Abstract
We describe a general computational method for designing proteins that bind a surface patch of interest on a target macromolecule. Favorable interactions between disembodied amino-acid residues and the target surface are identified and used to anchor de novo designed interfaces. The method was used to design proteins that bind a conserved surface patch on the stem of the influenza hemagglutinin (HA) from the 1918 H1N1 pandemic virus. After affinity maturation, two of the designed proteins, HB36 and HB80, bind H1 and H5 HAs with low-nanomolar affinity. Further, HB80 inhibits the HA fusogenic conformational changes induced at low pH. The crystal structure of HB36 in complex with 1918/H1 HA revealed that the actual binding interface is nearly identical to that in the computational design model. Such designed proteins may be useful for both diagnostics and therapeutics.
Introduction
Molecular recognition is central to biology, and high-affinity binding proteins, such as antibodies, are invaluable for both diagnostics and therapeutics(1). Current methods for producing antibodies and other proteins that bind a protein of interest involve screening of large numbers of variants generated by the immune system or by library construction(2). The computer-based design of high-affinity binding proteins is a fundamental test of the current understanding of the physical-chemical basis of molecular recognition and, if successful, would be a powerful complement to current library-based screening methods since it would allow targeting of specific patches on a protein surface. Recent advances in computational design of protein interactions have yielded switches in interaction specificity(3), methods to generate modest-affinity complexes(4, 5), two-sided design of a novel protein interface(6), and design of a high-affinity interaction by grafting known key residues onto an unrelated protein scaffold(7). However, the capability to target an arbitrarily selected protein surface has remained elusive.
Influenza presents a serious public-health challenge and new therapies are needed to combat viruses that are resistant to existing antivirals (8) or escape neutralization by the immune system. Hemagglutinin (HA) is a prime candidate for drug development as it is the major player in viral invasion of cells lining the respiratory tract. While most antibodies bind to the rapidly varying head region of HA, recently two antibodies, CR6261 and F10, were structurally characterized (9, 10) that bind to a region on the HA stem, which is conserved among all group 1 influenza strains (Fig. S1)(11). Here, we describe a computational method for designing protein-protein interactions de novo, and use the method to design high-affinity binders to the conserved stem region on influenza HA.
Computational design method
In devising the computational design strategy, we considered features common to dissociable protein complexes. During protein complex formation, proteins bury on average ~1,600Å2 of solvent-exposed surface area (12). Interfaces typically contain several residues that make highly optimized van der Waals, hydrogen bonding, and electrostatic interactions with the partner protein; these interaction hotspots contribute a large fraction of the binding energy (13).
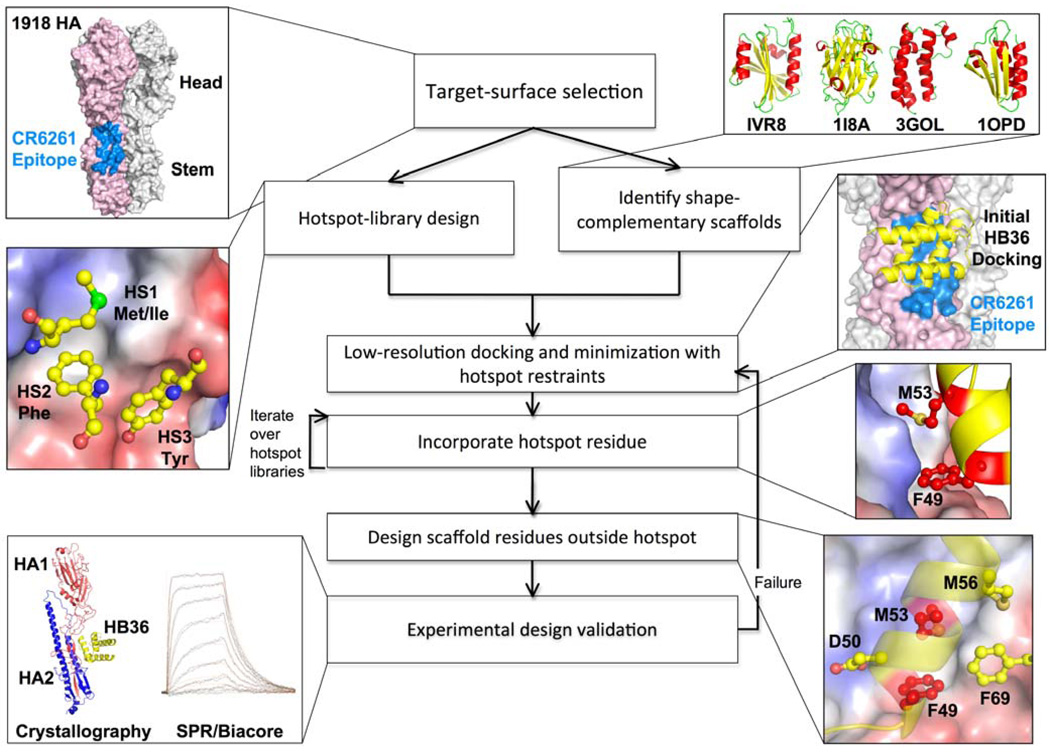
Our strategy thus centers on the design of interfaces that have both high shape complementarity and a core region of highly optimized, hotspot-like residue interactions. We engineer high-affinity interactions and high shape complementarity into scaffold proteins in two steps (see Fig. 1): (i) disembodied amino-acid residues are computationally docked or positioned against the target surface to identify energetically favorable configurations with the target surface; and (ii) shape-complementary configurations of scaffold proteins are computed that incorporate the key residues.
Fig. 1. Overview of the design process.
The flow chart illustrates key steps in the design process for novel binding proteins, with thumbnails illustrating each step in the creation of binders that target the stem of the 1918 HA.
Design of HA-binding proteins
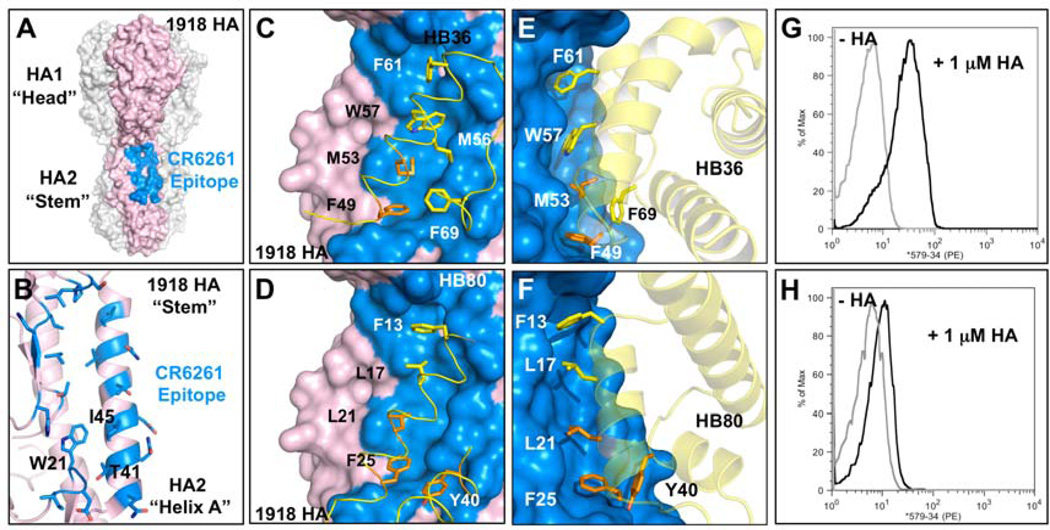
The surface on the stem of HA recognized by neutralizing antibodies consists of a hydrophobic groove that is flanked by two loops that place severe steric constraints on binding to the epitope (Fig. 2A–B)(14). In the first step of our design protocol (Fig. 1), the disembodied residues found through computational docking cluster into three regions (HS1, HS2, and HS3; Fig. 1). In HS1, a Phe side chain forms an energetically favorable aromatic-stacking interaction with Trp21 on chain 2 of the HA (HA2) (HA residue numbering corresponds to the H3 subtype sequence-numbering convention). In HS2, the nonpolar residues Ile, Leu, Met, Phe, and Val, make favorable van der Waals interactions with both the hydrophobic groove and HS1 (Fig. 1 and S2). In HS3, a Tyr side chain forms a hydrogen bond to Asp18 on HA2 and van der Waals interactions with the A-helix on HA2. The Tyr in HS3 resembles the conformation of a Tyr residue observed on the antibody in the structure of the HA and CR6261 Fab complex (Figs. S1 and S2); the HS1 and HS2 interactions are not found in the antibody structures (9, 10) (Fig. S1) (15).
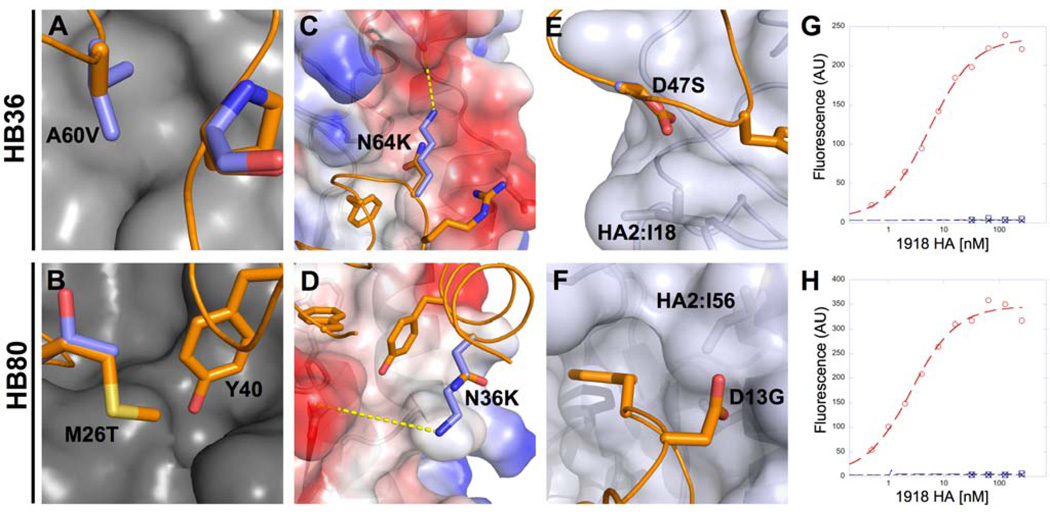
Fig. 2. Design of HB36 and HB80, targeting the stem of the 1918 HA.
(A) Surface representation of the trimeric HA structure (PDB 3R2X) from the 1918 pandemic virus, with one of the three protomers highlighted in pink. Broadly neutralizing antibody CR6261 binds a highly conserved epitope in the stem region (blue patch on surface), close to the viral membrane (bottom). (B) Enlarged view of the CR6261 epitope (blue region from part A), with CR6261 contact residues depicted as sticks. This target site on HA contains a groove lined by multiple hydrophobic residues. Loops on either side of this hydrophobic groove (above and below) constrain access to this region. Key residues on HA2 are noted in one-letter code. (C and D) Front view of the designed interaction between HB36 (C) and HB80 (D) and the target site on HA. HA is rotated approximately 60° relative to Fig. 2A. Contact segments of HB36 and HB80 are colored in yellow and residues are depicted as sticks, with hotspot residues highlighted all in orange (F49 and M53 for HB36 and L21, F25, and Y40 for HB80). Polar atoms of side chains are shown in red (oxygen) and blue (nitrogen). For clarity, the non-contacting regions from the designs have been omitted. (E and F) Further details of the designed interactions of HB36 (E) and HB80 (F) with 1918/H1 HA. (G and H) Initial binding data for HB36 (G) and HB80 (H) designs (before affinity maturation). When incubated with 1 µM 1918 HA, yeast displaying the two designed proteins show an increase in fluorescent phycoerythrin signal (x-axis) compared to the absence of 1918 HA. Coordinate files of models of 1918 HA in complex with HB36 and HB80 are available as Supporting Online Material.
In the second step, we searched a set of 865 protein structures selected for ease of experimental manipulation (16) (Table S1) for scaffolds capable of supporting the disembodied hotspot residues and shape complementary to the stem region. Each scaffold protein was docked against the stem region using the feature-matching algorithm PatchDock(17), identifying hundreds of compatible binding modes for each scaffold (260,000 in total). These coarse-grained binding modes were then refined using RosettaDock(18) with a potential function that favored configurations that maximized the compatibility of the scaffold protein backbone with as many hotspot residues as possible (see Supporting Online Material for details). Next, residues from the hotspot-residue libraries were incorporated on the scaffold. First, for each Phe conformation in HS1, scaffold residues with backbone atoms within 4Å of the hotspot residue were identified. For each of these candidate positions, the scaffold protein was placed to coincide with the backbone of the hotspot, the residue was modeled explicitly, and the rigid-body orientation was minimized. If no steric clashes were observed and the Phe was in contact with Trp21 and Thr41 of HA2 (Fig. 2B), the placement of the first hotspot was deemed successful; otherwise, another HS1 Phe conformation was selected and the process was repeated. For each success with HS1, nonpolar residues were incorporated at positions in the scaffold protein, from which the HS2 interactions could be realized, and the remainder of the scaffold protein surface was then redesigned using RosettaDesign(19).
Designing proteins also containing HS3 interactions was more challenging due to the large number of combinations of residue placements to be considered. To generate designs containing all three hotspot regions, we started by superimposing the scaffold protein on the backbone of the Tyr residue in HS3 (as for the Phe HS1 residue above). We then searched for two positions on the scaffold protein that were nearest to residues in HS1 and HS2 and were best aligned to them (see Supporting Online Material for details). These positions were then simultaneously designed to Phe in the case of HS1 and to nonpolar residues in the case of HS2. RosettaDesign(19) was then used to redesign the remainder of the interface on the scaffold protein, allowing sequence changes within a distance of 10 Å of the HA.
Experimental and structural characterization
A total 51 designs using the two hotspot-residue concept and 37 using the three-residue concept were selected for testing (Table S2 and supplemental coordinate files of all models). The designs are derived from 79 different protein scaffolds and differ from the scaffold by on average 11 mutations. Genes encoding the designs were synthesized, cloned into a yeast-display vector, and transformed into yeast strain EBY100(20, 21). Upon induction, the designed protein is displayed on the cell surface as a fusion between an adhesion subunit of the Aga2p yeast protein and a C-terminal c-myc tag. Cells expressing designs were incubated with 1 µM of biotinylated SC1918/H1 (A/South Carolina/1/1918 (H1N1)) HA ectodomain, washed, and dual-labeled with phycoerythrin-conjugated streptavidin and fluorescein-conjugated anti-c-myc antibody. Binding was measured by flow cytometry with the two fluorescent tags allowing simultaneous interrogation of binding to HA and surface display of the design.
73 designs were surface-displayed, and 2 showed reproducible binding activity towards the HA stem region(22)(Table S2) (for models, see Fig. 2C–F). One design, HA Binder 36 (HB36) used the two-residue hotspot, and bound to the HA with an apparent dissociation constant (Kd) of 200 nM(23)(Fig. 2G, Fig. S4). The starting scaffold, Structural Genomics target APC36109, a protein of unknown function from B. stearothermophilus (PDB entry 1U84), did not bind HA (Fig. S4), indicating that binding is mediated by the designed surface on HB36. A second design, HB80, used the three-residue hotspot and bound HA only weakly (Fig. 2H). The scaffold from which this design was derived, the MYB domain of the RAD transcription factor from A. Majus (PDB code: 2CJJ)(24), again did not bind the HA (Fig. S5).
In the computational models of the two designs (Fig. 2C–F), the hotspot residues are buttressed by a concentric arrangement of hydrophobic residues with an outer ring of polar and charged residues as often observed in native protein-protein interfaces. Both designs present a row of hydrophobic residues on a helix that fits into the HA hydrophobic groove. The complexes each bury approximately 1,550Å2 surface area (total), close to the mean value for dissociable protein interactions (12) and slightly larger than the total surface area buried by each of the two neutralizing antibodies (9, 10) (Fig. S1). The helical binding modes in these designs are very different from the loop-based binding observed in the antibody-bound structures.
Affinity maturation
The computational design protocol is far from perfect; the energy function that guides design contains numerous approximations (25) and conformational sampling is incomplete. We used affinity maturation to identify shortcomings in the design protocol. Libraries of HB36 and HB80 variants were generated by single site-saturation mutagenesis at the interface, or by error-prone PCR (epPCR), and subjected to two rounds of selection for binding to HA using yeast-surface display (21, 24).
For both designed binders, the selections converged on a small number of substitutions that increase affinity and provide insight into how to improve the underlying energy function. Among the key contributions to the energetics of macromolecular interactions are short-range repulsive interactions due to atomic overlaps, electrostatic interactions between charged and polar atoms, and the elimination of favorable interactions with solvent (desolvation). The affinity-increasing substitutions point to how each of these contributions can be better modeled in the initial design calculations.
Repulsive interactions
For HB36, substitution of Ala60 with the isosteres Thr/Val increased the apparent binding affinity 25-fold (apparent Kd’s for all design variants are listed in Table 1). These substitutions fill a void between the designed protein and the HA surface, but were not included in the original design because they were disfavored by steric clashes within HB36 (Fig. 3A). Backbone minimization, however, readily relieved these clashes resulting in higher predicted affinity for the substitutions. For HB80, a Met26Thr mutation significantly increased binding compared to the starting design. Modeling showed that Met26 disfavored the conformation of the Tyr hotspot residue, rationalizing the substitution to a smaller residue (Fig. 3B). More direct incorporation of backbone minimization in the design algorithm should allow identification of such favorable interactions from the start, whereas insuring that hotspot residues are fully relaxed in the design would eliminate unfavorable interactions.
Table 1.
Dissociation constants (Kd) for binding of design variants to SC1918 HA
| Design | Kd [nM]* |
|---|---|
| 1U84 (HB36 Scaffold) | NB (NB) |
| HB36 | 200 (> 2000) |
| HB36 Asp47Ser | 5 |
| HA36 Ala60Val | 8 |
| HB36.3 (HB36 Asp47Ser, Ala60Val) | 4 (29) |
| HB36.4 (HB36 Asp47Ser, Ala60Val, Asn64Lys) | 4 (22) |
| 2CJJ (HB80 Scaffold) | NB |
| HB80 | > 5000 |
| HB80 Met26Thr | 100 |
| HB80 Asn36Lys | 300 |
| HB80 Met26Thr Asn36Lys | 7.5 |
| HB80 Δ54–95, Met26Thr, Asn36Lys | 5 |
| HB80.3 (HB80 Δ54–95, Aspl2Gly, Ala24Ser, Met26Thr, Asn36Lys) | 3(38) |
Kd was determined using yeast surface display titrations.
Number in parentheses indicates Kd determined by SPR.
NB, no binding.
Fig. 3. Affinity Maturation of HB36 and HB80.
Substitutions that increase the affinity of the original designs can be classified as deficiencies in modeling the (A and B) repulsive interactions HB36 Ala60Val (A), HB80 Met26Thr (B); (C and D) electrostatics HB36 Asn64Lys (C), HB80 Asn36Lys (D); (E and F) and solvation HB36 Asp47Ser (E), HB80 Asp12Gly (F). Binding titrations of HB36.4 (G) and HB80.3 (H) to SC1918/H1 HA as measured by yeast surface display. Red circles represent the affinity-matured design, blue squares the scaffold protein from which the design is derived, and black crosses represent the design in the presence of 750nM inhibitory CR6261 Fab.
Electrostatics
In HB36, the substitution to Lys at position 64 places a complementary charge adjacent to an acidic pocket on HA near the conserved stem region (Fig. 3C); in HB80, an Asn36Lys substitution positions a positive charge 6.5Å from the negative Asp18 on HA2 (Fig. 3D). These substitutions all enhance electrostatic complementarity in the complex. The lysines were not selected in the design calculations because the magnitude of surface-electrostatic interactions between atoms outside of hydrogen-bonding range are largely reduced; improvement of the electrostatic model would evidently allow design of higher-affinity binders from the start.
Desolvation
In HB36, 8 different substitutions at Asp47 increased apparent affinity by over an order of magnitude compared to the original design (Table S3); the highest-affinity substitution was Asp47Ser that increased binding affinity circa 40-fold. The design of an unfavorable charged group in this position likely stems from underestimation of the energetic cost of desolvating Asp47 by the aliphatic Ile18 on HA2 (Fig. 3E); the substitutions remedy this error by replacing the Asp with residues that are less costly to desolvate upon binding. In HB80, an Asp12Gly substitution relieves the desolvation by the neighboring Ile56 on HA2 (Fig. 3F). With improvements in the solvation model, the deleterious Asp residues would not be present in starting designs.
The favorable substitutions were combined and the proteins were expressed with a His-tag in E. coli and purified by nickel affinity and size-exclusion chromatography. The variant HB36.3, incorporating the Asp47Ser and Ala60Val substitutions, bound to SC1918/H1 HA as confirmed by surface plasmon resonance (SPR; Fig. S6), ELISA, and co-elution on a size-exclusion column (Fig. S7). The HB36.4 variant, which incorporates Asp47Ser, Ala60Val, and Asn64Lys, bound to SC1918/H1 HA with a dissociation constant measured by SPR of 22nM and an off-rate of 7·10−3 s−1 (Table S4). Co-incubation with an excess of CR6261 Fab abolished binding to the HA (Fig. 3G), consistent with HB36.4 binding in close proximity to the same stem epitope on the HA. For the HB80 design, the combination of the affinity-increasing mutations reduced surface expression on yeast, indicative of poor stability. Therefore, we excised a C-terminal stretch (Δ54–95) greatly boosting surface expression of the design with no significant loss of binding affinity (Fig. S9). HB80.3, which incorporates the truncation as well as the Asp12Gly, Ala24Ser, Met26Thr, and Asn36Lys substitutions, has a Kd=38nM with off-rate of 4·10−2 s−1 by SPR. As with HB36.4, co-incubating HA with the CR6261 Fab completely abolished binding to HB80.3 (Fig. 3H), consistent with the designed binding mode.
Site-directed alanine mutagenesis of several core positions on each affinity-matured design partially or completely knocked out HA binding (Table S5, Fig. S10) supporting the computational model of the designed interfaces (26). Furthermore, no mutations were uncovered during selection for higher affinity that were inconsistent with the designed binding modes.
Crystal structure of the HB36.3-SC1918 HA complex
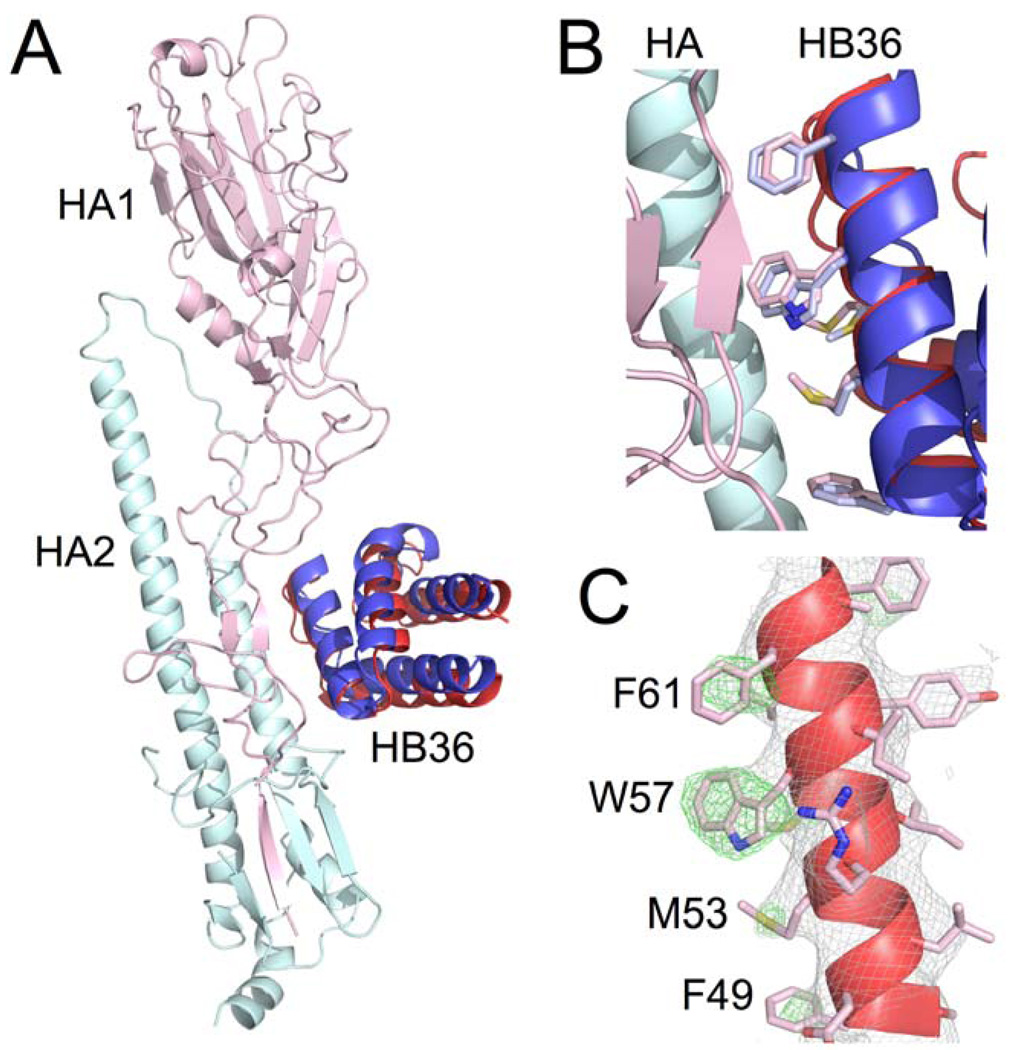
The crystal structure of HB36.3 in complex with the SC1918 HA ectodomain was determined to 3.1Å resolution. After molecular replacement using only the 1918/H1 HA structure as the search model (approximately 86% of the protein mass in the crystal asymmetric unit), clear electron density was observed for HB36.3 near the target surface in the HA stem region into which HB36.3 could be unambiguously placed. The orientation was essentially identical to the designed binding mode, with the modified surface of the main recognition helix packed in the hydrophobic groove on HA (Fig. 4A). To obtain unbiased density for the designed side chains, the native structure from which HB36.3 was derived (PDB entry: 1U86) was manually fit into the electron-density maps and contact side chains were pruned back to their β-carbon. After crystallographic refinement, electron density became apparent for the side chains of most of the contact residues on HB36.3, allowing the predominant rotamers to be assigned for Phe49, Trp57, Phe61, and Phe69. This unbiased density clearly shows that these four hydrophobic side chains are all positioned as in the designed model (Fig. 4B). The Met53 side chain is consistent with the design model (Fig. 4C), although other rotamers could also be fit to the map. For Met56, only very weak side-chain density was observed. Overall, the crystal structure is in excellent agreement with the designed interface, with no significant deviations at any of the contact positions.
Fig. 4. Crystal structure of HB36 in complex with SC1918 HA confirms designed interface.
(A) Superposition of the crystal structure of HB36.3-SC1918/H1 complex (HB36.3 in red with SC1918 HA1 subunit in pink and HA2 subunit in cyan) and the computational design (blue) reveals good agreement in the position of the main recognition helix, with a slight rotation of the rest of the protein domain. Superposition was performed using the HA2 subunits. For clarity, only the HA from the crystal structure is depicted here (the HA used for superposition of the design, which is essentially identical to the crystal structure, was omitted). (B) Close up of the SC1918 HA-HB36.3 interface, highlighting the close agreement between the design and the crystal structure. The main recognition helix is oriented approximately as in (A), with the HB36.3 crystal structure in red (pink side chains), the design in blue (light blue side chains), and HA in cyan and pink at left. (C) Unbiased 2Fo-Fc (gray mesh, contoured at 1σ) and Fo-Fc (green mesh, contoured at 3σ) electron-density maps for the main recognition helix of HB36.3. The helix is oriented as in (B), with key contact residues of the left face of the helix in this view labeled (the right surface faces and interacts with the core of the HB36.3 protein). Significant density was observed for most of the large side chains at the interface with HA, including F49, M53, W57, F61, and F69 (not visible in this view). While side chains are shown here to illustrate their agreement with the experimental electron density, maps were calculated after initial refinement of an HA-HB36.3 model with the following side chains truncated to alanine (no prior refinement with side chains present): F49, M53, M56, W57, F61, and F69.
Given the quite low (2 out of the 73 surface displayed proteins) design success rate and starting affinities, the atomic-level agreement between the designed and experimentally determined HB36.3-SC1918 HA complex is very encouraging and suggests that, despite their shortcomings, the current energy function and design methodology capture essential features of protein-protein interactions.
Cross-reactivity and inhibitory activity
The surface contacted by HB36.3 is accessible and highly conserved in the HAs of most group 1 influenza viruses, suggesting that it may be capable of binding not only other H1 HAs, but also other HA subtypes. Indeed, binding of HB36.3 to A/South Carolina/1/1918(H1N1) and A/WSN/1933(H1N1) is readily detectable in solution by gel filtration (Fig. S7), as well as high-affinity binding of HB36.4 to A/Vietnam/1203/2004 H5 subtype by yeast display (Fig. S11).
While a crystal structure of HB80 in complex with HA has not been obtained, the mutational data and the antibody-competition results suggest that HB80 also binds to the designed target surface, overlapping with HB36 and CR6261. Consequently, HB80.3 is also expected to be highly cross-reactive and binds with high affinity to A/Vietnam/1203/2004 H5 HA (Fig. S11), and to H1, H2, H5, and H6 subtypes by biolayer interferometry (Fig. 5 A,B). Overall, the pattern of HB80 binding mirrors that of CR6261 and binds most of the group 1 HAs tested, with no detectable binding to group 2 HAs.
Fig. 5. HB80 binds multiple HA subtypes and inhibits the conformational changes that drive membrane fusion.
(A) Phylogenetic tree depicting the relationship between the 16 influenza A hemagglutinin subtypes. These subtypes can be divided into two main lineages, groups 1 and 2. CR6261 has broad activity against group 1 viruses. HB80.3 has a similar cross-reactivity profile and binds multiple group 1 subtypes, including H1 and H5. (B) Binding data for HB80.3 and CR6261 Fab against a panel of HAs. “+”, “++”, and “+++” indicate relative degree of binding (approximately 10−7, 10−8, and 10−9 M, respectively), while “-“ indicates no detectable binding at the highest concentration tested (100nM). (C) HB80.3 inhibits the pH-induced conformational changes that drive membrane fusion. Exposure to low pH converts 1918 H1 HA (top panel) and the Viet04 H5 HA to a protease susceptible state (lane 1), while HAs maintained at neutral pH are highly resistant to trypsin (lane 3). Pre-incubation of HB80.3 with H1 and H5 prevents pH-induced conformational changes and retains the HAs in the protease-resistant, pre-fusion state (lane 2).
Antibody CR6261 inhibits influenza virus replication by blocking the pH-induced refolding of HA, which drives fusion of the viral envelope with the endosomal membrane of the host cell. Given extensive overlap between the HB80.3 and CR6261 binding sites and its high affinity for SC1918 HA, it seemed plausible that HB80.3 would also block this conformational change. Indeed, HB80.3 inhibits the pH-induced conformational changes in both H1 and H5 HAs (Fig. 5C, Fig. S12)(10), suggesting that this design may possess virus-neutralizing activity against multiple influenza subtypes (27). Further work will be needed to explore the potential utility of HB80.3 in a therapeutic or diagnostic setting, but these results suggest that de novo computational design of antiviral proteins is feasible.
Supplementary Material
Footnotes
Supporting Online Material
Materials and Methods
Supporting Discussion
Fig. S1 to S12
Tables S1 to S7
Coordinate files for models of complexes of HA with HB36 and HB80
References
References and Notes
- 1.Ledford H. Nature. 2008;455:437. doi: 10.1038/455437a. [DOI] [PubMed] [Google Scholar]
- 2.Lerner RA. Angew Chem Int Ed Engl. 2006;45:8106. doi: 10.1002/anie.200603381. [DOI] [PubMed] [Google Scholar]
- 3.Kortemme T, et al. Nat. Struct. Mol. Biol. 2004;11:371. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 4.Jha RK, et al. J Mol Biol. 2010;400:257. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang PS, Love JJ, Mayo SL. Protein Sci. 2007;16:2770. doi: 10.1110/ps.073125207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karanicolas J, et al. Mol. Cell. 2011 doi: 10.1016/j.molcel.2011.03.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, et al. Proc Natl Acad Sci U S A. 2007;104:5330. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bautista E, et al. N Engl J Med. 2010;362:1708. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 9.Sui J, et al. Nat Struct Mol Biol. 2009;16:265. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekiert DC, et al. Science. 2009;324:246. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Group 1 includes 10 of the 16 HA subtypes: H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16. Group 2 includes the remaining 6 subtypes: H3, H4, H7, H10, H14, and H15.
- 12.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 13.Clackson T, Wells JA. Science. 1995;267:383. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 14.Rossmann MG. J Biol Chem. 1989;264:14587. [PubMed] [Google Scholar]
- 15.The other hotspot residues (HS1 and HS2) differed from the sidechains observed in the crystal structures in their conformation or identity. Each hotspot residue was further diversified by constructing all conformations, the terminal atoms of which coincided with those modeled above. For instance, for HS3, these consisted of all Tyr conformations that matched the position of the aromatic ring and hydrogen bond. This diversification step produced a ‘fan’ of backbone positions for each residue in the hotspot libraries.
- 16.Proteins in the scaffold set contained no disulfides, were expressed in E. coli, and were predicted to form monomers (see Supplemental Information).
- 17.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Nucleic Acids Res. 2005;33:W363. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray JJ, et al. J Mol Biol. 2003;331:281. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 19.Kuhlman B, et al. Science. 2003;302:1364. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Skehel JJ, Wiley DC. Proc Natl Acad Sci U S A. 1999;96:8967. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao G, et al. Nat Protoc. 2006;1:755. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 22.A third design HB35 bound HA at apparent low µM affinity; however, binding was only partially abolished upon co-incubation of HA with the CR6261 Fab, indicating of at most partial contact with the target surface on the stem region of HA, and so this design was eliminated from further consideration (Fig. S7). A handful of other designs bound HA albeit weakly and with incomplete reproducibility (see supporting results).
- 23.We recorded dissociation constants using two main methods: by titration of HA against yeast surface-displayed designs, and by fitting both kinetic and equilibrium measurements using surface plasmon resonance. As there is a discrepancy in determining Kd's between the methods, measurements derived from yeast surface-display titrations are listed as apparent Kd and should be viewed qualitatively. (see Supporting Online Material).
- 24.Stevenson CE, et al. Proteins. 2006;65:1041. doi: 10.1002/prot.21136. [DOI] [PubMed] [Google Scholar]
- 25.Das R, Baker D. Annu Rev Biochem. 2008;77:363. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 26.The alanine-scan mutations were as follows: for HB36.3, Phe49, Met53, and Trp57; for HB80.1 Phe13, Phe25, and Tyr40 (Table S4 and supplemental resutls).
- 27.HB36.4 was not able to block the pH-induced conformational changes in the H1 HA under identical assay conditions, even though HB36.4 and HB80.3 have very similar dissociation constants and kinetic off-rates at pH 7.5 (Fig. S12 and supplemental results).
- 28.Computational designs were generated on resources generously provided by participants of Rosetta @ Home and the Argonne National Leadership Computing Facility. SJF was supported by a long-term fellowship from the Human Frontier Science Program, JEC was supported by the Jane Coffin Childs Memorial Fund, and EMS by career development award NIH/NIAID AI057141. Research in the Baker lab was supported by grants from the Defense Advanced Research Projects Agency, Defense Threat Reduction Agency, and HHMI and in the Wilson lab by NIH AI058113, predoctoral fellowships from the Achievement Rewards for College Scientists Foundation and the NIH Molecular Evolution Training Program GM080209 (D.C.E.), and the Skaggs Institute for Chemical Biology. X-ray diffraction datasets were collected at the Stanford Synchrotron Radiation Lightsource beamline 9-2 and at the Advanced Photon Source beamline 23ID-B (GM/CA-CAT). The GM/CA CAT 23-ID-B beamline has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. Coordinates and structure factors were deposited in the Protein Data Bank (PDB) as entry 3R2X.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.