Abstract
MEF2 is a MADS-box transcription factor required for muscle development in Drosophila. Here, we show that the bHLH transcription factor Twist directly regulates Mef2 expression in adult somatic muscle precursor cells via a 175-bp enhancer located 2245 bp upstream of the transcriptional start site. Within this element, a single evolutionarily conserved E box is essential for enhancer activity. Twist protein can bind to this E box to activate Mef2 transcription, and ectopic expression of twist results in ectopic activation of the wild-type 175-bp enhancer. By use of a temperature-sensitive mutant of twist, we show that activation of Mef2 transcription via this enhancer by Twist is required for normal adult muscle development, and reduction in Twist function results in phenotypes similar to those observed previously in Mef2 mutant adults. The 175-bp enhancer is also active in the embryonic mesoderm, indicating that this enhancer functions at multiple times during development, and its function is dependent on the same conserved E box. In embryos, a reduction in Twist function also strongly reduced Mef2 expression. These findings define a novel transcriptional pathway required for skeletal muscle development and identify Twist as an essential and direct regulator of Mef2 expression in the somatic mesoderm.
Keywords: Twist, MEF2, Drosophila, muscle development, adepithelial cells, imaginal discs
Studies in Drosophila and vertebrates have shown that MADS-box proteins of the myocyte enhancer factor-2 (MEF2) family and basic helix-loop-helix (bHLH) factors act in collaboration to direct differentiation of skeletal muscle (for review, see Molkentin and Olson 1996). Mef2 genes from fruit flies and vertebrates are expressed in precursors of all the muscle lineages early in development, and expression persists as the descendants of these cells differentiate (Edmondson et al. 1994; Lilly et al. 1994; Nguyen et al. 1994). The single Drosophila Mef2 gene is required for differentiation of skeletal, cardiac, and visceral muscles (Bour et al. 1995; Lilly et al. 1995; Ranganayakulu et al. 1995), and in mice, targeted inactivation of mef2c results in defects in heart morphogenesis and differentiation (Lin et al. 1997).
Understanding how these regulatory genes are themselves regulated is a central issue in the field. Because Mef2 genes are expressed in all myogenic lineages over a broad developmental time, multiple separate processes likely regulate Mef2 expression. Analyses of the Drosophila Mef2 5′-flanking region revealed a number of large regulatory regions spanning several kilobases (Lilly et al. 1995; Schulz et al. 1996) that, together, give rise to the complete pattern of Mef2 expression. No direct regulators of any Mef2 genes in somatic muscle, however, have been identified.
The bHLH protein Twist plays an important role in somatic muscle development in Drosophila. twist is expressed in the somatic mesoderm at high levels prior to differentiation (Dunin-Borkowski et al. 1995) and is required for differentiation of the somatic muscles (Baylies and Bate 1996). Although Twist activates a number of genes early in embryogenesis (Ip et al. 1992a,b), and tinman is a target of Twist in the visceral mesoderm (Lee et al. 1997), no Twist targets have been identified in the somatic lineage.
During the larval stage, twist and Mef2 are expressed in cells that give rise to adult somatic muscles (Bate et al. 1991; Ranganayakulu et al. 1995). These cells are specified in the embryo and are identifiable at the end of embryogenesis by their expression of high levels of twist. twist expression is maintained in proliferating adult muscle precursor cells throughout the larval and early pupal stages (Bate et al. 1991). The thoracic myoblasts are coincident with the adepithelial cells (Poodry and Schneiderman 1970) that give rise to the adult thoracic musculature (Reed et al. 1975; Fernandes et al. 1991). As the myoblasts migrate from the discs and begin to fuse and differentiate, twist expression is lost (Fernandes et al. 1991). How either twist or Mef2 are regulated in the adepithelial cells and the functional significance of this expression is unknown.
In this paper, we have analyzed Mef2 regulation in the precursors of the adult somatic muscles. Adepithelial cell expression of Mef2 is controlled by a 175-bp enhancer element 2245-bp upstream of the transcriptional start site. A single evolutionarily conserved E box is essential for enhancer activity, and Twist binds this site to activate Mef2 transcription directly. The enhancer is also active in embryonic somatic myoblasts prior to differentiation, and its activity during embryogenesis is dependent on the same E box required for later expression. To test the role of the enhancer in vivo, we used a temperature-sensitive twist mutant. Reduction in Twist function during the larval stage causes a loss of Mef2 expression in the adult muscle precursor cells, which results in adult muscle phenotypes similar to those observed for Mef2 hypomorphic mutants. Reduction in Twist function during the embryonic stage also affects MEF2 accumulation. These results show a central role for Twist in the development of the somatic mesoderm and identify Twist as the first known regulator of Mef2 expression in the somatic muscle lineage.
Results
Identification of the enhancer required for Mef2 expression in adult muscle precursors
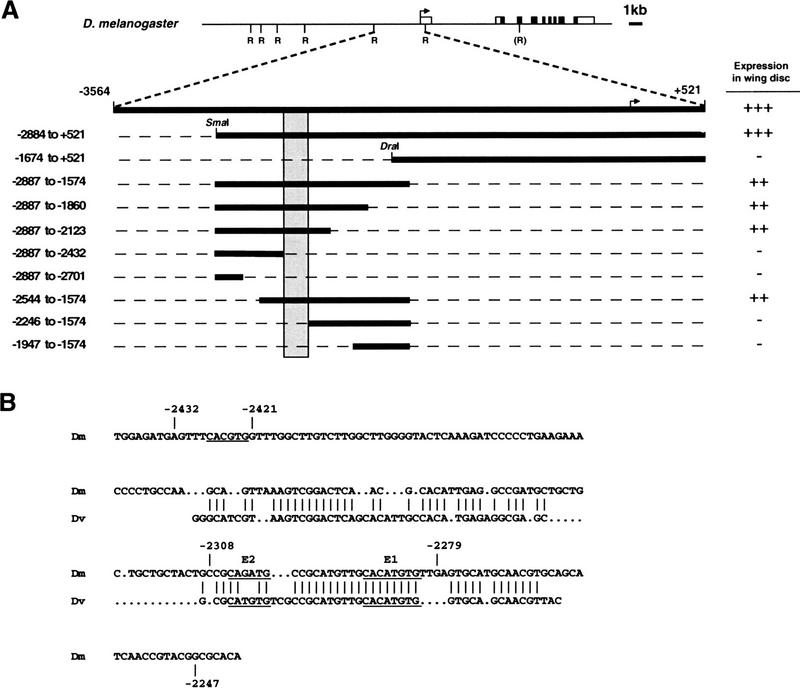
To identify the 5′ regulatory sequences responsible for Mef2 expression in adult muscle precursor cells, transgenic lines carrying different Mef2 enhancer/promoter fragments fused to lacZ were tested for β-galactosidase activity in the adepithelial cells of the wing imaginal disc. Schulz et al. (1996) reported that a 4-kb EcoRI fragment from −3564 to +521 is sufficient for expression in adepithelial cells (see Fig. 1A, top) as well as in other compartments of the embryonic mesoderm (Lilly et al. 1995). We have further scanned EcoRI fragments spanning 12 kb upstream and 10 kb downstream of the transcriptional start site for mesodermal enhancers. The 4-kb fragment was the only region capable of driving lacZ expression in the wing disc adepithelial cells (data not shown).
Figure 1.
Characterization of the Mef2 adepithelial cell enhancer. (A) The top line shows the genomic organization of the D. melanogaster Mef2 locus. Solid and open boxes represent coding and noncoding transcribed regions, respectively. Vertical ticks labeled R show the locations of EcoRI restriction sites (R in parentheses represents a synthetic site; see Schulz et al. 1996). Each of these EcoRI fragments, cloned upstream of the lacZ gene, was tested for expression in the wing disc adepithelial cells. Only the fragment from −3564 to +521 showed expression. Below is an expansion of this ∼4-kb region, within which the indicated fragments were tested. Summaries of their expression in wing imaginal discs are shown at right. The shaded bar denotes a 185-bp region that is required for expression in adepithelial cells. (B) Sequence similarity between D. melanogaster and D. virilis in the 185-bp region. Strong similarity is observed over a 100-bp segment. The D. virilis sequence shown is from −2448 to −2359. (Dm) D. melanogaster sequence; (Dv) D. virilis sequence. Three E boxes are underlined, and E1 and E2 are indicated.
The 4-kb fragment was sequenced, and smaller constructs were tested for activity in adepithelial cells (summarized in Fig. 1A). The adepithelial cell enhancer was initially localized between the SmaI site at −2884 and the DraI site at −1674. A fragment spanning this region (−2887 to −1574) was able to activate transcription of an heterologous promoter in wing imaginal discs.
To further localize the adepithelial cell Mef2 enhancer, deletion constructs within this region were generated and tested for expression in transgenic animals. These constructs identified a 185-bp region from −2431 to −2247, which was required for adepithelial cell expression of lacZ (shaded region in Fig. 1A).
In a second approach, the Mef2 gene from Drosophila virilis was cloned, and a 3.9-kb region upstream of the transcription start site (from −4248 to −358) was sequenced and tested for activity in transgenic larval discs. This fragment drove expression of lacZ in Drosophila melanogaster imaginal discs (data not shown), indicating that the adepithelial cell enhancer was present in this DNA, and that its regulation was conserved between these distantly related species. To identify elements that might be important for expression, the sequence of the 185-bp region from D. melanogaster was compared with the sequence of the 3.9-kb Drosophila virilis fragment. A block of 100 bp within the D. melanogaster 185-bp element was conserved in sequence and position from the transcriptional start site when compared with the D. virilis sequence (Fig. 1B). This suggested that the adepithelial cell enhancer element might lie within these 100 bp.
The 185 bp of D. melanogaster sequence (from −2431 to −2247) contains three E-box sequences (5′-CANNTG-3′), which are potential binding sites for bHLH proteins (Murre et al. 1989). Of these E boxes, two are conserved in position in D. virilis (Fig. 1B). One of these, E2, has a different core sequence, whereas the other, E1, is identical in core and flanking sequences. Note that the E1 sequence (5′-CACATGTG-3′) contains two overlapping E boxes. The third E box at the 5′ end of the 185-bp region, is not conserved in D. virilis.
To determine if this small region was sufficient for adepithelial cell enhancer activity, a 175-bp fragment from −2421 to −2247 was generated from the wild-type sequence of D. melanogaster. This sequence contained E1 and E2, but lacked the third E box because it was not conserved in D. virilis. In transgenic D. melanogaster, this region was capable of driving lacZ expression in the wing discs (Fig. 2A) in a pattern that recapitulated the expression observed for the largest −3564 to +521 construct. β-galactosidase activity was also detected in the haltere and leg imaginal discs, and groups of cells closely apposed to the abdominal body wall musculature, indicating that this enhancer is active in most, if not all, of the adult muscle precursor cells (data not shown).
Figure 2.
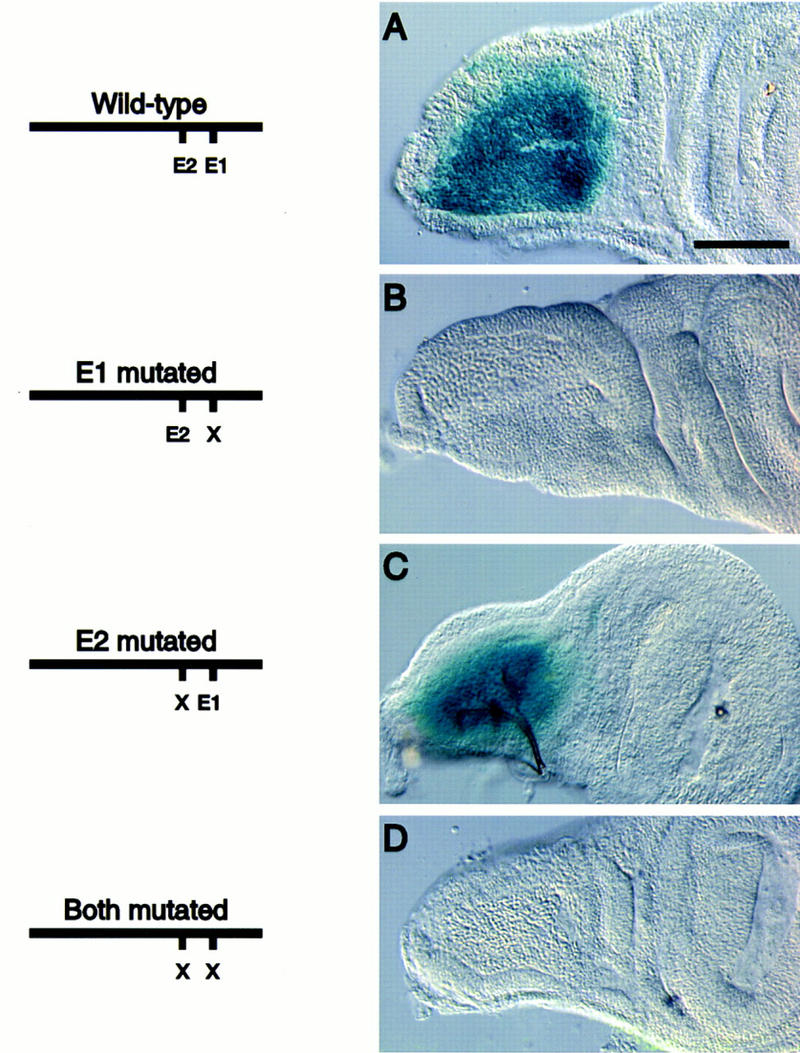
Expression of the 175-bp adepithelial cell enhancer in wing imaginal discs depends on the E1 E box. Transgenic lines carrying enhancer–lacZ constructs were tested for β-galactosidase expression in wing imaginal discs. (A) Wild-type 175-bp enhancer drives high levels of expression in the adepithelial cells; (B) enhancer with E1 mutated has no expression; (C) enhancer with E2 mutated drives lacZ expression at wild-type levels; (D) the enhancer with both E1 and E2 mutated has no activity. Bar, 50 μm.
The two E boxes were mutated singly and in combination to determine if these conserved sequence elements were required for activity of the 175-bp enhancer. Mutation of the most conserved E box, E1, ablated enhancer activity in the imaginal discs, whereas mutation of E2 had no effect upon expression (Fig. 2B,C). Mutation of both E boxes eliminated the activity of the enhancer (Fig. 2D). These results showed that only the most conserved E box (E1) was essential for enhancer activity.
An additional 30-bp construct (from −2308 to −2279) that contained both of the E boxes together with flanking sequence did not drive expression of lacZ in transgenic animals (data not shown), indicating that the E boxes alone are not sufficient for enhancer activity.
twist and Mef2 are coexpressed in adepithelial cells of the wing disc
To determine if twist and Mef2 are expressed in overlapping groups of cells, imaginal discs were reacted with antibodies raised against either Twist or MEF2 protein. Separately labeled discs were then embedded in paraffin and sectioned. Figure 3A shows a whole disc reacted with anti-MEF2 and sections are shown in Figure 3B and C. MEF2 was detected in a specific group of cells near the proximal end of the disc. Most, if not all, of the cells in this region were labeled. Sections of imaginal discs stained with an anti-Twist antibody showed a very similar distribution of signal in most, or all, of these same cells (Fig. 3D). These data indicate that Twist and MEF2 accumulate in overlapping groups of cells, which are the precursors of the adult thoracic muscles (Bate et al. 1991; Fernandes et al. 1991). Although there may be some cells in which only one of the two proteins accumulates, the majority of adepithelial cells express both proteins.
Figure 3.
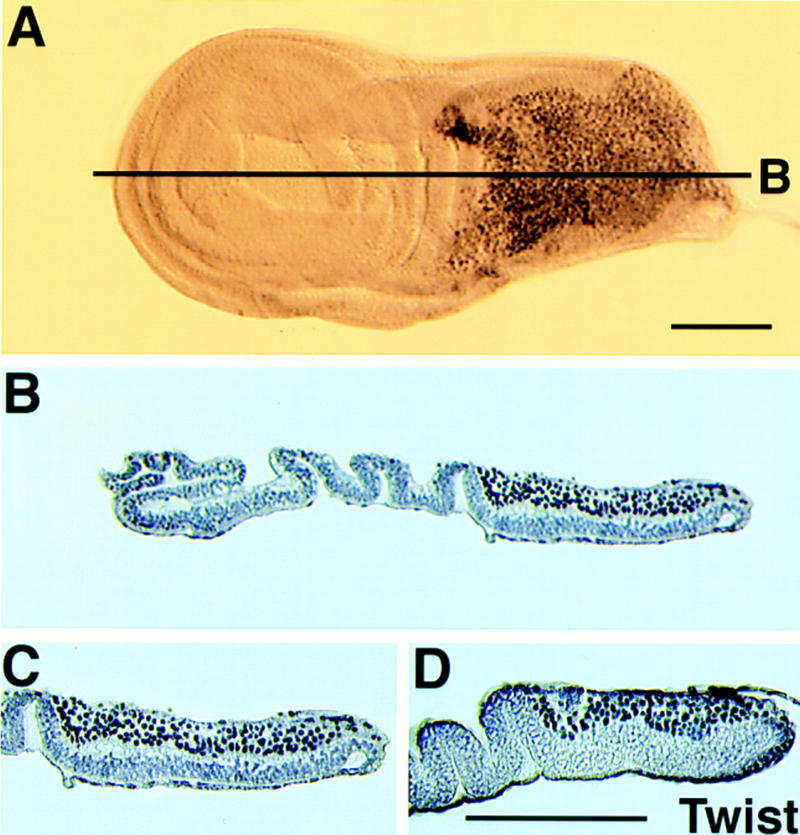
Mef2 and twist are expressed in overlapping domains in wing imaginal discs. Third larval instar wing imaginal discs were reacted with either anti-MEF2 (A) or anti-Twist (not shown) polyclonal antibodies. Discs were then embedded in paraffin and sectioned to localize the immunoreactive cells. Anti-MEF2 staining was localized to the proximal end of the disc (B), and closer observation (C) revealed that it was expressed in most or all of the cells in this region. Twist protein accumulated in a very similar pattern (D), showing that these two proteins are predominantly coexpressed in the adult muscle precursor cells. Bar, 50 μm.
Direct transcriptional activation by Twist through a single E box
To determine if Twist could directly activate the 175-bp enhancer in vivo, Drosophila Schneider line 2 (SL2) cells were cotransfected either with twist expression plasmid or with expression plasmid lacking a cDNA insert, along with the wild-type 175-bp enhancer–lacZ reporter and each of the mutant enhancer–lacZ constructs. Following transfection, cell lysates were assayed for β-galactosidase activity (Fig. 4A). Twist activated the wild-type enhancer several hundredfold. This activation occurred through the E1 E box, because mutation of that E box resulted in a complete loss of activity, whereas mutation of the E2 E box had no effect on enhancer activation.
Figure 4.
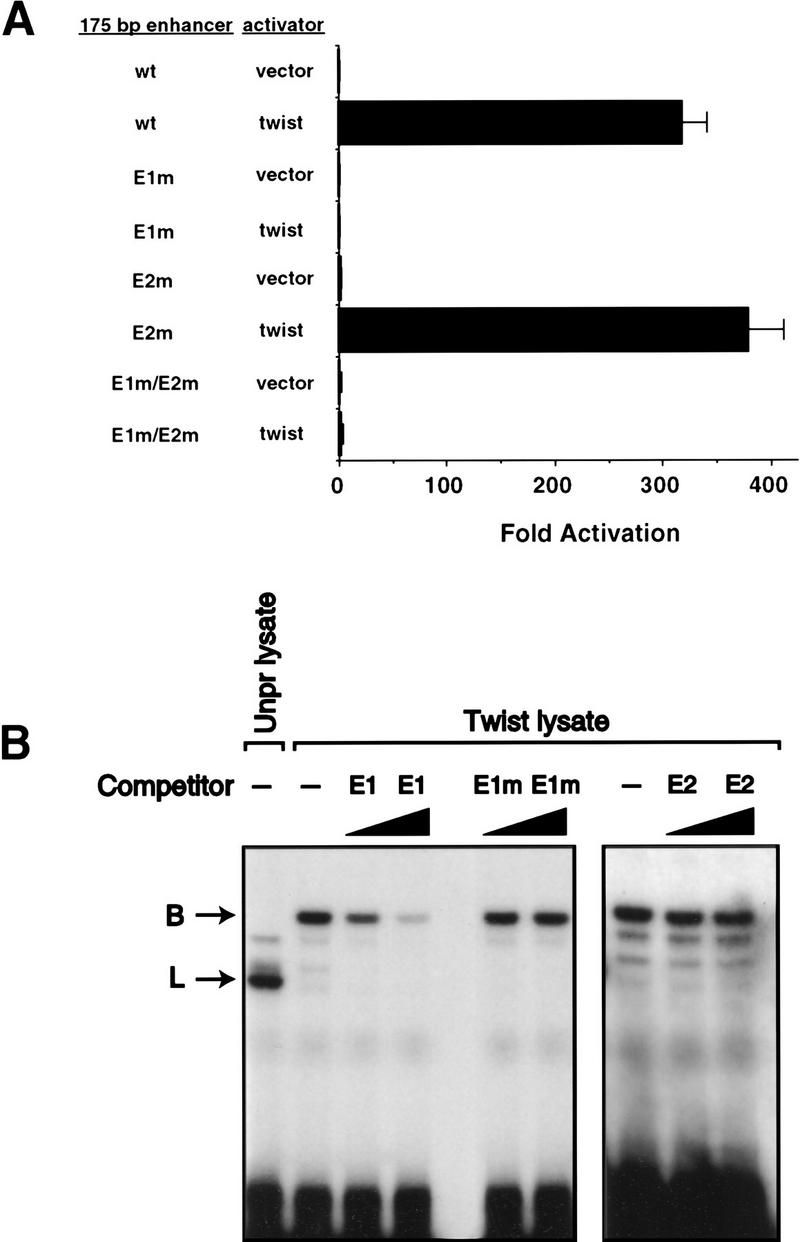
Twist activates the 175-bp enhancer in Drosophila SL2 cells. (A) SL2 cells were transfected with the indicated β-galactosidase reporter plasmids and either twist expression vector or expression vector without a cDNA insert. The data represent the mean values obtained in four independent transfections and are expressed as the fold-activation obtained in each sample over the activity generated by the wild-type enhancer construct in the absence of twist expression plasmid. Error bars represent the standard error of the means for the four experiments. The mean level of β-galactosidase activity produced by the wild-type enhancer construct in the absence of Twist was 3.4 × 10−6 units and in the presence of Twist was 1.1 × 10−3 units (one unit is defined as the amount of enzyme that will convert >95% of the β-d-galactose from 1 nmole of Galβ1–3GlcNAcβ1–3Galβ1–4Glc–AMC in a 10-μl reaction in 1 hr at 37°C and at pH 4.5). (B) Twist binding to the E1 E box. An oligonucleotide corresponding to the E1 E box was used as a probe in an electrophoretic mobility shift assay by use of in vitro-translated Twist. Unprogrammed lysate was included in a parallel lane. A 50- or 300-fold excess of unlabeled E1, unlabeled E1 mutant (E1m), or unlabeled E2 oligonucleotides were used as competitors. Competition by the wild-type E1 oligonucleotide but neither the E1 mutant nor E2 indicates a specific interaction between Twist and E1. B, probe bound with Twist; L, complex detected in unprogrammed lysate.
Activation of the enhancer by another mesoderm-specific bHLH transcription factor, Nautilus [(Nau) Michelson et al. 1990; Paterson et al. 1991], was also tested. Under conditions where Twist strongly activated the enhancer, Nau-expressing lysates showed no activation over background, indicating that Nau was unable to activate transcription through this enhancer (Twist, 127 ± 33-fold; vector alone: 1.00 ± 0.67-fold; Nau, 0.78 ± 0.20-fold).
To confirm that Twist was able to bind to the E1 sequence, electrophoretic mobility shift assays were performed with a radioactively labeled DNA probe corresponding to the E1 E box and Twist protein generated by transcription and translation in vitro (Fig. 4B). Addition of Twist-containing lysate generated a band (labeled B in Fig. 4B) not detected in control lysate. Binding to this site was competed by addition of unlabeled E1 probe, but not by addition of unlabeled E1 mutant DNA nor wild-type E2 DNA. These results indicate that Twist binds specifically to the E1 E box.
Ectopic Twist activates the 175-bp enhancer
To further investigate the responsiveness of the 175-bp enhancer to Twist, twist expression was ectopically induced in embryos to determine whether this resulted in ectopic activation of the 175-bp enhancer. Males carrying a UAS–twist construct (Baylies and Bate 1996) and a construct expressing GAL4 under the control of a heat-shock promoter (Brand and Perrimon 1990) were mated to females homozygous for different enhancer–lacZ constructs. Embryos were collected from this cross and following heat shock the induction of β-galactosidase reporter was determined.
Embryos carrying the wild-type 175-bp enhancer–lacZ construct showed expression of lacZ in the embryonic mesoderm in the absence of ectopic twist (Fig. 5A, also see below). After induction of twist expression, strong expression of the lacZ reporter was observed throughout the embryo (Fig. 5C). Embryos carrying the enhancer–lacZ in which E1 had been mutated showed no expression of lacZ in the absence (Fig. 5B) or presence (Fig. 5D) of ectopic Twist, indicating the single Twist-binding site in the enhancer was required for Twist responsiveness.
Figure 5.
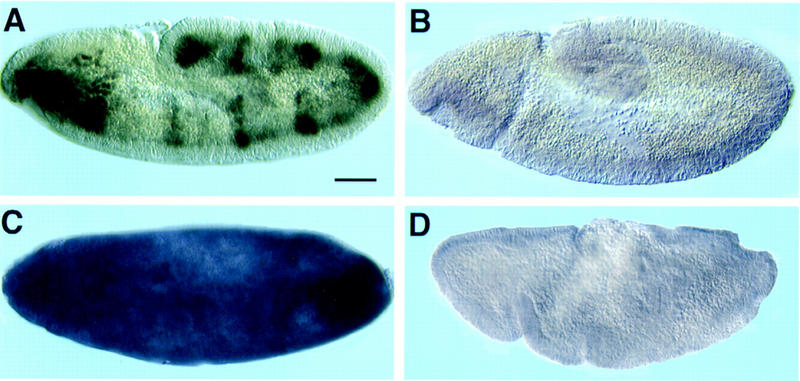
Twist activates the 175-bp enhancer in embryos. Embryos carrying either the wild-type 175-bp enhancer–lacZ construct (A,C) or the E1 mutated construct (B,D) were stained for β-galactosidase protein in the absence (A,B) or presence (C,D) of ectopically expressed twist. The 175-bp enhancer was active in wild-type embryos (A; see also Fig. 11), and activity was induced throughout the embryo after ubiquitous expression of twist (C). The E1 mutated enhancer was inactive in wild-type embryos (B) and was not induced by ectopic Twist (D), although the morphology of this embryo was affected. Bar, 100 μm.
Taken together, the results presented in Figures 2, 3, 4, and 5 strongly support the conclusion that Twist directly activates transcription of Mef2 in the adult muscle precursor cells of the wing imaginal discs. The activity of the 175-bp enhancer is absolutely dependent on the E1 E box, and in vivo and in vitro experiments show conclusively that Twist can interact with this sequence and activate transcription of reporter genes.
Twist activation of Mef2 is required for normal adult myogenesis
To determine whether twist is required for adult somatic myogenesis and if the effects are mediated by Mef2, a temperature-sensitive heteroallelic combination of two different twist alleles, twiRY50 and twiV50, was employed (Thisse et al. 1987). The RY50 and V50 alleles were crossed into a genetically marked background to permit identification of RY50/V50 mutant larvae (see Materials and Methods). Crosses were set up at the permissive temperature (18°C), and embryos were incubated at this temperature until hatching. Newly hatched mutant larvae were collected and subjected to two different incubation temperatures: To reduce Twist function for the rest of development, larvae were shifted to the restrictive temperature (30°C); as a control, mutants were maintained at 18°C for the rest of their development.
To determine the effects of reducing Twist function upon Mef2 expression in the adult muscle precursor cells, the expression of both Mef2 and a Mef2–lacZ transgenic construct was analyzed. Reduction in Twist function resulted in a loss of Mef2 expression in the adepithelial cells (Fig. 6A, top). Two other genes known to be expressed specifically in the adepithelial cells of the imaginal discs are the fibroblast growth factor receptor-1 homolog heartless (htl; Emori and Saigo 1993) and the homeobox gene cut (Blochlinger et al. 1990). Expression of these genes was unaffected by reducing Twist function (Fig. 6A), indicating that adepithelial cells had formed and proliferated normally.
Figure 6.
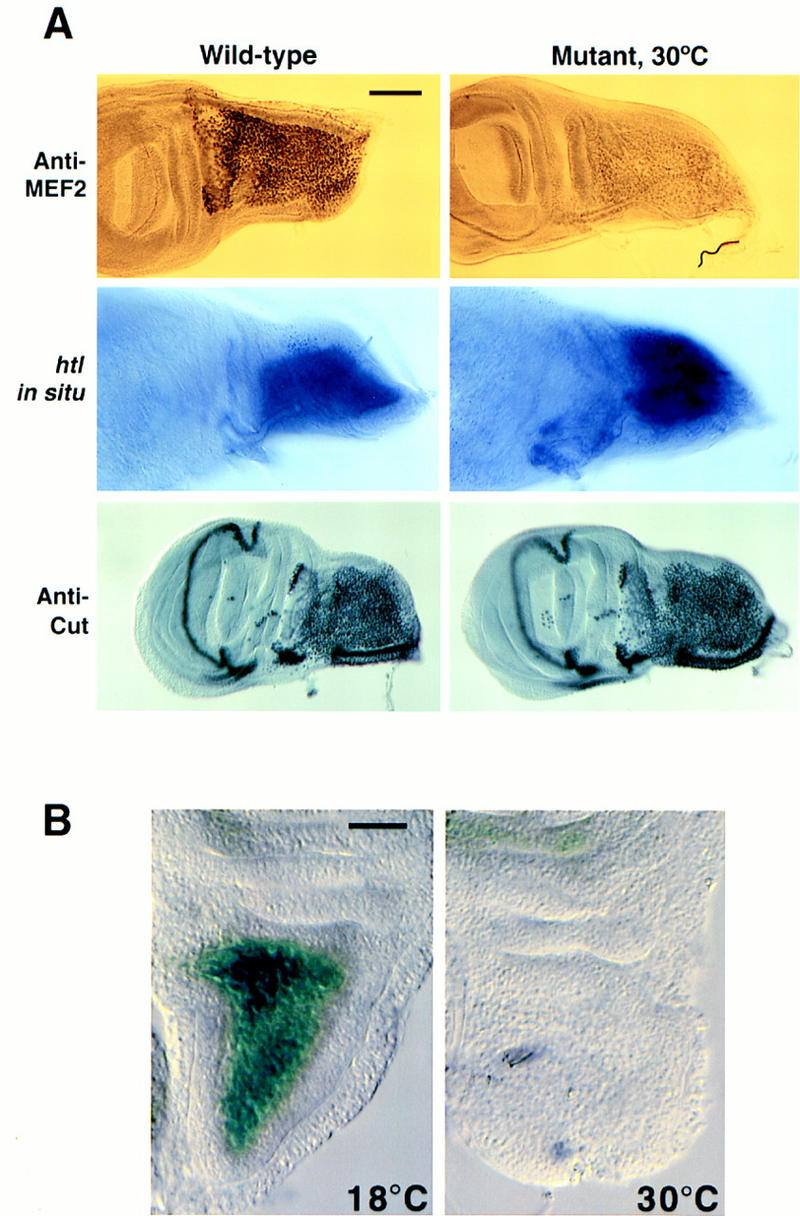
Expression of adepithelial cell markers in wild-type and RY50/V50 twist mutants. (A) (Left) Wild type. (Right) RY50/V50 raised at the restrictive temperature. (Top) Immunostaining by an anti-MEF2 polyclonal antibody. (Center) In situ hybridization with a digoxigenin-labeled antisense probe for the Drosophila fibroblast growth factor receptor homolog -1, htl. (Bottom) Immunostaining by an anti-Cut monoclonal antibody. Note that MEF2 accumulation is greatly reduced, yet the other markers are expressed at normal levels. Bar, 50 μm. (B) A ∼3.4-kb Mef2 enhancer/promoter fragment is sensitive to the levels of twist expression. Transgenic flies carrying the −2884 to +521 fragment cloned upstream of lacZ were crossed into the twist mutant backgrounds. RY50/V50 mutants raised at either the permissive (left, 18°C) or restrictive (right, 30°C) temperatures were analyzed for β-galactosidase activity in wing imaginal discs. Transgene expression was only observed at 18°C, indicating that Twist function is required for enhancer activity. Bar, 25 μm.
The expression of a Mef2–lacZ construct was also sensitive to reduction in Twist function (Fig. 6B): The transgene was active at the permissive temperature (18°C) but inactive at the restrictive temperature (30°C). This further shows that the adepithelial cell enhancer examined is responsive to the levels of Twist function as is the endogenous Mef2 gene.
Of the twist mutant larvae shifted to the restrictive temperature, 63% (n = 150) developed to adults, compared with 82% (n = 150) of control y w treated in the same manner. This indicates that there is a postembryonic requirement for Twist for normal viability. In addition, several of the resulting adults showed defects in wing positioning. This phenotype is common among flies in which the patterning or function of the indirect flight muscles is defective (see, e.g., Deak 1977; Mogami and Hotta 1981; for review, see Bernstein et al. 1993). twist mutants raised at the permissive temperature showed good viability (94%, n = 250) and no wing position abnormalities.
The adult musculature of wild type and RY50/V50 raised at the permissive or restrictive temperature was analyzed (Fig. 7, A–C). Within the thorax, there are two columns of dorsal longitudinal indirect flight muscles (DLMs), shown in cross section in panels A–C. In wild-type flies, there are six DLM fibers in each half thorax (panel A, numbered). There was a reduction to five fibers, however, in the twist mutants raised at the permissive temperature (panel B), and to three fibers in the mutants raised at the restrictive temperature (panel C). Analysis of a number of individuals showed that these DLM defects were highly reproducible (summarized in Fig. 7, D–F). Patterning defects were also observed in other thoracic and abdominal muscles including the tergal depressor of the trochanter muscles and the lateral transverse muscles of the abdomen (data not shown). These results show that twist is required for normal development of somatic muscle in the adult, and correlate with observations that twist is expressed in the precursor myoblasts of the adult somatic muscles (Bate et al. 1991; Currie and Bate 1991). The mild defects that are observed in RY50/V50 raised at the permissive temperature (five DLM fibers per hemithorax) indicate that even at this temperature, the mutant Twist protein is functionally compromised.
Figure 7.
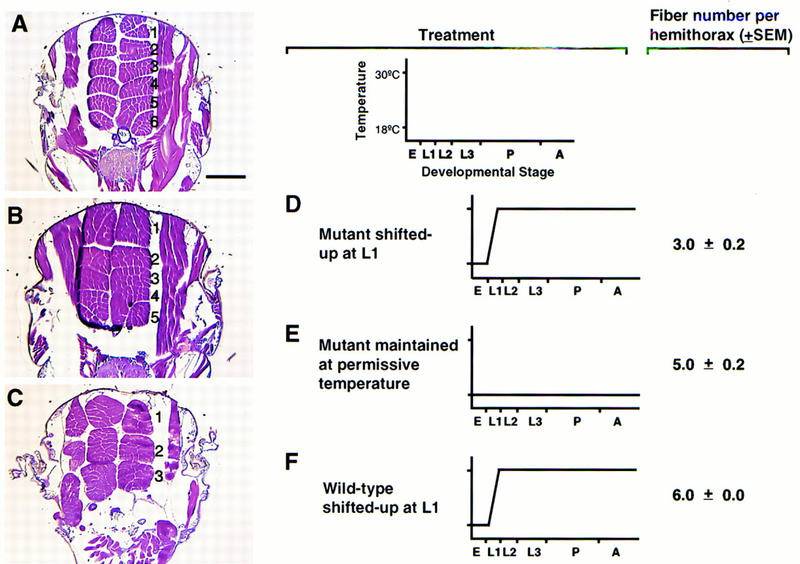
Adult muscle phenotypes resulting from reduced twist expression. (A–C) vertical transverse paraffin sections of thoraces to visualize the two rows of dorsal longitudinal indirect flight muscles (muscles are numbered 1–6 where present). (A) wild-type; (B) RY50/V50 raised at 18°C; (C) RY50/V50 raised at 30°C. Bar, 100 μm. (D–F) summary of temperature shifts performed and fiber defects observed; fiber numbers represent the average number of DLM fibers per hemithorax. (D) RY50/V50 raised at 30°C after embryogenesis; (E) RY50/V50 raised at 18°C; (F) wild-type raised at 30°C after embryogenesis. (s.e.m.) Standard error of the mean. Developmental stages: (E) embryonic stage; (L1–3) larval instars; (P) pupal stage; (A) adult stage.
Previously, Ranganayakulu et al. (1995) showed that Mef2 function is required for normal patterning of the DLMs, and the phenotypes observed here when Twist function is reduced are very similar to those observed previously. Because antibody staining showed that reduction in Twist function resulted in a loss of MEF2, it can be concluded that the patterning defects observed in this study are likely caused by a loss of MEF2 expression when Twist function is reduced.
Embryonic expression of the 175-bp enhancer
Because twist and Mef2 are also coexpressed at various times during embryogenesis (Lilly et al. 1994; Nguyen et al. 1994; Taylor et al. 1995), lines containing the 175-bp enhancer–lacZ construct were tested for lacZ expression in embryos. Early in embryogenesis, both twist and Mef2 are expressed at high levels in the developing mesoderm, however, there was only weak expression from the 175-bp enhancer at this stage and this was not uniform in the mesoderm (Fig. 8A). This indicates that at this stage Twist alone cannot activate the 175-bp enhancer, and it is likely that the genomic region governing the earliest expression of Mef2 is either outside of this enhancer or overlaps it.
Figure 8.
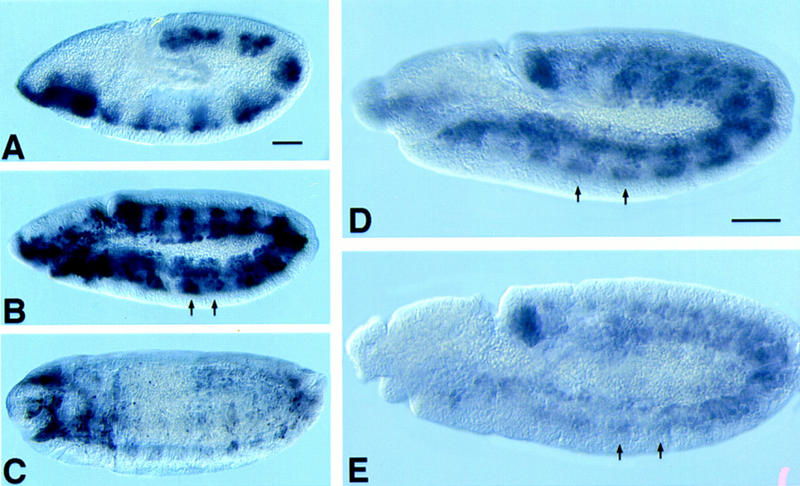
Expression of the 175-bp enhancer during embryogenesis. (A) At stage 9, the enhancer was only weakly active in the developing mesoderm and was segmentally modulated. (B) at late stage 11 the enhancer was active strongly in segmentally repeating groups of cells that are the precursors of the larval somatic muscles (arrows). (C) By stage 15, enhancer activity was waning. (D–E) Accumulation of MEF2 in the presence (D) or absence (E) of Twist function after gastrulation. (D) MEF2 acumulation in a late stage 11 twist mutant heterozygous embryo, showing normal expression in somatic muscle myoblasts (arrows). (E) MEF2 accumulation in a sibling temperature-sensitive twist mutant in which Twist function had been reduced after gastrulation. There is a general reduction in Mef2 expression in the somatic myoblasts (arrows), as well as in the precursors of the pharyngeal muscles (out of focus at anterior end). Bar, 100 μm.
The activity of the 175-bp enhancer increased during embryogenesis until stage 12, where it was expressed in segmentally repeating groups of cells corresponding to the unfused and undifferentiated myoblasts of the larval somatic muscles (Fig. 8B, arrows). These cells were identified as somatic myoblasts because some lines with high levels of expression had β-galactosidase protein that persisted during differentiation. At this stage, twist and Mef2 are coexpressed in many of these cells (Taylor et al. 1995). By stage 15, levels of β-galactosidase had declined in the differentiated somatic muscles, although protein persisted in some of the muscle fibers of the head and thorax (Fig. 8C).
The expression of lacZ in lines carrying the 175-bp enhancer with the E2 E box mutated was the same as for the wild-type enhancer, whereas both the E1 mutant enhancer and the double-mutant enhancer were inactive throughout embryogenesis. These results indicate that the 175-bp enhancer is active in a subset of mesodermal cells during embryogenesis, and that the activity of this element likely depends upon a bHLH factor.
Because ectopic Twist was able to activate Mef2 expression in embryos and because twist and Mef2 are coexpressed in somatic myoblasts by stage 12 (Taylor et al. 1995), the expression of Mef2 in the twist temperature-sensitive mutant background was studied. When twist was inactivated by temperature shift after gastrulation, there was a severe reduction in the level of MEF2 protein in the mutant embryos (Fig. 8E) as compared to heterozygous siblings (Fig. 8D).
Discussion
Members of the MEF2 family of transcription factors play central roles in muscle cell differentiation. In the absence of MEF2 function, myoblasts are properly specified in the embryo, but do not activate terminal muscle differentiation genes (Bour et al. 1995; Lilly et al. 1995; Ranganayakulu et al. 1995; Lin et al. 1997). Whereas an understanding of the mechanism for transcriptional activation by MEF2 proteins is emerging, little is known of the pathways that control Mef2 gene expression in different muscle cell types.
In this study, we have identified a novel enhancer that is responsible for Mef2 expression in somatic myoblasts prior to differentiation at two different developmental stages. A single conserved E box is essential for enhancer activity, and we show that the bHLH transcription factor Twist is capable of binding to this E box to activate transcription both in tissue culture and when ectopically expressed during embryogenesis. Furthermore, expression of the endogenous Mef2 gene is strongly reduced when Twist function is reduced, showing that one of the functions of Twist is to directly activate transcription of Mef2.
twist expression after gastrulation has been shown to be necessary for differentiation of the embryonic somatic mesoderm (Baylies and Bate 1996). On the basis of our results, the mutant phenotypes observed when Twist function is reduced could be caused by a loss of Mef2 expression in somatic myoblasts, although it is possible that somatic myoblasts do not develop normally when Twist function is reduced prior to this time. In adult muscle precursor cells, a reduction in Twist function in the larva clearly results in a loss of enhancer activity and of Mef2 expression in the imaginal myoblasts. These observations show an essential function in vivo for the 175-bp enhancer in the development of the skeletal musculature.
The 175-bp enhancer is one in a series of regulatory modules that control Mef2 expression during development of the somatic mesoderm. Early in embryogenesis, expression of twist and Mef2 are high throughout the mesoderm, yet the activity of the 175-bp enhancer is low and is not uniformly distributed. This indicates that sequences either outside the 175-bp enhancer or overlapping with it are required for normal Mef2 expression at this time, and that factors in addition to Twist may be required for enhancer function. It is likely that Mef2 is regulated by Twist at this stage (Taylor et al. 1995), but whether this activation is direct is unknown.
Later, as skeletal muscle cells differentiate, Mef2 expression persists in the somatic mesoderm (Bour et al. 1995; Lilly et al. 1995), despite a decline in both Twist levels (Dunin-Borkowski et al. 1995) and activity of the 175-bp enhancer. This indicates that Mef2 is regulated by factors other than Twist through a separate enhancer at this time. Enhancer activity responsible for Mef2 expression in differentiated muscles has been identified (Lilly et al. 1995; Schulz et al. 1996), however, the location and regulation of these enhancers has not been studied. Finally, a separate enhancer element has been recently defined that regulates Mef2 expression in some muscle founder cells in the embryo (R.A. Schulz, unpubl.; B. Zhao, R. Cripps, E. Olson, unpubl.). These observations underscore the complexity of regulatory mechanisms that contribute to Mef2 expression.
Ectopic expression of Twist in the embryo can induce reporter gene expression through the 175-bp enhancer, yet early in embryogenesis, when Twist is present uniformly in the mesoderm, the enhancer was not fully active. A possible explanation for this discrepancy is that Twist is over-expressed under the control of the hs–GAL4 driver. Perhaps high levels of Twist are sufficient to activate the enhancer throughout the embryo. In the mesoderm, lower levels of Twist alone cannot support enhancer activity.
A myogenic regulatory function for twist
The requirement for Drosophila twist in somatic muscle development contrasts with studies of the murine homolog M-Twist, which inhibits muscle cell differentiation in culture (Hebrok et al. 1994; Spicer et al. 1996). Thus, it has been unclear if Twist acts as an activator or a repressor in the myogenic lineage. Here, we show that Drosophila Twist is a direct and positive regulator of Mef2 expression, correlating with its requirement for muscle development in Drosophila and establishing it as a potent transcriptional activator in these cells. These results are consistent with other roles for Twist both early during mesoderm formation, where Twist activates snail, (Ip et al. 1992a) rhomboid (Ip et al. 1992b) and possibly other genes (Lai et al. 1991; Leptin 1991; Shishido et al. 1993), and later in the activation of tinman in the visceral mesoderm (Lee et al. 1997).
Twist function after gastrulation is important for the division of the mesoderm into somatic, cardiac, and visceral muscle lineages (Baylies and Bate 1996). Whereas our results show a role of Twist in the activation of Mef2 expression in somatic myoblasts, Twist must be involved in the activation or repression of additional genes to mediate these other functions. The identity of these genes is not yet known.
The role of twist in adult muscle development
One proposed function of twist during the larval stage might be to maintain adult muscle precursor cells in an undifferentiated state to control the proliferation of these cells prior to differentiation (Bate 1993). A reduction in twist function during the larval stage, however, does not result in major defects in the adepithelial cells. These cells exhibit many of the characteristics of wild-type cells, including expression of htl and cut, and the nuclear localization of Cut (Fig. 6) reveals that adepithelial cells apparently formed and proliferated normally. Thus, Twist must play a more subtle role in the development of the adult muscle cells.
We have shown that when Twist function is reduced during the larval stage, Mef2 is not expressed in the adult muscle precursor cells, resulting in abnormal patterning of the adult somatic muscles, most strikingly in the DLMs. Hypomorphic Mef2 mutant adults show very similar defects (Ranganayakulu et al. 1995), indicating that this phenotype, resulting from loss of twist expression, likely occurs through a requirement to maintain normal Mef2 levels.
It is striking that reducing Twist function during the embryonic stage has a severe effect on development of the musculature (Baylies and Bate 1996), yet, a reduction during the larval and pupal stages has a more minor effect on adult myogenesis. The basis for this difference in twist function at different stages is unclear. Perhaps the major function of twist in adult myogenesis occurs during the embryonic stage, at the same time as the critical role of twist in embryonic muscle development. Alternatively, it might be that adult myogenesis is initiated independently of twist, and twist expression in the adepithelial cells is only required for muscle patterning.
The DLM defects we observed can be explained in terms of DLM development during the pupal stage. DLMs develop when adepithelial cells from the wing disc surround and associate with three pairs of larval body wall muscles, the larval oblique muscles (LOMs; Fernandes et al. 1991). Once the adepithelial cells arrive, each LOM splits longitudinally to form templates for the six pairs of DLMs found in the adult. After splitting, adult myoblasts fuse with the LOMs and commence differentiation. Our results show that the DLM defects observed here and in Mef2 mutants likely result from a lack of Mef2 expression in the adepithelial cells, because twist is expressed in the adult myoblasts but not in the LOMs used as templates for DLM development (Fernandes et al. 1991).
The most likely cause of the DLM patterning defect is a failure of the LOMs to undergo splitting. Absence of LOM splitting would result in the observed reduction of final fiber number from six per hemithorax in wild-type flies to three in the mutants. Intermediate numbers of DLMs may result from the splitting of some LOMs but not others.
Conservation of regulatory pathways controlling Mef2 expression
Drosophila and vertebrates share the diversification of the mesoderm into skeletal, cardiac, and visceral muscle lineages, and there is conservation in both sequence and function in many of the major myogenic factors. It is, therefore, likely that there is also conservation in the mechanisms that regulate these factors during development. One enhancer required for Mef2 expression in a subset of cardial cells is regulated by Tinman (Gajewski et al. 1997), and it is tempting to speculate that the vertebrate tinman-like gene Nkx2.5 might regulate cardiac expression of mef2 genes in the mouse. Here, we have identified a second enhancer that is responsive to Twist and that is expressed in somatic myoblasts. It is likely that Drosophila Mef2 contains other enhancers that direct complex temporal and spatial expression patterns in other mesodermal compartments. Identification of these enhancers and the factors that regulate them should reveal the transcriptional networks governing Mef2 expression in multiple lineages.
Materials and methods
Generation of constructs for promoter/enhancer analysis
Constructs containing EcoRI genomic fragments are described in Lilly et al. (1995) and Schulz et al. (1996). For analysis of enhancer activity in transgenic flies, constructs devoid of an endogenous start site were cloned into the CaSpeR–hsp–AUG–β-gal vector (CHAB, which contains an hsp promoter upstream of lacZ; Thummel and Pirrotta 1992), whereas those including the transcriptional start site of Mef2 were cloned into a similar vector in which the heat shock promoter had been removed. Initial constructs were generated by use of appropriate restriction sites (see Fig. 1); all subsequent constructs were generated by PCR. PCR products were first cloned into the pCR2.1 vector (InVitrogen) and then cloned into CHAB by use of appropriate flanking sites.
Mutagenesis of the 175-bp enhancer was carried out by gene SOEing (Horton 1993). The E1 E box was changed from 5′-CACATGTG-3′ to 5′-CGGCCGTG-3′ that ablates both overlapping E boxes, and the E2 E box was mutated from 5′-CAGATG-3′ to 5′-GACGTC-3′. The smallest construct generated was a 30-bp fragment (−2306 to −2277); this was made by annealing two oligonucleotides to give the correct final sequence and this also generated 5′ overhangs at each end to permit insertion into the EcoRI site of CHAB.
Sequencing of genomic DNA and of PCR-generated mutant constructs was carried out with an ABI 373 automated sequencer.
P-element-mediated germ-line transformation was carried out as described (Rubin and Spradling 1982). y w embryos were coinjected with vector DNA and Δ2-3 helper plasmid (Robertson et al. 1988). G0 adults were crossed with y w, and transformants isolated in the G1 generation. For each new construct generated, at least three independent lines were analyzed.
Cloning of the D. virilis Mef2 gene
A Drosophila virilis genomic library made by J. Tamkun (University of California, Santa Cruz) and provided by S. Wasserman (University of Texas Southwestern Medical Center, Dallas) was screened with a probe made from the MADS and MEF2 regions of D. melanogaster Mef2 cDNA. From the clones isolated in the primary screen, secondary screening was performed on duplicate filters: one duplicate was probed with the same probe as the primary; the other duplicate was hybridized with a probe spanning the transcriptional start site of D. melanogaster Mef2 (from −140 to +440). Two independent clones hybridized with both probes. Sequencing both of these identified a region of strong sequence similarity to the transcriptional start site of D. melanogaster Mef2 (90% homology over 52 bp, from −15 to +37 in D. melanogaster). Upstream of the start site there is ∼4.2 kb of 5′ genomic sequence to the bacteriophage arm. A 3.9-kb fragment extending from a SalI site in the bacteriophage arm to an EcoRI site upstream of the transcriptional start (−4248 to −358) was cloned into CHAB and used to generate transgenic lines.
Cell culture and transfections
For cotransfection assays, the twist expression plasmid pPacTwi (Shirokawa and Courey 1997), was provided by J. Shirokawa and A. Courey (University of California, Los Angeles), and the nau expression plasmid was from B. Paterson [National Cancer Institute (NCI), National Institutes of Health (NIH), Bethesda, MD]. Reporter constructs were the same as those used for P-element mediated germ-line transformation of flies, which contain the 175-bp enhancer (wild type or mutant), and a minimal promoter upstream of the lacZ gene. SL2 cells were maintained at 25°C in Schneider’s Drosophila medium (GIBCO/BRL) supplemented with 10% heat-inactivated fetal calf serum (FCS). Two hours prior to transfection, cells were seeded at ∼80% confluence onto T25 flasks. Cells were transfected with the DOSPER reagent (Boehringer Mannheim). Six micrograms of DNA (3 μg of activator plasmid and 3 μg of reporter plasmid) were mixed with 18 μg of DOSPER in 2 ml of Dulbecco’s modification of minimal essential medium (DMEM) without serum or antibiotics and incubated for 15 min at room temperature. Schneider’s Drosophila medium was removed from the cells and the DMEM containing the DNA/lipid complex was added. After 5 hr, the DMEM was removed, 3 ml of fresh Schneider’s Drosophila medium containing 10% FCS was added, and the cells incubated for 43 hr prior to harvesting.
β-Galactosidase assays
Transfected cells were harvested by scraping the attached cells into the culture medium and centrifuging all adherent and nonadherent SL2 cells. Cell pellets were resuspended in 3 ml of PBS and centrifuged again. The cells were then resuspended in 0.1 ml of 0.1 m sodium phosphate (pH 7.0) and lysed by sonication. Debris was pelleted by centrifugation at 12,000g at 4°C for 15 min, and the supernatant was removed. The cell lysates were quantitated for total protein (Lowry et al. 1951), and 50 μg of lysate from each transfection was assayed for β-galactosidase activity as described previously (Sambrook et al. 1989). To allow for direct comparison of samples with dramatically different levels of β-galactosidase activity, reactions containing two 10-fold dilutions of extract from each sample were also conducted with 5 and 0.5 μg of lysate from each transfected sample. The total amount of cellular lysate in each reaction was held constant at 50 μg by adding the appropriate amount of untransfected SL2 cell lysate to each sample. Serial dilutions of purified β-galactosidase enzyme (Boehringer Mannheim) were also analyzed by mixing with 50 μg of lysate from untransfected SL2 cells to allow for direct comparison of the number of units of enzyme present in each transfected sample. Reactions were conducted at 37°C for 2 hr, then stopped by the addition of 1 m sodium carbonate. Samples were measured for absorbance at 420 nm to determine the amount of β-galactosidase activity in each sample. Only samples in the linear range of the assay (absorbance readings of 0.05 to 1.0) were used for comparison.
In vitro DNA binding assay
For in vitro transcription and translation of Twist, a 1.6-kb NdeI to EcoRI cDNA fragment containing the twist coding region was excised from the pAR 3040 plasmid (Ip et al. 1992b; kindly provided by M. Levine, University of California, Berkeley) and cloned into the unique NdeI–EcoRI sites of pCITE-2a (Novagen). Twist protein was generated from this construct with the TnT transcription and translation kit (Promega Corp.). To generate radiolabeled E1 probe DNA, two oligonucleotides of the sequence 5′-GGATGTTGCACATGTGTTGAGT-3′ and 5′-GGACTCAACACATGTGCAACAT-3′ were annealed. After annealing, the 5′ GG overhangs were filled in with Klenow enzyme (New England Biolabs) and [α-32P]dCTP. Probes were purified on a G25 spin column (Boehringer Mannheim), and 50,000–120,000 cpm were used in each assay. For competition experiments, wild-type E1 oligonucleotide, mutant E1m oligonucleotide, or wild-type E2 oligonucleotide was included in the reactions at 50× and 300× concentrations. The E1 mutant oligonucleotides had the sequences 5′-GGATGTTGCGGCCGTGTTGAG T-3′ and 5′-GGACTCAACACGGCCGCAACA T-3′ before annealing. The sequence of the mutated E box in this fragment is the same as that in the analyses of enhancer function in transgenic flies and in cotransfection assays. The E2 oligonucleotides had the sequences 5′-GGACTGCCGCAGATGCCGCATG-3′ and 5′-GGCATGCGGCATCTGCGGCAGT-3′. Each gel shift reaction consisted of probe, 1 μg of poly[d(I–C)]; 3–4 μl of TnT lysate and 1 μl of a 10× buffer consisting of 400 mm KCl, 150 mm HEPES at pH 7.9, 10 mm EDTA, 5 mm DTT, 50% glycerol. After incubation at room temperature for 20 min, reactions were electrophoresed on a 5% (wt/vol) polyacrylamide gel at 22 mA. Dried gels were subjected to autoradiography.
Histochemistry, immunohistochemistry, and in situ hybridization
Embryos were stained with antibodies as described by Patel et al. (1987). Imaginal discs for immunohistochemistry were dissected from larvae in PBS, fixed for 30 min on ice in 4% paraformaldehyde in PBS, washed in PBS and then incubated in 0.2% (vol/vol) Triton X-100 in PBS at room temperature for 20 min. Discs were then treated for antibody staining as for embryos. The anti-MEF2 antibody was from B. Paterson (NCI, NIH, Bethesda, MD; Lilly et al. 1995) and was used at a concentration of 1:1000. The anti-Twist polyclonal antibody was from C. Thisse and B. Thisse (University of Strasbourg, France) and was used at 1:500. These antibodies were detected by use of a peroxidase-linked goat anti-rabbit secondary antibody (Jackson Immunoresearch Laboratories, Inc.) at 1:1000. The anti-β-galactosidase monoclonal antibody (Promega) was used at 1:300, and the anti-Cut monoclonal antibody was used at 1:150. These antibodies were detected with the Vectastain Elite Staining Kit (Vector Laboratories, CA). The anti-Cut antibody, developed by I. Rebay, G. Dailey, K. Lopardo, and G. Rubin, was obtained from the Developmental Studies Hybridoma Bank maintained by the Department of Biological Sciences, the University of Iowa, Iowa City, under contract from the National Institute of Child Health and Development (NICHD). Stained discs were prepared for sectioning as described by Lyons et al. (1990).
Imaginal disc in situ hybridization was performed essentially as described by Tautz and Pfeiffle (1989) and O’Neill and Bier (1994). Briefly, imaginal discs were dissected from larvae and fixed in 4% formaldehyde, 0.5% Triton X-100 in PBS on ice for 30 min. After washing in 0.5% Tween-20 in PBS (PBTw), discs were treated with 12.5 μg/ml proteinase K in PBTw for 2 min. at room temperature, and then quenched in 2 mg/ml glycine in PBTw. Discs then were postfixed in 4% formaldehyde, 0.2% glutaraldehyde in PBTw for 20 min at room temperature. Remaining discs were then washed and subjected to prehybridization and hybridization as described. A digoxigenin (Boehringer Mannheim) labeled antisense riboprobe for the htl gene was generated from a plasmid provided by A. Michelson (Harvard University, MA). The riboprobe was detected by use of an alkaline phosphatase-conjugated sheep anti-digoxigenin antibody (1:1000), and BM purple AP substrate for the color reaction (Boehringer Mannheim).
Imaginal discs for X-gal staining were dissected from larvae, fixed for 20 minutes in 4% (vol/vol) formaldehyde in 1× PBS on ice, washed in 1× PBS and then stained in an X-gal solution at 37°C as described (Ashburner 1989).
Fly culture and temperature shifts
Flies were grown on standard culture medium at the indicated temperatures. All genetic symbols are described in Lindsley and Zimm (1992). The stocks SM1/twiRY50 bw and SM1/twiV50 bw sp; st e were provided by R. Reuter, (Universität zu Köln, Germany). Only the heteroallelic combination of these two alleles is temperature sensitive; neither allele alone is temperature sensitive (Thisse et al. 1987).
For larval temperature-shift experiments, crosses were set up in cages where flies laid eggs on agar plates darkened with grape juice. These plates were incubated at 18°C, and mutant larvae were collected from the plates shortly after hatching in groups of 50, and placed in well-yeasted vials for subsequent culture. The permissive temperature in these experiments was 18 ± 1.0°C. The restrictive temperature was 30 ± 1.0°C. For embryonic temperature-shift experiments, eggs were collected over a 2-hr period at 18°C and aged at the permissive temperature until after gastrulation. The incubation temperature was then shifted to 30°C by floating the agar plates in a water bath for the remainder of the experiment.
Identification of twist mutants
To separate the twist mutant larvae from nonmutant siblings at the larval stage, each mutant was crossed into a genetic background that was y w on the X chromosome and in which the mutants were balanced over a CyO balancer chromosome (designated CyO, y+) onto which the y+ gene had been introduced (see Mardahl et al. 1993). From the cross of y w; CyO, y+/twiRY50 bw × y w; CyO, y+/twiV50 bw sp heteroallelic mutant first instar larvae could be identified on the basis of the color of their mouth hooks: y− individuals have yellow mouth parts, compared with the black mouthparts and dark denticle belts of y+ individuals.
To identify twist mutant offspring during the embryonic stage, stocks were balanced over an SM6b chromosome that carries an even-skipped–lacz transgene. Mutant offspring were identified on the basis of their lack of reaction with anti-β-galactosidase antibody compared with siblings.
Analysis of adult phenotypes
For sectioning adult thoraces, the heads and abdomens were removed from 1- to 3-day-old adult flies. The thoraces were then fixed and embedded in paraffin by use of standard procedures (Lyons et al. 1990). Sections of 7–10 μm were stained with hematoxylin for 30 sec and eosin for 30 sec, then dehydrated and mounted in Permount. To quantify the muscle phenotype observed in the twist mutant adults, thoraces were dissected as described by Peckham et al. (1990) until the point at which thoraces were longitudinally bisected. Half-thoraces were observed under the dissecting microscope, and the number of DLM fibers was counted in each half-thorax. By convention, the average DLM count for flies in any class was determined by averaging the number of fibers in the right hemithorax, except where the right hemithorax was damaged, in which case the fiber number was taken from the left hemithorax for that individual fly. We have not seen any consistent left-right bias in any of the flies that were dissected. At least nine different adults were tested for each experiment.
Ectopic expression of twist
Ectopic expression of twist in embryos was under the control of a heat shock promoter via the GAL4 system (Brand and Perrimon 1993; Taylor et al. 1995). hs–GAL4 was from the Bloomington Drosophila Stock Center, and UAS–twi was provided by M. Baylies (Memorial Sloane-Kettering Cancer Center, New York, NY). These lines were used to generate UAS–twi; hs–GAL4/TM2, Ubx. Males of this genotype were crossed to females homozygous for enhancer–lacZ constructs, and progeny subjected to heat shock and immunohistochemistry (Taylor et al. 1995).
Acknowledgments
We are grateful to the following individuals for generously providing reagents: M. Baylies, A. Courey, M. Levine, A. Michelson, B. Paterson, R. Reuter, J. Shirokawa, C. Thisse, B. Thisse, and S. Wasserman. We thank J. Molkentin, F. Naya, G. Ranganayakulu, and A. Rawls for invaluable help during the course of this work, and T. Gemelli and K. Kunkle for excellent technical assistance. We thank A. Tizenor for assistance with graphics. We are grateful to D. McKearin and S. Wasserman for comments on the manuscript. Research was supported by grants from the NIH, the Muscular Dystrophy Association, the Human Frontiers Science Program and the American Heart Association to E.N.O. B.L.B. was supported by a postdoctoral fellowship from the American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eolson@hamon.swmed.edu; FAX (214) 648 1196.
References
- Ashburner M. Drosophila: A laboratory handbook and manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Bate M. The mesoderm and its derivatives. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1013–1090. [Google Scholar]
- Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M. twist: A myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- Bernstein SI, O’Donnell PT, Cripps RM. Molecular genetic analysis of muscle development, structure and function in Drosophila. Int Rev Cytol. 1993;43:63–152. doi: 10.1016/s0074-7696(08)61874-4. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Bodmer R, Jan LY, Jan YN. Patterns of expression of Cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes & Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes & Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Currie DA, Bate M. The development of adult abdominal muscles in Drosophila: Myoblasts express twist and are associated with nerves. Development. 1991;113:91–102. doi: 10.1242/dev.113.1.91. [DOI] [PubMed] [Google Scholar]
- Deak II. Mutations of Drosophila melanogaster that affect muscles. J Embryol Exp Morphol. 1977;40:35–63. [PubMed] [Google Scholar]
- Dunin-Borkowski OM, Brown NH, Bate M. Anterior-posterior subdivision and the diversification of the mesoderm in Drosophila. Development. 1995;121:4183–4193. doi: 10.1242/dev.121.12.4183. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- Emori Y, Saigo K. Distinct expression of two Drosophila homologs of fibroblast growth factor receptors in imaginal discs. FEBS Lett. 1993;332:111–114. doi: 10.1016/0014-5793(93)80494-f. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Bate M, Vijayraghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Gajewski K, Kim Y, Lee YM, Olson EN, Schulz RA. D-mef2 is a target for Tinman activation during Drosophila heart development. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Wertz K, Fuchtbauer E-M. M-twist is an inhibitor of muscle differentiation. Dev Biol. 1994;165:537–544. doi: 10.1006/dbio.1994.1273. [DOI] [PubMed] [Google Scholar]
- Horton RM. In vitro recombination and mutagenesis of DNA. In: White BA, editor. PCR protocols: Current methods and applications. Vol. 15. Totowa, NJ: Humana Press; 1993. pp. 251–261. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal–twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes & Dev. 1992a;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992b;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Lai Z, Fortini ME, Rubin GM. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- Lee YM, Park T, Schulz RA, Kim Y. Twist-mediated activation of the NK-4 homeobox gene in the visceral mesoderm of Drosophila requires two distinct clusters of E-box regulatory elements. J Biol Chem. 1997;272:17531–17541. doi: 10.1074/jbc.272.28.17531. [DOI] [PubMed] [Google Scholar]
- Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes & Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by the myogenic transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm G. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. . J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lyons GE, Schiaffino S, Barton P, Sassoon D, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardahl M, Cripps RM, Rinehart RR, Bernstein SI, Harris GL. Introduction of y+ onto a CyO chromosome. Dros Inf Serv. 1993;72:141–142. [Google Scholar]
- Michelson A, Abmayr SM, Bate M, Martinez-Arias A, Maniatis T. Expression of a MyoD family member prefigures muscle pattern in Drosophila embryos. Genes & Dev. 1990;4:2086–2097. doi: 10.1101/gad.4.12a.2086. [DOI] [PubMed] [Google Scholar]
- Mogami K, Hotta Y. Isolation of Drosophila flightless mutants which affect myofibrillar proteins of indirect flight muscle. Mol & Gen Genet. 1981;183:409–417. doi: 10.1007/BF00268758. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Defining the regulatory networks for muscle development. Curr Opin Genet & Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA. D-mef2: A Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JW, Bier E. Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. BioTechniques. 1994;17:870–875. [PubMed] [Google Scholar]
- Patel N, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: A glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Paterson BM, Walldorf U, Eldridge J, Dubendorfer A, Frasch M, Gehring W. The Drosophila homolog of vertebrate myogenic-determination genes encodes a transiently expressed nuclear protein marking primary myogenic cells. Proc Natl Acad Sci. 1991;88:3782–3786. doi: 10.1073/pnas.88.9.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham M, Molloy JE, Sparrow JC, White DCS. Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J Muscle Res Cell Motil. 1990;11:203–215. doi: 10.1007/BF01843574. [DOI] [PubMed] [Google Scholar]
- Poodry CA, Schneiderman HA. The ultrastructure of the developing leg of Drosophila melanogaster. Wilhelm Roux’ Archiv. 1970;166:1–44. doi: 10.1007/BF00576805. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Reed CT, Murphy C, Fristrom D. The ultrastructure of the developing pupal leg of Drosophila melanogaster. Wilhelm Roux’ Archiv. 1975;178:285–302. doi: 10.1007/BF00848064. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G M, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schulz RA, Chromey C, Lu M-F, Zhao B, Olson EN. Expression of the D-MEF2 transcription factor in the Drosophila brain suggests a role in neuronal cell differentiation. Oncogene. 1996;12:1827–1831. [PubMed] [Google Scholar]
- Shirokawa J, Courey AJ. A direct contact between the Dorsal rel homology domain and Twist may mediate transcriptional synergy. Mol Cell Biol. 1997;17:3345–3355. doi: 10.1128/mcb.17.6.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E, Higashijima S, Emori Y, Saigo K. Two FGF-receptor homologues of Drosophila: One is expressed in mesodermal primordium in early embryos. Development. 1993;117:751–761. doi: 10.1242/dev.117.2.751. [DOI] [PubMed] [Google Scholar]
- Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Taylor MV, Beatty KE, Hunter HK, Baylies MK. Drosophila MEF2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech Dev. 1995;50:29–41. doi: 10.1016/0925-4773(94)00323-f. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeiffle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thisse B, El Messal M, Perrin-Schmitt F. The twist gene: Isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS, Pirrotta V. New pCaSpeR P element vectors. Dros Inf Serv. 1992;71:150. [Google Scholar]