Abstract
Alpha/beta T cell receptors (TCRs) react with major histocompatibility complex proteins (MHC) plus peptides, a poorly understood phenomenon, probably because thymocytes bearing TCRs that manifest MHC-reactivity too well are lost by negative selection. Only TCRs with attenuated ability to react with MHC appear on mature T cells. Also, the interaction sites between TCRs and MHC may be inherently flexible and hence difficult to spot. Contacts between TCRs and MHC in the solved structures of their complexes were reevaluated with these points in mind. The results show that frequently used amino acids in TCR CDR1 and CDR2 regions are often used to bind MHC, in areas around small amino acids on the surfaces of MHC α helices that form a cup, allowing somewhat flexible binding of the TCRs. The TCR amino acids involved are specific to families of V regions and partially different rules govern recognition of MHC1 versus MHCII.
Terms: T cell receptor, MHC, evolution, conserved interactions, tolerance, selection, major histocompatibility complex
A HISTORICAL INTRODUCTION
It is almost 50 years since the discovery that the thymus had something to do with immune responses. Shortly thereafter, cells derived from the thymus were found to improve the ability of B cells to make antibodies, This was followed by the discovery of the carrier effect, i.e. the observation that the antibody response to haptens requires simultaneous recognition of both the hapten and epitopes on the attached protein carrier. These findings dovetailed unexpectedly well to give rise to the idea that T cells created in the thymus react with one portion of the antigen and then help B cells make antibody against a different determinant on the same molecule (1–4). This satisfying explanation for cell cooperation in immune responses left unresolved one key problem for immunologists, the problem of the major histocompatibility complex.
The fact that the major histocompatibility complex (MHC) represents a special case for immune responses had been recognized many years before, by people studying the rejection of grafts and tumors (5, 6). They found that differences at the MHC were recognized by the immune system extraordinarily rapidly. This phenomenon led Niels Jerne to propose that lymphocyte receptors, by which, at the time, he meant immunoglobulin proteins, had evolved evolutionary to react with alleles of the MHC (7). In a ground breaking theoretical paper, he suggested that lymphocytes developing in the thymus somatically mutate their evolutionarily generated receptors such that the receptors no longer react with the MHC of their host, but retain the ability to react well with MHC of others.
Meanwhile, others were beginning to realize that T and B cells do not react with the same determinants on antigens. Senyk et al., for example, showed that after immunization with bovine glucagons, rabbits and guinea pigs make antibodies against a glucagon N terminal peptide, whereas their lymphocytes divide in response to the C terminal part of the protein (8). This seemed to be due to different reactivities of T and B cells since we showed that mouse B and T cells cross react differently with red blood cells of different species and that, after immunization with sheep red blood cells, B cells can bind these cells directly, forming rosettes, but T cells cannot, even though the T cells are clearly able to respond to the antigen (9, 10). At about the same time a number of experiments showed that T cell were geared towards recognition of antigens that were cell associated rather than soluble. These kinds of experiments suggested that T and B cells recognize antigen in radically different ways and led to a lengthy search for the antigen receptor on T cells. The search was confused by reports that T cells secrete soluble factors that could bind antigen, by intermittent reports that T cells expressed immunoglobulin molecules and by the idea that genes coded by the MHC might themselves be part of T cell antigen receptors. On the other hand, a giant clue came from Ingenes, genes that control immune response against certain antigens and which map to the MHC (reviewed in (11).
A shift in perspective led to the concept that resolved all these apparent contradictions, with the discovery by Zinkernagel and Doherty that T cells must recognize not only antigen, but also to products of the MHC (12). The issue of whether these were properties of a single receptor, or two, one specific for antigen and the other for MHC, was cleared up by a “two hybrid experiment” which showed that cells able to react with two combinations of antigen and MHC, antigen a + MHC A and antigen b + MHC B, could not react with the mixed combinations, that is, antigen a + MHC B or antigen b + MHC A (13) and confirmed by the discovery of the polypeptides and genes of the alpha/beta T cell receptor (TCR) (14–20). T cell receptors are now known to be made up of two chains, alpha and beta, each composed of variable (Vα, Nα, Jα; Vβ, Dβ, Nβ, Jβ) and constant elements with all but the N regions germ line encoded (21). Biochemical and cellular immunological and, later, crystallography experiments, showed that TCRs usually react with a complicated ligand composed of the alpha helices of MHC proteins and peptides derived from antigen, bound to specially designed grooves of MHC molecules (22–26).
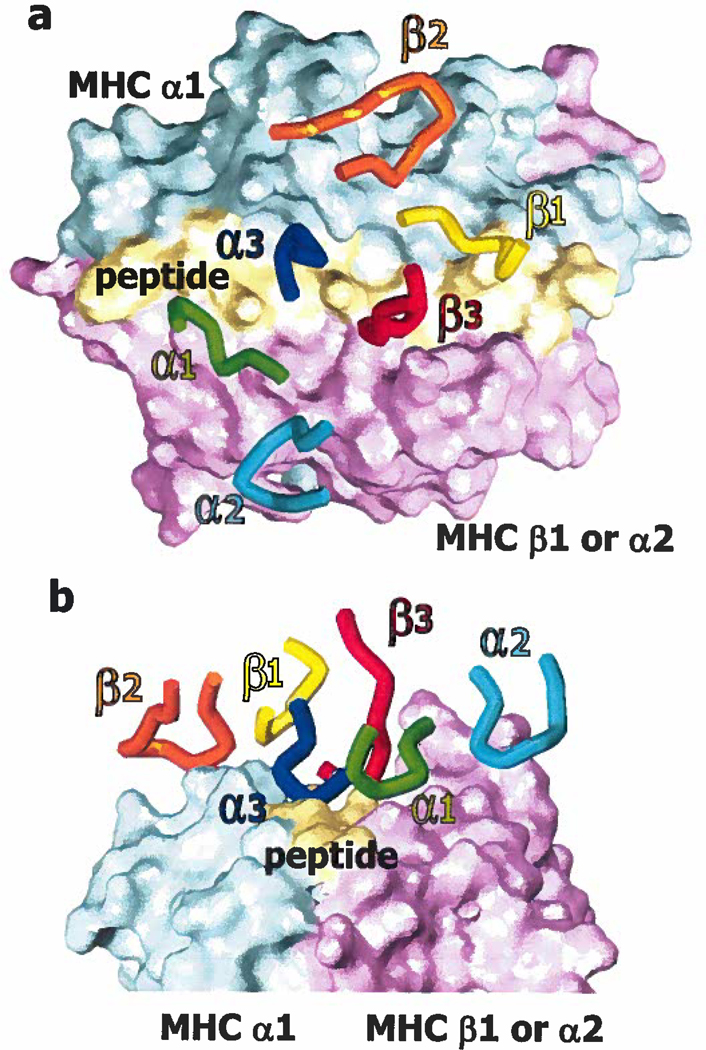
A number of structures of TCRs bound to their MHC/peptide ligands have been reported (27–46). T cell receptors bind MHC/peptide via their CDR loops, CDR1α, CDR2α, CDR1β and CDR2β, encoded in the germ line and CDR3α and CDR3β, made up at least partially of non germ line encoded residues and the C terminal and N terminal ends respectively of Vα or Vβ and Jα or Jβ. In these structures the TCRs often lie on a diagonal above the face of MHC/peptide (Figure 1). To varying degrees the 6 CDR loops of TCRs contact this face, usually with CDR1α and 2α over the α2 helix of MHC class I (MHCI) or the β helix of MHC class II (MHCII), and CDR1 β and 2β over the α1 helix of MHC class I or the α helix of MHC class II. CDR3α and CDR3β, on the other hand, are usually focused on amino acids of the peptide.
Figure 1. The TCR usually contacts MHC/peptide on a diagonal, via the loops of its CDR regions.
a. Shown is a plan of the contacts between the CDR1–3 loops of the α and β chains of a TCR and a space filling surface of MHC and peptide, exemplified here by a mouse TCR binding to the MHCII protein, IAu, engaged by a peptide from myelin basic protein (pdb 1U3H). The α1/α region of MHC is colored cyan, the α2/β region of MHC is in magenta and the peptide in yellow. The CDR loops are color coded and indicated in their corresponding colors on the diagram. b. As in a, shown in elevation.
We now know that an entirely different set of T cells exist, bearing receptors made up of gamma and delta chains. The structure of a γδTCR bound to its ligand has been solved (47). However, the interactions of such receptors with their ligands are not the subject of this review and, although they are of great interest, will not be discussed further.
EVIDENCE THAT TCRS HAVE NOT BEEN SELECTED EVOLUTIONARILY TO REACT WITH MHC
All these wonderful discoveries left unresolved Jerne's original hypothesis, the idea that what are now known to be T cell receptors have evolved evolutionary to react with MHC proteins (7). In fact the notion fell out of favor, after the discovery of positive selection, the phenomenon by which developing thymocytes are picked out to survive based on reaction between their receptors and self MHC + self peptides in the thymus (48, 49). The existence of positive selection allowed immunologists to account for the obsession of T cells with MHC without proposing that evolution had done the job. Nevertheless, some still hoped that solved structures of TCRs bound to their MHC/peptide ligands would shore up the evolutionary hypothesis. The thought was that conserved amino acids in the MHC helices and TCR CDR loops would serve as lynch pins for interaction and that other interactions amongst variable amino acids would determine specificity and affinity. However, the first few reported structures of TCRs bound to MHC/peptide did not reveal any obvious rules governing their interaction, at least at the fine structure level. Thus there didn’t seem to be any consistent pattern to the TCR amino acids that bound MHC and vice versa, and thus no evidence that evolution had created TCRs which reacted in some predictable way with MHC.
There are also other pieces of evidence suggesting that the evolutionary idea might not be right. For example, TCR genes and MHC genes map on different chromosomes, thus there is no obvious genetic mechanism that will maintain co-expression of particular TCR V region alleles and particular versions of the rapidly evolving MHC genes. While there are a few examples of TCR V region alleles or family members with a bias towards a particular MHC allele or class (50–52), in general most Vα and Vβ elements can be found in TCRs that recognize any of the extremely polymorphic alleles and isotypes of MHCI and MHCII. Worse still, in special cases, apparently quite conventional TCR alpha and beta chains react with MHC-like proteins (the CD1 proteins, for example CD1d) and their ligands that are quite different stereochemically from classical MHCI and MHCII (53–60). How could evolution select for TCR variable regions that could cope with all these differences? It’s easiest to conclude that it could not, and that the somatic process of positive selection in thymus must pick out from an immense collection of receptors with random specificities, those that can react with MHC.
The CDR sequences of TCR variable regions do not offer many clues to help us understand this problem either. T cell receptor genes do not usually mutate (61), so Jeme’s idea that somatic mutation might help solve the problem is not acceptable. If TCR CDR regions have been picked out evolutionary to react with MHC, then one might expect their sequences to offer some clue how that has happened. Clearly the evolutionary secret cannot lie in the CDR3 regions of TCRs, since CDR3 regions are created somatically and are only partly encoded in the genome. The secret, if there is one, probably lies in the CDR1 and CDR2 sequences of the V regions of the alpha and beta chains of the TCR. However, inspection does not offer much hope. There are many different sequences for TCRα and β CDR1s and CDR2s, and, although they can, and have been, assigned to families based on their amino acid contents and predicted structures, they are not particularly well conserved between mouse and man (62–64).
EVIDENCE THAT TCRs HAVE BEEN SELECTED EVOLUTIONARILY TO REACT WITH MHC
Despite the difficulty of proposing an evolutionary hypothesis based on the sequences of the TCR V genes or the initial structural data, several points suggest that Jerne might still, in principle, been right. First, in the solved structures, TCRs usually bind MHC/peptide in about the same orientation, angled across the MHC α helices with, as mentioned earlier, the TCR α chain over the α2 helix of MHCI or the β helix of MHCII, and the TCR β chain CDR regions over the MHC helices α1/α (Figure 1, reviewed in (65, 66)). If MHC recognition were simply a matter of positive selection why would the orientation never be reversed? It has been suggested that this may be because the accessory proteins CD4 or CD8, or perhaps CD3, lock the TCR/MHC/peptide complex into the observed positions, by binding simultaneously to both TCR and MHC (67) and this remains an untested possibility.
Secondly, the pivot point of the TCR on MHC is usually in about the same place, centered over peptide amino acids 4–6 in MHCI, and peptide amino acid 5 (P5) in MHCII complexes. If TCR obsession with MHC were simply a matter of positive selection, why wouldn’t the TCR slide drastically to one end or the other of MHC, or even interact with the side of the protein? Actually a dramatic shift of this nature is seen in the reaction of an NKT TCR with CD1d (see later) (68) but has only once been observed for TCR reacting with classical MHC (38). Again, the effects of CD4 and CD8 can be invoked (see above).
Thirdly, although TCR CDR regions vary between species, they are better conserved evolutionary than their counterparts in immunoglobulin (69, 70), suggesting that they have some required function.
Finally, there is a small amount of direct experimental evidence suggesting that evolution has played a role in the obsession of TCRs with MHC. For example, we showed that random combinations of TCR α and β chains react more frequently than expected with MHC (71), and others demonstrated that, even before positive selection has had a chance to have an impact, TCRs react with surprising frequency with MHC (72, 73).
NEGATIVE SELECTION WILL OBSCURE THE BIASES OF TCRS TO REACT WITH MHC
The specificity of the TCRs on developing thymocytes is tested in two ways, via both positive and negative selection. Positive selection allows the maturation of thymocytes bearing TCRs that react with self MHC + self peptides (48, 49), but negative selection deletes all thymocytes bearing TCRs that achieve this recognition too well (74, 75), We have reasoned that, if TCR V elements have been selected through evolution to react with MHC, this reactivity must be attenuated in the thymus to allow escape from negative selection. Thus, while the CDR1s and CDR2s of most Vα/Vβ combinations may produce an inherently MHC-reactive TCR, CDR3s generated via somatic recombination wiil sterically interfere to varying degrees with this reactivity, producing a repertoire of TCRs with a wide range of affinity for MHC. Only T cells with TCRs with reactivity for self MHC/peptides in a narrow affinity window will pass the tests of both positive and negative selection. Thus, in the resulting mature TCR repertoire only a few of the germ line encoded MHC interactions of the CDR1s and CDR2s may still be evident, although they still contribute to the overall interaction of the TCRs for MHC and their maintenance in the germ line is critical to provide a starting point for selection.
The consequence of this line of reasoning is that the germ line encoded MHC interactions may be difficult to spot in structural studies of T cells that have been through normal negative selection. The best place to look for these interactions would be in the window between positive and negative selection. Realizing this, we set out to examine the properties of TCRs in mice in which positive selection was relatively normal, but negative selection was limited. Several mice that fit the bill were available, however, we picked animals we had described previously, in which the MHCII protein has been replaced with an MHCII that is covalently bound to a single peptide (76). These animals were not perfect for the purposes of the experiment, they still expressed normal levels of MHCI, bound to the many different host peptides with which it is normally engaged, and they did of course express one MHCII/peptide combination. Therefore any thymocytes bearing TCRs that could react well with any of these combinations would still have been deleted. However, these animals had two advantages for the planned studies. They had previously been characterized and were known to contain high frequencies of T cells that could react very well with MHC, and indeed, cross react with MHC of many different alleles (76). These were properties we expected of the TCRs manifesting their evolutionarily selected abilities to bind MHC. Secondly, the animals did in fact contain mature, CD4+ T cells, that could be primed with MHC/peptide, and therefore at least one target MHC/peptide combination for these cells could be identified.
Mice expressing a single class II protein, IAb, bound to a single peptide, from Eα, (MHCI+ MHCII-li-IAb/Eα mice) and normal C57BL/6 (B6) animals were primed with IAb bound to another peptide, called 3K, because it contains 3 lysine residues that point straight up out of the IAb groove, and should engage TCRs that react with the combination. T cell hybridomas specific for IAb/3K were prepared from the two sets of animals. Only one of the IAb/3K specific TCRs from B6 mice had any alloreactivity, whereas many of the TCRs specific for the same MHC/peptide combination from MHCI+ MHCII- li- IAb/Eα animals were extravagantly alloreactive (Table 1) (77). Two of these TCRs were tested in detail and turned out to react both with MHCI and MHCII. This phenomenon didn’t seem to be a special feature of TCRs from the MHCI+ MHCII- li- IAb/Eα mice since similar results were obtained with TCRs from other mice in which negative selection was limited (78–83). Also, the degree of MHC reactivity of the TCRs appeared to be related to the degree to which negative selection was absent (Table 2). The few T cells in mice lacking almost all MHC proteins (MHCI- MHCII- li- in Table 2) had the highest frequency of reaction with allogeneic MHC proteins.
Table 1.
T cells that have gone through limited negative selection frequently react with MHC
| Mouse donor | Immunogen | Number of TCRs tested |
% of target MHC tested that stimulated |
|---|---|---|---|
| C57BL/6 | IAb/3K | 5 | 4.4 |
| B6→MHCl+ MHCll- li-IAb/Eα+ | IAb/3K | 2 | 0 |
| MHCl+ MHCll- li-IAb/Eα+ | IAb/3K | 19 | 40.4 |
MHCll- li- IAb/Eα+ mice express a single MHCll protein, IAb bound to the Eα peptide
Table 2.
T cell alloreactivity is controlled by the number of deleting MHC ligands in the animal
| Mouse donor | Number of TCRs tested |
% of target MHC tested that stimulated |
|---|---|---|
| C57BL/6 | 20 | 3.1 |
| MHCl+ MHCll- li- IAb/Eα+ | 19 | 44.1 |
| MHCl+ MHCll- li- IEk/99A+ | 8 | 52.2 |
| MHCl+ MHCll- li- | 3 | 18.2 |
| MHCl- MHCll- li- IAb/Eα+ | 45 | 41.1 |
| MHCl- MHCll- li- | 28 | 67.1 |
MHCl+ MHCll- li- IEk/99A mice express wild type MHCl of the k allele, and IEk bound to the 99A peptide (REF)
To check that the ability to produce very MHC-reactive TCRs on mature T cells was due to lowered opportunities for negative selection in the MHCI+ MHCII- li- IAb/Eα mice, chimeras were made in which the bone marrow derived cells were B6, but the thymus epithelial cells were MHCI+ MHCII- li- IAb/Eα. In these animals, positive selection of most conventional T cells occurs on thymus epithelium, whereas negative selection occurs on both the epithelial and bone marrow derived cells. IAb/3K specific T cells from these chimeric mice were not MHC-cross reactive (Table 1) (77). Therefore mature T cells bearing very MHC cross reactive TCRs are allowed to appear in MHCI+ MHCII- li- IAb/Eα mice because of the limited opportunities for negative selection in these animals.
Further studies showed that the MHC-cross reactive TCRs have several unexpected properties. Although they react with their immunogen, IAb/3K, with the same kinetics, affinities and footprints as normal TCRs do, they appear less specific for particular side chains of the amino acids of their ligand (77, 84). That is, they are more accepting of amino acid substitutions in both IAb and the 3K peptide than normal TCRs are. This is just another representation of the fact that these TCRs are more cross reactive, of course. If the TCRs can accept amino acid substitutions they are, indeed, more likely to be able to recognize different MHC alleles and classes than their wild type counterparts.
These results supported the view that the T cell repertoire is intrinsically MHC reactive, and that this reactivity is partially masked by negative selection in the thymus. To understand the results better we have recently solved the crystal structures of two TCRs both bound to the IAb/3K complex (Dai et al. submitted for publication). One TCR (B3K506) was from a T cell developing in normal mice and was extremely peptide and MHC specific. The other TCR (YAe62) was from one of the very cross reactive T cells described above. Both TCRs had about the same affinity for the IAb/3K ligand. The cross reactive TCR had a very concentrated footprint on IAb/3K with very strong contributions from several amino acids in Vα CDR1 (Y31) and Vβ CDR2 (Y48 and E54). These same amino acids contributed to the binding of the specific TCR. However, in this case these amino acids were less important in the overall footprint which was distributed over a much wider area. These results further supported our view that germ line encoded interactions might best be revealed by these cross reactive T cells that have avoided negative selection and that for normally developing T cells these interactions may be present in an attenuated form.
TCR V REGIONS HAVE BUILT IN BIASES FOR REACTION WITH MHC
The increasing numbers of solved structures of TCRs bound to MHC/peptide have been inspected many times in the hope that they will reveal the rules governing the reactions between TCRs and MHC (27–46). Apart from their usual diagonal mode of interaction and the usual placement of the TCR V region loops over the α helices of the MHC, such rules have not been apparent. Others have, however, noticed that particular TCR amino acids often make contact with MHC, and that, in the case of two structures involving the same TCR Vβ region, bound to different ligands, some of the V region amino acids used to bind MHC were identical and bound MHC in the same positions (42). Therefore, there was some hope that rules were present, although difficult to make out.
There are now more than 20 published structures for TCR/MHC. This is both an advantage and a disadvantage for people searching for the rules underlying the reactions. On the one hand, the increasing numbers of structures provide more data for analysis and a greater likelihood of spotting patterns. On the other hand, however, some of the new structures have been deliberately picked by their discoverers to illustrate unusual interactions, such as those with self antigens, or with very large peptides. They may therefore represent odd structures that are not illustrative of the general rules guiding recognition of MHC by TCRs.
Bearing in mind these issues, we were encouraged to revisit the published TCR/MHC structural information with new eyes and more flexible criteria in looking for conserved interactions. We were guided by several thoughts:
Almost all the structures published so far involve TCRs that have been through normal negative selection. Therefore these TCRs are unlikely to demonstrate all their built in abilities to react with MHC. Had they been able to do this, the cells bearing them would have been deleted in the thymus. Perhaps each TCR/MHC/peptide combination will manifest only a few of the evolutionarily selected interactions, with others prevented by the TCR CDR3 regions, or the peptide. Thus, different TCR/MHC/peptide combinations will use different evolutionarily determined interactions and some of the combinations may not use any at all.
Given the variability in TCR V region CDR1 and 2 sequences, the evolutionary selected interactions may be different for different TCR V regions.
The angle and pitch with which TCRs settle on to MHC varies because of differences in peptide and CDR3 sequences. The evolutionarily selected interactions may therefore have to have a built in flexibility, to accommodate shifts in the relative positions of the amino acids involved.
The evolutionary selected interactions may not be the same for MHCI and MHCII. Such an idea is suggested by the fact that some TCR V regions are preferentially used to react with one or other of the MHC classes (50–52).
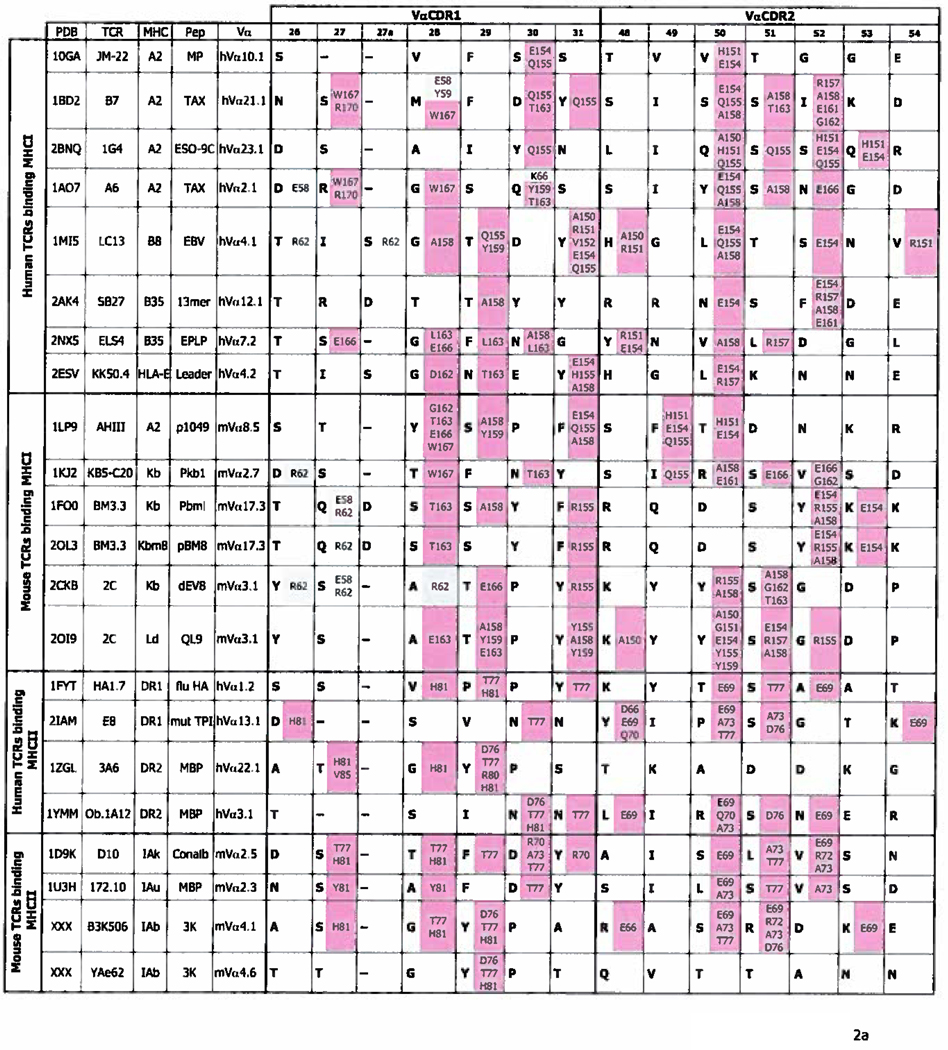
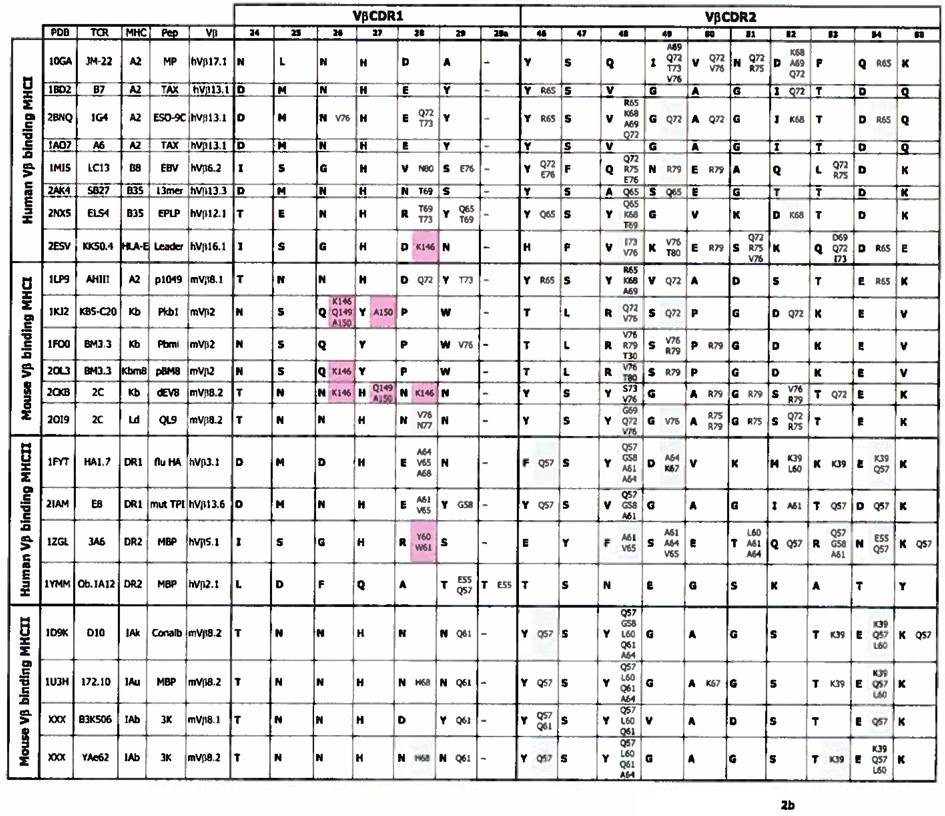
With these thoughts in mind we have re-examined the contacts between the CDR1 and CDR2s of Vα and Vβ regions and MHC in all the published structures of TCRs bound to MHC/peptide ligands. The analysis also included data from 2 structures we have recently solved, mentioned above. The only omissions from the analysis are TCR/MHC/peptide structures that are duplicates of particular TCR/MHC combinations. The results of these analyses are shown in Figures 2a and 2b.
Figure 2. Contacts between TCR CDR1 and 2 regions and MHC.
a. The figure shows the sequences (bolded) of CDR1α and CDR2α of TCRs whose structure, bound to the MHC/peptide ligands is known. In the same cell as each CDR amino acid are the MHC amino acids (not bolded) bound by that CDR residue, color coded to indicate the MHC region on which they lie. Amino acids from the α1/α regions of MHCI/MHCII are shaded blue, and those from the α2/β regions are shaded pink. Dashes indicate a gap in the CDR region, introduced according to (63, 64) to best align TCR sequences. The sequences are arranged such that TCRs binding MHCI are above, and those binding MHCII are below, in the Figure. Human and mouse TCRs are separated. TCR and MHC amino acids are numbered according to (63, 64). Structures are from (27–46) and Dai et al. submitted for publication. b. As in a except that the sequences of TCR CDRβ1 and β2 and their contacts are displayed.
The analyses are inherently incomplete because only a few structures of TCRs bound to their ligands have been reported, by comparison with the many TCR V region genes that exist in man and mouse. The results are also distorted by the fact that it seems that TCRs containing some V regions are better behaved in crystallographic studies than others, perhaps because the V regions in question allow the TCRs to fold more stably. Whatever the reason, the TCR/ligand structures reported so far are heavily biased in favor of TCRs using Vβ8 in the mouse or Vβ13, a related Vβ, in man, and a few other Vβ regions related to these two. There is less bias in the Vα regions used. These caveats limit our ability to make predictions about the universe of human and mouse TCR V regions. However, they may help our ability to see patterns of reactivity for those V regions that are overrepresented in the solved structures.
In fact some patterns are evident, even from the limited numbers of structures available. As previously reported, and mentioned earlier, Vα CDR amino acids usually contact amino acids in the α2 helix of MHCI, or the β helix of MHCII. Conversely, Vβ CDR amino acids bind amino acids in the α1 or α helices of MHCI or MHCII respectively (Figure 2a and b). (In this paper’s Figures, MHC amino acids are from the MHC α chain unless specifically marked “β”.} However, the Figure also suggests that the contacts between TCRs and MHC might follow more rules than just this one. For example, as previously noted, TCRs often use amino acids in positions 28, 29 and 31 of CDR1α; 50, 51 and 52 of CDR2α and positions 28 and 29 of CDR1β and 48 of CDR2β to bind MHC. (In this paper we will number TCR V regions and their amino acids as listed in (63, 64)). This usage is to some extent independent of whether the target MHC is MHCI or MHCII. A number of these positions have been discussed at length by ourselves and others in previous publication, so that in the discussion below we will concentrate on positions that have been less well covered.
Amino acids often used to bind MHC by Vαs
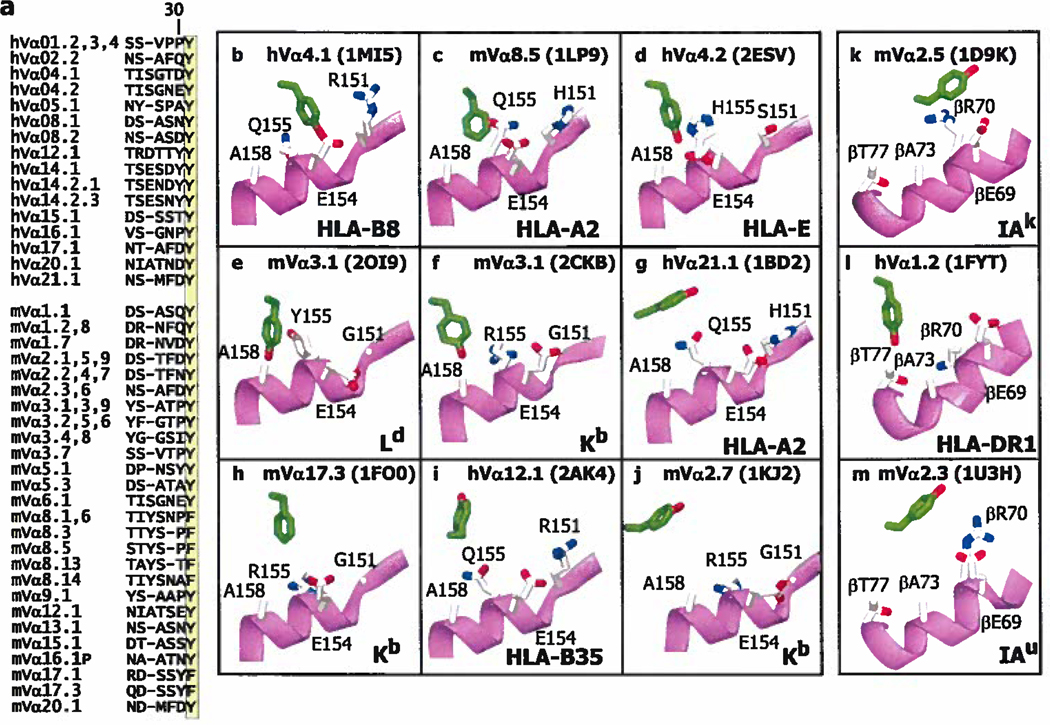
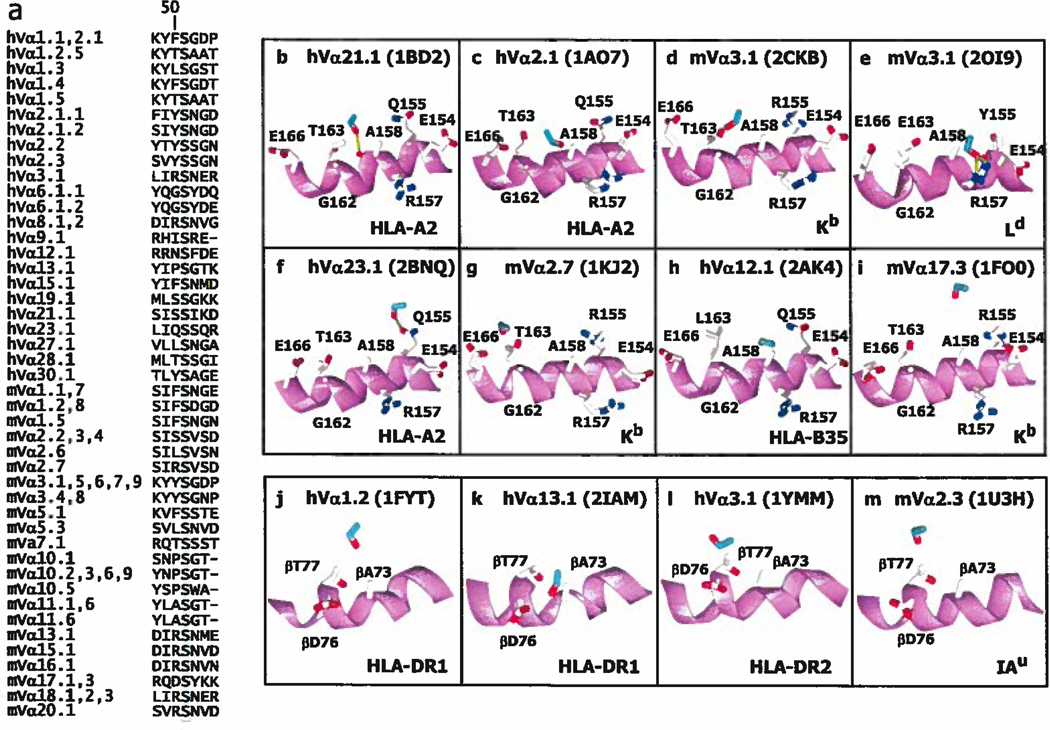
In Vα CDR1, the data in Figure 2a show that Y/F31 is very often bound to a site on MHC that includes amino acids around Q/H/R 155 of the α2 helix of MHCI or the equivalent position on MHCII, R70 on the MHCII β helix. About 40% of human and mouse TCRαs have a Y or F at position 31, at the C terminal end of their CDR1 regions (Figure 3a) so, if this were an interaction which routinely affects TCR/MHC binding, it could apply to many TCRs.
Figure 3. Y/F31 in CDR1α is often used in TCRs and often engages MHC at the same site.
a. The Figure lists the CDR1 sequences of the human (h) and mouse (m) Vα regions that contain an F or Y at position 31 (highlighted). V regions were omitted if the sequence of both their CDR1 and CDR2 regions were identical to one already displayed. The numbering of the V regions and their amino acids is according to (63, 64). The sequences were selected from those of 48 human Vαs and 75 mouse Vαs. b–m. Shown are the arrangements of MHC amino acids around Y/F31α (in green with red hydroxyl) in 9 (b–j) of the solved structures of TCRs bound to MHCI with the α2 helix of MHCI in magenta. Also indicated are the Vα region, MHC allele and pdb number. The structures are arranged from left to right and top to bottom roughly according the predicted strength of the interaction between Y/F31α and MHCI, with no predicted contact for Y/F31α in 2AK4 and 1KJ2, at the bottom of the Figure, k–m. Structures are shown as in b–j, but for TCRs bound to MHCII and with the β helix of MHCII in magenta. There is no predicted contact between Y/F31α and MHCII in 1U3H. Structures were selected from the references in Figure 2.
The structures of 13 TCRs which contain a VαY/F31, bound to different MHC ligands, have been solved. The relative positions of VαY/F31 and MHC/peptide in 12 of these are shown in Figure 3b–m. (One structure, 2OL3, was omitted because it involved that of a TCR already included, bound to a closely related MHC). VαY/F31 does not bind MHC in 3 of these structures (2AK4, 1KJ2 and 1U3H, Figure 3 i, j and m). In the one of the cases (2AK4), the entire TCR is lifted away from MHC by a very pronounced bulge in the engaged peptide, which in the structure is an unusual 13mer. In one of the other cases, the TCR (1KJ2) has a very long CDR3β and the total number of interactions between this TCR and MHC is less than normal (Figure 2).
The orientation of VαY/F31 is not identical in the 9 structures shown in which it binds MHC, sometimes adopting a vertical configuration, reacting with the C terminal side of α2H/Q/R155 or βR70 (in 1LP9, 2ESV, 2CKB, 1FO0,1FYT, Figure 3 c, d, f, h, l), and in other structures on the N terminal side of the same amino acid or horizontal relative to the axis of the MHC α helix (in 1M15, 1BD2, 1DK9, Figure 3b, g, k). For both MHCI and MHCII, the approach of VαCDR1 to the MHC in this region is often facilitated by the lack of a side chain on a highly conserved alanine on top of the MHC α helix (A158 for MHCI and βA73 for MHCII). This leads to a “cup” on the surface of the MHC α helix into which Y/F of TCRα31 can nestle in various orientations while still maintaining Van der Waals like interactions with some portion of the exposed helix backbone and surrounding amino acid side chains, in a way analogous to a ball and socket.
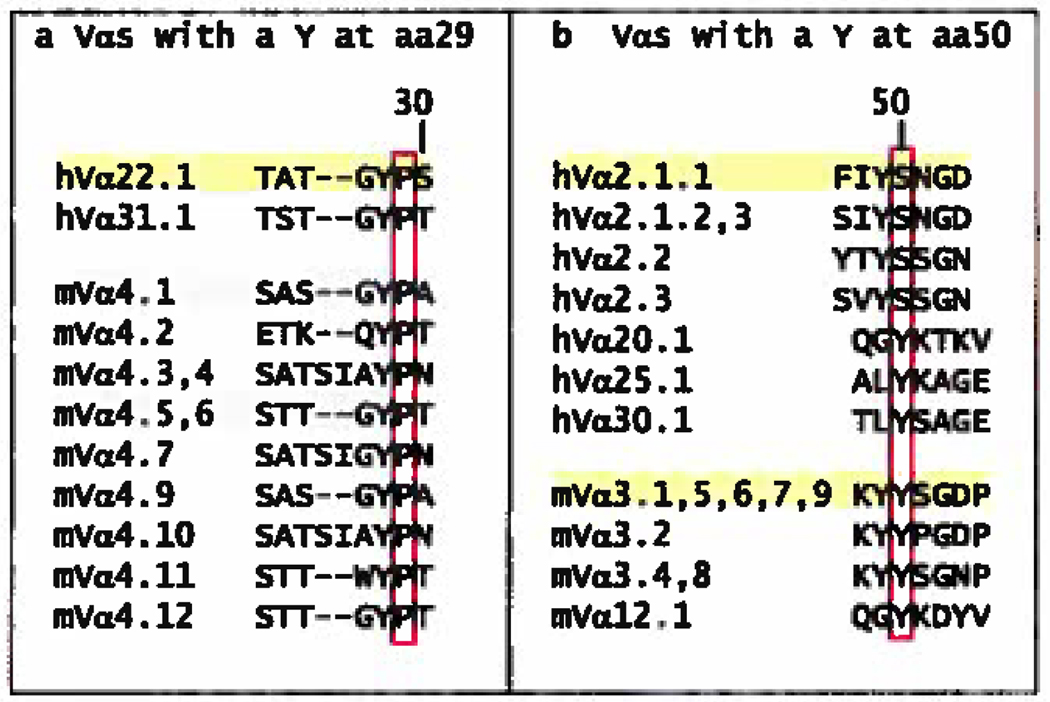
Almost as striking as the fact that VαY/F31 tends to bind to a particular site on MHC, is the fact that other amino acids at this same position do not. Alternate amino acids, in known structures, at position 31α are G, A, S, N and T. These amino acids engage MHC in only one of these structures (1YMM). In many cases, the interaction seems to be replaced by binding between amino acid Vα29 or Vα30 and the ligand (Figure 2a). In human Vα22 and Vα4, VαY29 seems to be a special case. As we have pointed out previously (Dai et al. submitted for publication), in the 3 structures available VαY29 makes a strong interaction with βH81 and βT77 of the MHCII β chain helix. An adjacent proline at position 30 of these Vαs orients VαY29 to point straight out from the tip of CDR1 making it particularly available for interaction. In the other structures whose Vα’s have a P30, the amino acid at p29 (P, T, S) also interacts with the MHC helix in the same region (Figure 2a). This may be a conserved structural feature of Va’s with this proline.
In VαCDR2, a serine at position 51 is the most conserved amino acid among the published structures (63, 64). In MHCI it most often interacts with the MHC helix slightly C-terminal to the site engaged by VαY/F31 (Figure 4b–g). Only in the occasional TCR does it not engage MHC (2AK4 and 1FO0, Figure 4 h,i). The approach of αCDR2 to the MHCI α helix is again facilitated by the conserved A158 (and G151 as well). In a few cases the hydroxyl of VαS51 makes an H-bond to the backbone or amino acid side chains of the MHC helix and other Van der Waals interactions can involve the participation of the backbones of VαS51 and the MHC helix. On MHCII the site for binding VαS51 is similar, but more fixed, engaging MHCII βT77 in various orientations (Figure 4 j–m). Again there is the impression that, particularly in the case of MHCI, S51 binds at a region of the MHC α helix with some flexibility in its exact site and pitch on the MHC protein.
Figure 4. S51 in CDR2α is often used in TCRs and often engages MHC at the same site.
a. The Figure lists the CDR2 sequences of the human (h) and mouse (m) Vα regions that contain an S at position 51 (highlighted). V regions were omitted and numbered as in Figure 3a. b–m. Shown are the arrangements of MHC amino acids around S51α (in blue with red hydroxyl) in 8 (b–i) of the solved structures of TCRs bound to MHCI with the α2 helix of MHCI in magenta. Also indicated are the Vα region, MHC allele and pdb number. The structures are arranged from left to right and top to bottom roughly according the predicted strength of the interaction or alignment between S51α and MHCI, with no predicted contact for S51α in 2AK4 and 1FO0. j–m. As in b–i, but for TCRs bound to MHCII and with the β helix of MHCII in magenta. Structures were selected from the references in Figure 2.
Others have pointed out the interaction of VαY50 in the interaction of several TCRs with MHCI (85). Va’s with this amino acid are not abundant (present in 12–16% of Vαs, Figure 5b), but in the VαCDR2s of the published structures, both VαY50, and other amino acids at this position very often interact with MHC, using the same area of MHCI (around A158) and MHCII (around βA73) (Figure 2a). Perhaps they are evolutionary selected to do this.
Figure 5. CDRα sequences that contain other amino acids that frequently bind MHC at the same position.
Listed are the human and mouse V region CDR1α sequences that include a Y at position 29 (a), or the human and mouse V region CDR2α sequences that contain a Y at position 50 (b). The amino acid of interest is in each case boxed in red. The sequences were selected and numbered as in Figure 3a and structures were selected from those in Figure 2.
Amino acids often used to bind MHC by Vβs
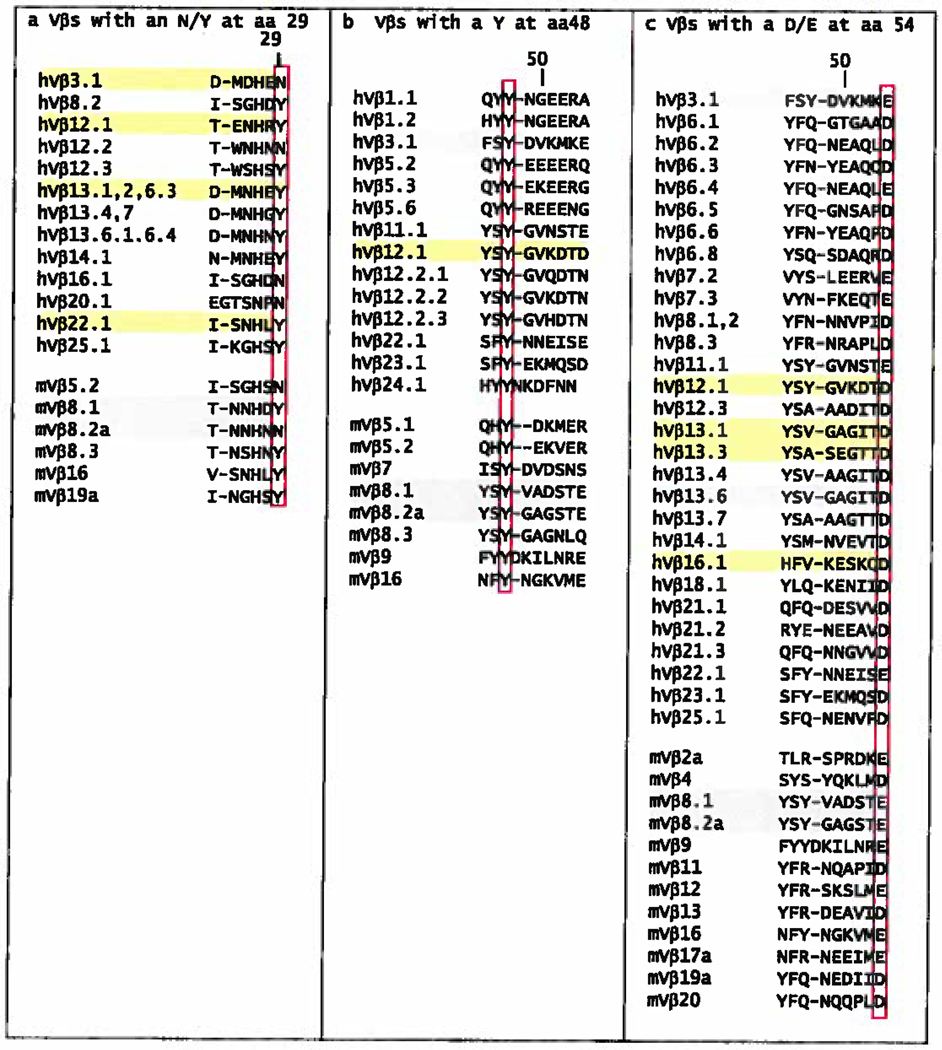
As pointed out above, the published TCR/MHC structures are dominated by TCRs using mVβ8 family members or the related human Vβ elements (hVβ3, hVβ12, hVβ13, hVβ17). With this large pool of data for a related set of Vβ's, a number of analyses have already pointed out particular amino acids that interact with MHC similarly from structure to structure (42, 67, 85). These include VβN/Y29 in CDR1 and VβY/F46, VβY48 and VβD/E54 in CDR2 of Vβ (Figure 2b). For example, in these Vβ families and a few others there is often a Y or N in position 29 of Vβ CDR1 (Figure 6). In the published structures these amino acids often make contact with the α1 MHC helix, especially in the structures with MHCII in which there is often an H-bond to Q61. Thus this may be an amino acid selected by evolution to achieve this task. Also in VβCDR1, many different amino acids at position 28 often contact the MHC. However, there is no obvious conserved pattern of recognition for amino acids at this position and in fact in some structures the rotation of the TCR has bought this amino acid over the peptide to contact the α2 helix of MHCI or the β1 helix of MHCII (Figure 2b), so this does not seem to be an evolutionary conserved interaction, were such to exist.
Figure 6. CDRβ sequences that contain amino acids that frequently bind MHC at the same position.
Listed are the human and mouse V region CDR1β sequences that include an N/Y at position 29 (a) or the human and mouse V region CDR2β sequences that contain a Y at position 48 (b) or a D/E at position 54 (c). The sequences were selected from those of 59 human Vβs and 23 mouse Vβs and were numbered as in Figure 3a.
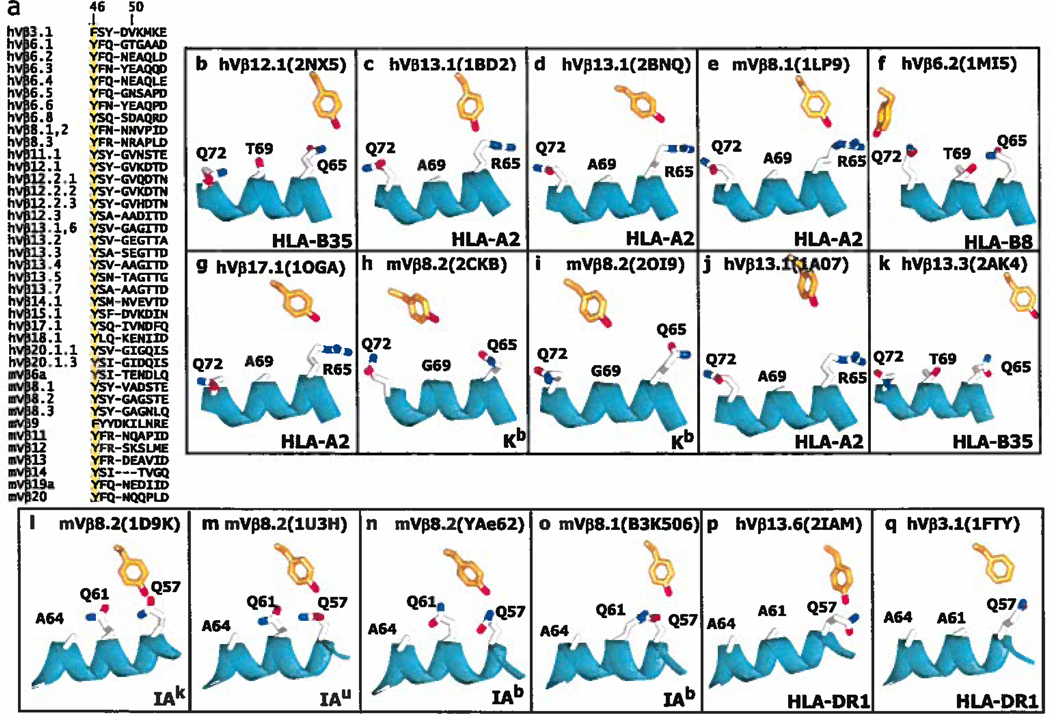
The most striking example of a conserved point of interaction is position 48 of VβCDR2, which in the mVβ8 family is a Y. Originally, Maynard et al. reported that this Y made very similar contacts with MHCII in the interactions of two different TCRs with 2 different MHC proteins (see 1U3H and 1D9K, (42)) and suggested that these residues might form an anchor point for interaction of TCRs using this Vβ, in this case mVβ8.2, with MHCII. We subsequently found that in the B3K506 and YAe-62 TCRs (using the closely related mVβs, 8.1 and 8.2) VβY48 bound to yet another MHCII molecule quite similarly. We also pointed out a similar interaction in a human TCR using the related hVβ3 (see 1FYT) bound to human HLA-DR1 and that in mVβ8 containing TCRs interacting with MHCI, VβY48 also interacts with the MHCI α1 helix in a similar location (Dai et al. submitted for publication). This case is similar to that discussed above for Vα in that the presence of a conserved small amino acid on the top of the α1 helix at position 69 of MHCI and 61 of MHCII creates an area for VβY48 to approach the helix where, via mainly Van der Waals interactions, it can pivot and slide somewhat on the helix without losing contact. In the Vβs of a number of the structures other amino acids (V, A, R, Q) at Vβ48 contact the MHC a1 helix in the same general area. It appears that this amino acid is a key determinant in anchoring the TCR to the MHC.
Another position in Vβ CDR2 worth considering is Vβ46. As shown in Figure 7a Vβ46 is a Y or F in a wide variety of Vβs, including 16 of the 22 structures analyzed in Figure 2b. In 5 of the structures with MHCI VβY/F46 contacts the MHC α1 helix, usually via α65 (Figure 7 b–f). In the other 5 MHCI structures VβY/F46 is too far away from MHC to bind, but lies in roughly the same position (Figure 7 g–k). In all 6 VβY/F46 containing TCR/MHCII structures this amino acid contacts Q57 of the α1 helix (Figure 7 l–q)
Figure 7. Y/F46 in CDR2β is often used in TCRs and often engages MHC at the same site.
a. The Figure lists the CDR2 sequences of the human (h) and mouse (m) Vβ regions that contain a Y/F at position 46 (highlighted). V regions were omitted and numbered as in Figure 3a. b–q. Shown are the arrangements of MHC amino acids around Y46β (in orange with red hydroxyl) in 10 (b–k) of the solved structures of TCRs bound to MHCI with the α1 helix of MHCI in cyan. Also indicated are the Vβ region, MHC allele and pdb number. The structures are arranged from left to right and top to bottom roughly according the predicted strength of the interaction or alignment between S51α and MHCI, with no predicted contact for Y46β in g–k, in the second row of the Figure. l–q. Structures are shown as in b–k, but for TCRs bound to MHCII and with the α helix of MHCII in cyan. Structures were selected from the references in Figure 2.
Finally Vβ CDR2 very frequently has an acidic D or E at position 54 (Figure 6). Four of the 14 of the TCR structures with MHCI that have a VβD/E54 contact R65 of the α1 MHC helix. Since so many different mouse and human Vβs have D/E54, it is a candidate for a conserved and sometimes used interaction site. Six of the 8 TCR structures with MHCII have VβD/E54, in all cases contacting Q57 of the MHCII α1 helix (Figure 2b). Also as pointed out previously, in the 5 cases with an E at Vβ54, there is a salt bridge to MHCII amino acid, αK39, that is not on the α helix of MHCII, but rather lies on one of the β strands of MHCII as it protrudes beyond the peptide binding site. This solvent exposed amino acid is highly conserved in MHCII, although not involved in the structural integrity of MHCII or in peptide binding. Thus, again, D/E54 could represent an TCR CDR2 amino acid that has been evolutionarily selected to react with MHCI and MHCII (in this case in different ways).
The biases for MHC reaction built into TCR Vαs and Vβs may control the orientation of TCRs on MHC
Our purpose in this analysis was to determine whether the existing TCR/MHC structures could be used to make a case for germ line encoded features of TCR Vα and Vβ elements that account for the obsession of TCRs with MHC ligands. It was clear that any analysis would have to account for both the conserved general diagonal orientation of the TCR on the MHC ligand and also for the considerable variation in angle and pitch of the TCR on the MHC surface. We were guided in this analysis by the idea that the need to avoid negative selection in the thymus might select for TCR CDR3s that attenuated these germ line interactions and the observation that in mice with limited negative selection highly MHC cross reactive T cells were common.
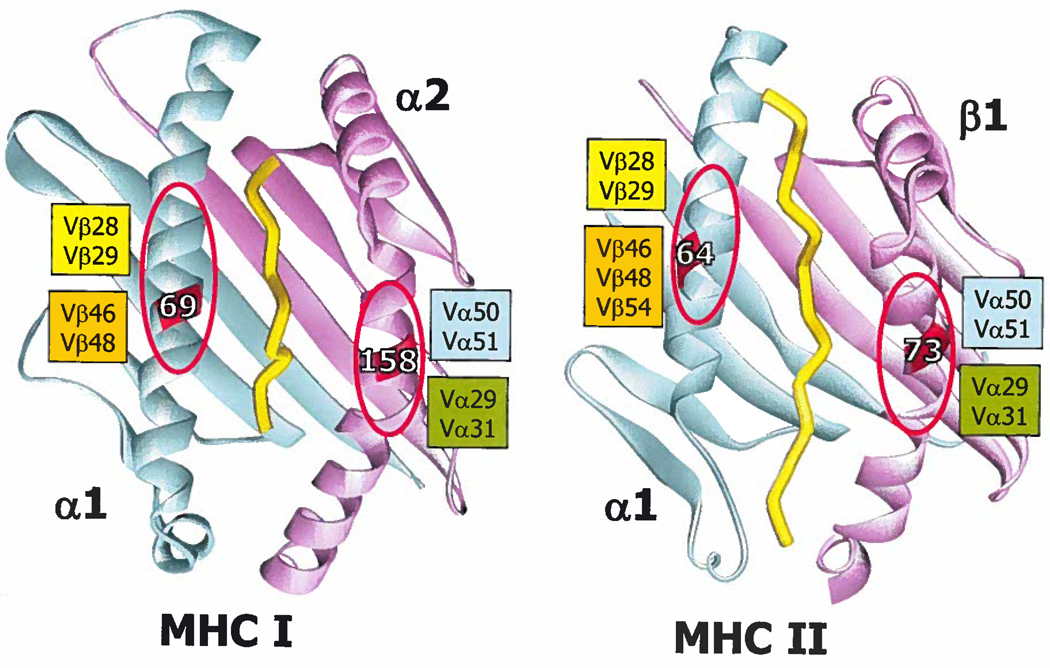
Therefore, in examining the structures we were looking for sites of interaction in which particular TCR amino acids were often, but not always found and for interactions that might be flexible and not necessarily lost by small shifts in orientation position. We conclude that such interactions can be identified. Our overall conclusions are summarized schematically in Figure 8. We propose that the anchor points on MHC molecules are not particular amino acid chains, but rather two exposed areas on the tops of the MHC α helices centered about α1 69 and α2 158 for MHCI and α1 64 and β1 73 for MHCII. These amino acids are highly conserved (mostly alanine and glycine) and create dish like areas that expose the backbones of the helices. The flanking amino acids on the helices determine the size of this area.
Figure 8. The amino acids in Vα and Vβ that frequently contact MHC in the same areas can determine the diagonal aspect of TCR binding to MHC.
Ribbon representations of HLA-A2/Tax peptide (pdb 1BD2) and IAb/3K (pdb 1LNU) are shown as an examples of an MHCI and MHCII molecule, respectively. The α1/α domains are colored cyan. The MHCI α2 domain and the MHCII β1 domains are colored magenta. The peptide is represented in yellow. The positions of 2 amino acids on the helices of each MHC molecule are labeled, α69 and α158 on MHC1 and α64 and β73 on MHCII. An area of around each of these is indicated with a red oval. TCR amino acids most often found in contact with the MHC within these areas are listed next to each oval. See text for further discussion.
In the majority of TCR/MHC structures these areas are major sites of interaction with specific amino acids in TCR CDR1 and CDR2 regions. Since these areas occur diagonally opposite one another on the helices, we propose that they determine the general diagonal orientation of the TCR on the MHC. The TCR amino acids involved in these interactions are often tyrosines that make many Van der Waals contacts with the backbones of helices and the flanking amino acid side chains. These types of interactions do not require a precise geometry and allow considerable flexibility in orientation and exact position along the helices, accounting for most of the variations in orientation of the TCRs.
Why aren’t all the evolutionary selected rules apparent in all TCR/MHC complexes?
If there are rules that govern the interactions between TCR V regions and MHC then one might expect that these would always be manifest, particularly when the V region is involved in recognition of the same MHC allele, regardless of the peptide. However, this is not always observed. For example, the structures of three human TCRs containing Vβ13.1, bound to the class I protein, HLA-A2 have been reported. One TCR (1G4 in 2NBQ) has many contacts between Vβ13.1 and HLA-A2, another (B7 in 1BD2), a few, and the last (A6 in 1AO7), none (Figure 2b), These differences are due to changes in the CDR3 regions of the TCR. A6 has a very long CDR3β, that lifts the TCR away from the MHC, so that contacts are not possible. We hypothesize that such phenomena may illustrate thymic selection at work and that these attenuations of the conserved interactions are sometimes needed to escape negative selection. In another case, a situation was described in which TCR contacts with MHC were limited by a very pronounced bulge in the surface of MHC, caused by the extraordinary length of the engaged peptide (in 2AK4, Figure 2). Thus the CDR3 regions of the TCR, and/or the peptide amino acids projecting out of the MHC groove determine whether or not the TCR will be lifted away from MHC, or pitched such that it can contact only a portion of the MHC amino acids.
Do the evolutionary selected interactions operate for both MHCI and MHCII recognition?
Our analysis of highly cross reactive T cells, such as YAe62, suggested that the same conserved MHC reactive features of a particular CDR1 and CDR2 might apply to both MHCI and MHCII recognition and we expected to find evidence for this in our analysis. In general, that is the case but there seem to be some differences. In fact the rules seem somewhat more apparent with MHCII than with MHCI. For example, in TCRs with mVβ8.2 the limits of area of contact between VβCDR2 Y48 and the α1 MHC helix appears to be more restricted on MHCII than on MHCI and the MHC αK39 that forms a unique salt bridge with VβCDR2 E54 does not exist in MHCI. One factor might be that a number of the TCR/MHCI structures involve MHC allo-antigens and therefore the TCRs did not come from a repertoire shaped by negative selection by that MHC during development. Thus, it is possible that as more TCR/MHCI structures appear involving conventional peptide antigens, some of these differences may fade.
A caveat
Engagement by a TCR amino acid of a particular site on MHC does not necessarily mean that the reaction is predetermined by the sequences of TCR and MHC. It may be forced by other elements, such as the docking sites of CD4 and CD8 or other phenomena (67) and, since there are limited numbers of amino acids in the CDR 1 and 2 regions of TCRs and on the alpha helices of MHC, some common reactions are bound to occur. We believe that this is unlikely for two reasons. First, the interactions are determined by the nature, not position, of the TCR amino acid (Figure 2). Second the prominent availability of particular TCR amino acids does not in itself predict a particular area of MHC contact as proposed in Figure 8. Moreover, there are some differences between MHCI and MHCII engagement (see above), suggesting that the system is built more subtly than suggested by the forced position argument. Ultimately, the hypothesis we propose simply focuses attention on particular amino acids whose function must be confirmed in proper straightforward experimental tests. Such can be done, by, for example, changing the relevant TCR amino acids in the mouse germ line, and observing the effects of the changes on TCR selection and function.
DO THE BIASES OF TCRs FOR RECOGNITION OF MHC APPLY TO NONCLASSICAL MHC MOLECULES?
In addition to the classical polymorphic MHC molecules, mammalian species express other relatively non-polymorphic MHCIb and MHC-like molecules, such as HLA-E, H2-M3, MR1 and the CD1 family (39, 86, 87)). These molecules are also recognized by αβTCR-bearing T cells and are important in fending off infectious diseases. For example, HLA-E restricted T cells have been identified in responses to bacteria and viruses, including Mycobacterium tuberculosis (Mtb) and cytomegalovirus (CMV) (88, 89). In mice, H2-M3 restricted T cells have a characteristic pre-activated phenotype, and mediate early T cell responses against Listeria monocytogenes (90). Similarly, T cells that recognize lipid antigens in the context of the CD1 family have been implicated in responses to Sphingomonas, Borrellia burgdorferi, and Mtb (91–96). One feature of these non-classical MHC proteins is the fact that they often present non-peptide antigens, such as glycolipids. The question then arises, how do TCRs that have, perhaps, evolved to react with classical MHC/peptide cope with these non-conventional MHCs and their bound ligands?
The non-polymorphic MHCIb molecule, HLA-E, presents peptides from the leader sequences of conventional MHCIa molecules (97). These MHC/self-peptide complexes are recognized by inhibitory NK receptors as surrogate markers for MHCI fidelity when MHCI expression is altered by specific pathogens (98, 99). Inhibitory NK receptors are quite tolerant of amino acid changes in HLA-E presented peptides (39), whereas αβTCRs have conventional specificities both for HLA-E and its engaged peptides. This is exemplified by the recent solution of the structure of a TCR, KK50.4, bound to HLA-E plus a cytomegalovirus-encoded mimic of an MHCI leader peptide (39). The overall recognition of HLA-E by the KK50.4 TCR has the same topology as T cells reacting with conventional MHC, with a similar diagonal binding mode, and uses the same TCR amino acids to dock with HLA-E (Figure 2a,b). Thus the evolutionary biases of TCRs for MHC apply, is they exist, to reaction with both classical MHC and HLA-E.
H2-M3 was originally identified as a minor histocompatibility molecule presenting a maternally linked factor (100), Like HLA-E, H2-M3 is relatively non-polymorphic, but is unique to murine species. The peptide binding groove of H2-M3 is unlike that of conventional MHCI molecules, accommodating a formylmethionine moiety at the NH2 terminus of the peptide that facilitates the presentation of bacterial and mitochondrial produced proteins (101, 102). The overall dimensions of the groove between the α-helices of H2-M3 are similar to those of conventional MHCI molecules, but the amino acids that line this pocket are primarily non-polar, facilitating the presentation of hydrophobic peptides. The molecular basis for TCR recognition of H2-M3 has not been determined, but the overall similarities to MHCIa suggest it may be receptive to conserved TCR interactions.
MHC-related protein 1 (MR1) is another β2m-associated, MHCIb molecule that is evolutionary conserved among mammals (103). MR1 is associated with stimulation of, and is required for the development of, mucosal associated invariant T (MAIT) cells. This population of T cells expresses, in mice, a TCR with an invariant Vα19-Jα33 TCRα (Vα19i) and, in human, the highly similar combination of Vα7.2 and Jα19 (Vα7.2i) (104). The natural antigens presented by MR1 in vivo are largely unknown, although a role for gut flora in the activation of these MAIT cells has been suggested. Surprisingly, while the amino-acid composition of the MR1 groove does not appear especially suited for glycolipid presentation, in contrast with CD1 molecules (see below), α-mannosylceramide was recently shown to stimulate Vα19i T cells in a MR1-dependent manner (105). The structure of MR1 is currently unknown, but based on the overall similarity to MHCIa and MHCIb a computational analysis suggested an MHC-like fold and allowed a mutational analysis of the α-helices and putative antigen binding groove. The data from the response of several T cell hybridomas to these mutants suggested both an antigen presentation function as well as an orthogonal TCR docking mode similar to that of conventional T cells (106).
TCR recognition of CD1d
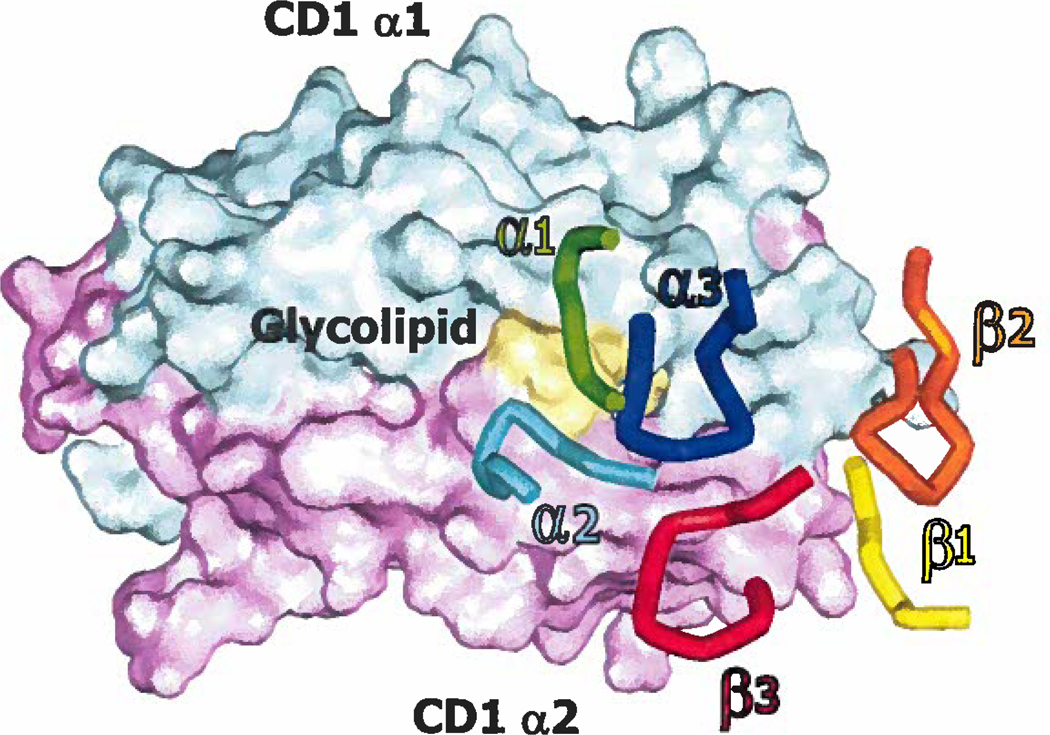
The human CD1 family and mouse CD1d are non-polymorphic, β2m-associated, MHCI-like molecules expressed predominately on hematopoetic cells. Compared to classical MHCI and MHCII molecules, the antigen binding groove of CD1 family members is narrower, has a more pronounced bulge in its α2 helix, and is composed primarily of non-polar amino acids to facilitate the presentation of diverse hydrophobic ligands, including lipids, glycolipids, and lipopeptides. While the lipid tails serve to “anchor” these antigens in the groove, the polar head groups extend out of the groove, for recognition by T cells (reviewed in (58)).
Group I CD1 isoforms (CD1a, b and c) present many different antigens, including lipids and lipopeptides that vary in size and shape (58,107–111). Most human T cells reactive to CD1a, CD1b, and CD1c express diverse Vα and Vβ gene segments (108,109, 111), suggesting that group I CD1-reactive T cells may have a broad range of unique antigen specificities. Indeed, CD1a and CD1b reactive T cell lines have been shown to be highly specific for particular peptide moieties on didehydroxymycobactin antigens (for CD1a) and for carbohydrate moieties of mycolyl lipid antigens (for CD1b) (112,113). While the precise molecular basis for αβTCR recognition of CD1a, CD1b, or CD1c has not yet been resolved by crystallography, mutational analysis of the TCR interacting face of CD1b suggested a diagonal orientation of TCR contacts similar to the binding mode observed in conventional T cells (114). There remains the interesting question how TCRs with apparently normal geometry can contact the group I CD1 proteins and classical MHC proteins in the saem way, given the quite different geometry of the two types of MHC molecules.
The situation is entirely different for the group II isoform of CD1d, the only CD1 isoform conserved between mouse and human (58). In contrast to the highly diverse TCR repertoire expressed by T cells reactive to other MHC molecules and group I CD1s, most CD1d reactive cells in humans and mice are natural killer (NK) T cells (115, 116). These express an invariant TCRα composed of Vα24-Jα18 (Vα24i TCRα) in humans and the nearly identical Vα14-Jα18 (Vα14i TCRα), in mice (117, 118). The peripheral NKT population uses a diverse, but limited number of TCRβ gene segments, with the majority of the population expressing Vβ11 in humans and Vβs 2, 7 and 8.2 in mice (115, 116). The invariant TCRα chain, and the complete lack of NKT cells in Jα18 deficient mice, strongly suggests a critical role for the TCRα in the recognition of glycolipid/CD1d complexes (119). On the other hand, the role of the more diverse TCRβ is less clear. NK T cells react with a number of glycolipid antigens (116). Perhaps different TCRβs allow reaction with different glycolipids.
We recently assessed the role of the Vα14i TCRα and mTCRβ chains in recognition of different glycolipid/CD1d complexes by mutagenesis analysis (Scott-Browne et al. submitted for publication). We showed that mouse Vα14i TCRα recognition of multiple α-linked glycolipids is conferred by a functional “hot-spot” composed of germ line encoded amino acids within CDR3α, CDR1α and CDR2β. This functional "hot-spot" does not differ between structurally distinct antigens, suggesting that the Vα14i TCRα functions as a pattern recognition receptor. The mutagenesis identified the critical role of germ line encoded residues in the Vα14i TCRα, and the requirement for these residues provided a basis for the extremely biased TCR repertoire of NKT cells. While the mutagenesis alone could not provide the molecular basis of the interaction between the TCR and CD1d, the addition of structural data, facilitated a precise definition of glycolipid recognition by NKT cells.
Because CD1d has an MHC-like fold and TCRβ chains from conventional T cells could provide glycolipid/CD1d specificity, early models of the TCR-CD1d interaction postulated that the TCR would dock on CD1d as they do on MHCI/MHCII (55, 56). However, this hypothesis has been shown to be wrong by the recently solved crystal structure of the Vα24i TCR bound to hCD1d/αGC (68). In this structure the Vα24i TCR is oriented parallel to, rather than diagonally, over CD1d (Figure 9). The interaction of the Vα24i TCRα chain with CD1d is dominated by CDR3α residues (encoded by Jα18) binding to many conserved residues of CD1d, while glycolipid specificity is contributed primarily by CDR1α (encoded by Vα24/14).
Figure 9. TCRs bind CD1d/glycolipid in an orientation that is completely different from that used to bind classical MHC/peptide.
Shown is a plan of the contacts between the CDR1-3 loops of the α and β chains of the hVα24i iTCR and a space filling surface of CD1d and α-galactosylceramide (data taken from (68), pdb2PO6). The α1 region of CD1d is colored cyan, the α2 region of CD1d is in magenta and the glycolipid in yellow. The CDR loops are color coded and indicated in their corresponding colors on the diagram.
Followers of the evolutionary arguments (above) might ask what controls the ability of the Vα24i/14i TCRα to bind preferentially CD1d. The phenomenon probably occurs for several reasons. First, hJα18 residues in CDR3α interact with a large surface on CD1d and are needed for the specificity of the NKT TCRs since, while some Vα24- human CD1d/αGC TCRs have been isolated, these TCRs still use Jα18 (55, 120). Secondly, residues in Vα24/14 CDR1α are probably also important. Overall the CDR1α are nearly identical between mouse Vα14 and human Vα24 (63, 64), suggesting evolutionary pressure has conserved these sequences and their V’s at 26α and P’s at 28α are unique in mouse Vαs, and human and mouse Vαs respectively. These amino acids are crucial for the ability of NKTCRs to react with CD1d/glcolipid (Scott-Browne et al. submitted for publication) and may be among features that control the function of theVα24i/14iTCRα.
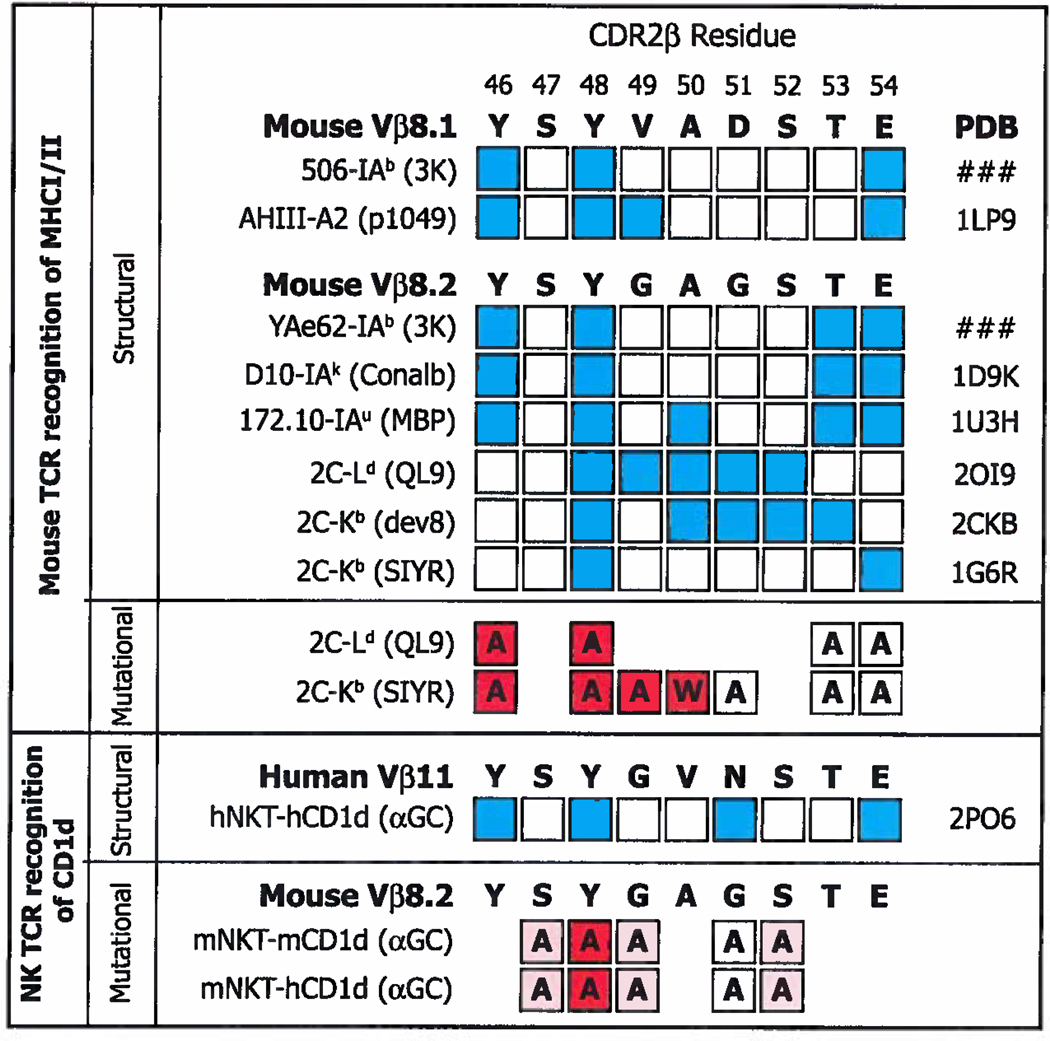
As far as the NK TCR β chain residues are concerned, these are focused on CD1d, rather than the antigen (68). Although the residues that contact CD1d, Y46β, Y48β, and E54β, also bind classical MHC (Figures 2 and 7) they do so at very different sites (compare Figures 1 and 9). The case is most striking for Vβ8.2 Y48β, which has been shown by both structural and mutational experiments to be absolutely required for Vβ8.2+ NKT TCRs to bind CD1d/glycolipid (Figure 10) ((68), Scott-Browne et al. submitted for publication) and which is often crucial to the interaction of conventional TCRs with classical MHC (Figure 2, (27, 30, 32, 34, 36, 42, 46,121), Dai et al. submitted for publication). This amino acid binds classical MHCs at a relatively conserved site (Figure 2b, (27, 30, 32, 34, 36, 42, 46) and Dai et al. submitted for publication). Nevertheless, it binds CD1d at a totally different site (Figure 9, (68))
Figure 10. Some of the CDR2β amino acids used to contact classical MHCI/MHCII and CD1d are identical.
Above. The amino acid sequences of mVβ8.1 and mVβ8.2 in bold. Amino acids that have been shown in sturtures to bind MHC are indicated in blue, together with the names of the TCRs and pdb files used. Amino acids in white squares do not contact MHC in the indicated structure. Below this are shown the results of mutational analyses, in which the amino acid in question was mutated as shown in each square. The influence of each mutation is indicated by particular shades of red, red filled squares indicate >1 log change in reactivity, while white indicates a <0.5 change in log reactivity (122, 123). Below Contact points identified by structural analysis (68) between a hVα24i TCR and hCD1d/αGC and identified by mutational analyses, using staining with CD1d/αGC tetramers, between a mVα14i TCR and mCD1d/αGC or hCD1d/αGC (Scott-Browns et al. submitted for publication). Red filled squares indicate >50% loss in tetramer mean fluorescent intensity (MFI), pink indicates a change between 10% and 50% change in tetramer MFI, while white indicates a <10% change in tetramer MFI.
Thus Vα24i/14iTCRs behave quite differently from most TCRs recognizing other MHC/ligands. They use invariant TCRα chains and a restricted number of TCRβ chains. They solve the problem of the different geometry of CD1d versus classical MHC by binding the two types of protein in quite different ways. Nevertheless, it is remarkable that some of the same TCR Vb residues are crucial to binding both ligands, an example, of evolutionary exploitation by the TCRs on NKT cells
SUMMARY POINTS.
Certain amino acids in TCR CDR1 and CDR2 regions often contact MHC. These contact points are in similar positions on MHC in the complexes of different TCRs bound to different MHCs.
The target positions on MHC are often cups on the MHC α helices, containing small amino acids. The TCR CDR amino acids can bind at variable positions within the cups, contacting MHC backbone and the side chains of adjacent amino acids.
This arrangement allows flexibility in the pitch of the CDR1/2 amino acid as it contacts MHC, thus allowing the TCR to accommodate CDR3 regions and peptide ligands of different sequences and lengths and yet still bind the MHC/peptide ligand in approximately the same orientation.
TCRs do not usually use all of their built-in abilities to react with MHC because negative selection in the thymus removes from mature repertoires, TCRs that react too well with MHC.
The CDR1/2 amino acids that are often involved in binding MHC are not the same for all families of V regions.
The CDR1/2 amino acids that are frequently used may not always be the same in reactions with MHCI and MHCII.
The positions of the frequently used CDR1/2 amino acids and their targets on MHC suggest that these contacts may impose the usual diagonal mode of binding of TCRs on MHC.
The Vα24i/14i TCRs binding CD1d, and conventional TCRs, binding classical MHC, use some of the same CDR2β residues to contact their ligands, even though the mode of binding in the 2 cases is quite different. Vα241/14i TCRs appear to have adopted evolutionarily selected TCR residues for their own purposes.
ACKNOWLEDGMENTS
The authors thank Frances Crawford, Janice White and Rachel Frugge for their technical help in the work that led to this analysis and Drs. Whitney MacDonald and K.C. Garcia for many helpful discussions. This work was supported in part by USPHS grants AI-17134, AI-19785, AI-22295, AI-057485 and Cancer Center CA-046934.
Acronyms
- TCR
alpha beta T cell receptor
- MHC
major histocompatibility complex
- CDR
complementarity determining region
- CD1
cluster of differentiation
- V region
variable region
- NKT cell
natural killer T cell
Contributor Information
Philippa Marrack, Email: marrackp@njc.org.
James P. Scott-Browne, Email: james.scott-browne@uchsc.edu.
Shaodong Dai, Email: dais@njc.org.
Laurent Gapin, Email: gapinl@njc.org.
John W. Kappler, Email: kapplerj@njc.org.
LITERATURE CITED
- 1.Claman HN, Chaperon EA, Triplett RF. Thymus-marrow cell combinations. Synergism in antibody production. Proc Soc Exp Biol Med. 1966;122:1167–1171. doi: 10.3181/00379727-122-31353. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell GF, Miller JF. Cell to cell interaction in the immune response. II. The source of hemolysin-forming cells in irradiated mice given bone marrow and thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:821–837. doi: 10.1084/jem.128.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchison NA. The carrier effect in the secondary response to hapten-protein conjugates. V. Use of antilymphocyte serum to deplete animals of helper cells. Eur J Immunol. 1971;1:68–75. doi: 10.1002/eji.1830010204. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky K. The carrier effect and cellular cooperation in the induction of antibodies. Proc R Soc Lond B Biol Sci. 1971;176:385–392. doi: 10.1098/rspb.1971.0002. [DOI] [PubMed] [Google Scholar]
- 5.Gorer PA. Genetic and antigenic basis of tumor transplantation. J. Path. and Bact. 1937;44:691–697. [Google Scholar]
- 6.Demant P, Graff RJ. Transplantation analysis of the H-2 system. Transplant Proc. 1973;5:267–270. [PubMed] [Google Scholar]
- 7.Jerne NK. The somatic generation of immune recognition. Eur J Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 8.Senyk G, Nitecki D, Goodman JW. Immunogenicity of glucagon: determinants responsible for antibody binding and lymphocyte stimulation. Science. 1971;171:407–408. doi: 10.1126/science.171.3969.407. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann M, Kappler JW. Regulation of the immune response. II. Qualitative and quantitative differences between thymus- and bone marrow-derived lymphocytes in the recognition of antigen. J Exp Med. 1973;137:721–739. doi: 10.1084/jem.137.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter P, Munro A, McConnell I. Properties of educated T cells for rosette formation and cooperation with B cells. Nat New Biol. 1972;236:52–53. doi: 10.1038/newbio236052a0. [DOI] [PubMed] [Google Scholar]
- 11.McDevitt HO. Discovering the role of the major histocompatibility complex in the immune response. Annu Rev Immunol. 2000;18:1–17. doi: 10.1146/annurev.immunol.18.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- 13.Kappler JW, Skidmore B, White J, Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981;153:1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allison JP, McIntyre BW, Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. 1982. J Immunol. 2005;174:1144–1151. [PubMed] [Google Scholar]
- 15.Meuer SC, Fitzgerald KA, Hussey RE, Hodgdon JC, Schlossman SF, Reinherz EL. Clonotypic structures involved in antigen-specific human T cell function. Relationship to the T3 molecular complex. J Exp Med. 1983;157:705–719. doi: 10.1084/jem.157.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haskins K, Kubo R, White J, Pigeon M, Kappler J, Marrack P. The major histocompatibility complex-restricted antigen receptor on T cells. I. Isolation with a monoclonal antibody. J Exp Med. 1983;157:1149–1169. doi: 10.1084/jem.157.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 18.Yanagi Y, Yoshikai Y, Leggett K, Clark SP, Aleksander I, Mak TW. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984;308:145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- 19.Chien Y, Becker DM, Lindsten T, Okamura M, Cohen DI, Davis MM. A third type of murine T-cell receptor gene. Nature. 1984;312:31–35. doi: 10.1038/312031a0. [DOI] [PubMed] [Google Scholar]
- 20.Sim GK, Yague J, Nelson J, Marrack P, Palmer E, Augustin A, Kappler J. Primary structure of human T-cell receptor alpha-chain. Nature. 1984;312:771–775. doi: 10.1038/312771a0. [DOI] [PubMed] [Google Scholar]
- 21.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 22.Shimonkevitz R, Colon S, Kappler JW, Marrack P, Grey HM. Antigen recognition by H-2-restricted T cells. II. A tryptic ovalbumin peptide that substitutes for processed antigen. J Immunol. 1984;133:2067–2074. [PubMed] [Google Scholar]
- 23.Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to la histocompatibility molecules. Nature. 1985;317:359–361. doi: 10.1038/317359a0. [DOI] [PubMed] [Google Scholar]
- 24.Morrison LA, Lukacher AE, Braciale VL, Fan DP, Braciale TJ. Differences in antigen presentation to MHC class I-and class II-restricted influenza virus-specific cytolytic T lymphocyte clones. J Exp Med. 1986;163:903–921. doi: 10.1084/jem.163.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 26.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 27.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 28.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 29.Ding YH, Smith KJ, Garboczi DN, Utz U, Biddison WE, Wiley DC. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 30.Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey RE, Smolyar A, Hare B, Zhang R, Joachimiak A, Chang HC, Wagner G, Wang J. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 31.Reiser JB, Darnault C, Guimezanes A, Gregoire C, Mosser T, Schmitt-Verhulst AM, Fontecilla-Camps JC, Malissen B, Housset D, Mazza G. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 32.Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. Embo J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiser JB, Gregoire C, Darnault C, Mosser T, Guimezanes A, Schmitt-Verhulst AM, Fontecilla-Camps JC, Mazza G, Malissen B, Housset D. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- 34.Buslepp J, Kerry SE, Loftus D, Frelinger JA, Appella E, Collins EJ. High affinity xenoreactive TCR:MHC interaction recruits CD8 in absence of binding to MHC. J Immunol. 2003;170:373–383. doi: 10.4049/jimmunol.170.1.373. [DOI] [PubMed] [Google Scholar]
- 35.Chen JL, Stewart-Jones G, Bossi G, Lissin NM, Wooldridge L, Choi EM, Held G, Dunbar PR, Esnouf RM, Sami M, Boulter JM, Rizkallah P, Renner C, Sewell A, van der Merwe PA, Jakobsen BK, Griffiths G, Jones EY, Cerundolo V. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 37.Deng L, Langley RJ, Brown PH, Xu G, Teng L, Wang Q, Gonzales MI, Callender GG, Nishimura MI, Topalian SL, Mariuzza RA. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat Immunol. 2007;8:398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- 38.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoare HL, Sullivan LC, Pietra G, Clements CS, Lee EJ, Ely LK, Beddoe T, Falco M, Kjer-Nielsen L, Reid HH, McCluskey J, Moretta L, Rossjohn J, Brooks AG. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat Immunol. 2006;7:256–264. doi: 10.1038/ni1312. [DOI] [PubMed] [Google Scholar]
- 40.Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. Embo J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst AM, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? Embo J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 45.Tynan FE, Burrows SR, Buckle AM, Clements CS, Borg NA, Miles JJ, Beddoe T, Whisstock JC, Wilce MC, Silins SL, Burrows JM, Kjer-Nielsen L, Kostenko L, Purcell AW, McCluskey J, Rossjohn J. T cell receptor recognition of a 'super-bulged' major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 46.Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, Burrows SR, McCluskey J, Rossjohn J. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 47.Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- 48.Fink PJ, Bevan MJ. H-2 antigens of the thymus determine lymphocyte specificity. J Exp Med. 1978;148:766–775. doi: 10.1084/jem.148.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zinkernagel RM, Callahan GN, Althage A, Cooper S, Klein PA, Klein J. On the thymus in the differentiation of "H-2 self-recognition" by T cells: evidence for dual recognition? J Exp Med. 1978;147:882–896. doi: 10.1084/jem.147.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sim BC, Aftahi N, Reilly C, Bogen B, Schwartz RH, Gascoigne NR, Lo D. Thymic skewing of the CD4/CD8 ratio maps with the T-cell receptor alpha-chain locus. Curr Biol. 1998;8:701–704. doi: 10.1016/s0960-9822(98)70276-3. [DOI] [PubMed] [Google Scholar]
- 51.Bill J, Appel VB, Palmer E. An analysis of T-cell receptor variable region gene expression in major histocompatibility complex disparate mice. Proc Natl Acad Sci U S A. 1988;85:9184–9188. doi: 10.1073/pnas.85.23.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 53.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 54.Sullivan BA, Kronenberg M. TCR-mediated recognition of glycolipid CD1 complexes. Curr Top Microbiol Immunol. 2007;314:165–193. doi: 10.1007/978-3-540-69511-0_7. [DOI] [PubMed] [Google Scholar]
- 55.Gadola SD, Koch M, Maries-Wright J, Lissin NM, Shepherd D, Matulis G, Harlos K, Villiger PM, Stuart DI, Jakobsen BK, Cerundolo V, Jones EY. Structure and binding kinetics of three different human CD1d-alpha-gatactosylceramide-specific T cell receptors. J Exp Med. 2006;203:699–710. doi: 10.1084/jem.20052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, Bharadwaj M, Godfrey DI, Rossjohn J, McCluskey J. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 59.Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 60.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- 61.Marrack P, Shimonkevitz R, Hannum C, Haskins K, Kappler J. The major histocompatibility complex-restricted antigen receptor on T cells. IV. An antiidiotypic antibody predicts both antigen and I-specificity. J Exp Med. 1983;158:1635–1646. doi: 10.1084/jem.158.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark SP, Arden B, Kabelitz D, Mak TW. Comparison of human and mouse T-cell receptor variable gene segment subfamilies. Immunogenetics. 1995;42:531–540. doi: 10.1007/BF00172178. [DOI] [PubMed] [Google Scholar]
- 63.Arden B, Clark SP, Kabelitz D, Mak TW. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 64.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 65.Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 66.Hennecke J, Wiley DC. T cell receptor-MHC interactions up close. Cell. 2001;104:1–4. doi: 10.1016/s0092-8674(01)00185-4. [DOI] [PubMed] [Google Scholar]
- 67.Mazza C, Malissen B. What guides MHC-restricted TCR recognition? Semin Immunol. 2007 doi: 10.1016/j.smim.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 69.Jaeger EE, Bontrop RE, Lanchbury JS. Structure, diversity, and evolution of the T-cell receptor VB gene repertoire in primates. Immunogenetics. 1994;40:184–191. doi: 10.1007/BF00167078. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka T, Nei M. Positive darwinian selection observed at the variable-region genes of immunoglobulins. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- 71.Blackman M, Yague J, Kubo R, Gay D, Coleclough C, Palmer E, Kappler J, Marrack P. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47:349–357. doi: 10.1016/0092-8674(86)90591-x. [DOI] [PubMed] [Google Scholar]
- 72.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 73.Merkenschlager M, Graf D, Lovatt M, Bommhardt U, Zamoyska R, Fisher AG. How many thymocytes audition for selection? J Exp Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 75.von Boehmer H, Teh HS, Kisielow P. The thymus selects the useful, neglects the useless and destroys the harmful. Immunol Today. 1989;10:57–61. doi: 10.1016/0167-5699(89)90307-1. [DOI] [PubMed] [Google Scholar]
- 76.Ignatowicz L, Kappler J, Marrack P. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 77.Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 78.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 79.Fung-Leung WP, Surh CD, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H2-M-deficient mice. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 80.Laufer TM, DeKoning J, Markowitz JS, Lo D, Glimcher LH. Unopposed positive selection and autoreactivity in mice expressing class II MHC only on thymic cortex. Nature. 1996;383:81–85. doi: 10.1038/383081a0. [DOI] [PubMed] [Google Scholar]
- 81.Fukui Y, lshimoto T, Utsuyama M, Gyotoku T, Koga T, Nakao K, Hirokawa K, Katsuki M, Sasazuki T. Positive and negative CD4+ thymocyte selection by a single MHC class II/peptide ligand affected by its expression level in the thymus. Immunity. 1997;6:401–410. doi: 10.1016/s1074-7613(00)80283-6. [DOI] [PubMed] [Google Scholar]
- 82.Gapin L, Fukui Y, Kanellopoulos J, Sano T, Casrouge A, Malier V, Beaudoing E, Gautheret D, Claverie JM, Sasazuki T, Kourilsky P. Quantitative analysis of the T cell repertoire selected by a single peptide-major histocompatibility complex. J Exp Med. 1998;187:1871–1183. doi: 10.1084/jem.187.11.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huseby ES, Crawford F, White J, Kappler J, Marrack P. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc Natl Acad Sci U S A. 2003;100:11565–11570. doi: 10.1073/pnas.1934636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 85.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 86.Stroynowski I, Forman J. Novel molecules related to MHC antigens. Curr Opin Immunol. 1995;7:97–102. doi: 10.1016/0952-7915(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 87.Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat Immunol. 2007;8:563–568. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- 88.Heinzel AS, Grotzke JE, Lines RA, Lewinsohn DA, McNabb AL, Streblow DN, Braud VM, Grieser HJ, Belisle JT, Lewinsohn DM. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J Exp Med. 2002;196:1473–1481. doi: 10.1084/jem.20020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazzarino P, Pietra G, Vacca P, Falco M, Colau D, Coulie P, Moretta L, Mingari MC. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur J Immunol. 2005;35:3240–3247. doi: 10.1002/eji.200535343. [DOI] [PubMed] [Google Scholar]
- 90.Seaman MS, Wang CR, Forman J. MHC class Ib-restricted CTL provide protection against primary and secondary Listeria monocytogenes infection. J Immunol. 2000;165:5192–5201. doi: 10.4049/jimmunol.165.9.5192. [DOI] [PubMed] [Google Scholar]
- 91.Wu D, Xing GW, Poles MA, Horowitz A, Kinjo Y, Sullivan B, Bodmer-Narkevitch V, Plettenburg O, Kronenberg M, Tsuji M, Ho DD, Wong CH. Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc Natl Acad Sci U S A. 2005;102:1351–1356. doi: 10.1073/pnas.0408696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 93.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 94.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 95.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 96.Behar SM, Porcelli SA. CD1-restricted T cells in host defense to infectious diseases. Curr Top Microbiol Immunol. 2007;314:215–250. doi: 10.1007/978-3-540-69511-0_9. [DOI] [PubMed] [Google Scholar]
- 97.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 98.Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 99.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang CR, Loveland BE, Lindahl KF. H-2M3 encodes the MHC class I molecule presenting the maternally transmitted antigen of the mouse. Cell. 1991;66:335–345. doi: 10.1016/0092-8674(91)90623-7. [DOI] [PubMed] [Google Scholar]
- 101.Vyas JM, Shawar SM, Rodgers JR, Cook RG, Rich RR. Biochemical specificity of H-2M3a. Stereospecificity and space-filling requirements at position 1 maintain N-formyl peptide binding. J Immunol. 1992;149:3605–3611. [PubMed] [Google Scholar]
- 102.Wang CR, Castano AR, Peterson PA, Slaughter C, Lindahl KF, Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 103.Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phytogeny of the MHC class I-related MR1 gene. J Immunol. 1998;161:4066–4077. [PubMed] [Google Scholar]
- 104.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, Lantz O. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shimamura M, Huang YY, Okamoto N, Suzuki N, Yasuoka J, Morita K, Nishiyama A, Amano Y, Mishina T. Modulation of Valpha19 NKT cell immune responses by alpha-mannosyl ceramide derivatives consisting of a series of modified sphingosines. Eur J Immunol. 2007;37:1836–1844. doi: 10.1002/eji.200636689. [DOI] [PubMed] [Google Scholar]
- 106.Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, Fremont DH, Hansen TH. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. 2005;280:21183–21193. doi: 10.1074/jbc.M501087200. [DOI] [PubMed] [Google Scholar]
- 107.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 108.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli SA, Brenner MB. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grant EP, Beckman EM, Behar SM, Degano M, Frederique D, Besra GS, Wilson IA, Porcelli SA, Furlong ST, Brenner MB. Fine specificity of TCR complementarity-determining region residues and lipid antigen hydrophilic moieties in the recognition of a CD1-lipid complex. J Immunol. 2002;168:3933–3940. doi: 10.4049/jimmunol.168.8.3933. [DOI] [PubMed] [Google Scholar]
- 110.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 111.Young DC, Moody DB. T-cell recognition of glycolipids presented by CD1 proteins. Glycobiology. 2006;16:103R–112R. doi: 10.1093/glycob/cwj111. [DOI] [PubMed] [Google Scholar]
- 112.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 113.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O'Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 114.Melian A, Watts GF, Shamshiev A, De Libero G, Clatworthy A, Vincent M, Brenner MB, Behar S, Niazi K, Modlin RL, Almo S, Ostrov D, Nathenson SG, Porcelli SA. Molecular recognition of human CD1b antigen complexes: evidence for a common pattern of interaction with alpha beta TCRs. J Immunol. 2000;165:4494–4504. doi: 10.4049/jimmunol.165.8.4494. [DOI] [PubMed] [Google Scholar]
- 115.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 116.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 117.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4-8- T cells. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 120.Brigl M, van den Elzen P, Chen X, Meyers JH, Wu D, Wong CH, Reddington F, Illarianov PA, Besra GS, Brenner MB, Gumperz JE. Conserved and heterogeneous lipid antigen specificities of CD1d-restricted NKT cell receptors. J Immunol. 2006;176:3625–3634. doi: 10.4049/jimmunol.176.6.3625. [DOI] [PubMed] [Google Scholar]
- 121.Manning TC, Parke EA, Teyton L, Kranz DM. Effects of complementarity determining region mutations on the affinity of an alpha/beta T cell receptor: measuring the energy associated with CD4/CD8 repertoire skewing. J Exp Med. 1999;189:461–470. doi: 10.1084/jem.189.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Manning TC, Schlueter CJ, Brodnicki TC, Parke EA, Speir JA, Garcia KC, Teyton L, Wilson IA, Kranz DM. Alanine scanning mutagenesis of an alphabeta T cell receptor: mapping the energy of antigen recognition. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- 123.Lee PU, Churchill HR, Daniels M, Jameson SC, Kranz DM. Role of 2CT cell receptor residues in the binding of self- and allo-major histocompatibility complexes. J Exp Med. 2000;191:1355–1364. doi: 10.1084/jem.191.8.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]