Abstract
Background
Previous studies have identified evidence of genetic influence on alcohol use in samples selected to be informative for alcoholism research. However, there are a growing number of genome-wide association studies (GWAS) using samples unselected for alcohol consumption (i.e. selected on other traits and forms of psychopathology), which nevertheless assess consumption as a risk factor. Is it reasonable to expect that genes contributing to variation in alcohol consumption can be identified in such samples?
Methods
An exploratory approach was taken to determine whether linkage analyses for heaviness of alcohol consumption, using a sample collected for heterogeneous purposes, could replicate previous findings. Quantity and frequency measures of consumption were collected in telephone interviews from community samples. These measures, and genotyping, were available for 5441 individuals (5067 quasi-independent sibling pairs). For 1533 of these individuals, data were collected on two occasions, about 8.2 years apart, providing two datasets that maximize data collected at either a younger or an older age. Analyses were conducted to address the question of whether age and heavier levels of alcohol consumption effects outcome. Linkage results were compared in the younger and older full samples, and with samples in which approximately 10%, 20%, and 40% of drinkers from the lower end of the distribution of alcohol consumption were dropped.
Results
Linkage peaks varied for the age differentiated samples and for percentage of light drinkers retained. Larger peaks (LOD scores > 2.0) were typically found in regions previously identified in linkage studies and/or containing proposed candidate genes for alcoholism including AGT, CARTPT, OPRD1, PIK3R1, and PDYN.
Conclusions
The results suggest that GWA studies assessing alcohol consumption as a covariate for other conditions will have some success in identifying genes contributing to consumption-related variation. However, sample characteristics, such as participant age, and trait distribution, may have substantial effects on the strength of the genetic signal. These results can inform forthcoming GWA studies where the same restrictions apply.
Keywords: alcohol consumption, linkage analysis
INTRODUCTION
Genome-wide association studies (GWAS) are a powerful new tool for identifying genes influencing common diseases, conditions, and traits (Kingsmore et al., 2008; Lango and Weedon, 2008). As alcohol consumption is considered to influence risk for many diseases and conditions (e.g. cancer (La Vecchia et al., 2008; Toriola et al., 2008), low bone density (Berg et al., 2008), and hypertension (Chen et al., 2008)), many ongoing GWAS will assess alcohol consumption as a risk factor. Such datasets may offer the possibility of identifying genes and polymorphisms affecting alcohol use.
There is substantial evidence from latent variable genetic approaches using twin samples that genes contribute to variation in alcohol consumption levels. In general, studies find that approximately 30% to 60% of variance in alcohol consumption is influenced by genetic factors (Heath and Martin, 1994). Linkage and GWA studies have also identified chromosomal regions (Edenberg and Foroud, 2006; Long et al., 1998; Prescott et al., 2005; Reich et al., 1998; Wyszynski et al., 2003) and genes (Johnson et al., 2006; Uhl et al., 2008) associated with alcohol-related traits. However, they have typically used samples ascertained for a family history of substance abuse. Is it therefore reasonable to expect to find similar evidence of genetic influence in samples unselected for problems related to substance use?
A problem with samples unselected for consumption, is that low consumption levels may reflect a mixture of biological and social causes, thereby obscuring evidence of genetic influence. Furthermore, in many studies focussed on chronic medical conditions, participants will typically be older, at an age where current drinking patterns may not reflect past heavy use, and recall of lifetime history may be imperfect or unavailable. Nevertheless, characterisation of genetic causes of variation in alcohol use in the general population is important.
To explore these issues, the current study used linkage analyses of heaviness of alcohol consumption. The samples examined were ascertained for multiple purposes and included data from some individuals who were assessed on two occasions, approximately 8 years apart on average. Thus, we are able to compare linkage results for somewhat younger versus older samples. Will the trend for heavier drinking among younger participants enhance linkage in the sample maximising data collected at the younger age? Or conversely, will stronger environmental influences on consumption in younger individuals, as suggested by the lower heritabilities found in our younger cohort (Hansell et al., 2008), dampen linkage signals?
To further assess the effect of having light drinkers in samples (i.e. samples unselected for heaviness of consumption), post-hoc analyses will examine the effects of truncating heaviness of consumption by excluding the lowest 10%, 20%, and 40% of the distribution of consumption. Cultural causes of low consumption will be disproportionately represented in the extreme lower tail of the distribution of consumption and may dampen linkage signals.
Thus, the present study had three aims. First, in a sample ascertained for heterogeneous purposes, the aim was to determine whether linkage was confirmed in gene regions previously associated with alcohol-related traits. The second aim was to examine whether linkage is influenced by participant age. Finally, the effect of including light drinkers in analyses was examined by comparing full-sample results with linkage obtained using truncated samples that dropped individuals at the lower end of the consumption distribution. Linkage analyses have some limitations, not least of which is low power to detect linkage for complex traits influenced by multiple genes. Accordingly, evidence of linkage in gene regions with known associations to alcohol-related traits would be a promising finding.
MATERIALS AND METHODS
Sample
The core of this community sample was drawn from the Australian Twin Registry (ATR) volunteer twin panel, which was formed in 1978–79 and supported by the Australian National Health and Medical Research Council. Multiple collection phases of alcohol-related data ensued over the next 25 years, as described in detail in Hansell et al. (2008). Briefly, data were drawn from two phases of collection via mailed questionnaire and three subsequent phases of collection through telephone interview. The first mailed questionnaire survey (1988–1989) targeted twins born up to and including 1964 and their relatives, while the second (1989–1992) targeted twins born from 1964 through 1972 and their relatives. Follow-up telephone interviews, which were adapted from the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA, Bucholz et al., 1994), were conducted for both the older twin cohort (1992–1993) and the younger twin cohort (1996–2000). The third phase of telephone interviews (2001–2005) indexed smokers, drinkers, and controls from the older and younger cohorts. For 53% of our sample, self-reported ancestry for all grandparents was available. Ancestry was predominantly European with 93% having full European ancestry. Of these, 65% had full British and/or Irish ancestry and 94% had at least 2 grandparents of British and/or Irish decent.
For the current analyses, both alcohol phenotypes and genotypes suitable for linkage analysis were available for 5441 individuals (2110 males, 3331 females) from 2134 families. The sample comprised 3554 twins (including 1609 complete pairs, of which 95 were MZ), 1684 non-twin siblings, and 203 children of twins (including 1 DZ pair). The number of quasi-independent sibling pairs (QISPs) in the dataset was 5067. Lifetime abstainers (i.e. individuals who had never tried alcohol) were excluded from the study and are not included in these sample numbers. Genotyping was available for 1642 parents (including 599 parental pairs).
As data were collected on more than one occasion for some individuals, the composition of the dataset could vary. Two compositions were examined. In both cases, telephone interview data were selected over mailed questionnaire data. Telephone interview data collected on two occasions were available for 1533 individuals, with a mean time between interviews of 8.2 years (SD = 3.1 years, range 1–13 years). One dataset maximised data collected from older individuals by selecting the most recent telephone interview data, whereas data from earlier interviews were selected for the alternative dataset, thus maximising data from younger individuals. Table 1 shows participant numbers and age for the different data collection phases and for each dataset composition. Test-retest correlations for the 1533 retested individuals were 0.51 for Quantity × Frequency, 0.48 for Quantity, and 0.58 for Frequency.
Table 1.
Participant Numbers and Age by Study and Dataseta
| Study | Older Dataset | Younger Dataset | ||
|---|---|---|---|---|
| No. | Age Range in Years (Mean, SD) |
No. | Age Range in Years (Mean, SD) |
|
| Mailed Questionnaire 1 (1988–1992) | 707 | 19–76 (36.6, 10.2) | 707 | 19–76 (36.6, (10.2) |
| Mailed Questionnaire 2 (1989–1992) | 126 | 19–42 (27.4, 4.5) | 126 | 19–42 (27.4, 4.5) |
| Telephone Interview 1 (1992–1993) | 1514 | 29–80 (43.2, 11.4) | 2309 | 28–80 (41.6, 10.5) |
| Telephone Interview 2 (1996–2000) | 408 | 25–36 (29.8, 2.3) | 1142 | 24–36 (30.0, 2.4) |
| Telephone Interview 3 (2001–2005) | 2686 | 19–80 (43.8, 9.4) | 1157 | 19–80 (45.1, 9.5) |
| TOTAL | 5441 | 19–80 (41.3, 10.7) | 5441 | 19–80 (39.0, 10.6) |
|
SUBSET - those interviewed twice (included in TOTAL) |
1533 | 29–77 (42.8, 9.1) | 1533 | 24–65 (34.6, 7.3) |
The datasets differ only for 1533 individuals for whom telephone interview data were collected on two occasions (on average 8 years apart). For these individuals, data collected on the second occasion were included in the Older Dataset, while data collected on the first occasion were included in the Younger Dataset. Therefore differences in linkage results for the two datasets are due entirely to data being collected at different ages for this subset of individuals.
Informed oral consent was received from participants in the first two phases of telephone interviews and informed oral and written consent was received from phase three telephone interview participants. Ethics approval was received from the institutional review boards (Queensland Institute of Medical Research (QIMR) and Washington University, School of Medicine) appropriate to each study.
Measures
Total alcohol intake, derived from questions regarding the number of drinks consumed in a typical drinking day and drinking frequency (i.e. Quantity × Frequency), was the primary phenotype of interest. However, its individual quantity and frequency components were also examined. See means with standard deviations and medians in Supplementary Table 2.
Quantity
In the mailed questionnaires, quantity was assessed by the item “On average how many drinks would you have ON EACH DAY THAT YOU HAVE SOME ALCOHOL”, and responses for weekend days were examined. The phase 1 (older cohort) telephone interview question was “Think of the times when you’ve had alcohol during the past 12 months. How many drinks do you typically drink on these days when you had an alcoholic drink?”. For the second (younger cohort) and third (combined cohort) phases of telephone interviews, the question was “In the past 12 months, how many alcoholic drinks would you have on a typical day when you had any alcoholic drinks?”. In the mailed questionnaires and first phase of telephone interviews, the actual number of drinks was recorded. However, for the second and third phases of telephone interviews, participants were given response choices. Subsequently, all responses were coded into the following categories: 1 = zero drinks, 2 = 1–2 drinks, 3 = 3–4 drinks, 4 = 5–6 drinks, 5 = 7–8 drinks, 6 = 9–11 drinks, 7 = 12–15 drinks, 8 = 16–18 drinks, 9 = 19–24 drinks, 10 = 25–30 drinks, 11 = 31 or more drinks.
Frequency
In the mailed questionnaires, drinking frequency was assessed by the item “Write in below the number which best describes how often the following people have had alcoholic drinks DURING THE PAST 12 MONTHS”. The telephone interview question regarding drinking frequency was “During the past 12 months, how often have you had alcoholic drinks?”. Response choices varied slightly between studies. They were recoded into the following 6 categories that are common to both the telephone interview and earlier mailed questionnaire studies (6 = at least once daily, 5 = 3–6 days per week, 4 = 1–2 days per week, 3 = 1–3 days per month, 2 = less often, 1 = never).
The Quantity categories were recoded to reflect the number of drinks per day. Similarly, the Frequency categories were recoded to reflect a “times per week” measure. Thus the Quantity × Frequency measure reflected the number of drinks per week.
Zygosity Determination and Genotyping
Zygosity was initially determined by self-report questionnaire with standard questions regarding physical similarity and degree to which others could tell co-twins apart. If co-twins gave inconsistent answers, they were followed-up by telephone, and if inconsistency or uncertainty was still apparent, they were asked to send in photographs at various ages, from which a zygosity assignment was made by project staff. Zygosity assignment based on self-report and responses to standard informative questions has been shown to be approximately 97% accurate (Reed et al., 2005). Subsequently, 347 pairs were genotyped at nine independent DNA microsatellite polymorphisms plus the sex marker amelogenin using the ABI Profiler PlusT multiplex marker set (AmpFLSTRR Profiler PlusT, Applied Biosystems, Foster City, CA). The probability of dizygotic twins being concordant for both alleles at all of the polymorphic loci examined when using this kit, is reported to be less than 10−4 (i.e., resulting in 99.99% zygosity certainty) (Nyholt, 2006). Additionally, most pairs were genotyped, as part of ongoing linkage studies, at a minimum of 20 (maximum of 1369) microsatellite markers. Only 16 pairs had fewer than 20 markers for one or both co-twins and consequently have not had zygosity confirmed through microsatellite genotyping. Of these, 6 pairs have been checked for blood group.
Participants have typically participated in multiple studies, and consequently, genotype data have come from a compilation of microsatellite genome scans performed for ~15 different studies at the Queensland Institute of Medical Research (QIMR). These were conducted at a number of facilities: (a) Sequana Therapeutics, 437 markers; (b) Gemini Genomics, 222 markers; (c) the Marshfield Clinic's Mammalian Genotyping Service, 777 markers (only 393 markers for some studies) (d) the University of Leiden, 430 markers; (e) the Australian Genome Research Facility, 394 markers; and (f) the Finnish Genome Centre, 400 markers. Allowing for overlap between the various sets (most centres used the ABI2 set), there were in total 1461 unique markers, although no individual was genotyped for all of these. Note that additional markers in these datasets were available, but were excluded due to concerns about unreliability given earlier comparisons of allele calls between the genome scans (Cornes et al., 2005).
Each individual genome scan was checked prior to inclusion using the following procedure. First, checks for Mendelian errors were performed using the 'pedtool' software for genetic data manipulation (written by author Scott Gordon). The algorithm used was a modified Lange-Goradia genotype elimination algorithm (see O'Connell and Weeks, 1999). Counts of errors, both per marker and per family, were used to identify problems either with the marker set, or particular families. Problematic markers (i.e. those producing exceptionally high numbers of Mendelian inconsistencies, or with significant differences in allele frequencies relative to other datasets) were excluded. Second, the heterozygosity of each individual for X-chromosome markers was examined and used to confirm recorded gender. Third, the autosomal markers were used to test the correctness of intra-family relationships and to test for inter-family relatedness. Version 2 of the program RELPAIR (Epstein et al., 2000), as well as other software such as GRR (Abecasis et al., 2001), were used. Fourth, MERLIN (version 0.10.1)(Abecasis et al., 2002) was used to identify unlikely recombination events, indicating problems with data such as miscoded pedigree structure. Fifth, the needed alterations to family structure, and deletion/relabelling of problem samples, were applied and tested by repeating the same procedure.
The various genome scans were integrated using the custom 'pedtool' software. The input to this integration consisted of (a) the raw data, (b) a list of target markers (those from different genotyping labs were kept distinct), (c) cM positions using an integrated genetic map based on NCBI build 35.1 (Duffy, 2006), and (d) a set of data-correction rules, derived from earlier cleaning of the component genome scans, that code for corrections to assumed pedigree structure and the dropping of bad DNA samples.
For each chromosome, all genotypes for that chromosome were merged into a data structure after applying the relevant correction rules. Mendelian inheritance checks were then applied automatically, and error genotypes removed across whole families. These data were then further cleaned using MERLIN. Relationship checks on the merged dataset were also performed to check for problems not detectable from individual genome scans.
For the 5441 individuals with alcohol phenotypes, the number of unique autosomal markers per individual ranged from 1 to 1359 (mean = 553.7 ±232.3, median = 394), with 103 individuals having fewer than 300 markers (65 having fewer than 200, 39 having fewer than 100 and 18 having fewer than 50). Intermarker distances for all chromosomes are reported for this sample in Supplementary Table 3 (means range 7.0–8.9cM, standard deviations range 2.7–3.8, and medians range 6.7–10.4cM). The number of unique autosomal markers for the 1642 parents ranged from 3 to 1236 (mean = 490.0 ±213.9, median = 389), with 91 individuals having fewer than 300 markers. These samples were further checked for genotyping errors using MERLIN (1.0.1) and flagged markers were dropped from the dataset (single markers were dropped for each of 104 participants and two markers were dropped for each of 7 participants). Average marker heterozygosity of 75.1% was found using MERLIN (1.0.1) and PEDSTATS (Wigginton and Abecasis, 2005). This cleaned dataset was the one used for analyses.
Statistical Analyses
Pedigree-wide regression analyses were conducted for the 22 autosomal chromosomes using MERLIN-REGRESS version 1.1.2 (Abecasis et al., 2002). The method, an extension of the Haseman-Elston procedure (Haseman and Elston, 1972), implements a regression-based process for linkage analysis using trait-squared sums and differences to predict identity-by-descent (IBD) sharing between any non-inbred relative pairs (Sham et al., 2002). For allelic regions containing influential genes, greater phenotype similarity (i.e. smaller squared trait differences) is expected for relative pairs with more alleles in common. MINX (Abecasis et al., 2002) was used to examine linkage for the X chromosome.
Multipoint linkage analyses were examined for the older and younger datasets. The position and order of markers was established using a locally weighted linear regression map (http://www/qimr.edu.au/davidD) based on NCBI Build 35.1 physical map positions, deCODE and Marshfield maps. Both co-twins from MZ pairs were included in analyses (with zygosity identified), although the MZ relationship per se does not contribute to linkage. The result is similar to including one co-twin with phenotypes averaged for the pair. However, including their individual phenotypes makes clear to the model that there are two measurements, which provides added confidence. Post hoc linkage analyses examined the effects of truncating heaviness of consumption by excluding the lowest 10%, 20%, and 40% of the distribution of consumption (exact proportions, based on the Quantity × Frequency score, were 11.3%, 23.3% and 37.3% for the older dataset and 10.9%, 23.5%, and 38.3% for the younger dataset).
Preliminary analyses were performed using SPSS (version 15.0 for Windows, 2006). Quantity × Frequency and Quantity were log transformed (log10(x + 1) and Frequency was square root transformed (Tabachnick and Fidell, 1989) to correct for positive skew. The effects of sex, age, age2, sex × age, and sex × age2 were examined using stepwise linear regression, with significant terms (p < .05) regressed out and residuals used in the linkage analyses (for significant effects, see supplementary material, Table 1).
Normalized residuals (i.e. mean 0, variance 1) were used for all variables to facilitate analyses in MERLIN-REGRESS (Sham et al., 2002). Univariate outliers with z-score values exceeding ±3.3 (less than 0.1% of the dataset) and bivariate outliers (less than 0.4% of dataset) were excluded from the linkage analyses. Bivariate, or within-family, outliers are defined as sibling pairs, or QISPs, for whom the squared phenotypic difference is much larger than expected from the population correlation. The method used for identifying these outliers is described in detail in Benyamin et al. (2008). For outlying QISPs, data for the individual with the largest deviation from the mean was removed.
RESULTS
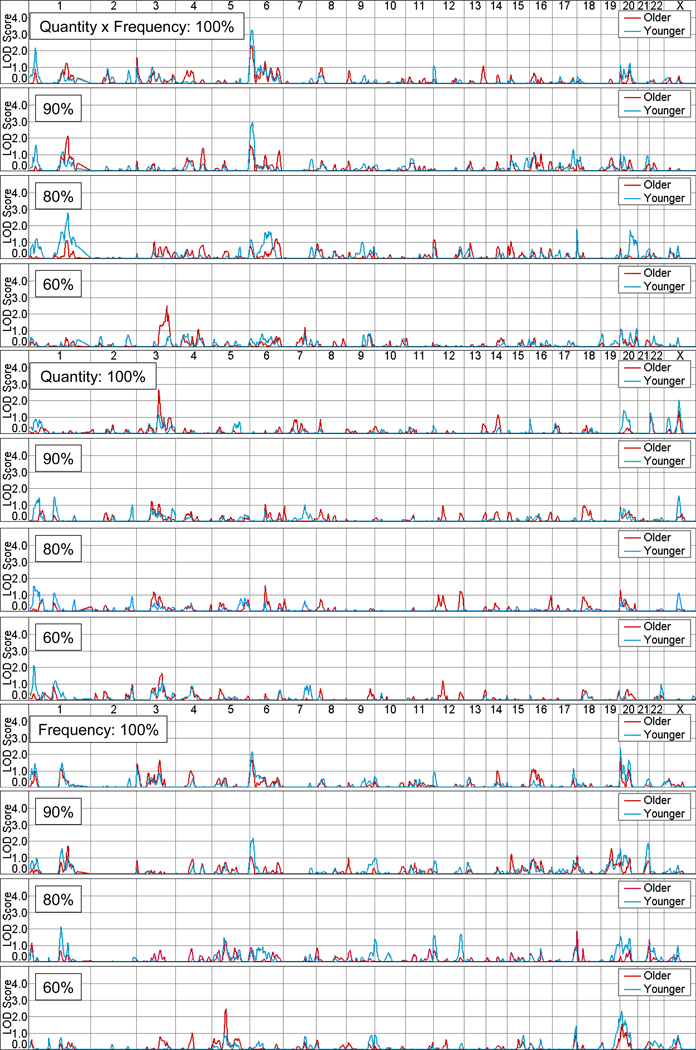
The plotted results of univariate multipoint linkage analyses are shown in Figure 1 for the older and younger datasets for Quantity × Frequency and for its component parts, Quantity and Frequency. Linkage is also shown for truncated samples, in which increasing proportions of lighter drinkers were dropped, resulting in samples of approximately 90%, 80%, and 60% of the full sample. Linkage peaks with LOD scores greater than 1.5 are reported in Table 2.
Figure 1.
Linkage plots for Quantity × Frequency, Quantity, and Frequency with full and truncated samples (100%, 90%, 80%, & 60%), for the older and younger datasets, are shown for the 22 autosomal chromosomes and the X chromosome.
Table 2.
All LOD Scores Peaks >1.5 (>2.0 are shown in bold)
| Chr | Marker | Position (cM) |
Sample (%) & Dataset |
Q × F | Quantity | Frequency | |||
|---|---|---|---|---|---|---|---|---|---|
| LOD | p | LOD | p | LOD | p | ||||
| 1 | D1S2667 | 29.932 | 80 Younger | 1.515 | .004 | ||||
| 60 Younger | 2.104 | .0009 | |||||||
| 1 | D1S2697 | 38.303 | 100 Younger | 2.189 | .0007 | ||||
| 1 | D1S3669 | 42.290 | 90 Younger | 1.580 | .003 | ||||
| 1 | D1S218 | 189.213 | 80 Younger | 2.136 | .0009 | ||||
| 1 | AAT200 | 194.299 | 80 Younger | 1.640 | .003 | ||||
| 90 Younger | 1.581 | .003 | |||||||
| 1 | GATA88F03 | 227.289 | 90 Older | 2.157 | .0008 | ||||
| 80 Younger | 2.803 | .0002 | |||||||
| 1 | D1S1602 | 229.785 | 90 Older | 1.710 | .003 | ||||
| 3 | D3S1307 | 3.323 | 100 Older | 1.590 | .003 | ||||
| 3 | D3S1267 | 131.771 | 100 Older | 2.682 | .0002 | ||||
| 3 | GATA152F04 | 137.183 | 100 Older | 1.655 | .003 | ||||
| 3 | ATA85B10 | 149.064 | 60 Older | 1.651 | .003 | ||||
| 3 | GATA92B06 | 177.444 | 60 Older | 2.503 | .0003 | ||||
| 3 | AAC030 | 186.928 | 60 Older | 2.011 | .0012 | ||||
| 5 | GATA141B10 | 82.539 | 60 Older | 2.498 | .0003 | ||||
| 6 | D6S344 | 3.086 | 90 Older | 1.556 | .004 | ||||
| 6 | ATA109H09 | 8.658 | 100 Older | 2.296 | .0006 | 1.674 | .003 | ||
| 6 | D6S1574 | 14.381 | 100 Younger | 2.178 | .0008 | ||||
| 100 Older | 1.588 | .003 | |||||||
| 6 | SE30 | 15.120 | 100 Younger | 3.265 | .00005 | ||||
| 90 Younger | 2.993 | .0001 | |||||||
| 6 | D6S309 | 19.326 | 90 Younger | 2.220 | .0007 | ||||
| 6 | ATA28B11 | 89.222 | 80 Younger | 1.610 | .003 | ||||
| 80 Older | 1.563 | .004 | |||||||
| 6 | ATA11D10 | 109.806 | 80 Younger | 1.651 | .003 | ||||
| 12 | GATA101G01 | 2.981 | 80 Younger | 1.582 | .003 | ||||
| 12 | ATA29A06 | 163.818 | 80 Younger | 1.680 | .003 | ||||
| 18 | D18S59 | .002 | 80 Younger | 1.814 | .002 | ||||
| 80 Older | 1.860 | .002 | |||||||
| 19 | GATA156F11 | 62.969 | 90 Older | 1.555 | .004 | ||||
| 19 | AAT249 | 106.179 | 60 Younger | 1.856 | .002 | ||||
| 20 | D20S103 | 2.134 | 100 Older | 1.603 | .003 | ||||
| 20 | AAAT007 | 2.448 | 100 Younger | 2.399 | .0004 | ||||
| 90 Younger | 1.549 | .004 | |||||||
| 80 Younger | 1.555 | .004 | |||||||
| 20 | D20S889 | 11.111 | 60 Younger | 2.358 | .0005 | ||||
| 20 | GATA51D03 | 13.019 | 60 Older | 1.570 | .004 | ||||
| 20 | AATTC013 | 30.549 | 60 Younger | 1.817 | .002 | ||||
| 20 | GATA29F06 | 51.678 | 100 Younger | 1.675 | .003 | ||||
| 20 | GATA90E02 | 59.888 | 80 Younger | 1.730 | .002 | ||||
| 21 | UT1355 | 59.932 | 90 Younger | 1.891 | .002 | ||||
| X | DXS6800 | 88.237 | 100 Younger | 2.050 | .0011 | ||||
| 90 Younger | 1.560 | .004 | |||||||
Note: Chr = Chromosome, Q × F = Quantity × Frequency, p-values are derived from multipoint analyses (i.e. they are not corrected for multiple testing).
Quantity × Frequency
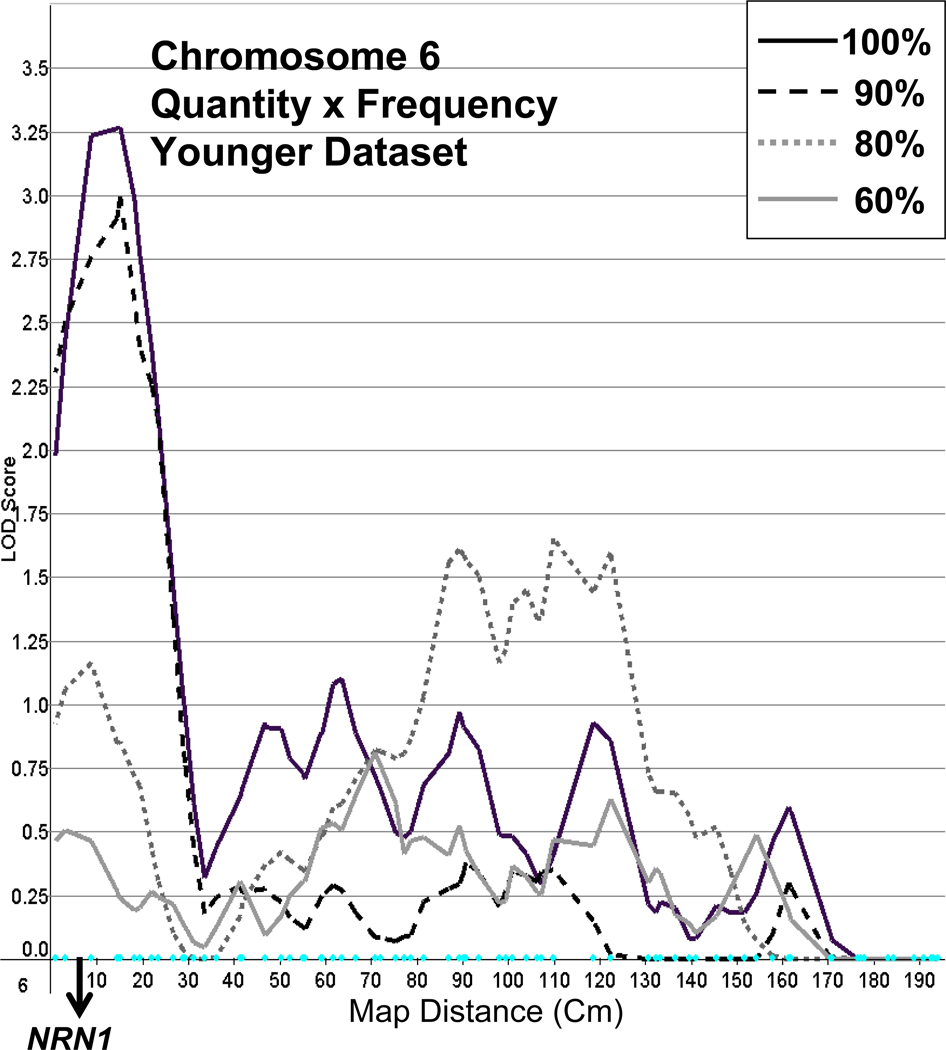
This, the primary outcome measure, provided the best linkage results, peaking with a LOD of 3.3 on chromosome 6 at marker SE30 (between D6S344 and D6S470), when using the younger dataset. A less substantial peak (LOD = 2.3) was found in the same region at marker ATA109H09, using the older dataset. In both datasets, this peak declined as lighter drinkers were progressively dropped (see Figure 2).
Figure 2.
Linkage on chromosome 6 for Quantity × Frequency with full and truncated samples (100%, 90%, 80%, 60%), using the younger dataset, and with the location of the candidate gene NRN1 (neuritin 1) indicated.
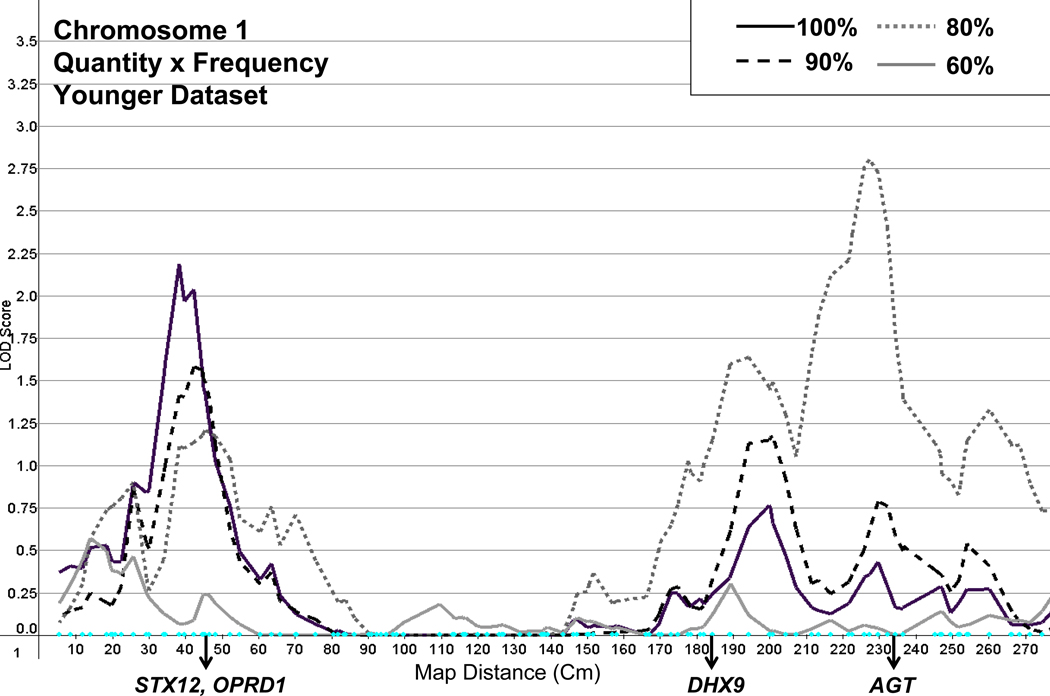
A number of regions of linkage activity were found on chromosome 1 (Figure 3). The first peaked with a LOD of 2.2 at marker D1S2697 (between D1S1612 and D1S199), using the younger dataset. This peak declined as an increasing number of lighter drinkers were dropped. All linkage peaks in this region for the older dataset had a LOD of less than 1.0. Activity peaked with a LOD of 2.8 at marker GATA88F03 (between D1S491 at and D1S225), also using the younger dataset. Linkage in this region for the younger dataset increased as lighter drinkers were dropped, with the exception of the most concentrated sample of heavier drinkers, for which no linkage was observed. The older dataset also showed evidence of linkage in this region, peaking with a LOD of 2.2 at the same marker for the sample with 10% of drinkers at the low end of the consumption distribution dropped.
Figure 3.
Linkage on chromosome 1 for Quantity × Frequency with full and truncated samples (100%, 90%, 80%, 60%), using the younger dataset, and showing the location of candidate genes STX12 (syntaxin 12-binding protein), OPRD1 (opioid receptor, delta-1), DHX9 (DEAH (Asp-Glu-Ala_His) box Polypeptide 9), and AGT (angiotensinogen).
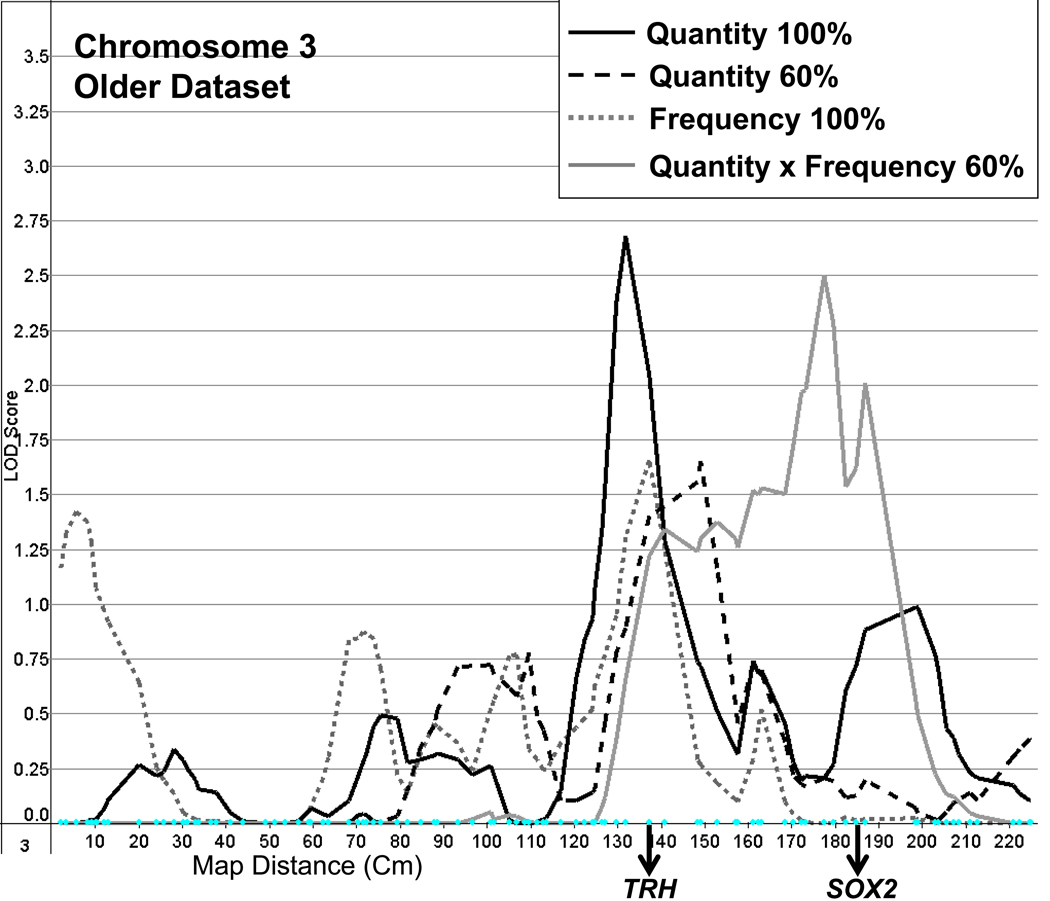
A peak with a LOD of 2.5 was observed on chromosome 3 at marker GATA92B06 for the older dataset in its most truncated form (Figure 4). Other small linkage peaks, with LOD scores greater than 1.5, were found in other regions of chromosomes 3 and 6, and on chromosomes 18 and 20.
Figure 4.
Linkage on chromosome 3 for quantity and frequency measures with full and/or truncated samples (100%, 60%), using the older dataset, and showing the location of the candidate genes TRH (thyrotropin-releasing hormone) and SOX2 (SRY (sex determining region Y) – box 2).
Quantity
The most notable peak for Quantity, with a LOD of 2.7, was found on chromosome 3 at D3S1267 (between D3S2460 and D3S1292) using the older dataset (Figure 4). Linkage was maximised in the full sample, while the most concentrated sample peaked nearby with a LOD of 1.7 at ATA85B10. Peak LOD scores for the intermediate samples ranged from 0.7 to 1.1. In the same region, peaks with LOD scores ranging from 0.5 to 1.2 were found for the younger dataset.
In the younger dataset, a peak with a LOD of 2.1 was found on chromosome 1 at marker D1S2667 (between D2S1612 and D1S199). This peak was maximal for the sample containing fewest light drinkers and declined as more light drinkers were included in the sample. No linkage was found when analysing the older dataset. Also in the younger dataset, a LOD of 2.05 was found on the X chromosome at marker DXS6800 (between DXS8029 and DXS6789). This peak was maximal for the full sample and decreased as increasing numbers of light drinkers were dropped from the sample. A small peak (LOD = 1.4) was also found using the older dataset, full sample. In addition, a minor peak, with a LOD of 1.6, was found on chromosome 6 at ATA28B11, using the older dataset and reduced light drinkers.
Frequency
Peaks found for Frequency echoed those found for Quantity × Frequency on chromosomes 1 and 6. Further, a minor peak (LOD = 1.7) was found in the same region on chromosome 3 (GATA152F04) as the best peak for Quantity, using the older dataset and the full distribution of drinkers (Figure 4).
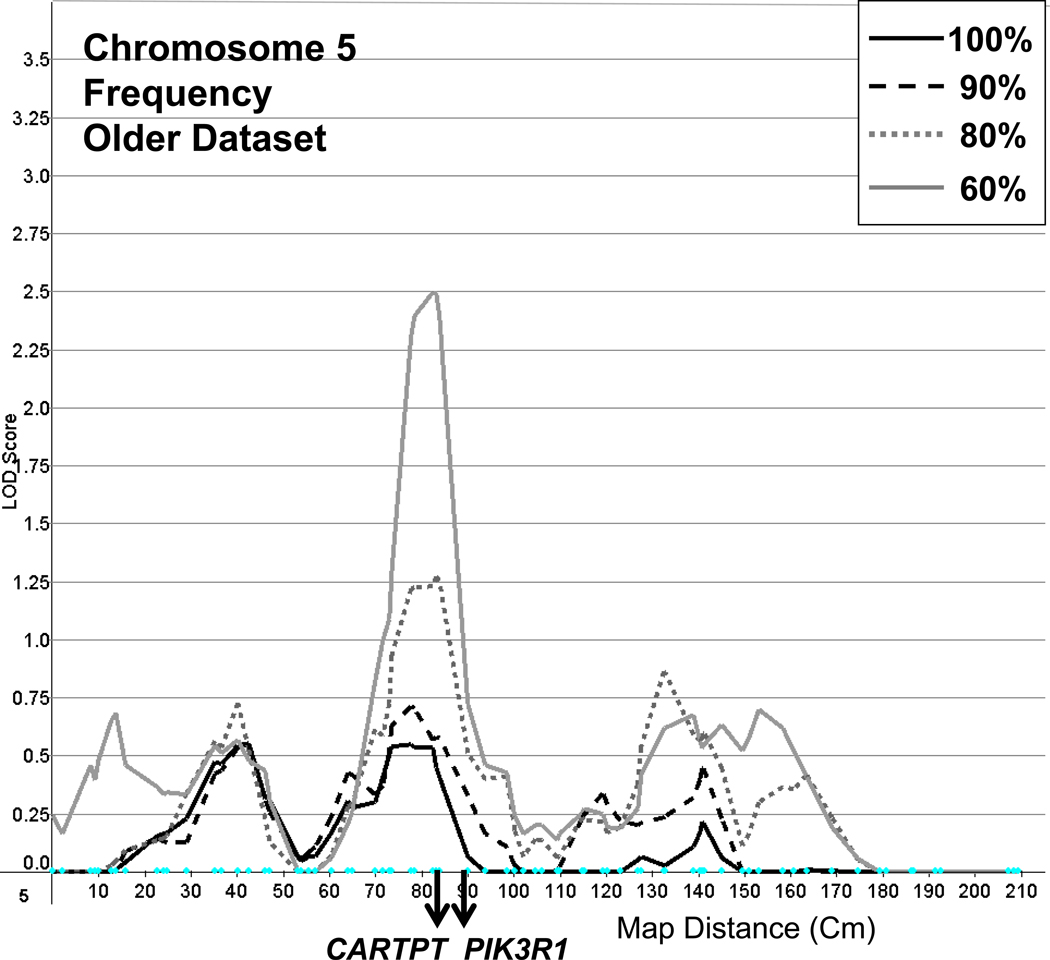
A linkage peak with a LOD of 2.5 was found on chromosome 5 at marker GATA141B10, for the older dataset in its most truncated form, with declining peaks as larger numbers of light drinkers were included in analyses (Figure 5). In addition, a series of peaks were found on chromosome 20, particularly for the younger dataset, as reported in Table 2. Further, minor peaks with LOD scores greater than 1.5 were also found on chromosomes 12, 18, 19 and 21.
Figure 5.
Linkage on chromosome 5 for Frequency with full and truncated samples (100%, 90%, 80%, 60%), using the older dataset and showing the genes CARTPT (cocaine- and amphetamine-regulated transcript (CART) prepropeptide) and PIK3R1 ((phosphoinositide-e-kinase, regulatory subunit 1 (alpha)).
DISCUSSION
Our study took an exploratory approach to examine the feasibility of identifying genes influencing alcohol consumption in samples selected for heterogeneous purposes. A number of linkage peaks with LOD scores greater than 2.0, and peaking at 3.3, were found in candidate gene regions for alcohol-related traits. Peaks were found to vary for the same sample when data collected at different time points (with a mean interval of 8 years) were used for a subset of the sample. Further, linkage varied as lighter drinkers were progressively excluded from the dataset being analysed. Thus, plausible linkage for alcohol consumption can be identified in samples selected for heterogeneous purposes, but sample characteristics can have a substantial influence on the strength of the finding.
Linkage peaks with LOD scores greater than 2.0 were found on chromosomes 1p, 1q, 3q, 5q, 6p, 20p, and Xq. The major peaks, with the exception of those found on 20p and Xq, were found in regions where previous studies have reported linkage for alcohol-related traits (see Table 3), with our best finding confirming linkage reported for chromosome 6 by Hill et al (2004). Similarly, a number of smaller linkage peaks on 12q and 21q (with LOD scores between 1.5 and 2.0), were found to be in regions previously associated with measures of alcohol dependence (Ma et al., 2005). In contrast to the present study, the earlier studies examined samples selected for family history of drinking problems, with most utilizing data from the Collaborative Study on the Genetics of Alcoholism (COGA) sample (Agrawal et al., 2007; de Andrade et al., 2005; Dick et al., 2002; Ma et al., 2005). As shown in Table 3, proposed candidate genes for alcoholism (e.g. AGT, CARTPT, OPRD1, PIK3R1, and PDYN ) can be found in the regions of interest.
Table 3.
Best Linkage Peaks (LODs > 2) Shown with Convergent Findings
| Chr | Marker (Position/cM) |
LOD | Number of Genes in Region (~±1 LOD) & Candidate Gene Examples |
Independent Linkage Results in Humans |
|---|---|---|---|---|
| 1 1 |
D1S2667 (29.9) D1S2697 (38.3) |
2.104 2.189 |
Total genes in region = 497 with 49 being ethanol-related*, including: STX12 (syntaxin 12-binding protein) (Rodd et al., 2006) 1p35.3; separates for ethanol preference in rats (Treadwell, 2006) and down-regulated in alcoholic vs. control human brains (Sokolov et al., 2003). OPRD1 (opioid receptor, delta-1) 1p36.1-p34.3; reported to modulate substance dependence risk , but null findings also reported (Xuei et al., 2007; Zhang et al., 2008). |
De Andrade et al. (2005): LOD=2.15 @ 52 cM for maximum number of drinks in 24 hours. |
| 1 | D1S218 (189.2) | 2.136 | Total genes in region = 238 with 33 being ethanol-related*, including: DHX9 (DEAH (Asp-Glu-Ala-His) box polypeptide 9) 1q25: down-regulated in the temporal cortex of alcohol abuse indivudals versus controls (Sokolov et al., 2003). |
Hill et al (2004): LOD=3.46 @ 169 cM (D1S196) for alcoholism. |
| 1 | GATA88F03 (227.3) | 2.803 | Total genes in region = 237 with 20 being ethanol-related*, including: AGT (angiotensinogen) 1q42.2; may mediate alcohol consumption as precursor of angiotensin II, which correlates with voluntary alcohol intake in mice (Maul et al., 2001); baseline levels are higher in prefrontal cortex of alcohol-preferring vs. non- alcohol preferring rats (Rodd et al., 2006); down-regulated in frontal cortex of alcoholic vs. control human brains (Lewohl et al., 2000). |
Agrawal et al. (2007): LOD=2.0 @ 213 cM for DSM-IV alcohol dependence symptoms. Dick et al. (2002): LOD=2.3 @ 235 cM for a factor age of onset of regular drinking and harm avoidance. |
| 3 | D3S1267 (131.8) | 2.682 | Total genes in region = 187 with 11 being ethanol-related*, including: TRH (thyrotropin-releasing hormone) 3q13.3-q21; In mice, thyrotropin-releasing hormone appears to modulate various parameters related to ethyl alcohol consumption (e.g. French et al., 1993) . |
Ma et al. (2005): LODs up to 1.3 @ ~130 cM (3p21) for alcohol dependence. Ehlers et al. (2005): LOD=2.2 @ 143 cM for alcohol craving. |
| 3 | GATA92B06 (177.4) AAC030 (186.9) |
2.503 2.011 |
Total genes in region = 187 with 10 being ethanol-related*, including: SOX2 (SRY (sex determining region Y) – box 2) 3q26.3-q27; up-regulated in the prefrontal cortex of alcoholic versus control subjects (Iwamoto et al., 2004). |
|
| 5 | GATA141B10 (82.5) | 2.498 | Total genes in region = 169 with 52 being ethanol-related*, including: PIK3R1 (phosphoinositide-3-kinase, regulatory subunit 1 (alpha)) 5q13.1; associated with patterns of risky alcohol consumption in male adolescents (Desrivieres et al., 2008). CARTPT (cocaine- and amphetamine-regulated transcript (CART) prepropeptide) 5q13.2; associated with alcoholism in Korean males (Jung et al., 2004). |
Hill et al (2004): LOD=3.54 @ 108 cM (D5S644) for alcoholism. |
| 6 6 6 6 |
ATA109H09 (8.7) D6S1574 (14.4) SE30 (15.1) D6S309 (19.3) |
2.296 2.178 3.265 2.220 |
Total genes in region = 80 with 23 being ethanol-related*, including: NRN1 (neuritin 1) (Rodd et al., 2006) 6p25.1; implicated in the development of schizophrenia (Moises et al., 2002) and differential expression profiles for bipolar (Jurata et al., 2004); cross-matched with alcoholism due to clinical comorbidity that may in part be due to genetic overlap (Nurnberger Jr. et al., 2004). |
Hill et al (2004): LOD=4.25 @ 15 cM (D6S1574) for alcoholism. Ma et al. (2005): LODs up to 1.57 @ 6p24 for definitions of alcohol dependence. |
| 20 20 |
AAAT007 (2.4) D20S889 (11.1) |
2.399 2.358 |
Total genes in region = 91 with 7 being ethanol-related*, including: PDYN (prodynorphin) 20pter-p12; associated with risk for alcohol dependence (Xuei et al., 2006) and cocaine/alcohol codependence (Williams et al., 2007). |
|
| X | DXS6800 (88.2) | 2.050 | Total genes in region = 230 with 15 being ethanol-related*, including: ITGB1BP2 (integrin beta 1 binding protein (melusin) 2) Xq12-q13.1; down- regulated in the nucleus accumbens, a brain region thought to mediate the rewarding effects of addictive substances, in alcoholics versus controls (Flatscher-Bader et al., 2005). |
as listed at ERGR (Ethanol-Related Gene Resource - http://bioinfo.vipbg.vcu.edu/ERGR)
Some linkage peaks detected in previous studies have coincided with candidate genes, most obviously with the GABA receptor and alcohol dehydrogenase genes on chromosome 4. Results from multiple association studies from our own laboratory (Macgregor et al., in press) and others (Edenberg, 2007; Edenberg et al., 2004), particularly on the ADH1B and GABRA2 genes, confirm that variation in these regions does affect alcohol consumption and dependence risk. Our largest linkage peaks on chromosome 4 were around 1.0 for Frequency. One may therefore ask why more substantial linkage to these regions was not found in our study. The likely reason is that the proportion of variance in consumption explained by polymorphisms in these genes is small, probably only a few percent, and so the power of even quite large linkage studies to detect these loci will be low. With low power, some studies will find significant or near-significant linkage and others will not. In addition, some of our negative findings may be due to uneven marker distribution leading to gaps in coverage.
In general, linkage found in the current study was stronger when the dataset maximised data collected at a younger age, as was the case for peaks on 1p and 6p for Quantity × Frequency. This supported the proposal that the trend for heavier drinking among younger individuals could enhance linkage for consumption. However, peaks found on 3q (Quantity) and 5q (Frequency) were maximal for the dataset maximising data collected at an older age. These results suggest that factors under genetic control, which influence drinking habits, may change as people age, although previous analyses suggest that heritability itself is stable over periods of approximately 5 to 10 years (Hansell et al., 2008). Nonetheless, our earlier analyses also showed consumption to be less heritable in a younger versus older cohort (23–39 years vs. 28–90 years), consistent with other findings (Carmelli et al., 1993), and consistent with the view that environmental and genetic influences on consumption may change over time.
Linkage results were also found to vary when comparing the full sample to truncated samples in which varying proportions of light drinkers were dropped. Linkage that is maximal in the full sample, and drops as the sample is reduced, is consistent with the influence of genes influencing consumption in all individuals or with decreased power to detect linkage in the smaller sub-samples. In contrast, linkage that is maximal in a sub-sample, by definition, suggests the influence of genes that are expressed only in a proportion of the population. Both modes of influence were in evidence, for example, with linkage maximised in the larger samples on 1p and 6p for Quantity × Frequency and for a sub-sample on 5q for Frequency.
Convergent findings suggest that the current analyses were successful in identifying chromosomal regions containing genes contributing to variation in alcohol consumption traits. However, the study had insufficient power to identify significant linkage. This problem is exacerbated by the issue of multiple testing. A further limitation is the not inconsiderable number of previously reported linkage results, covering virtually all chromosomes, which would increase the likelihood of chance convergence. Accordingly, the results require confirmation by demonstration of genetic association for plausible genes under the suggestive linkage peaks. Confirmation by demonstration of genetic association is also necessary as linkage and GWAS employ different methods, which usually produce different results. Nonetheless, the results of the current study are encouraging for the use of datasets collected for other purposes in ongoing GWAS to identify genes that may contribute to individual differences in alcohol consumption behaviors. Further, the growing trend for large meta-analyses may hold the key to positive findings in the future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all participating families, the project coordinators and interviewers under the supervision of Dixie Statham, the data managers under the supervision of John Pearson and David Smyth, and laboratory personnel under the supervision of Megan Campbell and Anjali Henders. For genome scans of twins and siblings, we acknowledge and thank the Mammalian Genotyping Service, Marshfield WI (Director: Dr James Weber) for genotyping under a grant to Dr Daniel O’Connor; Drs Eline Slagboom and Dorret Boomsma for the Leiden genome scan; Dr Peter Reed for the Gemini genome scan; and Dr Jeff Hall for the Sequana genome scan. National Institute of Health grants have supported this work, including grants to Drs Andrew Heath (AA07728, AA10248, AA11998, AA13321), Pamela Madden (DA12854), Nick Martin (AA13326), Michele Pergardia (DA019951), Richard Todd (AA13320), and John Whitfield (AA014041). Dr Arpana Agrawal receives support from DA023668 and the Alcoholic Beverages Medical Research Foundation and Dr Katherine Morley from an NHMRC Public Health Fellowship (520452). Harry Beeby and Sarah Medland designed and wrote the software used for plotting linkage results (Beeby et al., 2006).
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genetics. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, Saccone NL, Grucza RA, Wang JC, Cloninger CR, Edenberg HJ, Foroud T, Hesselbrock V, Kramer J, Bucholz KK, Kuperman S, Nurnberger JIJ, Porjesz B, Schuckit MA, Goate AM, Bierut LJ. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2007;93(1–2):12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeby HN, Medland SE, Martin NG. ViewPoint and ViewDist: utilities for rapid graphing of linkage distributions and identification of outliers. Behav Genet. 2006;36(1):7–11. doi: 10.1007/s10519-006-9045-z. [DOI] [PubMed] [Google Scholar]
- Benyamin B, Perola M, Cornes BK, Madden PAF, Palotie A, Nyholt DR, Montgomery GW, Peltonen L, Martin NG, Visscher PM. Within-family outliers: segregating alleles or environmental effects? A linkage analysis of height from 5815 sibling pairs. Eur J Hum Genet. 2008;16:516–524. doi: 10.1038/sj.ejhg.5201992. [DOI] [PubMed] [Google Scholar]
- Berg KM, Kunins HV, Jackson JL, Nahvi S, Chaudhry A, Harris KAJ, Malik R, Arnsten JH. Association between alcohol consumption and both esteoporotic fracture and bone density. Am J Med. 2008;121(5):406–418. doi: 10.1016/j.amjmed.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Carmelli D, Heath AC, Robinette D. Genetic analysis of drinking behavior in World War II verteran twins. Genet Epidemiol. 1993;10:201–213. doi: 10.1002/gepi.1370100306. [DOI] [PubMed] [Google Scholar]
- Chen L, Davey Smith G, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5(3):e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornes BK, Medland SE, Ferreira MAR, Morley KI, Duffy DL, Heijmans BT, Montgomery GW, Martin NG. Sex-limited genome-wide linkage scan for body mass index in an unselected sample of 933 Australian Twin Families. Twin Res Hum Genet. 2005;8(6):616–632. [PubMed] [Google Scholar]
- de Andrade M, Olswold CL, Slusser JP, Tordsen LA, Atkinson EJ, Rabe KG, Slager SL. Identification of genes involved in alcohol consumption and cigarettes smoking. BMC Genet. 2005;6 Suppl 1:S112. doi: 10.1186/1471-2156-6-S1-S112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres S, Krause K, Dyer A, Frank J, Blomeyer D, Lathrop M, Mann K, Banaschewski T, Laucht M, Schumann G. Nucleotide sequence variation within the P13K p85 alpha gene associates with alcohol risk drinking behaviour in adolescents. PLoS ONE. 2008;3(3):e1769. doi: 10.1371/journal.pone.0001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Nurnberger J, Jr, Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL, Porjesz B, Begleiter H, Hesselbrock V, Foroud T. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res. 2002;26(10):1453–1460. doi: 10.1097/01.ALC.0000034037.10333.FD. [DOI] [PubMed] [Google Scholar]
- Duffy DL. An integrated genetic map for linkage analysis. Behav Genet. 2006;36(1):4–6. doi: 10.1007/s10519-005-9015-x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li T-K, Nurnberger JI, Jr, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha-2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67(5):1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- French TA, Masserano JM, Weiner N. Influence of thyrotropin-releasing hormone and catecholaminergic interactions on CNS ethanol sensitivity. Alcohol Clin Exp Res. 1993;17(1):99–106. doi: 10.1111/j.1530-0277.1993.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Hansell NK, Agrawal A, Whitfield JB, Morley KI, Zhu G, Lind PA, Pergadia ML, Madden PAF, Todd RD, Heath AC, Martin NG. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Res Hum Genet. 2008;11(3):287–305. doi: 10.1375/twin.11.3.287. [DOI] [PubMed] [Google Scholar]
- Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Heath AC, Martin NG. Genetic influences on alcohol consumption patterns and problem drinking: results from the Australian NH&MRC twin panel follow-up survey. Ann N Y Acad Sci. 1994;708:72–85. doi: 10.1111/j.1749-6632.1994.tb24699.x. [DOI] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004;49:379–385. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu Q-R, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the Collaborative Study on the Genetics of Alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SK, Hong MS, Suh GJ, Jin SY, Lee HJ, Kim BS, Lim YJ, Kim MK, Park HK, Chung JH, Yim SV. Association between polymorphism in intron 1 of cocaine- and amphetamine-regulated transcript gene with alcoholism, but not with bipolar disorder and schizophrenia in Korean population. Neurosci Lett. 2004;365(1):54–57. doi: 10.1016/j.neulet.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Bukhman YV, Charles V, Capriglione F, Bullard J, Lemire AL, Mohammed A, Pham Q, Laeng P, Brockman JA, Altar CA. Comparison of microarray-based mRNA profiling technologies for identification of psychiatric disease and drug signatures. J Neurosci Methods. 2004;138:173–188. doi: 10.1016/j.jneumeth.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Lindquist IE, Mudge J, Gessler DD, Beavis WD. Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov. 2008;7(3):221–230. doi: 10.1038/nrd2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Vecchia C, Zhang ZF, Altieri A. Alcohol and laryngeal cancer: an update. Eur J Cancer Prev. 2008;17(2):116–124. doi: 10.1097/CEJ.0b013e3282b6fd40. [DOI] [PubMed] [Google Scholar]
- Lango H, Weedon MN. What will whole genome searches for susceptibility genes for common complex disease offter to clinical practice? J Intern Med. 2008;263(1):16–27. doi: 10.1111/j.1365-2796.2007.01895.x. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–1882. [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an american indian population. Am J Med Genet Neuropsychiatr Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Ma Q, Yu Y, Meng Y, Farrell J, Farrer LA, Wilcox MA. Genome-wide linkage analysis for alcohol dependence: a comparison between single-nucleotide polymorphism and microsatellite marker assays. BMC Genet. 2005;6 Suppl 1:S8. doi: 10.1186/1471-2156-6-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PAF, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. doi: 10.1093/hmg/ddn372. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul B, Siems W-E, Hoehe MR, Grecksch G, Bader M, Walther T. Alcohol consumption is controlled by angiotensin II. FASEB J. 2001;15:1640–1642. doi: 10.1096/fj.00-0797fje. [DOI] [PubMed] [Google Scholar]
- Moises HW, Zoega T, Gottesman II. The glial growth factors deficiency and synaptic destabilization hypothesis of schizophrenia. BMC Psychiatry. 2002;2:8. doi: 10.1186/1471-244X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Wiegand R, Bucholz K, O'Connor S, Meyer ET, Reich T, Rice J, Schuckit M, King L, Petti T, Bierut L, Hinrichs AL, Kuperman S, Hesselbrock V, Porjesz B. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch Gen Psychiatry. 2004;61(12):1246–1256. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- Nyholt D. On the probability of dizygotic twins being concordant for two alleles at multiple polymorphic loci. Twin Res Hum Genet. 2006;9(2):194–197. doi: 10.1375/183242706776382383. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. An optimal algorithm for automatic genotype elimination. Am J Hum Genet. 1999;65(6):1733–1740. doi: 10.1086/302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Caldwell CB, Carey G, Vogler GP, Trumbetta SL. The Washington University Twin Study of Alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:48–55. doi: 10.1002/ajmg.b.30124. [DOI] [PubMed] [Google Scholar]
- Reed T, Plassman BL, Tanner CM, Dick DM, Rinehart SA, Nichols WC. Verification of self-report of zygosity determined via DNA testing in a subset of the NAS-NRC twin registry 40 years later. Twin Res Hum Genet. 2005;8(4):362–367. doi: 10.1375/1832427054936763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–215. [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, Jerome RE, Lumeng L, Nurnberger JI, Jr, Edenberg HJ, McBride WJ, Niculescu AB. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics. 2006;J:1–35. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72:756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Tabachnick BG, Fidell LS. Using multivariate statistics. 2nd ed. New York: Harper Collins; 1989. [Google Scholar]
- Toriola AT, Kurl S, Laukanen JA, Mazengo C, Kauhanen J. Alcohol consumption and risk of colorectal cancer: the Findrink study. Eur J Epidemiol. 2008;23(6):395–401. doi: 10.1007/s10654-008-9244-4. [DOI] [PubMed] [Google Scholar]
- Treadwell JA. Integrative strategies to identify candidate genes in rodent models of human alcoholism. Genome. 2006;49:1–7. doi: 10.1139/g05-083. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Johnson C, Fatusin OO, Liu Q-R, Contoreggi C, Li C-Y, Buck K, Crabbe J. "Higher order" addiction molecular genetics: Convergent data from genome-wide association in humans and mice. Biochem Pharmacol. 2008;75:98–111. doi: 10.1016/j.bcp.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- Williams TJ, LaForge KS, Gordon D, Bart G, Kellogg S, Ott J, Kreek MJ. Prodynorphin gene promoter repeat associated with cocaine/alcohol codependence. Addict Biol. 2007;12(3–4):496–502. doi: 10.1111/j.1369-1600.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Wyszynski DF, Panhuysen CI, Ma Q, Yip AG, Wilcox M, Erlich P, Farrer LA. Genome-wide screen for heavy alcohol consumption. BMC Genet. 2003;4 Suppl 1:S106. doi: 10.1186/1471-2156-4-S1-S106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr, Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry. 2006;11(11):1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Xuei X, Flury-Wetherill L, Bierut L, Dick D, Nurnberger J, Jr, Foroud T, Edenberg HJ. The opioid system in alcohol and drug dependence: family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(7):877–884. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelerntner J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13(5):531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.