160 subunit has structural and functional similarities to NusG, which is involved in the λ N antitermination system. Our results suggest that DSIF is a eukaryotic antiterminator protein that may be involved in Tat activation of HIV-1 transcription. Most recent studies support this idea because they suggest a factor(s) that is required in addition to P-TEFb for Tat transactivation (Jones 1997; Mancebo 1997; Zhu et al. 1997).
In summary, we have identified a new negative transcription elongation factor, DSIF, that is conserved between humans and yeast. The discovery of DSIF should help us to understand transcription elongation in eukaryotes. Specifically, the ability of purified DSIF to recapitulate the DRB sensitivity observed in crude extracts should facilitate the establishment of a purified in vitro system in which DRB inhibition depends on DSIF. Such studies will help to identify both the kinase and the target of the kinase involved in the regulation of DSIF activity. In addition, the conservation of DSIF with Spt4–Spt5 means that genetic and molecular studies in yeast can be used to identify other factors that interact with DSIF and Spt4–Spt5 and to understand their roles in vivo.
Materials and methods
Cell culture
HeLa spinner cells were grown in minimal essential medium (MEM) containing 5% horse serum as described (Wada et al. 1991).
Preparation of HeLa cell nuclear extracts
Nuclear extracts were prepared as described (Dignam et al. 1983).
Construction of DNA template
pTF3-6C2AT-100 was constructed by PCR methods with the following primer; 5′-CCAAGCTTAGATTTGGGAAATATAA-3′. PCR reactions contained pTF3-6C2AT (Wada et al. 1991) as DNA template in the presence of universal primer and the above primer. The amplified DNA fragment was cloned between the EcoRI and HindIII sites of pTF3-6C2AT.
Kinetically synchronized transcription assays
Transcription reactions were performed essentially as described (Wada et al. 1991), except for minor modifications. Reactions (8 μl) were carried out in TRX buffer [25 mm HEPES–NaOH (pH 7.9), 10% (vol/vol) glycerol, 50 mm KCl, 6 mm MgCl2, 0.5 mm DTT, and 0.5 mm EDTA] containing 250 ng of DNA template (pTF3-6C2AT, unless otherwise indicated) and 32 μg of protein of HeLa nuclear extract that had been treated with hexokinase to deplete endogenous ATP (Wang et al. 1992). Reactions were assembled on ice and incubated at 30°C for 45 min. Seventeen microliters of TRX buffer containing 5 μCi of [α-32P]UTP (800 Ci/mmole), A/C/UTP mixture (final concentration: 60 μm ATP; 600 μm CTP; 5 μm UTP; 80 μm OMe–GTP), and 50 units of RNase T1 (GIBCO BRL) was then added. After a further incubation for 10 min, the reactions were terminated and G-less transcripts were isolated and analyzed by urea–PAGE as described (Wada et al. 1991). For DRB- or H8-inhibition experiments, DRB (at the final concentration of 50 μm, unless otherwise indicated, Sigma) or H8 (50 μm, Research Biochemicals International) was added to reactions after the 45-min preincubation.
Where the partially purified transcription system was used, 2 μl of concentrated 0.3–1.0 m KCl phosphocellulose eluate was added in place of nuclear extract to transcription reactions. Reactions (20 μl) were carried out in TRX buffer containing 250 ng of DNA template, concentrated 0.3–1.0 m KCl phosphocellulose eluate and the protein faction indicated. Reactions were assembled on ice and incubated at 30°C for 45 min. Five microliters of TRX buffer containing 5 μCi of [α-32P]UTP (800 Ci/mmole), A/C/UTP mixture (final concentration: 60 μm ATP; 600 μm CTP; 5 μM UTP; 80 μm OMe–GTP), 50 units of RNAse T1 (GIBCO BRL), with or without DRB or H8, was then added and incubated further for 10 min. G-less transcripts were analyzed as described above.
Where the minimally reconstituted transcription system was used, 10 ng of purified recombinant histidine-tagged fusion human TBP, 30 ng of purified recombinant histidine tagged fusion human TFIIB, 16 ng of purified recombinant human TFIIF subunits RAP30 and RAP74, and 0.5 μl of highly purified RNA Pol II (Usuda et al. 1991) were added to transcription reactions.
Transcription assays were quantified by AMBIS radioanalytic system or Quantity One Software (PDI).
Purification of DSIF
HeLa cell nuclear extracts (120 ml, 840 mg) were prepared as described previously (Dignam et al. 1983) and diluted with an equal volume of HGE [20 mm HEPES–NaOH (pH 7.9), 20% glycerol (vol/vol), 0.2 mm EDTA, 0.5 mm dithiothreitol (DTT)] containing 0.5 mm PMSF. The diluted extracts were applied to a P11 phosphocellulose column (Whatman) equilibrated with HGE containing 0.05 m KCl (HGE.05; the number following HGE denotes the molar concentration of KCl) and the column was washed with 360 ml of HGE.05 and step eluted with HGE.3, HGE.5, and HGE.85. These fractions were tested for their abilities to synthesize transcripts that were 380 nucleotides long. We usually omitted the step elution with HGE.5 and HGE.85; therefore, after eluting with HGE.3, we added HGE containing 1.0 m KCl for elution. The 0.3–1.0 m KCl step was further concentrated 10-fold with precipitation with (NH4)2SO4, followed by centrifugation at 13,600g in a F0650 rotor (Beckman) for 30 min at 4°C. The precipitate was resuspended and dialyzed against 10 mm HEPES-NaOH (pH 7.9), 10% (vol/vol) glycerol, 50 mm KCl, 6 mm MgCl2, 0.1 mm EDTA, 0.25 mm DTT. This fraction was used as a partially purified transcription system to test DRB sensitivity.
DSIF activity was found in the 0.05–0.3 m KCl step from P11. This eluate (270 mg of protein) was diluted with an equal volume of HGE and applied to a 60-ml DEAE–Sepharose fast-flow column (Pharmacia) equilibrated with HGE.15. After loading, the column was washed with three column volumes of HGE.15 and then eluted with one column volume of HGE1.0. The DSIF activity was eluted at 1.0 m KCl (120 mg of protein). The DSIF fractions were dialyzed against HGE.15 and then fractionated on hydroxylapatite (Bio-Rad). The column was washed with HGE.05 and step eluted with 0.2 and 0.4 m potassium phosphate (pH 7.5). DSIF activity was eluted at 0.2 m potassium phosphate. The DSIF fractions (50 mg of protein) were dialyzed against HGE containing 0.6 m (NH4)2SO4, followed by loading onto a phenyl–Superose column (Pharmacia). DSIF activity was eluted between 0.3 and 0 m (NH4)2SO4. The fractions containing DSIF were adjusted to a conductivity equivalent to HGE.1 and loaded onto a MonoS column (Pharmacia; 0.1 ml). The column was then eluted with a gradient from 0.1 to 0.5 m KCl. The DSIF fractions (eluting at 0.2 m KCl) were pooled (0.47 mg of protein) and applied to a Mono Q column (Pharmacia; 0.1 ml) equilibrated with HGE.2. The column was washed with 10 column volumes of HGE.35 and eluted with a gradient from 0.35 to 0.5 m KCl. DSIF activity was eluted between 0.4 and 0.45 m KCl and further applied to a Superose 12 column (Pharmacia; 2.4 ml) equilibrated with HGE.1. From 840 mg of HeLa cell nuclear extracts, 2 μg of the purified DSIF polypeptides could be obtained.
Renaturation of DSIF
Purified DSIF was subjected to 12.5% SDS-PAGE and proteins were recovered from the gel, acetone precipitated, denatured, and renatured as described previously (Wada et al. 1991). In the case of rSupt4h or recombinant DSIF p160, bacterial strain BL21(DE3) was transformed with a plasmid encoding histidine-tagged fusion Supt4h protein or DSIF p160 protein and induced with IPTG, and the expressed fusion protein was purified as described (Wada et al. 1996). Then, 2 μg of purified recombinant protein was subjected to 10% SDS-PAGE and renatured as described previously (Wada et al. 1991).
cDNA cloning of DSIF p160
For peptide microsequencing analysis of p160, ∼600 ml of HeLa cell nuclear extracts were fractionated as described above except that the MonoQ peak fraction of DSIF containing ∼10 μg of p160 was TCA precipitated and resolved by SDS-PAGE and a band corresponding to p160 was excised from the gel. Four peptides sequences, DWFAK, SWVRLK, DMLEFPAQELRK, and DNRFAVALDSEQNNIHVK were obtained after Lysyl endopeptidase digestion. Two degenerate oligonucleotides 5′-TT(AGCT)ACATG(AG)AT(AG)TT(AG)TT(CT)TG(CT)TC-3′ and 5′-GA(CT)ATG(CT)T(AGT)GA(AG)TT(CT)CC(AGT)GC(AGCT)CA(AG)GA-3′ were synthesized on the basis of the peptides DNRFAVALDSEQNNIHVK and DMLEFPAQELRK, and used for internal PCR with HeLa cDNA as the template. After 28 cycles, a 416-bp fragment was amplified. This probe was then used to screen a λgt-10 HeLa cDNA library (Watanabe et al. 1993). Three positive clones were isolated from ∼2 × 106 phages, and found to contain ∼2.0 kb of a 3′-partial cDNA of p160. RACE–PCR was used to obtain the 5′-end of the cDNA (Frohman et al. 1988). First-strand cDNA was synthesized from HeLa mRNA with a p160-specific primer 5′-CGGCACAATGAGGCCTGTGTCG-3′. Two rounds of PCR (28 cycles each) with an anchor primer and gene-specific nested primers yielded a discrete band of ∼1.8 kb. The overlapping cDNAs were sequenced and found to encode a single ORF of 1087 amino acids, containing all four peptide sequences obtained by microsequence analysis.
Construction of rSupt4h protein expression vector
The histidine-tagged fusion Supt4h protein expression vector was constructed by inserting a PCR-generated fragment that contains SUPT4H sequences from 68 to 421 nucleotides (Hartzog et al. 1996) into pET14b (Novagen) between NdeI and BamHI sites. The PCR-amplified SUPT4H cDNA was confirmed by DNA sequencing.
Construction of recombinant p160 expression vector
To amplify a DNA fragment encoding the amino-terminal region of the p160 subunit, two oligonucleotides 5′-Pr, 5′-CAGTCAGGCGTCGTGCGAACAG-3′ and rev-0, 5′-CGCACGATGACACCCACAGTCTG-3′ were synthesized and used for internal PCR with DSIF p160 cDNA as the template, and then the amplified DNA fragment was subcloned in the SmaI site in pBluescriptSK(+) [pBSSK(+); Stratagene] to generate pBS–DSIFNterm. An EcoRI DNA fragment-encoded carboxy-terminal region of the p160 subunit was isolated from the λgt10 clone and subcloned in to the EcoRI site in pBSSK(+) to generate pBS-DSIFCterm. A BamHI–EcoRV fragment of p160 cDNA in pBS–DSIFCterm was subcloned between the BamHI and NotI (filled-in) sites in pBS–DSIFNterm to generate pBS–DSIFp160. For expression of DSIF p160 protein, two oligonucleotides, FLAGp160pr, 5′-CACCATGGACTACAAGGACGACGATGACAAGCATATGTCGGACAGCGAGGACA-3′, and rev-4, 5′-CTCAGAGAGTGTTGGCTTCACACC-3′, were synthesized and used for internal PCR with DSIF p160 cDNA as the template, and then the amplified DNA fragment was subcloned in the SmaI site of pBSSK(+) to generate pBS–FLAGNterm. A XhoI–XbaI fragment of p160 cDNA in pBS–FLAGNterm was subcloned between the XhoI and XbaI sites in pBS–DSIFp160 to generate pBS–FLAGp160. A NdeI–EcoRV fragment of p160 cDNA in pBS–FLAGp160 was subcloned between the NdeI and BamHI (filled-in) sites in pET14b (Novagen) to generate pET–DSIFp160.
Preparation of recombinant proteins
Recombinant human TFIIB and TBP with a histidine tag were expressed in E. coli strain BL21 (DE3), and purified essentially as described previously (Malik et al. 1991; Takeda et al. 1992). Purified human TFIIF subunits RAP30 and RAP74 were kindly provided by Dr. S. Kitajima (Tokyo Medical and Dental University, Japan).
Preparation of DSIF p160 fusion protein and antibody
A plasmid that expresses glutathione S-transferase (GST)–DSIF p160 was generated by inserting the DSIF p160 NcoI–EcoRV fragment (encoding the carboxy-terminal 250 residues) into the bacterial expression vector pGEX-5X-3 (Pharmacia). Fusion protein was produced in E. coli and purified as described previously (Wada et al. 1996). The purified GST–DSIFp160250 fusion protein was dialyzed against and used to immunize rats and a monoclonal antibody (α-p160) was prepared as described by Harlow and Lane (1988).
Immunodepletion of DSIF
Purified monoclonal p160 antibodies (4.5 mg) were coupled to 0.1 ml of protein G–Sepharose (Pharmacia) with 20 mm dimethylpimelimidate as described (Harlow and Lane 1988). The resin was incubated for 1 hr at 4°C with HeLa cell nuclear extracts (0.2 ml, 0.8 mg/ml). An unbound fraction was separated from the resin and incubated with the new p160 antibody affinity resin for 1 hr at 4°C. The unbound fraction was then collected and used as DSIF-depleted nuclear extracts. Immunoadsorbed complexes in the first and second incubations were washed with 0.2 ml of HGE.1 and eluted by addition of 200 μl of 4× protein dye solution (Wada et al. 1996) following incubation for 5 min at 98°C. The protein G–Sepharose was used as a control material.
Immunoprecipitation
Anti-RNA Pol II CTD antibodies (4.5 mg, Promega) were coupled to 25 μl of protein G–Sepharose for 1 hr at 4°C. The resin was incubated for 1 hr at 4°C with HeLa cell nuclear extracts (0.15 ml, 0.8 mg/ml). Immunoadsorbed complexes were then washed with 0.75 ml of HGE.1 and eluted by addition of 25 μl of 4× protein dye solution (Wada et al. 1996) following incubation for 5 min at 98°C.
Western blotting
Proteins were separated by SDS-PAGE and transferred to PVDF membranes according to standard procedures. After a 1 hr incubation with Tris-buffered saline containing 0.1% Tween 20 and 10% skim milk, the membranes were incubated for 1 hr at room temperature with specific antibodies. Following extensive washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody and developed by the ECL system (Amersham). Anti-TBP polyclonal antibody, anti-TFIIB polyclonal antibody, anti-Rap30 polyclonal antibody, and anti-Cdk7 polyclonal antibody were purchased from Santa Cruz Biotechnology, Inc., anti-TFIIE α subunit polyclonal antibody was provided by Dr. Ohkuma (Osaka University, Japan).
Acknowledgments
We are indebted to Drs. M. Dahmus, P. Sharp, A. Berk, R. Weinmann, J. Parvin, R. Kingston, and H. Watanabe for many helpful discussions and we thank Dr. M. Goto for help with cDNA cloning, Dr. R. Roeder for TBP and TFIIB expression vectors, Dr. S. Kitajima for purified TFIIF subunits, and Dr. Ohkuma for anti-TFIIE α subunit antibody. This work was supported by a Grant-in Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan to H.H. and by a National Institutes of Health (NIH) grant GM32967 to F.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The nucleotide sequence data of DSIF p160 reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence data bases under accession number AB000516.
Footnotes
E-MAIL hhanda@bio.titech.ac.jp; FAX 81-45-924-5834.
References
- Akoulitchev S, Makela TP, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Bradsher JN, Jackson KW, Conaway RC, Conaway JW. RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J Biol Chem. 1993a;268:25587–25593. [PubMed] [Google Scholar]
- Bradsher JN, Tan S, McLaury HJ, Conaway JW, Conaway RC. RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J Biol Chem. 1993b;268:25594–25603. [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Buermeyer AB, Thompson NE, Strasheim LA, Burgess RR, Farnham PJ. The HIP1 initiator element plays a role in determining the in vitro requirement of the dihydrofolate reductase gene promoter for the C-terminal domain of RNA polymerase II. Mol Cell Biol. 1992;12:2250–2259. doi: 10.1128/mcb.12.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Richardson JP. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci. 1995;92:4738–4742. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang P-W, Fogel E, Jackson CL, Lieuallen K, Lennon G, Qu X, Wang S-Q, Kurnit DM. Isolation, sequencing, and mapping of the human homologue of the yeast transcription factor, Spt5. Genomics. 1996a;38:421–424. doi: 10.1006/geno.1996.0646. [DOI] [PubMed] [Google Scholar]
- Chiang P-W, Wang SQ, Smithivas P, Song WJ, Crombez E, Akhtar A, Im R, Greenfield J, Ramamoorthy S, Van Keuren M, et al. Isolation and characterization of the human and mouse homologues (SUPT4H and Supt4h) of the yeast SPT4 gene. Genomics. 1996b;34:368–375. doi: 10.1006/geno.1996.0299. [DOI] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem. 1989;264:2250–2257. [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- Egyhazi E. A tentative initiation inhibitor of chromosomal heterogeneous RNA synthesis. J Mol Biol. 1974;84:173–183. doi: 10.1016/0022-2836(74)90220-4. [DOI] [PubMed] [Google Scholar]
- ————— Inhibition of Balbiani ring RNA synthesis at the initiation level. Proc Natl Acad Sci. 1975;72:947–950. doi: 10.1073/pnas.72.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Initiation inhibition and reinitiation of the synthesis of heterogenous nuclear RNA in living cells. Nature. 1976;262:319–321. doi: 10.1038/262319a0. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granick D. Nucleolar necklaces in chick embryo fibroblast cells. I. Formation of necklaces by dichlororibobenzimidazole and other adenosine analogues that decrease RNA synthesis and degrade preribosomes. J Cell Biol. 1975;65:398–417. doi: 10.1083/jcb.65.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartzog GA, Basrai MA, Ricupero-Hovasse SL, Hieter P, Winston F. Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2848–2856. doi: 10.1128/mcb.16.6.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog, G.A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- Jones KA. Taking a new TAK on tat transactivation. Genes & Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- Kato H, Sumimoto H, Pognonec P, Chen CH, Rosen CA, Roeder RG. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes & Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes & Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR, Ouzounis CA. KOW: A novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage λ. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- Li J, Mason SW, Greenblatt J. Elongation factor NusG interacts with termination factor ρ to regulate termination and antitermination of transcription. Genes & Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- Makela TP, Parvin JD, Kim J, Huber LJ, Sharp PA, Weinberg RA. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Hisatake K, Sumimoto H, Horikoshi M, Roeder RG. Sequence of general transcription factor TFIIB and relationships to other initiation factors. Proc Natl Acad Sci. 1991;88:9553–9557. doi: 10.1073/pnas.88.21.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone EA, Fassler JS, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV tat transcriptional activation in vivo and in vitro. Genes & Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Meggio F, Shugar D, Pinna LA. Ribofuranosyl-benzimidazole derivatives as inhibitors of casein kinase-2 and casein kinase-1. Eur J Biochem. 1990;187:89–94. doi: 10.1111/j.1432-1033.1990.tb15280.x. [DOI] [PubMed] [Google Scholar]
- Mogridge J, Mah TF, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the λ N protein. Genes & Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Lis JT. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Sluder AE, Greenleaf AL. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Conaway JW, Conaway RC. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. The role of general transcription factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Samuels M, Fire A, Sharp PA. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982;257:14419–14427. [PubMed] [Google Scholar]
- Sehgal PB, Darnell JE, Tamm I. The inhibition by DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell. 1976a;9:473–480. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science. 1976b;194:431–433. doi: 10.1126/science.982026. [DOI] [PubMed] [Google Scholar]
- Sekimizu K, Kobayashi N, Mizuno D, Natori S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry. 1976;15:5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- Serizawa H, Conaway JW, Conaway RC. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature. 1993;363:371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- Stachora AA, Schafer RE, Pohlmeier M, Maier G, Ponstingl H. Human Supt5h protein, a putative modulator of chromatin structure, is reversibly phosphorylated in mitosis. FEBS Lett. 1997;409:74–78. doi: 10.1016/s0014-5793(97)00486-9. [DOI] [PubMed] [Google Scholar]
- Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: Effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes and acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada R, Nakatani Y, Hoffman A, Kokubo T, Hasegawa S, Roeder RG, Horikoshi M. Identification of human TFIID components and direct interaction between a 250-kDa polypeptide and the TATA box-binding protein (TFIIDt) Proc Natl Acad Sci. 1992;89:11809–11813. doi: 10.1073/pnas.89.24.11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Hand R, Caliguiri LA. Action of dichlorobenzimidazole riboside on RNA synthesis in L-929 and HeLa cells. J Cell Biol. 1976;69:229–240. doi: 10.1083/jcb.69.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda Y, Kubota A, Berk AJ, Handa H. Affinity purification of transcription factor IIA from HeLa cell nuclear extracts. EMBO J. 1991;10:2305–2310. doi: 10.1002/j.1460-2075.1991.tb07767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Watanabe H, Usuda Y, Handa H. Different biological activities of the hetero- and homodimers formed by the 47- and 43-kilodalton proteins of transcription factor ATF/E4TF3. J Virol. 1991;65:557–564. doi: 10.1128/jvi.65.2.557-564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Kawase H, Hiramoto M, Ferdous A, Takayama M, Lee KA, Hurst HC, Handa H. Copurification of casein kinase II with transcription factor ATF/E4TF3. Nucleic Acids Res. 1996;24:876–884. doi: 10.1093/nar/24.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Carey M, Gralla JD. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Sawada J, Yano K, Yamaguchi K, Goto M, Handa H. cDNA cloning of transcription factor E4TF1 subunits with Ets and notch motifs. Mol Cell Biol. 1993;13:1385–1391. doi: 10.1128/mcb.13.3.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. Transcriptional regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. Analysis of SPT genes: A genetic approach toward analysis of TFIID, histones and other transcription factors of yeast; pp. 1271–1293. [Google Scholar]
- Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Yankulov K, Yamashita K, Roy R, Egly JM, Bentley DL. The transcriptional elongation inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- Yankulov KY, Pandes M, McCracken S, Bouchard D, Bentley DL. TFIIH functions in regulating transcriptional elongation by RNA polymerase II in Xenopus oocytes. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandomeni R, Bunick D, Ackerman S, Mittleman B, Weinmann R. Mechanism of action of DRB. III. Effect on specific in vitro initiation of transcription. J Mol Biol. 1983;167:561–574. doi: 10.1016/s0022-2836(83)80098-9. [DOI] [PubMed] [Google Scholar]
- Zandomeni R, Zandomeni MC, Shugar D, Weinmann R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem. 1986;261:3414–3419. [PubMed] [Google Scholar]
- Zhou Q, Sharp PA. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes & Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]