Abstract
The nicotinic acetylcholine receptor and related Cys-loop receptors are ligand-gated ion channels that mediate fast synaptic transmission throughout the central and peripheral nervous system. A highly conserved aspartate residue (D89) that is near the agonist binding site, but does not directly contact the ligand, plays a critical part in receptor function. Here we probe the role of D89 using unnatural amino acid mutagenesis coupled with electrophysiology. Homology modeling implicates several hydrogen bonds involving D89. We find that no single hydrogen bond is essential to proper receptor function. Apparently the side chain of D89 establishes a redundant network of hydrogen bonds; these bonds preorganize the agonist binding site by positioning a critical tryptophan residue that directly contacts the ligand. Earlier studies of the D89N mutant led to the proposal that a negative charge at this position is essential for receptor function. However, we find that receptors with neutral side chains at position 89 can function well, if the side chain is less perturbing than the amide of asparagine (nitro or keto groups allow function), or if a compensating backbone mutation is made to relieve unfavorable electrostatics.
Neuroreceptors are central players of synaptic transmission, receiving and interpreting chemical signals between neurons in the nervous system. Neuroreceptors of the ligand-gated ion channel (LGIC) family convert incoming chemical signals into electrical output. In the LGIC gating process, neurotransmitters are recognized by ligand-binding domains, and binding triggers conformational changes within the structure to form an ion-conducting pore.
The nicotinic acetylcholine receptor (nAChR) has served as a prototype for understanding the structure and function of the Cys-loop family of LGICs (also known as pentameric LGICs). In this superfamily of receptors, which also includes γ-aminobutyric acid, glycine, and serotonin receptors, the five subunits are symmetrically or pseudosymmetrically arranged around a central ion-conducting pore. Each subunit contains a four-helix transmembrane domain that contains the ion channel gate and an extracellular ligand-binding domain. Members of nAChR family are expressed at the neuromuscular junction and in the electric organ of eels and rays (muscle-type receptors), as well as in the central nervous system (neuronal receptors). The muscle-type receptor is the best characterized, and the form studied here is the embryonic muscle nAChR, with a subunit stoichiometry of two α1 subunits and one each of β1, γ, and δ subunits(1). The nAChR has two agonist binding sites located at the α/γ and α/δ subunit interfaces(2–4). The α subunits contribute the primary binding site components, termed loops A, B, and C, while the γ and δ subunits contribute the complementary components, primarily loop D, with possible contributions from loops E and F. The focus of the present work is on loops A and B of the α subunit.
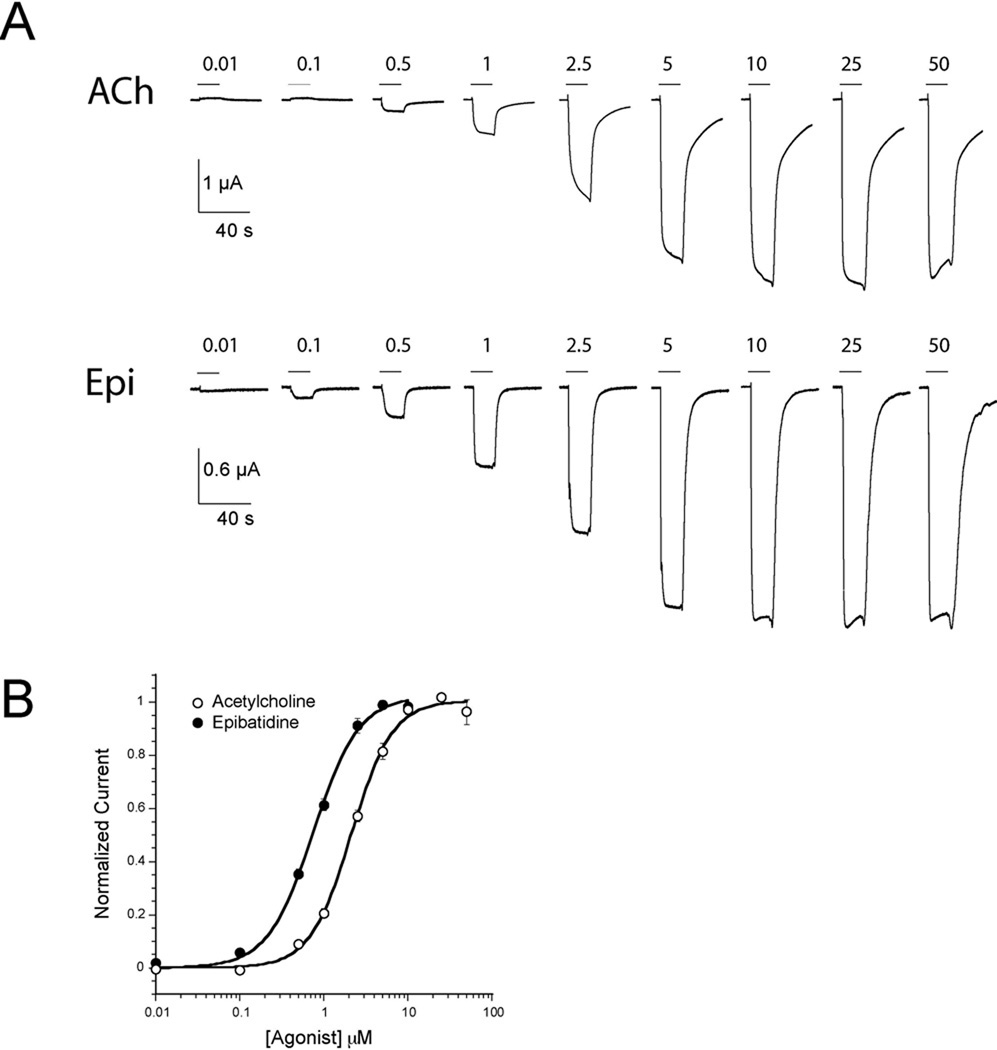
Work over the past several years on acetylcholine binding protein (AChBP) orthologs from mollusks has led to important new insights into the structures of Cys-loop receptor ligand-binding domains(5–8). AChBP is a soluble, homopentameric protein produced in glial cells that is homologous to the nAChR ligand-binding domain. Crystal structures of AChBP with various agonists bound have established that the nAChR binding site is comprised of a box of conserved aromatic residues. One of these conserved aromatic residues is a tryptophan on loop B, W149 (Figure 1, 2). Previous studies by this lab established that this tryptophan makes a strong cation-π interaction with ACh in the muscle-type receptor (9), and its role as a component of the AChBP “aromatic box” confirmed those findings. Subsequent work showed that the potent nicotinic agonist epibatidine also makes a cation-π interaction with W149. (9, 10). Nicotine is a quite weak agonist at the muscle-type receptor; its actions at the neuronal receptors are more substantial. As such, epibatidine is a better candidate for studying interactions between non-ACh agonists and the muscle-type receptor.
Figure 1.
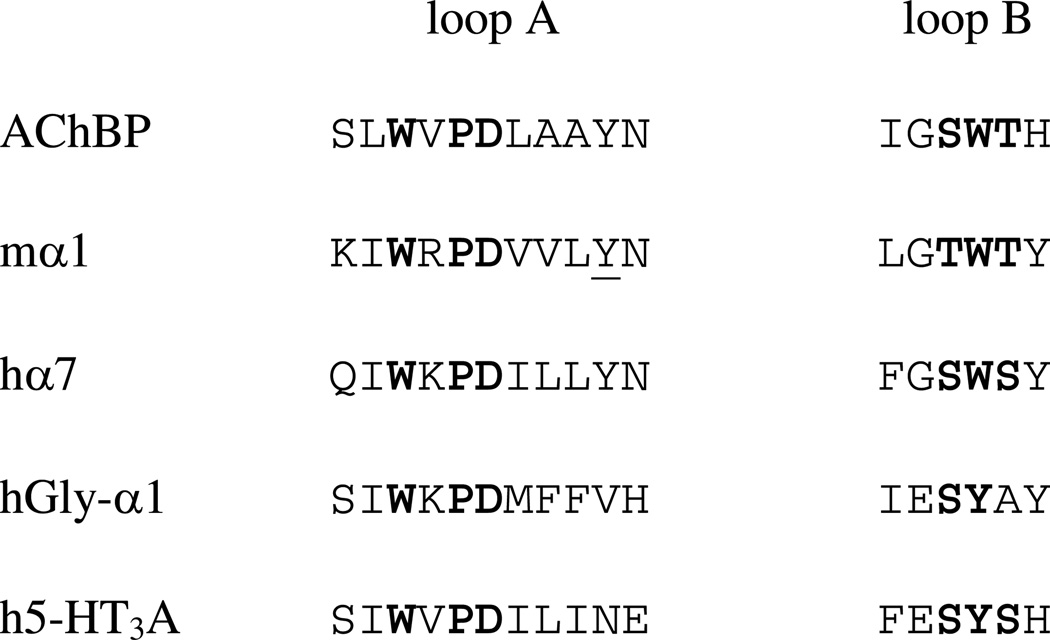
Alignments of portions of loops A and B for several Cys-loop receptors. The WxPD motif of loop A and the region around W149 of loop B are highlighted. AChBP: ACh binding protein from Limnaea stagnalis; mα1: nAChR α1 subunit of mouse muscle (studied here); hα7: human nAChR α7 subunit; hGly-α1: human glycine receptor α1 subunit; h5-HT3A: human 5-HT3 A subunit.
Figure 2.
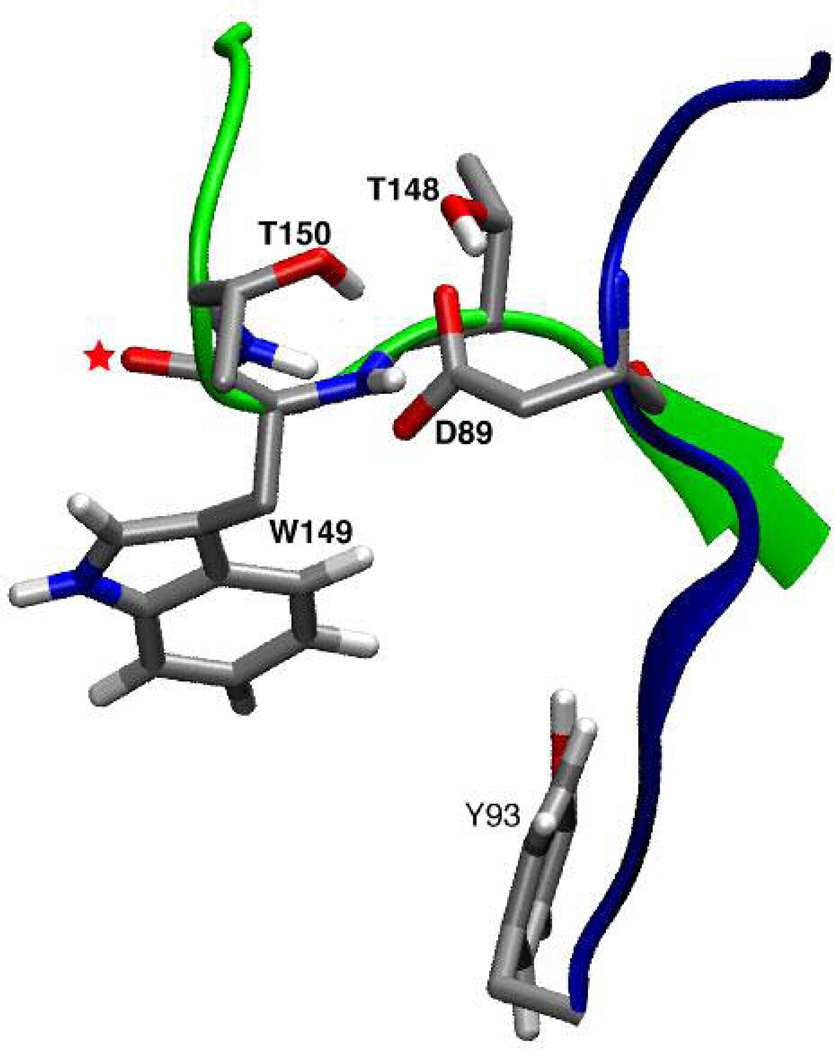
Region investigated in the present work. Loop A is shown in blue; loop B in green. Highlighted are the side chains of D89, T148, and T150, which together can form a network of hydrogen bonds. Also shown are contributors to the agonist binding site: Y93 from loop A; W149 side chain and W149 backbone carbonyl (red star) from loop B. This carbonyl and the side chain of Trp 149 point directly at the agonist, which in this view lies “behind” loop B. This image results from MD simulations, as discussed in text.
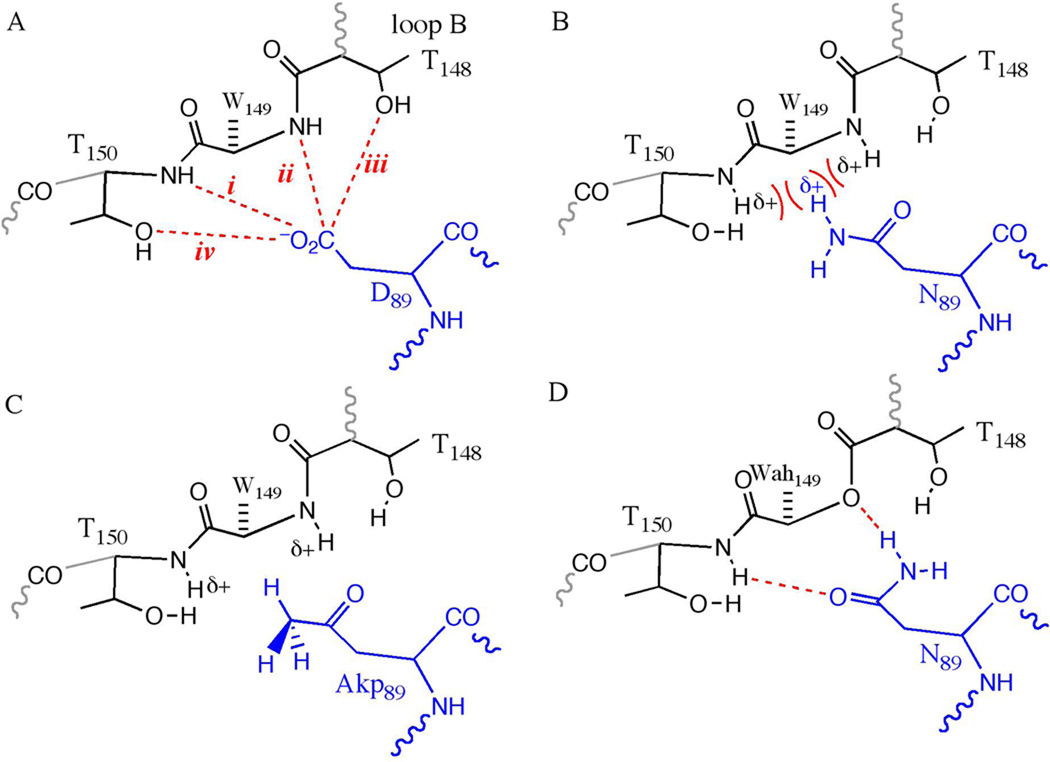
The AChBP crystal structures suggest other important ligand-binding domain interactions that require verification through experiment. One structurally interesting interaction involves a conserved aspartate on loop A, D89 (we use the residue numbering that corresponds to the nicotinic receptor, not the AChBP numbers)(6). This residue is part of a highly conserved WxPD motif exhibited across the entire Cys-loop superfamily (Figure 1). In the crystal structures of AChBP, D89 is positioned to interact with loop B through any of a number of hydrogen bonds between the aspartate carboxylate side chain and loop B residues T148, W149, and T150 (Figure 2). For the purposes of discussion, a schematic of the putative hydrogen bonding network, labeling potential hydrogen bonds, is shown in Figure 3A. The high conservation of the WxPD motif, and the clear interaction of D89 with a known component of the agonist binding site (loop B/W149) have generated considerable interest in D89. Note that loop A also contains a canonical contributor to the aromatic box, Y93
Figure 3.
Schematics of potential hydrogen bonding interactions between loops A (blue) and B (black). A. The wild type receptor. B. D89N, highlighting the potential electrostatic clash. C. D89Akp; note the lack of an electrostatic clash. D. The double mutant D89N/W149Wah; note how hydrogen bond ii could, in principle, be restored.
Recently, Lee and Sine(11) have investigated D89 and its role in agonist binding by combining site-directed mutagenesis with single channel kinetic analyses. Neutralization of the negative charge at position 89 through a mutation to asparagine or threonine (D89N and D89T, respectively; Figure 3B) substantially compromised receptor function, producing significant decreases in the bimolecular forward rate constant for agonist-receptor binding. In contrast, mutating residues T148 and T150, singly and in combination, did not seriously impact function. From these results, Lee and Sine concluded that D89 plays a structural role in stabilizing loop B, in particular W149, for agonist association. They concluded that essential structural features of the D89/loop B network were the negative charge of D89 and hydrogen bonds between the aspartate carboxylate and the amide backbones of T150 and T149 (hydrogen bonds i and ii of Figure 3A). They also proposed that a possible polarization of the backbone carbonyl of W149 contributes to ACh binding(6).
In the present study, we sought to further probe the role of the conserved D89 in the nAChR ligand-binding domain. Through the incorporation of unnatural amino acids using the nonsense suppression methodology(12–14), we introduced more subtle modifications to the side chain of D89, allowing what we have termed “chemical-scale” studies of such complex receptors. By chemical scale we mean, in effect, the distance scale to which chemists are accustomed: the functional group; the specific bond rotation or local conformational change; the precise noncovalent interaction. We have also incorporated amide-to-ester backbone mutations in loop B to probe proposed hydrogen bonds to this region. We conclude that the significantly disruptive D89N mutation affects receptor function in several ways, including: a distortion of the hydrogen bonding network; the introduction of an electrostatic clash between the asparagine amide side chain and the backbone amides of loop B; and, to a lesser extent, a neutralization of charge. We also find that, of the network of hydrogen bonds implied by the AChBP structure (Figure 3A), no one hydrogen bond is singularly important. Rather, maintaining the overall network of hydrogen bonds and avoiding electrostatic and/or steric clashes are essential for proper function. To support our unnatural amino acid mutagenesis studies, we conducted molecular dynamics simulations of the wild-type and D89N mutant ligand-binding domains to evaluate the hydrogen bonding network.
Materials and Methods
Synthesis of Wah cyanomethyl ester (3-(3-Indolyl)-2-hydroxypropanoic acid cyanomethyl ester)
α-hydroxytryptophan (Wah) (3-(3-Indolyl)-2-hydroxypropanoic acid) cyanomethyl ester was synthesized according to previously published methods(10, 15). The hydroxy acid (255 mg, 1.24 mmol) was dissolved in 1.9 ml of ClCH2CN (30 mmol) and 514 µl Et3N (3.65 mmol). Upon stirring under Ar for 45 min, the solution turned pale yellow. The reaction mixture was concentrated and dried under vacuum. The material was dry loaded onto a flash silica gel column and run in 9:1 methylene chloride/ethyl acetate to give 242 mg (80 % yield) of hydroxyl-tryptophan cyanomethyl ester: 1H NMR (DMSO) δ 3.04 (m, 2H), 3.32 (broad s, 1H), 4.38 (broad s, 1H), 4.94 (s, 2H), 6.96 (t, 1H, J = 7.2 Hz), 7.05 (t, 1H, J= 6.9 Hz), 7.12 (d, 1H, J = 2 Hz), 7.32 (d, 1H, J = 7.8 Hz), 7.51 (d, 1H, J = 7.5 Hz) , 10.85 (s, 1H); 13C NMR 30.0, 48.9, 70.7, 109.7, 111.3, 115.8, 118.3, 118.3, 120.8, 123.8, 127.3, 136.0, 172.7: Electrospray MS Calcd for C13H12N2O3 plus H: 245.08. Found m/z (M+H): 245.0.
Synthesis of dCA-Wah
Hydroxytryptophan cyanomethyl ester (11 mg, 45 µmol) was dissolved in 315 µl dry DMF in a flame-dried 5 ml round bottom flask with a stir bar. The dinucleotide dCA (20 mg, 16.7 µmol) was added, and the reaction stirred under Ar for 9 hours. Upon completion of the reaction, the pure compound was obtained by preparative HPLC. Electrospray MS Calcd for C30H35N9O15P2 minus H: 823.17. Found m/z (M-H): 822.0.
Synthesis of t-Butyl 2-Diphenylmethyleneimino-4-nitro-butanoate
Lithium diisopropylamide (6.8 mL 0.5 M in THF, 3.4 mmol) was added to a solution of N-diphenylmethyleneglycine t-butyl ester (1 g, 3.4 mmol) in THF (6.8 mL) at −78°C. After 1 h, nitroethylene (4.25 mL 0.8 M in THF, 3.4 mmol), prepared from 2-nitroethanol as reported previously(16), was added to the mixture and stirred for an additional 1 h. The reaction was then brought to room temperature and a 1:1 mixture of ethyl acetate and water (20 mL) was added. The organic layer was separated, washed with brine, dried (Mg2SO4), and evaporated. Crude product was purified on a flash silica column using a 1:1 ethyl acetate and hexane solvent system to yield 690 mg pure product (53%): 1H NMR (CDCl3) δ 1.44 (s, 9H), 2.58 (m, 2H), 4.08 (t, 1H), 4.53 (m, 2H), 7.20 (m, 2H), 7.36 (m, 3H), 7.47 (m, 3H), 7.67 (m, 2H); ES-MS calculated mass for C21H24N2O4: 368.17, Found m/z (M+H+): 369.0, (M+Na+): 390.8, (M+K+): 407.0.
Synthesis of nitrohomoalanine
t-Butyl 2-Diphenylmethyleneimino-4-nitrobutanoate was deprotected by adding 1 N HCl and stirring for 15 h at room temperature. The reaction was lyophilized and taken onto the next reaction.
Synthesis of (NVOC)-nitrohomoalanine
Nitrohomoalanine (230 mg, 0.6 mmol) and Na2CO3 (132 mg, 1.2 mmol) were dissolved in water (10 mL). To this solution, NVOC-Cl (171 mg, 0.6 mmol) in dioxane (10 mL) was added and stirred for 4 h at room temperature. The reaction was evaporated to half of the reaction volume and then diluted with 20 mL of water. The solution was extracted with ether (20 mL) until the organic was no longer colored. The aqueous layer was acidified with HCl to a pH ~2 (solution became cloudy) and extracted with dichloromethane until organic layer was clear. The organic layers were dried and evaporated to yield 209 mg of (NVOC)-nitrohomoalanine (90%); ES-MS calculated for C14H17N3O10: 387.09, Found m/z (M-H−): 387.0. Crude product was taken on directly to the next step.
Synthesis of (NVOC)-nitrohomoalanine cyanomethyl ester
(NVOC)-nitrohomoalanine (50 mg, 0.13 mmol) was dissolved in 5 mL ClCH2CN. Et3N (18.3 µL, 0.13 mmol) was added and the solution was stirred under Ar for 4 h. The reaction was evaporated and purified on a flash silica column with a 1:1 mixture of ethyl acetate and hexanes solvent system to yield 40 mg of product (72%). 1H NMR (CDCl3) δ 2.95 (m, 2H), 3.18 (m, 2H), 3.96 (s, 3H), 4.02 (s, 3H), 4.33 (m, 1H), 4.96 (d, 2H), 5.56 (d, 2H), 5.72 (d, 1H), 6.91 (s, 1H), 7.72 (s, 1H). ES-MS calculated for C16H18N4O10: 426.10, Found m/z (M+Na+): 449.0, (M+K+): 465.0.
Synthesis of Nha-dCA
(NVOC)-nitrohomoalanine cyanomethyl ester (10 mg, 2.3 µmol) was dissolved in 0.5 mL dry DMF. The dinucleotide dCA was added (10 mg, 8.4 µmol) and stirred under Ar overnight. The reaction was purified by reverse phase HPLC. ES-MS calculated for C33H38N11O22P2: 1005.9, Found m/z (M-H−): 1004.2.
Synthesis of Akp-dCA
The synthesis of 2-amino-4-ketopentanoic acid and preparation of Akp-dCA was described previously(17).
Unnatural Amino Acid Suppression
Synthetic amino acids and α-hydroxy acids were conjugated to the dinucleotide dCA and ligated to truncated 74 nt tRNA as previously described.(15, 18) Aminoacyl tRNA was deprotected by photolysis immediately prior to co-injection with mRNA, as described.(18, 19) Typically, 25 ng of tRNA were injected per oocyte along with mRNA in a total volume of 50 nl/cell. mRNA was prepared by in vitro runoff transcription using the Ambion (Austin, TX) T7 mMessage mMachine kit. The site of interest was mutated to the amber stop codon by standard means, verified by sequencing through both strands. Mouse muscle embryonic nAChR in the pAMV vector was used. A total of 4.0 ng of mRNA was injected in the subunit ratio of 10:1:1:1 α:β:γ:δ. In all cases, a Leu-to-Ser mutation at a site 50 Å from the nAChR ligand-binding domain in the M2 helix, known as 9’, was used to lower EC50 values to a measurable range(10, 20). Previous work on this mutation has shown that a Leu9’Ser mutation in the β subunit lowers EC50 values 40-fold without changing trends in EC50 values(13, 20). In addition, the α subunits contain an HA epitope in the M3–M4 cytoplasmic loop for western blot studies. Control experiments show that this epitope does not detectably alter EC50. As a negative control for suppression, truncated 74 nt tRNA or truncated tRNA ligated to dCA was co-injected with mRNA in the same manner as fully charged tRNA. At the positions studied here, no current was ever observed from these negative controls. The positive control for suppression involved wild-type recovery by co-injection with 74 nt tRNA ligated to dCA-Thr or dCA-Trp. Frameshift suppression at αD89 was utilized as described by Rodriguez et. al.(21)
Electrophysiology
Stage VI oocytes of Xenopus laevis were employed. Oocyte recordings were made 24 to 48 h post injection in two-electrode voltage clamp mode using the OpusXpress™ 6000A (Axon Instruments, Union City, California). Oocytes were superfused with Ca2+-free ND96 solution at flow rates of 1 ml/min before application, 4 ml/min during drug application, and 3 ml/min during wash. Holding potentials were −60 mV. Data were sampled at 125 Hz and filtered at 50 Hz. Drug applications were 15 s in duration. Acetylcholine chloride was purchased from Sigma/Aldrich/RBI. Epibatidine was purchased from Tocris as (±) epibatidine dihydrochloride. All drugs were prepared in sterile ddi water for dilution into calcium-free ND96. Dose-response data were obtained for a minimum of 10 concentrations of agonists and for a minimum of 3 cells. Dose-response relations were fitted to the Hill equation to determine EC50 and Hill coefficient values.
Generation of Mouse Muscle nAChR Heteropentamer Computational Model
A model of the mouse muscle ligand-binding domain was created by first aligning the mouse muscle nAChR sequence with the sequence of the Torpedo marmorata acetylcholine receptor whose structure was solved using electron microscopy by Unwin(22) and resolved to 4 Å resolution(23). An alignment was generated using the T-Coffee website. A homology model was then built with this alignment using Prime(24), within the Schrödinger suite of programs. The individual chains of the T. marmorata structure 2BG9 were used as templates for each subunit type. Chain A of the T. marmorata structure was used as a template for both α subunits of the mouse muscle nAChR. Each subunit was exported as a PDB file and aligned in Swiss PDB Viewer(25). This structure was imported back to Prime where a sidechain prediction algorithm was used.
This heteropentamer was converted to GROMACS(26) format and inserted in a periodic box with 7 Å gaps between the protein and the box edge for molecular mechanics minimizations and simulations. SPC water molecules(26) were used to add explicit solvation to the model followed by the insertion of sodium and chloride ions to bring the molarity of the box to 150 mM. An excess of sodium ions was added to neutralize the charge of the protein.
Ligand Incorporation into Mouse Muscle nAChR Model
Using this heteropentamer, another structure was generated containing carbamylcholine (CCh) in the two ligand-binding pockets of the mouse muscle nAChR. This was performed by aligning the box residues of the mouse muscle model with the D binding site of the AChBP structure containing CCh (1UV6). Gromacs parameters for the ligand (CCh) were initially generated using ProDRG (http://davapc1.bioch.dundee.ac.uk/programs/prodrg/)(27). The charges generated by ProDRG for CCh were modified due to a large positive charge that was inaccurately placed on the nitrogen atom of the ammonium. Instead, ChelpG charges from HF/6–31G calculations were used with some attenuation of the partial charges between carbamoyl protons and oxygens to fit within GROMACS MD parameters.
Generation of the D89N Mutant Structures
D89N mutant structures were made from the homology model PDB file by mutating D89 of both α subunits to Asn using Swiss PDB. The two structures differed in their orientations of the NH2 of the side chain: D89N1 placed the NH2 in a position analogous to OD1 of D89 in the wild type model, while D89N2 placed the NH2 in a position analogous to OD2. The mutant models were then placed in a hexagonal periodic box and treated similarly to the wild type model.
Molecular Dynamics Simulations. All four nAChR structures (agonist free wild type, CCh bound wild type, D89N1, and D89N2) underwent one minimization step. Then, under the GROMACS forcefield, MD simulations were begun. The MD simulations started at 0 K and warmed up to 310 K over the first 25 ps. The protein and drug (CCh bound structure) were highly restrained during this warmup followed by 100 ps of slowly releasing the restraints. All simulations after this point continued unrestrained for 5000 ps (wild type structures) or 7500 ps (D89N mutant structures).
Analysis of Molecular Dynamics Simulations
All molecular dynamics trajectories were analyzed using the tools included in the GROMACS suite(26). All characterizations were performed on the final 500 ps of the simulations. Each trajectory file contained data at 0.5 ps intervals, yielding 1000 frames of analysis per simulation. Distances and hydrogen bonds were analyzed using the g_dist and g_hbond programs, respectively. The default g_hond hydrogen bond structural cut-offs (a donor/acceptor distance of 3.5 Å and an acceptor/hydrogen/donor angle of 30°) were used when monitoring hydrogen bonds.
RMSD values for the CCh bound wild type, D89N1, and D89N2 simulations were calculated using the g_rms program. All comparisons were made with respect to the average loop B structure of the final 500 ps of the agonist free wild type simulation obtained from g_cluster, using the gromos method and an rmsd cut-off of 0.14 Å. The α carbons of the two structures to be compared were aligned prior to the rmsd calculation. Figure 8 was produced from PDB files generated by g_cluster, using the above conditions on the agonist free and CCh bound wild type simulation trajectories.
Figure 8.
Two views of hydrogen bonding interactions for the receptor without (A) and with (B) CCh bound. Generated with the g_cluster progran of the GROMACS suite from the final 500 ps of the 5 ns nAChR ligand binding domain simulations.
Results
Conventional Mutants, D89N and D89E
The present work evaluates receptors using the macroscopic parameter EC50, the effective concentration of agonist necessary to achieve half-maximal response, rather than the more information rich, but more time consuming, single-channel analyses of Lee and Sine(11). We made this choice both in order to examine a large number of mutants and to avoid the additional challenges of performing single-channel studies at the low expression levels often associated with unnatural amino acid mutagenesis. Of course, EC50 is a composite value that could be influenced by changes in agonist affinity or in gating. Since the mutations we are evaluating are in close proximity to the agonist binding site and are quite remote from the gate of the channel, we interpret changes in EC50 as affecting binding more than gating. Consistent with this view, single channel analyses of several D89 mutants reveal much more substantial changes in binding parameters than in gating parameters(11).
For comparison, we first studied two conventional mutants, D89N and D89E that were also studied by Lee and Sine. The D89E mutant produced a modest 4-fold increase in EC50 for both ACh and epibatidine. However, the D89N mutant produced much more substantial 23 and 28-fold increases in EC50 for ACh and epibatidine, respectively (Table 1). Our results parallel the single channel work, in which D89N produced a significant decrease in rate constants for ACh association while D89E resulted in a modest decrease in association rates and barely any change in the overall agonist binding equilibria.
Table 1.
D89 Mutantsa
| Wild Typeb | D89N | D89E | D89Nha | D89Akp | ||
|---|---|---|---|---|---|---|
| ACh | EC50 | 0.83 ± 0.04 | 19 ± 1 | 3.4 ± 0.3 | 14 ± 1 | 8.0 ± 0.7 |
| Hill Coeff. | 1.8 ± 0.1 | 1.6 ± 0.1 | 1.6 ± 0.1 | 1.3 ± 0.1 | 1.7 ± 0.2 | |
| n | 22 | 8 | 9 | 8 | 5 | |
| Epi | EC50 | 0.60 ± 0.04 | 13 ± 1 | 2.4 ± 0.1 | 11 ± 1 | 5.0 ± 0.5 |
| Hill Coeff. | 1.6 ± 0.1 | 2.0 ± 0.2 | 1.7 ± 0.2 | 1.7 ± 0.3 | 1.5 ± 0.2 | |
| n | 22 | 5 | 3 | 8 | 5 |
EC50 (µM) and Hill coefficient ± standard error of the mean. The receptor has a Leu9’Ser mutation in M2 of the β subunit.
Data reported previously.
Unnatural Mutants, D89Nha and D89Akp
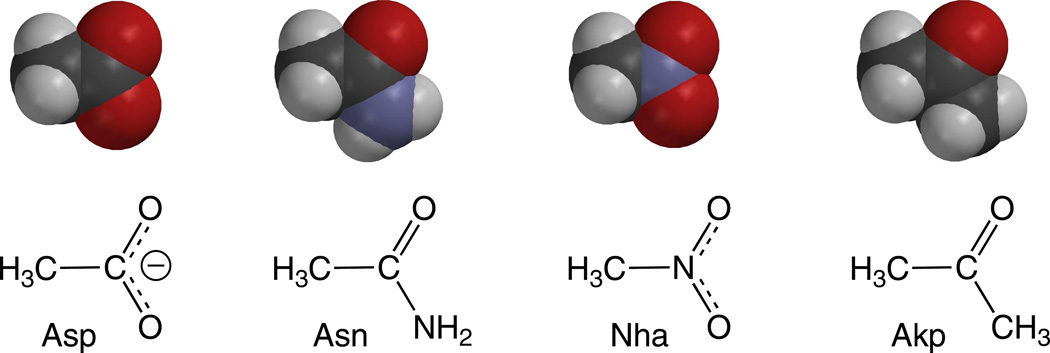
The essential role of D89 is confirmed by the large perturbation of the D89N mutant. While generally considered a subtle change, an Asp-to-Asn mutation does more than simply neutralize charge. It also replaces a hydrogen bond accepting O with a hydrogen bond donating NH2, and in the present context this change introduces a possible electrostatic clash between position 89 and loop B (Figure 3B). The amide side chain of asparagine places an Nδ−–Hδ+ bond dipole in close proximity to the Nδ−–Hδ+ bond dipoles of the loop B amide backbone at T150 and W149. This possible repulsive interaction between the Nδ−–Hδ+ dipoles could contribute to the deleterious effects of the D89N mutant.
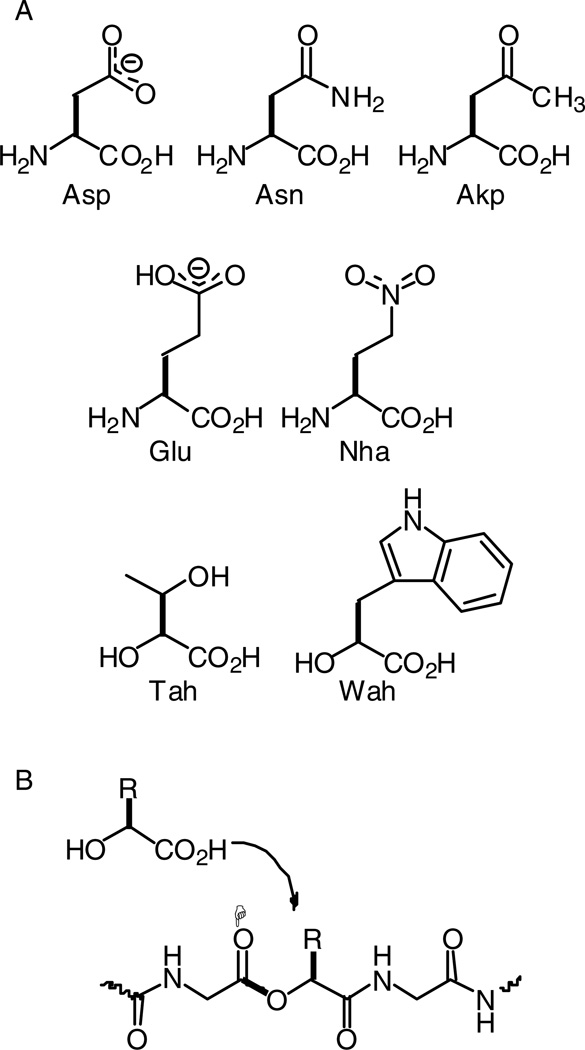
In an attempt to dissect apart these two features of the D89N mutant, we incorporated two unnatural amino acids that neutralize the negative charge of Asp without introducing an electrostatic clash. A nitro group (NO2) is isoelectronic and isosteric to a carboxylate, but it has no negative charge (Figure 4). Also, a nitro group is a substantially weaker hydrogen bond acceptor than carboxylate(28); the measured difference in energetics of carboxylate and nitro hydrogen bonding is 1.5 – 2.0 kcal/mol(29), corresponding to a factor of 10–20 in an equilibrium constant. The ideal residue would be nitroalanine, the nitro analog of Asp, but it is not chemically compatible with the nonsense suppression methodology. Therefore, we studied nitrohomoalanine (Nha), the nitro analog of Glu (Figure 5). Since the D89E mutant produces only a modest change in receptor function, comparing the Nha mutant to the Glu mutant was deemed meaningful.
Figure 4.
Functionalities of the side chains considered here, shown as space-filling models. Note the greater steric similarity between Asp and Nha and between Asn and Akp.
Figure 5.
A. Natural and unnatural amino (and hydroxy) acids considered here. B. Consequences of incorporating an α-hydoxy acid into a protein. The critical ester linkage is highlighted in bold; the carbonyl that is modulated is noted with a star.
Incorporation of Nha at position 89, D89Nha, resulted in a modest 4-fold increase in EC50 when compared to the isosteric D89E receptor for both ACh and epibatidine (Table 1). This change is comparable to that of the original D89E mutation. The D89Nha mutant behavior suggests that charge neutralization is no more deleterious than an increase in side chain length at position 89.
The second neutral unnatural amino acid we incorporated was 2-amino-4-ketopentanoic acid (Akp, Figure 5), producing D89Akp. Akp is a direct analog of Asp, and the ketone side chain of Akp is sterically similar to the Asp and especially the Asn side chains (Figure 4). However, Akp lacks the Nδ−–Hδ+ bond dipole (Figure 3C), and so does not contribute an electrostatic clash. When Akp was incorporated at position 89, 8- and 10-fold increases in EC50 were observed for ACh and epibatidine, respectively, relative to wild type (Table 1). The ~2.5-fold difference in EC50 between the D89Akp and D89N receptors can be attributed to the electrostatic clash produced by the Asn Nδ−-Hδ+ dipole.
Loop B Backbone Mutations
Of the four possible hydrogen bonds in the D89/loop B network, two involve side chain-to-side chain interactions, and two involve hydrogen bonds with the loop B amide backbone (Figure 3A). The side chain interactions (iii and iv) have been probed by conventional mutagenesis(11). T148L, T150A, and T148L/T150A mutants were not substantially disruptive, suggesting hydrogen bonds iii and iv are not crucial. By inference, the backbone hydrogen bonds i and ii have been proposed to be especially critical to receptor function(11).
Probing backbone hydrogen bonds requires the power of unnatural amino acid mutagenesis. Appropriate amide-to-ester mutations remove the backbone NH that can contribute to the hydrogen bond (Figure 5B). In the present system, the T150Tah mutation disrupts hydrogen bond i, and the W149Wah mutation disrupts hydrogen bond ii. The W149Wah mutation produced very modest effects (Table 2), suggesting that hydrogen bond ii is nonessential.
Table 2.
Loop B and Double Mutantsa
| Wild Typeb | T150Tahb | W149Wah | D89N | D89N / T150Tah |
D89N / W149Wah |
||
|---|---|---|---|---|---|---|---|
| ACh | EC50 | 0.83 ± 0.04 | 0.25 ± 0.01 | 0.81 ± 0.03 | 19 ± 1 | 15 ± 1 | 2.2 ± 0.1 |
| Hill Coeff. | 1.8 ± 0.1 | 1.4 ± 0.04 | 1.6 ± 0.1 | 1.6 ± 0.1 | 1.4 ± 0.2 | 1.8 ± 0.1 | |
| n | 22 | 17 | 7 | 8 | 7 | 6 | |
| Epi | EC50 | 0.60 ± 0.04 | 2.2 ± 0.2 | 1.6 ± 0.1 | 13 ± 1 | 2.9 ± 0.3 | 0.76 ± 0.05 |
| Hill Coeff. | 1.6 ± 0.1 | 1.3 ± 0.1 | 1.6 ± 0.1 | 2.0 ± 0.2 | 1.2 ± 0.1 | 1.7 ± 0.1 | |
| n | 22 | 16 | 5 | 5 | 6 | 6 |
EC50 (µM) and Hill coefficient ± standard error of the mean. The receptor has a Leu9’Ser mutation in M2 of the β subunit.
Data reported previously.
The T150Tah mutation has been studied previously(10). It is unique among the mutations considered here in that the results for ACh and epibatidine are qualitatively different; the EC50 for ACh decreased ~3-fold, while that for epibatidine increased ~4-fold. Ester backbones not only eliminate a hydrogen bond donor in the backbone; they also weaken the corresponding (adjacent) carbonyl as a hydrogen bond acceptor. The carbonyl perturbed by the T150Tah mutation, the W149 backbone carbonyl (H in Figures 2, 5B), points away from the region being probed here and directly into the agonist binding site. Epibatidine can make a hydrogen bond to this carbonyl, and the ester mutation weakens the hydrogen bond accepting ability of the carbonyl, accounting for the rise in EC50. No such hydrogen bond is possible for ACh. As such, we consider ACh the better gauge of the importance of hydrogen bond i, and we ascribe a nonessential role for it.
D89N and ester double mutants
Backbone ester mutations in loop B were also produced in an attempt to recover wild-type receptor function from the D89N mutant. If indeed a Nδ−–Hδ+•••Hδ+–Nδ− dipole-dipole clash is introduce by the D89N mutation, an appropriate backbone ester would not only alleviate the clash, but would replace it with a potentially favorable hydrogen bond (Figure 3D). Two double mutants were evaluated, D89N/T150Tah and D89N/W149Wah (Table 2); representative traces for the latter are shown in Figure 6. The D89N/T150Tah double mutant (attempting to rescue hydrogen bond i) did not fully recover wild-type receptor function: 18- and 5-fold increases in EC50 were observed for ACh and epibatidine, respectively. In contrast, the D89N/W149Wah double mutant (attempting to rescue hydrogen bond ii; Figure 3D) produced near wild-type activity, with EC50 increases of only 2.7-fold and 1.3-fold for ACh and epibatidine, respectively. These results suggest that there is an asymmetry in the D89/loop B network that allows the electrostatic clash of the Asn side chain to be relieved through an ester at position 149 but not at position 150. Note that the D89N/W149Wah mutant receptor is an example of a receptor that contains no negative charge in the vicinity of position 89, but retains nearly wild-type activity.
Figure 6.
Representative dose-response relations (A) and fits to the Hill Equation (B). Where errors bars are not evident in the fits, they are smaller than the markers. The receptor is the D89N/W149Wah double mutant.
Molecular Dynamics Simulation of nAChR Ligand-binding Domain
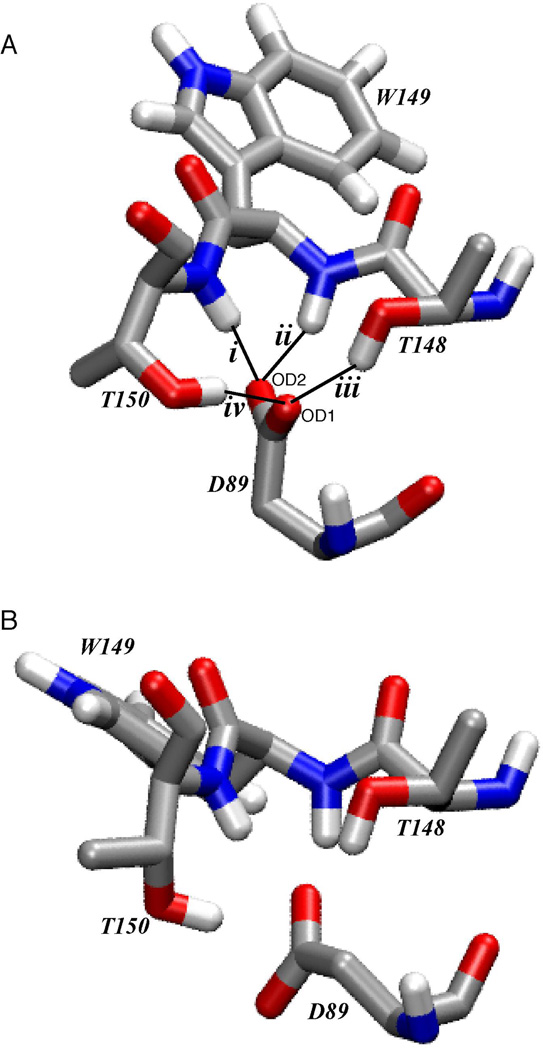
We performed two simulations of the mouse muscle nAChR ligand-binding domain, one without and one with the agonist carbamylcholine (CCh) bound. Other simulations of AChBP and variants of the nAChR have appeared(30–35). The agonist free structure is based on Unwin’s model of the receptor from T. marmorata, which is nearly identical in sequence to the mouse muscle receptor. CCh was then docked into this structure in a manner compatible with the crystal structure of AChBP that contains CCh in the agonist binding site(6). We monitored hydrogen bonds i through iv, considering both oxygens of the D89 carboxylate (referred to as OD1 and OD2, Figure 8), for a total of eight possible interactions. Along with the D89/loop B hydrogen bonds, we monitored two “control” hydrogen bonds that are part of a well-defined α-helix in the ligand binding domain. The results summarized in Figure 7 and Table 3 are calculated from the final 500 ps of the 5 ns simulations.
Figure 7.
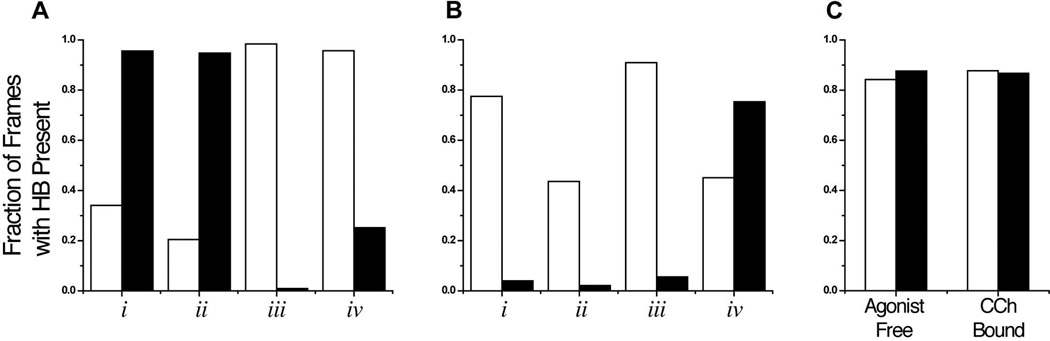
Analysis of hydrogen bonding in the molecular dynamics simulations of the nAChR ligand-binding domain without (A) and with (B) CCh bound. Hydrogen bonds were monitored between both D89 carboxylate oxygens (OD1, white bars, and OD2, black bars) and the four loop B hydrogen bond donors. C. Control hydrogen bonds between the backbone amide of K10 and the backbone carbonyl of R6 (K10R6, white bar) and the backbone amide of L11 and the backbone carbonyl of L7 (L11L7, black bar) were also monitored in both structures. All observations were made during the final 500 ps of the 5 ns simulations. Data expressed as the percentage of the 1000 observed frames where a given hydrogen bond was present.
Table 3.
Molecular Dynamics Simulationsa
| D89-Loop B Distance (Å)b | Number of D89-LoopB Hydrogen Bondsc |
RMSDd | |
|---|---|---|---|
| WT Agonist Free | 4.3 ± 0.2 | 4.7 ± 0.8 | -- |
| WT CCh Bound | 5.2 ± 0.3 | 3.4 ± 0.8 | 1.5 ± 0.2 |
| D89N1 | 5.3 ± 0.3 | 1.6 ± 0.6 | 2.5 ± 0.1 |
| D89N2 | 8.1 ± 0.5 | 1.5 ± 0.9 | 3.8 ± 0.3 |
All statistics averaged over the final 500 ps of the given simulation (1000 frames) and presented as mean ± standard deviation of the mean.
Distance measured from D89 or N89 γ carbon to the W149 α carbon.
All hydrogen bonds between position 89 and loop B residues, T148, W149, and T150. Includes both oxygens of D89 in wild type simulations.
RMSD calculated in reference to the average structure from the final 500 ps of the agonist free wild type simulation. Average structure from g_cluster program of GROMACS suite.
The agonist-free structure shows a very well defined hydrogen bonding network. In a representative structure (Figure 8A), one carboxylate oxygen (OD2) makes hydrogen bonds to the two backbone NHs (i and ii), while the other carboxylate oxygen makes hydrogen bonds to the two side chain OHs (iii and iv). These are strong hydrogen bonds, being present more frequently than the reference hydrogen bonds of the α helix. Occasionally, one carboxylate oxygen simultaneously makes three hydrogen bonds, and so on average there are 4.7 hydrogen bonds between D89 and loop B. Note that the orientation of the carboxylate in this simulation (Figure 2) differs from previous models, the side chain having rotated to enable formation of four hydrogen bonds.
Addition of the agonist CCh leads to a reduced interaction between D89 and loop B. In the agonist-free simulation, D89 interacts with all four loop B hydrogen bond donors in 94% of the frames, but that number drops to 34% of the frames in the CCh-bound simulation. Hydrogen bonds to the loop B backbone, interactions i and ii, are present less frequently in the CCh-bound structure (decreases of 17% and 50%, respectively, in the number of frames with a hydrogen bond present), while the side chain interactions, hydrogen bonds iii and iv, remain. The average number of hydrogen bonds drops from 4.7 to 3.4 when agonist binds, and the distance between D89 and W149 increases from 4.3 Å to 5.1 Å (measured from the D89 γ carbon to the W149 α carbon; Table 3). In the CCh-bound structure, OD1 makes the majority of the interactions with loop B, while OD2 interacts only with the T150 hydroxyl side chain (Figure 8B). Structurally, the changes between empty and occupied agonist binding sites arise because the D89 side chain rotates to a less symmetrical arrangement that favors one carboxylate oxygen over the other in hydrogen bonding (Figure 8). Also the T150 side chain reorients, but the OH group ends up in a similar location. In addition, as others have noted, the side chain of W149 reorients to make a cation-π interaction with the agonist.
Simulations of the D89N mutant were performed on the agonist-free structure only, because the Asn mutant mainly affected the kinetics of agonist association to the unbound nAChR(11). Two simulations that differ in the initial orientation of the Asn side chain were considered. In the D89N1 simulation, the N89 amide nitrogen was placed in a position comparable to OD1 of the agonist-free wild type structure, while in D89N2, the nitrogen was placed in the OD2 position.
As summarized in Table 3, both simulations show that the D89N mutant substantially disrupts the interaction between position 89 and loop B. The average distance between N89 and loop B (measured from the N89 γ carbon to the W149 α carbon) over the final 500 ps was larger for both D89N1 and D89N2 simulations (5.3 Å and 8.1 Å, respectively) than either wild type simulation. Also, in the D89N mutant simulations the 4.7 hydrogen bonds seen in the agonist free wild type simulation are reduced to ~1.5, and many of these hydrogen bonds do not correspond to hydrogen bonds i–iv, but are new hydrogen bonds involving the backbone of N89. The substantial disruption of the D89N mutant structures can also been observed in the loop B region. A comparison between the final loop B structure of the agonist free wild type simulation and comparable structures from the D89N1 and D89N2 simulations yielded RMSDs of 2.5 Å and 3.8 Å, respectively (Table 3).
Discussion
The study of the nAChR ligand-binding domain has been transformed by information gained from the AChBP crystal structures. Structural interactions found in the AChBP crystal structures have served as starting points for new mutation studies in the nAChR and other Cys-loop family receptors(36, 37). However, since the AChBP is not an actual LGIC and has < 25% homology to the closest nAChR relative, α7, experiments are necessary to test the relevance of interactions found in the crystal structures. The use of unnatural amino acids has allowed us to probe the relevance of these AChBP interactions at a chemical scale unavailable with conventional mutagenesis(10, 12, 13, 20).
In the present study, we have evaluated the structural role of the highly conserved residue, D89. According to the AChBP crystal structures, D89 provides the contact point between loop A, which contains D89 and agonist binding site residue Y93, and loop B, which contains the critical agonist binding site residue W149 (Figure 2). In the AChBP structures the D89 carboxylate forms hydrogen bonds with the backbone amides of loop B residues T150 and W149, while also interacting with the hydroxyl side chains of T148 and T150. This network is conserved among the primary ligand-binding subunits of the Cys-loop family of receptors (Figure 1). As noted before, D89 is part of a highly conserved WxPD motif found in essentially all known Cys-loop receptors. The residues aligning with T148 are conserved as hydroxyl side chains, serine or threonine. W149 is part of the conserved aromatic box that comprises the agonist-binding site. In three different Cys-loop receptors – the nAChR considered here, the 5-HT3 (serotonin) receptor, and the GABAC receptor – the aromatic residue that aligns with W149 makes direct contact with bound agonist through a cation-π interaction. (9, 10) (20, 38).
The essential role of D89 was established by the severe consequences of the relatively modest mutation D89N, an effect reported previously(11). Such a mutation could disrupt any or all of the 4 potential hydrogen bonds suggested by the AChBP structures (Figure 3). Conventional mutagenesis previously showed that hydrogen bonds iii and iv could be removed without significant disruption of receptor function. Using unnatural amino acid mutagenesis, we have now ablated hydrogen bonds i (T150Tah) and ii (W149Wah), and neither change is seriously disruptive. Clearly, no single hydrogen bond between loop B and loop A is critical to receptor function.
Along with disrupting hydrogen bonding, the D89N mutation neutralizes the negative charge of the wild type Asp. Others have concluded that binding of cationic agonists such as ACh requires a negative charge in the loop A/loop B interface region. However, more subtle charge-neutralization strategies, such as incorporation of Nha or Akp, do not reproduce the full D89N effect. This suggests that another factor is operative.
Along with disrupting hydrogen bonds and neutralizing charge, we propose that the D89N mutation introduces a destabilizing Nδ−–Hδ+•••Hδ+–Nδ− dipole-dipole clash. The other charge neutralizing mutations, D89Nha and D89Akp, do not experience such a clash and so are less disruptive. Also, the introduction into the D89N construct of a second, backbone mutation that removes one of the offending Nδ−–Hδ+ dipoles (D89N/W149Wah) restores near wild type behavior. This double mutant has no negative charge at the loop A/loop B interface yet is near wild type in behavior.
Another proposed role for D89 is a polarization of the W149 carbonyl (H in Figure 2), which points into the agonist binding site and can directly contact agonists(6). An amide carbonyl is highly polarized Cδ+=Oδ−, and the partial negative charge on oxygen could contribute to binding cationic agonists. It has been proposed that the negative charge of D89 could enhance this polarization(6), although we are unaware of any precedent for this type of effect. Our results do not appear to support this suggestion. In the mutant T150Tah the key carbonyl is much less polarized, as it is now an ester carbonyl rather than an amide carbonyl. However, this mutation decreases ACh EC50, opposite to expectations from the polarization suggestion.
The picture that emerges for the role of D89 is not simple. It seems certain that hydrogen bonding between the side chain of D89 and loop A is involved. However, no one interaction is singularly important. A subset of the full complement of interactions between D89 and loop B is required to stabilize the nAChR ligand-binding site.
Another possible indication of the disruptive nature of the D89N mutation can be found from a pair of Cys-loop receptors that are gated by serotonin. The 5-HT3 receptor has a Trp that aligns with the nAChR Trp α149, and it makes a cation-π interaction with the agonist serotonin.(20) Interestingly, the MOD-1 receptor from C. elegans naturally contains the D89N substitution, a rare exception to the highly conserved WxPD motif. Also, even though MOD-1 is highly homologous to the 5-HT3 receptor, and it binds the same neurotransmitter, the cation-π interaction in MOD-1 has moved away from the loop B aromatic reside to a different side chain of the aromatic box that is on loop C. (39) This could reflect the disruption of loop B caused by the D89N substitution in MOD-1.
Molecular dynamics simulations generally support the experimental data of others and ourselves. In our simulations, a sharp reduction of hydrogen bonding in the position 89/loop B network suggests that the D89N mutation severely disrupts interactions between position 89 and loop B. In fact, in one of the D89N simulations, the Asn side chain ceases to interact at all with loop B. Both D89N mutant simulations produced loop B structures that differed from their wild type counterparts. From these observations we conclude that loop B adjusts in structure to accommodate the Asn.
From our molecular dynamics simulations of the mouse muscle nAChR ligand-binding domain, it appears that D89 interacts with loop B more in the agonist-free state than in the agonist-bound state. Relative to the agonist-free structure we find an average of ~one less hydrogen bond between D89 and loop B; and D89 is almost 1 Å further from loop B in the CCh-bound structure. Because loop B itself does not relocate substantially on agonist binding (as revealed in the comparison of our agonist free and CCh bound wild type simulations), the role of D89 is to preorganize the empty agonist binding site into a conformation that favors binding, an effect that would enhance binding affinity(40). Apparently, once the agonist is bound, the stabilization of loop B provided by D89 is no longer necessary, and the interaction between the two weakens. This is consistent with an earlier conclusion that the D89N mutation affects agonist association but not subsequent steps in the channel activation mechanism(11).
In conclusion, chemical-scale studies of the D89/loop B network have further refined our understanding of this interesting and highly conserved structural feature. D89 and loop B form a redundant network of hydrogen bonding interactions, no one of which is essential. In addition, the charge on D89 is not essential for receptor function. The D89N mutation both disrupts the hydrogen bonding network and also introduces a repulsive electrostatic interaction, significantly destabilizing the D89/loop B network. These results, along with molecular dynamics simulations and earlier single channel studies(11), indicate that the role of the D89/loop B network is to preorganize the agonist binding site for ligand binding, with no significant contribution to the gating mechanism.
Acknowledgment
We thank Tingwei Mu for providing the unnatural amino acid Akp. This work was supported by the National Institutes of Health (NS 34407 and NS 11756) and by the George Hoag Family Foundation.
Abbreviations
- ACh
acetylcholine
- AChBP
acetylcholine binding protein
- LGIC
ligand gated ion channel
- nAChR
nicotinic acetylcholine receptor
- CCh
carbamylcholine
- Nha
nitrohomoalanine
- Akp
2-amino-4-ketopentanoic acid
- Wah
α-hydroxytryptophan
- Tah
α-hydroxythreonine
References
- 1.Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel. Journal of Molecular Biology. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- 2.Corringer P-J, Novere NL, Changeux J-P. Nicotinic receptors at the amino acid level. Annual Review of Pharmacology and Toxicology. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 3.Grutter T, Changeux J-P. Nicotinic receptors in wonderland. Trends in Biochemical Sciences. 2001;26:459–463. doi: 10.1016/s0968-0004(01)01921-1. [DOI] [PubMed] [Google Scholar]
- 4.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nature Reviews Neuroscience. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 5.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 6.Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 7.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfacres and conformations. EMBO Journal. 2005;24:3625–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smit AB, Syed NI, Schaap D, van Minnen J, Klumperman J, Kits KS, Lodder H, van der Schors RC, van Elk R, Sorgedrager B, Brejc K, Sixma TK, Geraerts WPM. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 9.Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: A cation-pi binding site in the nicotinic receptor. PNAS. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashin AL, Petersson EJ, Lester HA, Dougherty DA. Using physical chemistry to differentiate nicotinic from cholinergic agonists at the nicotinic acetylcholine receptor. J. Am. Chem. Soc. 2005;127:350–356. doi: 10.1021/ja0461771. [DOI] [PubMed] [Google Scholar]
- 11.Lee WY, Sine SM. Invariant aspartic acid in muscle nicotinic receptor contributes selectively to the kinetics of agonist binding. J. Gen. Physiol. 2004;124:555–567. doi: 10.1085/jgp.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougherty DA. Unnatural amino acids as probes of protein structure and function. Current Opinion in Chemical Biology. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- 13.Kearney PC, Nowak MW, Zhong W, Silverman SK, Lester HA, Dougherty DA. Dose-response relations for unnatural amino acids at the agonist binding site of the nicotinic acetylcholine receptor: tests with novel side chains and with several agonists. Mol Pharmacol. 1996;50:1401–1412. [PubMed] [Google Scholar]
- 14.Nowak MW, Gallivan JP, Silverman SK, Labarca CG, Dougherty DA, Lester HA, Conn PM. Methods in Enzymology. Academic Press; 1998. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system; pp. 504–529. [DOI] [PubMed] [Google Scholar]
- 15.England PM, Lester HA, Dougherty DA. Incorporation of esters into proteins: Improved synthesis of hydroxyacyl tRNAs. Tetrahedron Letters. 1999;40:6189–6192. [Google Scholar]
- 16.Ranganathan D, Rao B, Ranganathan S, Mehrotra AK, Iyengar R. Nitroethylene: A Stable, Clean, and Reactive Agent for Organic Synthesis. Journal of Organic Chemistry. 1980;45:1185–1189. [Google Scholar]
- 17.Mu T. Division of Chemistry and Chemical Engineering. Pasadena: California Institute of Technology; 2006. p. 126. [Google Scholar]
- 18.Nowak MW, Gallivan JP, Silverman SK, Labarca CG, Dougherty DA, Lester HA. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Ion Channels, Pt B. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- 19.Li LT, Zhong WG, Zacharias N, Gibbs C, Lester HA, Dougherty DA. The tethered agonist approach to mapping ion channel proteins toward a structural model for the agonist binding site of the nicotinic acetylcholine receptor. Chemistry & Biology. 2001;8:47–58. doi: 10.1016/s1074-5521(00)00055-7. [DOI] [PubMed] [Google Scholar]
- 20.Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-π interactions in ligand recognition by serotonergic (5-HT3) and nicotinic acetylcholine receptors: The anomalous binding properties of nicotine. Biochemistry. 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez EA, Lester HA, Dougherty DA. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. PNAS. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 23.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 24.Schrodinger I. Portland. OR: [Google Scholar]
- 25.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 26.van der Spoel D, Lindahl E, Hess B, van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Feenstra KA, R vD, Berendsen HJC. 2004 [Google Scholar]
- 27.Schuettelkopf AW, F vADM. Acta Crystal D60. 2004:1355–1363. [Google Scholar]
- 28.Kelly TR, Kim MH. Relative binding affinity of carboxylate and its isosteres: nitro, phosphate, phosphonate, sulfonate, and δ-lactone. Journal of the American Chemical Society. 1994;116:7072–7080. [Google Scholar]
- 29.Thorson JS, Chapman E, Schultz PG. Analysis of hydrogen bonding strengths in proteins using unnatural amino acids. Journal of the American Chemical Society. 1995;117:9361–9362. [Google Scholar]
- 30.Gao F, Bern N, Little A, Wang H-L, Hansen SB, Talley TT, Taylor P, Sine SM. Curariform antagonists bind in different orientations to acetylcholine-binding protein. J. Biol. Chem. 2003;278:23020–23026. doi: 10.1074/jbc.M301151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao F, Bren N, Burghardt TP, Hansen S, Henchman RH, Taylor P, McCammon JA, Sine SM. Agonist-mediated conformational changes in acetylcholine-binding protein revealed by simulation and intrinsic tryptophan fluorescence. J. Biol. Chem. 2005;280:8443–8451. doi: 10.1074/jbc.M412389200. [DOI] [PubMed] [Google Scholar]
- 32.Henchman RH, Wang H-L, Sine SM, Taylor P, McCammon JA. Asymmetric structural motions of the homomeric α7 nicotinic receptor ligand binding domain revealed by molecular dynamics simulation. Biophys. J. 2003;85:3007–3018. doi: 10.1016/S0006-3495(03)74720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henchman RH, Wang H-L, Sine SM, Taylor P, McCammon JA. Ligand-induced conformational change in the α7 nicotinic receptor ligand binding domain. Biophys. J. 2005;88:2564–2576. doi: 10.1529/biophysj.104.053934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Novere N, Grutter T, Changeux J-P. Models of the extracellular domain of the nicotinic receptors and of agonist- and Ca2+binding sites. PNAS. 2002;99:3210–3215. doi: 10.1073/pnas.042699699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schapira M, Abagyan R, Totrov M. Structural model of nicotinic acetylcholine receptor isotypes bound to acetylcholine and nicotine. BMC Structural Biology. 2002;2:1. doi: 10.1186/1472-6807-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Criado M, Mulet J, Bernal JA, Gerber S, Sala S, Sala F. Mutations of a conserved lysine residue in the N-terminal domain of α7 nicotinic receptors affect gating and binding of nicotinic agonists. Mol Pharmacol. 2005;68:1669–1677. doi: 10.1124/mol.105.015446. [DOI] [PubMed] [Google Scholar]
- 37.Mukhtasimova N, Free C, Sine SM. Initial coupling of binding to gating mediated by conserved residues in the muscle nicotinic receptor. J. Gen. Physiol. 2005;126:23–39. doi: 10.1085/jgp.200509283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lummis SCR, L. Beene D, Harrison NJ, Lester HA, Dougherty DA. A cation-π binding interaction with a tyrosine in the binding site of the GABAC receptor. Chemistry & Biology. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Mu TW, Lester HA, Dougherty DA. Different binding orientations for the same agonist at homologous receptors: A lock and key or a simple wedge? J. Am. Chem. Soc. 2003;125:6850–6851. doi: 10.1021/ja0348086. [DOI] [PubMed] [Google Scholar]
- 40.Cram DJ. Preorganization - from Solvents to Spherands. Angewandte Chemie-International Edition in English. 1986;25:1039–1057. [Google Scholar]