Abstract
A high-resolution primer extension technique was used to study the relationships between repair, transcription, and mutagenesis in RNA polymerase III transcribed genes in Saccharomyces cerevisiae. The in vivo repair of UV-induced DNA damage by nucleotide excision repair (NER) and by photoreactivation is shown to be preferential for the nontranscribed strand (NTS) of the SNR6 gene. This is in contrast to RNA polymerase II genes in which the NER is preferential for the transcribed strand (TS). The repair-strand bias observed in SNR6 was abolished by inactivation of transcription in a snr6Δ2 mutant, showing a contribution of RNA polymerase III transcription in this phenomenon. The same strand bias for NER (slow in TS, fast in NTS) was discovered in the SUP4 gene, but only outside of the intragenic promoter element (box A). Unexpectedly, the repair in the transcribed box A was similar on both strands. The strand specificity as well as the repair heterogeneity determined in the transcribed strand of the SUP4 gene, correlate well with the previously reported site- and strand-specific mutagenesis in this gene. These findings present a novel view regarding the relationships between DNA repair, mutagenesis, and transcription.
Keywords: Cyclobutane pyrimidine dimers, nucleotide excision repair, photoreactivation, RNA polymerase III transcription, SNR6, Saccharomyces cerevisaie
UV light is an efficient DNA damaging agent, mainly responsible for the formation of pyrimidine dimers (PDs). These lesions are mostly eliminated by photoreactivation (PR) and/or nucleotide excision repair (NER) (Friedberg et al. 1995). The first process is a direct unistep DNA repair mechanism that reverses cyclobutane pyrimidine dimers (CPDs) by reversing the linkage between the adjacent pyrimidines with a light-initiated electron transfer reaction (Sancar 1990, 1996b; Wood 1996). It was shown recently that the PR of active genes is modulated by chromatin structure and transcription (Livingstone-Zatchej et al. 1997; Suter et al. 1997). The Saccharomyces cerevisiae photolyase preferentially repairs the nontranscribed strands (NTSs) of RNA polymerase II (RNAP II)-transcribed genes, whereas the PR of the transcribed strand (TS) is inhibited by a stalled RNA polymerase (Livingstone-Zatchej et al. 1997; Suter et al. 1997). This provides an explanation for the previous observation that the photorepair of the Escherichia coli galactokinase-forming capacity is inhibited when the gene is transcriptionally active (Kolsch and Starlinger 1965).
The NER is a multistep mechanism that copes with a large range of DNA damage including CPDs (Sancar 1996a; Wood 1996). During the last decade, a link was observed between the NER process and transcription. It is clear that the transcriptionally active genes are more rapidly repaired and that their TSs are preferentially repaired (Hanawalt 1995; Friedberg 1996a,b; Sancar 1996a). NER and transcription are linked in two different ways:
First, the presence of specific cellular factors assures preferential repair of the template strands of active genes. In E. coli, this process is known as transcription-coupled repair (TCR) and is under the control of the mfd gene product also called TRCF (transcription repair coupling factor) (Selby and Sancar 1993, 1994). In human and S.cerevisiae cells, the strand-specific repair of active genes requires the products of CSA and CSB/RAD26 genes (Bhatia et al. 1996), however, the biochemical coupling of transcription and repair has not been shown as yet.
The second connection is the dual function of TFIIH components in transcription and NER (Feaver et al. 1993; Drapkin et al. 1994; Wang et al. 1994; Svejstrup et al. 1995; Friedberg 1996b). The role of TFIIH in connecting these two processes is still puzzling because it is only involved in promoter clearance (Goodrich and Tjian 1994) and seems to dissociate from the elongating RNAP II machinery once the nascent transcript becomes longer than ∼30 nucleotides (Zawel et al. 1995). The high TFIIH affinity for the RNAP II complex, however, may play a role in rapid loading of the NER apparatus on the damaged site in the vicinity of a stalled RNAP II (Chalut et al. 1994). Does strand-specific repair of active genes only concern the RNAP II-transcribed genes?
It is well known that in addition to RNAP II, the transcription of eukaryotic genomes requires RNAP I and RNAP III, which transcribe different sets of genes (Zawel and Reinberg 1995). RNAP III is responsible for the transcription of several cellular and viral RNAs. Most RNA transcribed by RNAP III correspond to very short transcription units that are extensively covered with transcription factors binding to intragenic promoter elements. The genes transcribed by RNAP III fall into three different classes depending on the promoter structures and their cognate transcription factors. RNAP III involves the accessory transcription factors TFIIIA, TFIIIB, and TFIIIC, which interact with the promoter elements (A, B, and C boxes) to form a stable preinitiation complex (Hernandez 1993; Geiduscheck and Kassavetis 1995; Zawel and Reinberg 1995).
The first and second class promoters are intragenic and TATA boxless, and could be exemplified by the 5S RNA and tRNA gene promoters. In both classes of promoters, the binding of TFIIIC is followed by the recruitment of TFIIIB that can directly contact RNAP III and initiates several rounds of transcription (Hernandez 1993; Geiduscheck and Kassavetis 1995; Zawel and Reinberg 1995).
The U6 snRNA exemplifies the third class of RNAP III transcribed genes. The yeast SNR6 promoter contains a canonical TATA box at −30, an internal degenerate box A at +21 and a downstream box B at 202 bp from box A (Brow and Guthrie 1990). SNR6 is an essential gene coding for a nontranslated small RNA involved in RNA splicing in yeast and human cells (Brow and Guthrie 1988, 1990). A 2-bp deletion at the B box dramatically inhibits the SNR6 transcription and alters the nucleosome arrangement in the flanking regions (Marsolier et al. 1995).
The relationship between excision repair of RNAP III genes and their transcription has not been explored in depth. It was stated in a recent report that, in human cells, the transcription by RNAP III of tRNASec and tRNAVal genes is uncoupled to NER (Dammann and Pfeifer 1996). In S. cerevisiae, however, the tRNA suppressor gene SUP4-o showed a preferential mutation induction occurring at sites in which the dipymidine was on the TS (Armstrong and Kunz 1990). This might be explained by a possible transcription of the NTS by RNAP II from a cryptic promoter within the plasmid vector used in that study (Armstrong and Kunz 1995). Alternatively, the strand preferential mutagenesis could imply a repair strand bias in the S. cerevisaie RNAP III transcribed genes. To investigate this question, a detailed analysis is required.
In this study, a high-resolution technique was used to investigate the effect of transcription on the repair of UV-induced DNA damage in two different RNAP III-transcribed genes, SUP4 and SNR6. We report a preferential repair of the NTS of the SNR6 gene by both NER and PR. This strand bias of NER and PR was abolished by transcriptional inactivation of the SNR6 gene, showing the contribution of the transcription by RNAP III in this strand-specific repair. Moreover, the same strand bias was observed by analysis of NER in the SUP4 gene. Surprisingly, the nucleotide excision repair in the SUP4 intragenic promoter element box A was not strand specific. These results provide important insight into the mechanisms relating DNA repair to transcription.
Results
DNA repair analysis at nucleotide resolution of UV-induced DNA damage
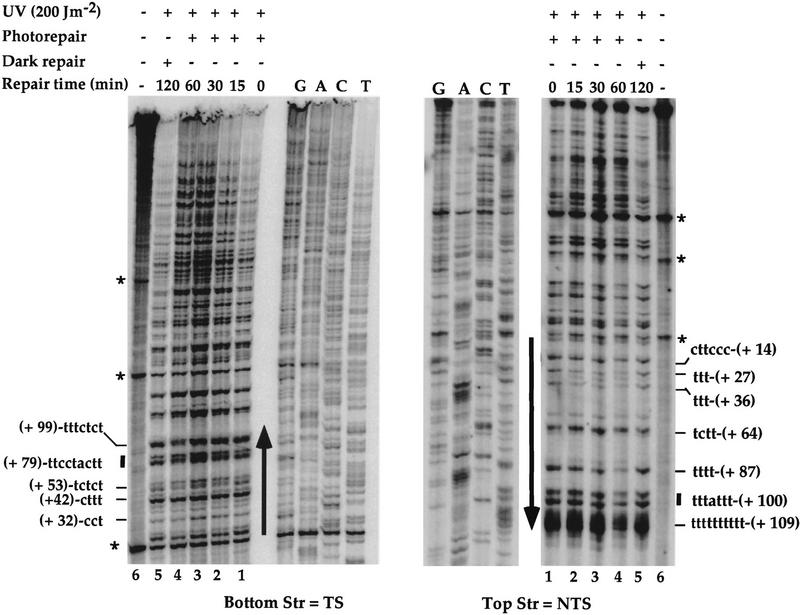
In this study, a primer extension assay was used to investigate the repair of photodimers formed in yeast genomic DNA. Yeast cells were UV-irradiated in suspension in water with 200 J/m2. At this UV dose, ∼0.3 PD is formed in each kilobase and up to 40% of the cells survive. The irradiated cells were then reincubated for different repair times, either in a growth medium, under yellow light at 30°C to allow nucleotide excision repair (dark repair, from 0 to 4 hr), or in water under the photoreactivating light (predominantly at 366 nm) (from 0 to 1 hr). The UV-damaged and repaired genomic DNA were purified, cut with EcoRI, denatured, and annealed to appropriate radiolabeled primers. PDs were then mapped by primer extension with Taq polymerase. Efficient blockage of Taq polymerase elongation occurs almost exclusively at PDs, producing radiolabeled DNA fragments of different sizes (Wellinger and Thoma 1996). Once separated on a polyacrylamide gel, these fragments give rise to different bands representing the PD positions. The intensities of these bands at the repair time (0 hr) correspond to the frequency of PD formation at particular sites. The repair is visualized by a time-dependent decrease in the intensities of these different bands (Fig. 1A, lanes 1–5). Genomic DNA purified from nonirradiated cells was used both for DNA sequencing by use of the same primers, allowing a precise localization of PDs, and as a control for nonspecific Taq polymerase blockage (Fig. 1A, lane 6). This sensitive and direct technique is apropos for investigating the PD formation and repair at high resolution in any region of S. cerevisiae genomic DNA, in particular, when the region to be analyzed is short.
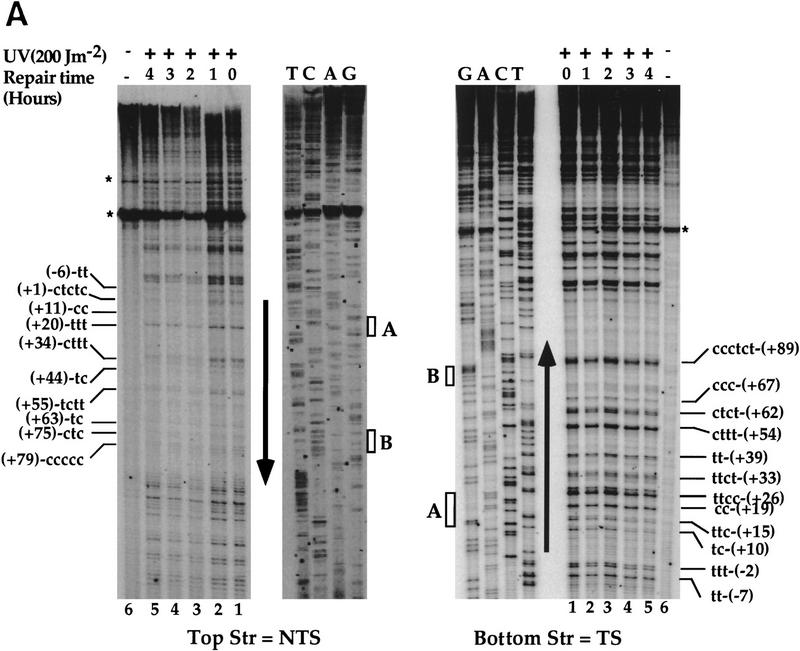
Figure 1.
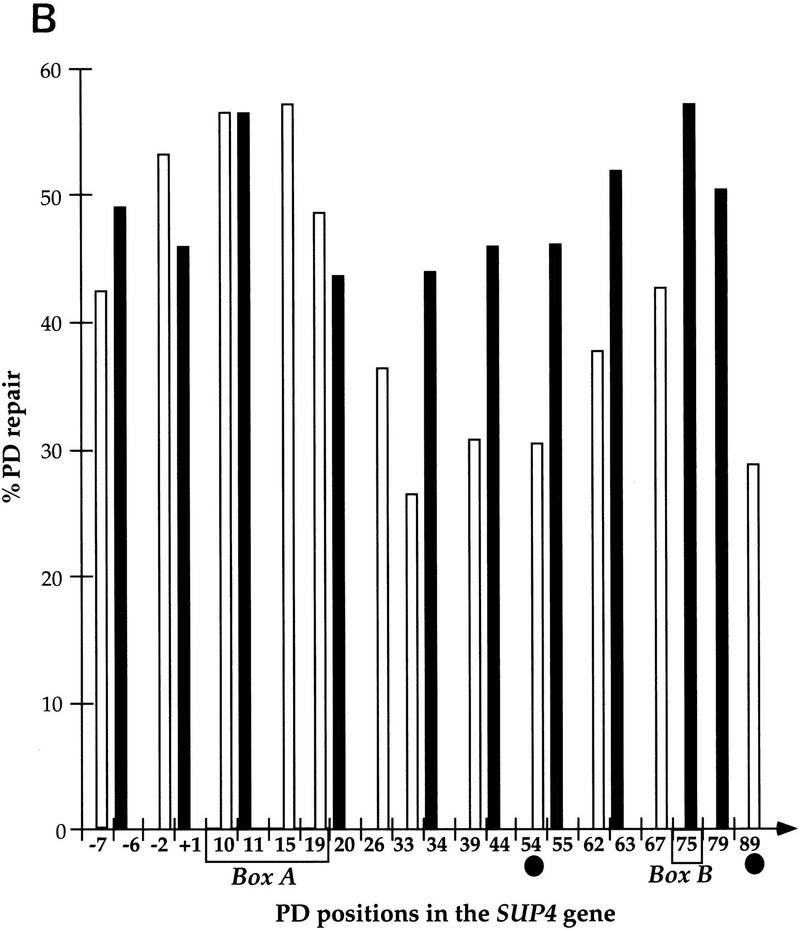
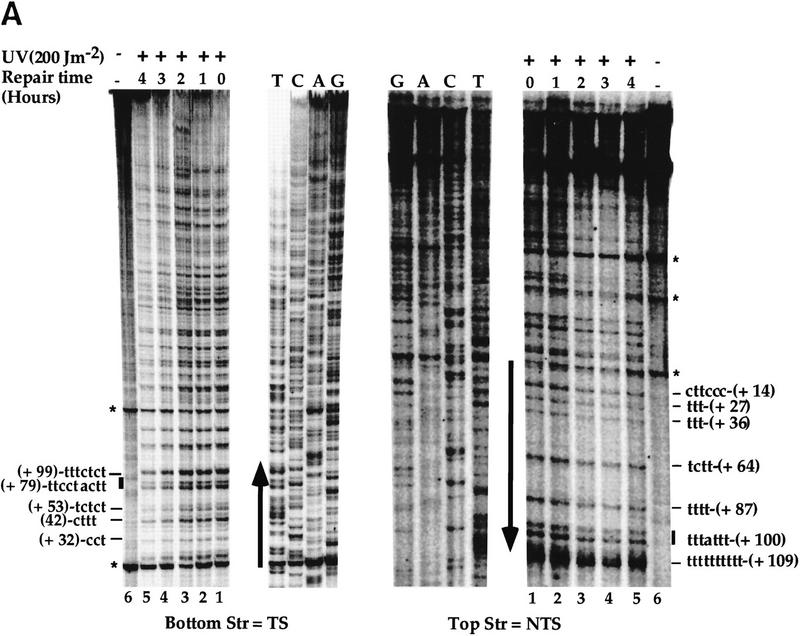
High-resolution analysis of nucleotide excision repair in the SUP4 gene. Yeast cells were UV-irradiated with 200 J/m2 and reincubated under yellow light for the indicated repair times. PD repair was analyzed by primer extension. (A) Primer extension products in the TSs and NTSs. UV-irradiated DNA (lanes 1–5), nonirradiated DNA (lane 6), DNA sequencing (lanes T,C,A,G). The letters on the left and the right sides represent pyrimidine clusters; the numbers refer to the 5′ nucleotide of the pyrimidine cluster in the SUP4 gene sequence (Knapp et al. 1978). Asterisks indicate nonspecific Taq polymerase arrests; (arrows) transcribed part; (boxes) the intragenic promoter elements box A and B. The top strand is NTS; the bottom strand is TS. (B) Quantitative analysis of PD removal from SUP4 TSs and NTSs. The fraction of PDs (%) removed after 4 hr from the TS (open bars) and NTS (solid bars). The numbers on the x-axis represent the positions of PD clusters. (•) Hot spot of mutagenesis determined previously (Armstrong and Kunz 1990). The arrow indicates the direction of transcription. The data are averages of two experiments.
The NTS of the SUP4 gene is preferentially repaired by NER, but only outside of the intragenic promoter element box A
To investigate nucleotide excision repair in RNAP III genes, the strain FTY113 was UV-irradiated and reincubated at 30°C for dark repair, and the genomic DNA was isolated at different repair times. The SUP4 gene, coding for a tRNATyr was chosen for this aim because it is a well-studied RNAP III gene. SUP4 contains the intragenic promoter elements A and B but lacks a TATA box (Knapp et al. 1978). DNA repair of the SUP4 gene was then investigated by primer extension. A detailed analysis of the autoradiographs indicated a generally faster removal of the lesions formed in the NTS (Fig. 1A, top strand) compared with those of the TS (Fig. 1A, bottom strand). Figure 1B shows quantitative results after 4 hr of dark repair. The repair of the lesions formed in the NTS seems more homogeneous, with repair levels of ∼50%. On the other hand, the excision repair on the TS appears more heterogeneous, depending on the position of the lesions in the gene (Fig. 1B). In box A, the lesions were removed with a rate similar to that of the lesions formed in the NTS, and in the upstream nontranscribed promoter region, ∼50% in 4 hr. In contrast, the photodimers formed in the TS, outside of box A, were more slowly repaired around 35% (ranging from 28% to 42%). Thus, in this part of the gene, and unlike RNAP II genes, NER seems to be preferential for the NTS. These results show a site- and strand-specific repair in concordance with the high and site-specific mutagenesis found in the TS of the SUP4-o gene (Armstrong and Kunz 1990).
The NTS of the SNR6 gene is preferentially repaired by NER
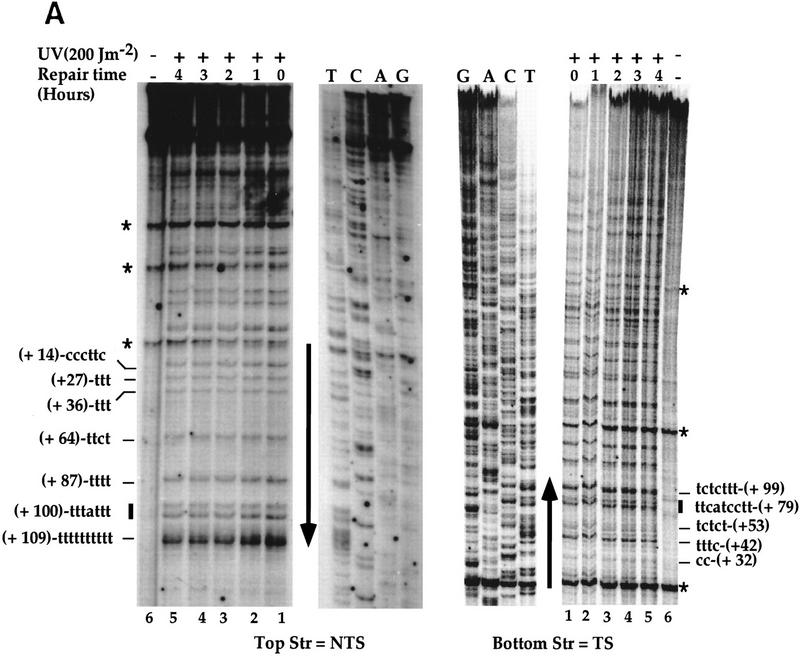
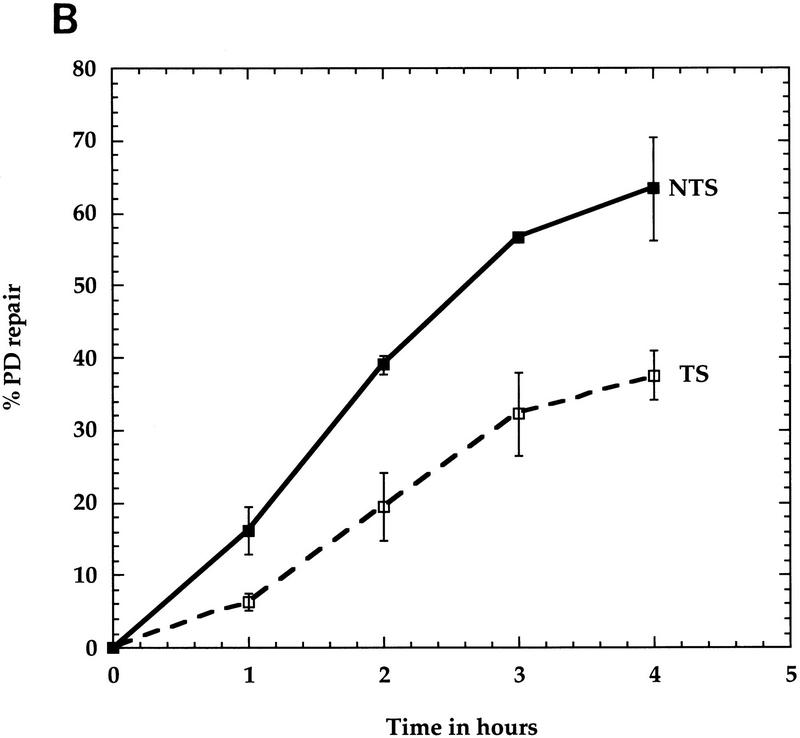
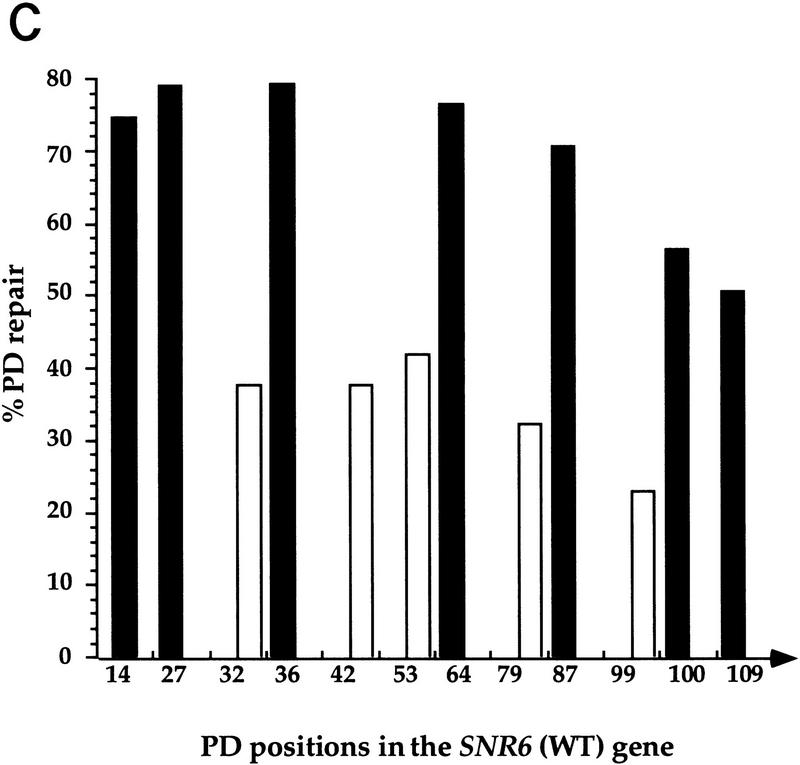
The NTS-specific repair observed in the SUP4 gene was unexpected based on the well-established preferential repair of RNAP II TSs. To test whether this observation could be extended to other types of RNAP III transcribed genes, NER was investigated in the SNR6 gene. Figure 2A indicates a faster decrease in the intensities of the bands in the NTS (top strand) compared with those in the TS (bottom strand). This observation was substantiated by PhosphorImager quantification of the different bands of these gels, taking into account the loading differences (see Materials and Methods). DNA repair is presented in Figure 2B as repair averages of all the PDs removed from the TS and NTS, respectively. Figure 2B shows a more efficient excision repair of the NTS. During 2 hr of dark repair, ∼40% of the PDs were removed from the NTS, whereas only 20% was removed from the TS. After 4 hr of repair, ∼70% of the lesions were excised from the NTS, but only 35% were removed from the TS. Compared with the SUP4 gene, a slightly higher repair was noticed in the NTS of the SNR6 gene. Site-specific repair is shown for 4 hr (Fig. 2C). The repair rates at individual sites in each individual strand were similar, with a slight decrease toward the 3′ end of the gene in both strands (Fig. 2C). This figure also shows that the repair strand bias concerns all the PDs formed along the SNR6 gene. The SNR6 and SUP4 results together show that the preferential repair of the NTS is not gene specific, suggesting that the reduced repair rate in the TSs could be general for the RNAP III transcribed genes. It implies a role of RNAP III transcription in this phenomenon.
Figure 2.
High-resolution analysis of nucleotide excision repair in the SNR6 gene. DNA was used as in Fig. 1. (A) Primer extension reactions on the TSs and NTSs. The legends are as in Fig. 1A. Indicated are pyrimidine clusters (the numbers refer to the 5′ nucleotide in the SNR6 gene; Marsolier et al. 1995); pyrimidine clusters separated by a purine (solid bars); nonspecific Taq polymerase arrests (asterisks); the transcribed part (arrow). The top strand is NTS; the bottom strand is TS. (B) Quantitative analysis of PD removal in the TSs and NTSs. Represented is the fraction of PD (%) removed from each strand at each repair time. Each data point corresponds to an averaged value of all the PDs removed from the transcribed part of the gene in the TS (□) and NTS (▪), respectively. Averages with standard deviations of three experiments are shown for NTS (line) and TS (broken line). (C) Site-specific removal of PD after 4 hr of NER is shown. The numbers on the x-axis represent the PD positions in the SNR6 gene. (Open bars) TS; (solid bars) NTS. The arrow indicates the direction of transcription.
The preferential repair of the NTS in the SNR6 is dependent on RNAP III transcription
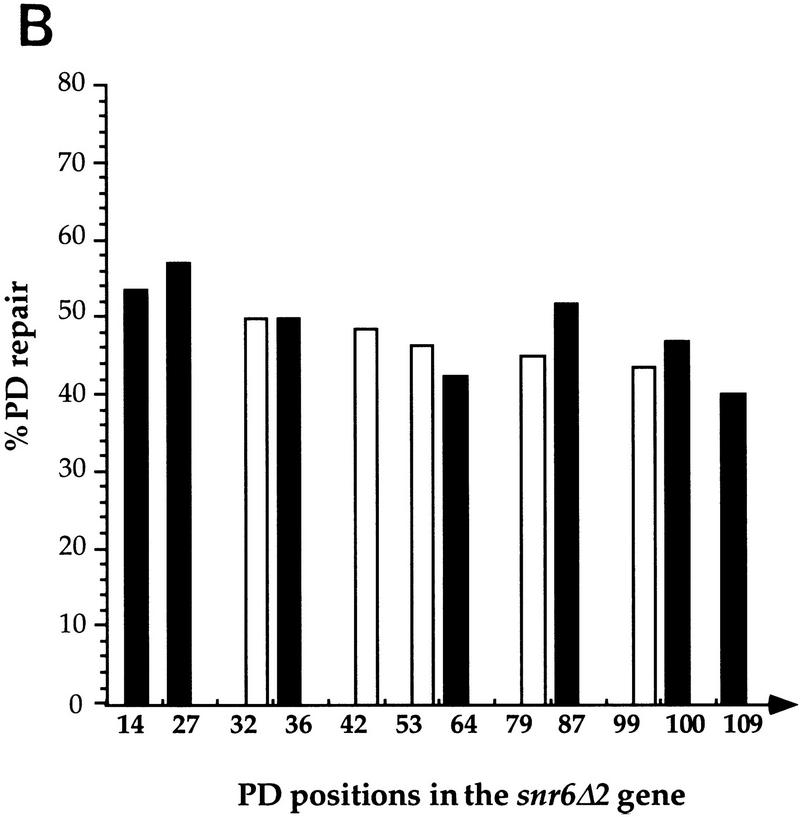
To test whether the slow repair of the SNR6 TS is the result of transcription by RNAP III, repair was analyzed in the FTY115 strain in which the transcription of the SNR6 gene was abolished by a 2-bp deletion in the box B (snr6Δ2). Because the SNR6 gene is essential for cell survival, the FTY115 cells contain a plasmid bearing a wild-type copy of the gene (Marsolier et al. 1995). Primers that allow for analysis of the genomic snr6Δ2 mutant were used (Marsolier et al. 1995). In the Δ2 mutant, the PDs seem to be removed with similar kinetics from both snr6Δ2 strands (Fig. 3A). Quantitative results showed that ∼50% of the lesions were excised after 4 hr of repair (Fig. 3B). The lesions formed in the TS were repaired more efficiently in the snr6Δ2 mutant (50%), in which SNR6 is transcriptionally inactive (Fig. 3B), than in the wild-type cells (35%; Fig. 2C). In both strands of the nontranscribed snr6Δ2 gene, the repair rate was intermediate between the repair rates of the TS and NTSs of the wild-type SNR6 gene (Fig. 3B). This indicates that the inactivation of RNAP III transcription abolished the difference in repair rates observed between the TSs and NTS of the SNR6 gene. These data show a role of the transcription by RNAP III in the repair strand bias observed in the wild-type SNR6 gene and eliminate any hypothetical effect of DNA sequence in this phenomenon.
Figure 3.
Repair analysis of pyrimidine dimers formed in the transcriptionally silent snr6Δ2 gene. snr6Δ2 mutant cells were treated as described in Fig. 1. (A) The same primers were used for primer extension in the top strand (NTS) and in the bottom strand (TS). (B) PD repair after 4 hr. (Open bars) TS; (solid bars) NTS. (See legend to Fig. 1 for details.)
RAD1 gene deletion abolishes the repair of the SNR6 gene
To see whether the repair inhibition observed in the TS of SNR6 concerns the NER process or another DNA metabolism process, the RAD1 gene that codes for a component of the NER 5′ endonuclease Rad1p/Rad10p (Wood 1996) was deleted in the FTY113 and FTY115 strains constructing the strains AAY1 and AAY2, respectively. The rad1Δ cells were UV-irradiated and reincubated for repair in the dark. The damaged genomic DNA was purified and the repair in the SNR6 gene was analyzed by primer extension. The results presented in Figures 4 and 5 show that PDs formed in both strands of the SNR6 gene persisted during the 2 hr of incubation (Fig. 4 and 5, cf. lanes 1 and 5). The quantitative analysis of these gels confirmed that <10% of the dimers were removed from each strand of the gene (Fig. 4B). A similar result was obtained after 4 hr of incubation (data not shown). This shows that the repair of the RNAP III SNR6 gene is RAD1-dependent, and thereby, the observed preferential repair of the NTS was carried out by NER.
Figure 4.
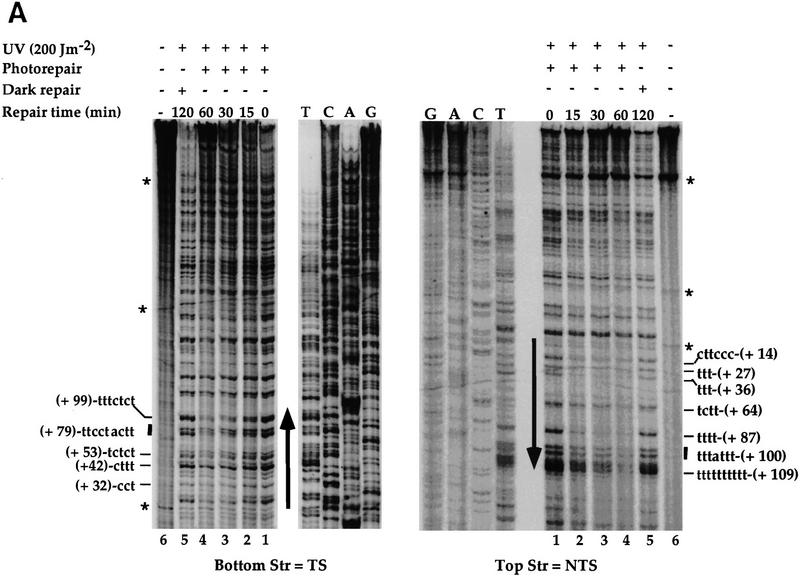
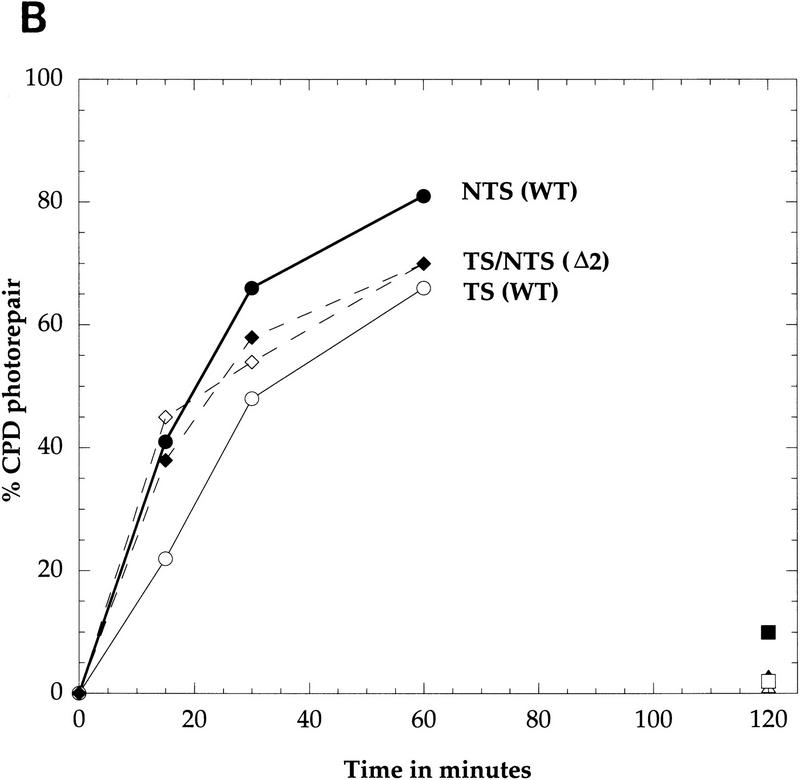
Repair of the SNR6 gene by photolyase. AAY1 (rad1Δ) cells were UV-irradiated with 200 J/m2 and exposed to photoreactivating light for different times as indicated (lanes 1–4). As a control, an aliquot was kept in the dark for 2 hr (lane 5) and another aliquot was not irradiated (lane 6). DNA was extracted, and CPD repair was analyzed by primer extension. (A) The legends are as for Fig. 1A. (B) Quantitative analysis of CPD repair in SNR6 and snr6Δ2. The CPD fraction (%) repaired at each time was determined as a repair average of all the CPD formed in the TS and in the NTS of SNR6 (line) and snr6Δ2 (broken line); values are averages of two primer extension experiments. TS (Open symbols); NTS (solid symbols); PR (diamonds and circles); NER of Snr6Δ2 (squares); NER of SNR6 (triangles); triangles and squares overlap.
Figure 5.
Analysis of the PR in the transcriptionally silent snr6Δ2 gene. AAY2 (rad1Δ, snr6Δ2) was treated as the AAY1, and the repair experiment was performed as described in Fig. 4A (A).
The NTS of the SNR6 gene is preferentially repaired by photolyase
It was discovered previously that photolyase preferentially repairs the NTS of transcribed RNAP II genes, whereas the TS is slowly repaired (Suter et al. 1997). To study the photorepair of a RNAP III gene and to see whether the preferential repair of the SNR6 NTS is confined to the NER mechanism or can be extended to other DNA repair processes, the PR process was studied in the SNR6 gene. AAY1 strain (rad1Δ) in which NER was abolished by deletion of the RAD1 gene was used. The cells were UV-irradiated in suspension in water at a dose of 200 J/m2 and then exposed to the photoreactivating light for 15, 30, and 60 min. As a control, one aliquot of UV-irradiated cells was incubated in the dark. DNA was isolated, treated as described previously, and the PD repair was analyzed by primer extension. The results presented in Figure 4 reveal a preferential photorepair of the NTS. As for NER, most of the CPDs formed in the NTS were more rapidly repaired than those formed in the TS. In the dark, no repair was detected (Fig. 4A, lane 5), indicating that the repair observed is light dependent. The quantitative analysis of DNA repair is represented in Figure 4B. Each data point represents an average of all the CPDs formed in each strand, respectively. During the first 15 min of repair, ∼20% of CPDs were repaired from the TS, whereas up to 40% were photoreversed from the NTS. After 60 min of repair, >80% of CPDs were repaired from the NTS, but only ∼65% were repaired from the TS. Hence, PR, as well as NER, are both slower in the TS of the SNR6 gene. Because the PR is a very rapid process (Suter et al. 1997), the repair difference observed is more pronounced during the first 15 min. In addition, the results show that photolyase has the same strand bias in RNAP II (Suter et al. 1997) and RNAP III genes.
The NTS-specific photorepair is dependent on transcription by RNAP III
To investigate the relationships between photorepair and RNAP III transcription, the AAY2 strain (rad1Δ, snr6Δ2) in which the genomic snr6Δ2 is transcriptionally silent, was used. AAY2 cells were UV-irradiated under the same conditions as the AAY1 cells, and reincubated for photorepair from 0 to 60 min. The time course analysis of the PR presented on Figures 4B and 5 show that both snr6Δ2 strands are repaired with similar rates. Approximately 40% of CPDs were photoreactivated from both strands of the gene after 15 min under the photoreactivating light. Thus, the photorepair rate of both strands was similar to that obtained for the NTS of the transcriptionally active SNR6, but two-fold higher than that of the TS (Fig. 4B). This shows that the photorepair of the TS is more efficient in the absence of transcription. After 60 min of repair, ∼70% of CPDs were photoreversed in both strands of the silent snr6Δ2 gene. For the samples incubated in the dark, <10% of the lesions were repaired (Fig. 4B). This result shows that the photorepair-strand bias observed in the wild-type SNR6 gene is abolished when the transcription is inactivated by a mutation, indicating that the strand bias of PR in the wild-type SNR6 gene is dependent on transcription by RNAP III.
Discussion
The NTSs of RNAP III genes are preferentially repaired by NER and PR
It is shown in this study that in yeast S. cerevisiae and in contrast to RNAP II transcribed genes, the RNAP III TSs are more slowly repaired by NER and PR than the NTSs. This phenomenon was discovered in two genes belonging to different RNAP III subclasses, SNR6 and SUP4 (Figs. 1 and 2). These results present the first example of a preferential repair of the NTS by NER. Hence, two different repair pathways show preferential repair of the NTS of a gene transcribed by RNAP III. What could be the cause of this phenomenon?
The NTS-specific repair is RNAP III transcription dependent
Because the strand bias for NER has been shown in two different genes, it can be excluded that the strand-specific repair results from a sequence difference between the two strands of these genes. Moreover, the results presented in Figures 3, 4, and 5 showed that the inhibition of SNR6 transcription led to similar excision repair and PR efficiencies in the TSs and NTSs of this gene. These data constitute a solid indication toward a role of transcription by RNAP III in the repair-strand bias, and rule out the dependency of this phenomenon on DNA sequence. A plausible explanation for this preferential repair of the NTS is that the RNAP III, like the RNAP II, could be arrested in vivo by a UV-induced DNA damage in the TS (Donahue et al. 1994; Selby et al. 1997). The arrested RNAP III may cover the lesion, delaying the repair processes either by NER or by PR in the TS. For the NER process, this hypothesis implies that RNAP III is not connected to the NER machinery neither by TFIIH [which is not required for RNAP III transcription (Geiduscheck and Kassavetis 1995)] nor by strand-specific repair factors. For RNAP II genes, Rad26p and CSA/CSB are good candidates for having the role of the E. coli TRCF, respectively, in S. cerevisiae and human cells (Friedberg 1996b)—the role being to accelerate the excision repair of the TS, which allows the RNAP II to resume transcription rapidly. In the absence of these links between transcription and repair, even RNAP II TSs became repaired slowly. It was shown recently that PR preferentially repairs the NTSs of the three RNAP II genes, URA3, HIS3, and GAL10 (Livingstone-Zatchej et al. 1997; Suter et al. 1997). This strand bias is dependent on RNAP II transcription (Livingstone-Zatchej et al. 1997). Moreover, RNAP I (Vos and Wauthier 1991; Christians and Hanawalt 1993; Fritz and Smerdon 1995) and RNAP III (Dammann and Pfeifer 1996) transcription in mammalian cells was shown not to be connected to NER. These results point out the fact that the relationships between DNA repair and transcription seem to be confined to NER and RNAP II transcription. This emphasizes an important role of TFIIH in this connection, because it is the only RNAP II transcription factor that is essential for NER as well (Friedberg 1996b). The difference between yeast and mammalian cells may be the result of the behavior of the stalled RNAP III at the lesion. It is possible that the S. cerevisiae RNAP III ternary complex is more stable than the mammalian RNAP I or RNAP III ternary complexes, leading to a strong protection of the lesions and, thereby, to the repair obstruction in the TS.
The disappearance of the repair strand bias in the snr6Δ2 mutant was accompanied by a slight diminution of DNA repair rate at the NTS and a slight increase of repair efficiency in the TS, either by PR or by NER (Figs. 3B and 4B). If we assume that slow repair of the TS is caused by blocked polymerases, then a release of polymerases by gene inactivation is expected to yield high repair rates equivalent to that of the NTS of the wild-type SNR6 gene, which was not observed. The intermediate levels of repair determined in snr6Δ2 may therefore be explained by the presence of nucleosomes and other DNA-binding factors in the snr6Δ2 gene. These nucleosomes may not be positioned, because they were not detected in the snr6Δ2 in a previous study (Marsolier et al. 1995).
DNA repair and transcription: RNAP II and RNAP III systems
A major question that arises from these data is why the excision repair of the TS of RNAP III genes is not enhanced as for RNAP II genes. Several reasons could explain the difference between RNAP II and RNAP III transcription regarding their relationships with DNA repair. First, RNAP III genes are very short, thus less frequently hit by DNA-damaging agents than the much longer RNAP II genes. Second, most of the RNAP III genes are multicopy (Percudani et al. 1997). Hence, very high doses of damaging agents would be required to inactivate all of them in the same time. Third, the half-life of RNAP III transcripts is rather long, therefore, cells may survive a long period without transcription of the damaged gene.
The NER is not strand-specific at the SUP4 intragenic box A
No NER strand bias was observed in the SUP4 intragenic promoter element box A, because the lesions of both strands were repaired with similar rates (50% in 4 hr). This element is important for an accurate initiation of transcription, because it is one of the binding sites of TFIIIC complex (Geiduscheck and Kassavetis 1995). Unlike RNAP II promoters, this region is transcribed, and that raises an interesting question as to why TS and NTS are repaired with similar rates. If we admit that the repair strand bias is the result of transcription, it is reasonable to assume that the absence of this strand bias is caused by the inhibition of RNAP III transcription. This inhibition could result from a very weak or no binding of TFIIIC on the UV-damaged box A element. The TFIIIC–box A complex is essential for transcription initiation, because it is TFIIIC that directs TFIIIB/RNAP III to the transcription start site (Geiduscheck and Kassavetis 1995; Zawel and Reinberg 1995). In a recent report, Tommasi et al. (1996) have shown that UV-induced PDs can inhibit the binding of some transcription factors (not TFIIIC) to their cognate elements in vitro.
Mutagenesis is linked to nucleotide excision repair
It was reported previously that the mutagenesis frequency in the SUP4-o gene is higher in the TS than in the NTS (Armstrong and Kunz 1990, 1992). This higher mutagenesis could be explained by a lower DNA repair in the TS. The results presented in this paper support this hypothesis. In the box A region in which the mutagenesis is very low, the excision repair efficacy is the highest in this strand (Fig. 1B). Outside of this promoter region, however, in which slower repair was found, several mutagenesis hot spots were detected as in the pyrimidine clusters CTTT and CCCTCT at positions 54 and 89, respectively (Armstrong and Kunz 1990). Another example of higher mutagenesis in the TS was observed previously in mfd-deficient E. coli lacking the TCR process (Oller et al. 1992). These results are in contrast with those of the TSs of RNAP II genes that are preferentially repaired and, consequently, show a lower mutagenesis as compared with the NTSs (Vrieling et al. 1989; McGregor et al. 1991; Sage et al. 1993). In conclusion, these data establish a direct link between NER and mutagenesis, and indicate that RNAP III transcription is not coupled to NER and that, in opposition to RNAP II genes, it is the NTSs of RNAP III genes that are preferentially repaired.
Materials and methods
Yeast strains
FTY113: MATα, ade2-102, ura3-52, lys2-801, hisΔ200, leuΔ1, trp1Δ63/pRS314+U6 TRP1, SNR6; FTY115: MATα, ade2-102, ura3-52, lys2-801, hisΔ200, leuΔ1, trp1Δ63/pRS314+U6 TRP1, snr6Δ2 (Marsolier et al. 1995). AAY1 and AAY2 are rad1Δ deletion strains derived from FTY113 and FTY115, respectively. RAD1 deletion was generated by gene replacement technique (Rothstein 1983).
Culture and UV irradiation of yeast cells
Three liters of yeast cells were grown at 30°C in minimal medium [2% dextrose, 0.67 yeast nitrogene base without amino acids (Difco)] (Sherman et al. 1986) supplemented with the appropriate amino acids to a final density of ∼107 cells/ml. The cells were then collected by centrifugation and resuspended in 1 liter water to a concentration of 1.5 × 107 to 3 × 107 cells/ml. Two hundred fifty-milliliter aliquots were transferred to a 22 × 31.5-cm plastic tray and UV-irradiated at room temperature with a dose of 200 J/m2, with four Sylvania G15T8 germicidal lamps (predominantly 254 nm) at 1 mW/cm2 (measured with a UVX radiometer UVP Inc., San Gabriel, CA).
PR
After UV irradiation, samples of 250–500 ml were photoreactivated in water with Sylvania type F15 T8/BLB bulbs (emission peak at 366 nm) at 1.5 mW/cm2 for 15–60 min. Two hundred fifty-milliliters of cells were collected and chilled in ice.
Dark repair
After UV irradiation, minimal medium supplemented with the appropriate amino acids was added, and the cells were incubated at 30°C (in the dark) or under yellow light (Sylvania GE Gold fluorescent light) for various repair times. Repair was arrested by collecting cells (by centrifugation) and chilling them immediately in ice. All postirradiation steps were done in yellow light to prevent PR.
DNA preparation and enzyme digestions
Genomic DNA preparation was carried out with Qiagen tips and protocols (QIAGEN Genomic DNA Handbook). Pellets containing 1.5 × 109 to 3 × 109 cells were resuspended in 12 ml of buffer Y1 (1 m sorbitol, 0.1 m EDTA, 14 mm βMEtOH). One milligram per milliliter of Zymolyase was added (100,000 U/gram; Seikagaku Kogyo Co., Tokyo, Japan) and the cells were incubated at 30°C for ∼30 min. Spheroplasts were harvested by centrifugation and resuspended in 15 ml of buffer G2 (800 mm GuHCL, 30 mm EDTA, 30 mm Tris, 5% Tween-20, 0.5% Triton X-100 at pH 8.0) supplemented with 100 ml of proteinase K (10 mg/ml) and 100 ml RNase A (10 mg/ml). The suspension was then incubated for 2 hr at 60°C. Following the cell lysis, the cellular debris was spun down at 10,000 rpm at 4°C, and the supernatant was loaded on a pre-equilibrated Qiagen genomic tip for DNA purification. Pelleted by centrifugation at 10,000 rpm, DNA was dissolved in 200 ml of TE at pH 8.0. DNA was then digested to completion with EcoRI (Böhringer Mannheim), precipitated with ethanol, and redissolved in the same volume of TE (pH 8).
Primer extension analysis
Primer labeling and primer extension were done as described in Wellinger and Thoma (1996) with some modifications. The 10-pmole primer was 5′ end-labeled with T4 nucleotide kinase (Biolabs) in the presence of 10 pmoles of radioactive [γ-32P]ATP. The mixture was incubated for 30 min at 37°C, the primer was then separated from the nonincorporated ATP with a G50 Sephadex column (Boehringer) and dissolved in an appropriate volume of TE (pH 8). Approximately 1–5 μg of EcoRI-digested genomic DNA was mixed with (1–5 ng) of radiolabeled primer and was subjected to 30 cycles of repeated denaturation (94°C for 45 sec), annealing (60°C for 4 min, 30 sec) and extension (72°C for 2 min) reactions, with 0.2 unit of Taq DNA polymerase (Perkin Elmer) for each reaction. The reaction products were ethanol precipitated and analyzed on a 4% or 5% polyacrylamide, urea (50%) gel. The gels were then dried on a Whatman DE 81 paper and either autoradiographed (Fuji RX films) or analyzed with PhosphorImager (Molecular Dynamics). DNA sequencing was done in parallel by use of Sanger chain termination method utilizing the same primers. The primers used were PAGE purified. For the SNR6 gene, top strand (no. 716), 5′-CGTACCATTGCATAGCTGTAACAATATTC-3′; bottom strand (no. 717a), 5′-TATATTGCTACCATGACTGTCTGAG-3′. For the SUP4 gene, top strand (no. 844), 5′-GCAATATGTCACAATTTGATAATA-3′; bottom strand (no. 845), 5′-CACTCTGAACCATCTTGGAAGGA-3′. For the SNR6 gene, the primers were chosen to anneal with regions that map outside of the SNR6 fragment present in the plasmid (Marsolier et al. 1995).
Quantifications
The sequencing gels were used to calculate the relative repair of the lesions. First, a volume box was layed around each lesion. The value corresponding to the gel background was subtracted from the measured value. The obtained number was then divided by the value obtained by a volume box that covered the whole lane. This most accurately corrects for loading differences (see Figure 1A; Wellinger and Thoma 1997). The values obtained for the nonirradiated DNA were then subtracted to correct for unspecific background signal and Taq polymerase blockage. For standardization, the corrected values obtained at t = 0 (no repair) were defined as 100% damage.
Acknowledgments
We are grateful to Dr. R.E. Wellinger, Dr. U. Schieferstein, and all the members of the F. Thoma group for their help and interesting discussions. We thank Dr. U. Suter for continous support and Dr. C. Weissmann for access to the PhosphorImager. This work was supported by grants from the Swiss National Science Foundation and by the ETH–Zürich, to (F.T.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL thoma@cell.biol.ethz.ch; FAX 41 1 633 10 69.
References
- Armstrong JD, Kunz BA. Site and strand specificity of UVB mutagenesis in the SUP4-o gene of yeast. Proc Natl Acad Sci. 1990;87:9005–9009. doi: 10.1073/pnas.87.22.9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Excision repair influences the site and strand specificity of sunlight mutagenesis in yeast. Mutat Res. 1992;274:123–133. doi: 10.1016/0921-8777(92)90059-c. [DOI] [PubMed] [Google Scholar]
- ————— Excision repair and gene orientation modulate the strand specificity of UV mutagenesis in a plasmid-borne yeast tRNA gene. Environ Mol Mutagen. 1995;25:12–22. doi: 10.1002/em.2850250104. [DOI] [PubMed] [Google Scholar]
- Bhatia PK, Wang Z, Friedberg EC. DNA repair and transcription. Curr Opin Genet Dev. 1996;6:146–150. doi: 10.1016/s0959-437x(96)80043-8. [DOI] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- ————— Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes & Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- Chalut C, Moncollin V, Egly JM. Transcription by RNA polymerase II: A process linked to DNA repair. BioEssays. 1994;16:651–655. doi: 10.1002/bies.950160910. [DOI] [PubMed] [Google Scholar]
- Christians FC, Hanawalt PC. Lack of transcription-coupled repair in mammalian ribosomal RNA genes. Biochemistry. 1993;32:10512–10518. doi: 10.1021/bi00090a030. [DOI] [PubMed] [Google Scholar]
- Dammann R, Pfeifer GP. Lack of gene- and strand-specific DNA repair in RNA polymerase III-transcribed human tRNA genes. Mol Cell Biol. 1996;17:219–229. doi: 10.1128/mcb.17.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn KJ, Sancar A, Reinberg D. Dual Role of TFIIH in DNA excision repair and in transcription by RNA Polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Bardwell L, Bardwell AJ, Buratowski S, Gulyas KD, Donahue TF, Friedberg EC, Kornberg RD. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- Friedberg EC. Cockaine syndrome-a primary defect in DNA repair, transcription, both or neither. BioEssays. 1996a;18:731–738. doi: 10.1002/bies.950180908. [DOI] [PubMed] [Google Scholar]
- ————— Relationships between DNA repair and transcription. Annu Rev Biochem. 1996b;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, DC.: ASM Press; 1995. [Google Scholar]
- Fritz LK, Smerdon MJ. Repair of UV damage in actively transcribed ribosomal genes. Biochemistry. 1995;34:13117–13124. doi: 10.1021/bi00040a024. [DOI] [PubMed] [Google Scholar]
- Geiduscheck EP, Kassavetis GA. Comparing transcriptional initiation by RNA polymerases I and III. Curr Opin Cell Biol. 1995;7:344–351. doi: 10.1016/0955-0674(95)80089-1. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. DNA repair comes of age. Mutat Res DNA Repair. 1995;336:101–113. doi: 10.1016/0921-8777(94)00061-a. [DOI] [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor. Genes & Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Knapp G, Beckman JS, Johnson PF, Fuhrman SA, Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978;14:221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Kolsch E, Starlinger P. A difference in the photoreactivation of UV-damage between genes in the repressed and genes in the de-repressed state. Z Vererbungsl. 1965;96:304–306. doi: 10.1007/BF00895046. [DOI] [PubMed] [Google Scholar]
- Livingstone-Zatchej M, Meier A, Suter B, Thoma F. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 1997;25:3795–3800. doi: 10.1093/nar/25.19.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier MC, Tanaka S, Livingstone-Zatchej M, Grunstein M, Thoma F, Sentenac A. Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes & Dev. 1995;9:410–422. doi: 10.1101/gad.9.4.410. [DOI] [PubMed] [Google Scholar]
- McGregor WG, Chen RH, Lukash L, Maher VM, McCormick JJ. Cell cycle-dependent strand bias for UV-induced mutations in the transcribed strand of excision repair-proficient human fibroblasts but not in repair-deficient cells. Mol Cell Biol. 1991;11:1927–1934. doi: 10.1128/mcb.11.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oller AR, Fijalkowska IJ, Dunn RL, Schaaper RM. Transcription-repair coupling determines the strandedness of ultraviolet mutagenesis in Escherichia coli. Proc Natl Acad Sci. 1992;89:11036–11040. doi: 10.1073/pnas.89.22.11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S. Transfer RNA gene redandancy and transcriptional selection in Saccharomyces cerevisiae. J Mol Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sage E, Drobetsky EA, Moustacchi E. 8-Methoxypsoralen induced mutations are highly targeted at crosslinkable sites of photoaddition on the non-transcribed strand of a mammalian chromosomal gene. EMBO J. 1993;12:397–402. doi: 10.1002/j.1460-2075.1993.tb05671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. DNA excision repair. Annu Rev Biochem. 1996a;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- ————— No “End of History” for photolyases. Science. 1996b;272:48–49. doi: 10.1126/science.272.5258.48. [DOI] [PubMed] [Google Scholar]
- Sancar GB. DNA photolyases—Physical properties, action mechanism, and roles in dark repair. Mutat Res. 1990;236:147–160. doi: 10.1016/0921-8777(90)90002-m. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- ————— Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: Footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Suter B, Livingstone-Zatchej M, Thoma F. Chromatin structure modulates DNA repair by photolyase. EMBO J. 1997;16:2150–2160. doi: 10.1093/emboj/16.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ, Wang Z, Feaver WJ, Wu X, Bushnell DA, Donahue T, Friedberg EC, Kornberg RD. Different forms of TFIIH for transcription and DNA repair: Holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Swiderski PM, Tu Y, Kaplan BE, Pfeifer GP. Inhibition of transcription factor binding by ultraviolet-induced pyrimidine dimers. Biochemistry. 1996;35:15693–15703. doi: 10.1021/bi962117z. [DOI] [PubMed] [Google Scholar]
- Vos JMH, Wauthier EL. Differential introduction of DNA damage and repair in mammalian genes transcribed by RNA polymerase-I and polymerase-II. Mol Cell Biol. 1991;11:2245–2252. doi: 10.1128/mcb.11.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieling H, Vanrooijen ML, Groen NA, Zdzienicka MZ, Simons JWIM, Lohman PHM, Zeeland AAV. DNA strand specificity for UV induced mutations in mammalian cells. Mol Cell Biol. 1989;9:1277–1283. doi: 10.1128/mcb.9.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Svejstrup JQ, Feaver WJ, Wu X, Kornberg RD, Friedberg EC. Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature. 1994;368:74–76. doi: 10.1038/368074a0. [DOI] [PubMed] [Google Scholar]
- Wellinger RE, Thoma F. Taq DNA polymerase blockage at pyrimidine dimers. Nucleic Acids Res. 1996;24:1578–1579. doi: 10.1093/nar/24.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Nucleosome structure and positioning modulate nucleotide excision repair in the non transcribed strand of an active gene. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- Zawel L, Kumar PP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes & Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]