Abstract
The Abbott RealTime HIV-1 assay is a real-time nucleic acid amplification assay available for HIV-1 viral load quantitation. The assay has a platform for automated extraction of viral RNA from plasma or dried blood spot samples, and an amplification platform with real time fluorescent detection. Overall, this study found no clinically relevant differences in viral load, if samples were extracted manually.
Keywords: HIV, Viral load monitoring
HIV viral load testing is used for monitoring treatment, determining prognosis and risk of disease progression, and for ascertaining treatment failure (Yilmaz, 2001). The newer laboratory viral load assays are based on real time detection and can detect viral load values as low as 20 copies/ml (Scott et al., 2009a). The FDA approved Abbott RealTime HIV-1 assay uses the automated extraction and detection m2000 system platform (Abbott Molecular Inc., Des Plaines, IL, USA). This assay targets the pol/IN region of HIV-1 using partially double-stranded probes (Huang et al., 2007) with a dynamic range of 40–1.0E+7 copies/ml. This fully automated system has a reporting capacity of 93 patient results per an 8-h day and has been fully validated against other real time assays (Scott et al., 2009b). Laboratories also have the option of performing manually the front end extraction for reasons such as lower sample throughput (21 reported patient results per 8-h day), the m2000sp purchase cost, lack of space for the m2000sp or a desire to minimise the use of complex equipment requiring service and maintenance (Crump et al., 2009).
This study therefore investigates any differences in viral load values between samples processed manually and using the m2000sp automated platform. K3 EDTA blood samples were recruited from 147 participants with written informed consent in Moshi, Tanzania (approval obtained from the Kilimanjaro Christian Medical Centre (KCMC) Research Ethics Committee approval number 156). Forty milliliters of blood was collected from each participant and the plasma separated within 4 h. This was divided into four aliquots, of 5ml each, and frozen at −80°C. Frozen plasma was shipped to the University of the Witwatersrand, Johannesburg, South Africa, on dry ice, where replicates and dilutions for another method validation plan were prepared (Crump et al., 2009) and then shipped back to the KCMC in Moshi, Tanzania, on dry ice. Storage at both sites was at −80°C before testing. In Johannesburg, the 1ml plasma extraction protocol (including additional for the ‘dead’ volume) was performed on neat samples using the automated m2000sp platform according to the manufacturer's instructions. In Moshi, samples were extracted manually from 1ml plasma samples using the CE marked Abbott Sample Preparation System (Abbott Laboratories, Abbott Park, IL), as per the manufacturer's instructions. Extracted samples from both methods were amplified and detected on the m2000rt platforms at each site according to the manufacturer's instructions. Agreement in viral load values between the different extraction methods was measured using the Bland–Altman and percentage similarity (Bland and Altman, 1995; Scott et al., 2003).
The total 147 samples (neat plasma tested at both sites) yielded 62 samples that were quantified by both extraction methods. The remaining 85 were either <40 copies/ml (n = 12 {14.1%}) or ‘target not detected’ by both methods of extraction. Manual extraction yielded two additional quantifiable samples (46 copies/ml and 83 copies/ml) that were <40 copies/ml after automated extraction, and therefore there was 97.7% concordance between the two extraction methods. The mean viral load, for the automated extraction samples yielding quantifiable results, was log 4.4 copies/ml (25,118 copies/ml) with a range of log 2.0 (100 copies/ml) to log 7.0 copies/ml (10 million copies/ml). The mean difference between the automated and the manual extraction values was log 0.0015 copies/ml (confidence interval of the mean difference −0.076; 0.079), with a standard deviation of this difference being log 0.306 copies/ml. The limits of agreement (0.536; −0.688 copies/ml) and the mean percentage similarity (100.4% with 4.0% standard deviation and 4.0% percentage similarity CV) showed good overall agreement between the methods of extraction. The Bland–Altman plot (Fig. 1) shows no values >log 1.0 difference and therefore no clinically relevant outliers. Seven (11.3%) values had differences in viral load values >log 0.5 copies/ml. Fig. 1 also suggests that the majority of values >log 4.5 copies/ml, generate higher values after automated extraction, and similarly viral load values <log 4.0 copies/ml generate higher values after manual extraction. This observation may also explain why manual extraction could quantify an additional two samples in the lower range than automated extraction, but this would have to be confirmed on a larger sample size.
Fig. 1.
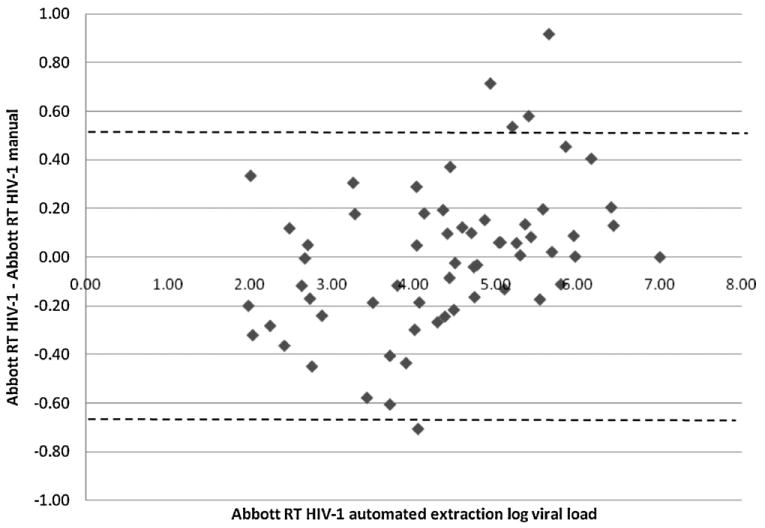
Bland–Altman scatter plot of the difference in viral load values between samples extracted by automated and manual protocols followed by quantitation on the Abbott RT HIV-1 m2000rt. The vertical axis represents the difference between viral load values from the automated minus values from the manually extracted samples. The dotted lines are the limits of agreement.
In summary, samples extracted manually or by the automated protocol on the m2000sp platform for further quantitation on the Abbott RealTime HIV-1, show no clinically relevant differences in viral load values. Although the manual extraction was performed here by a single skilled technical laboratory operator, inter-operator variability may occur with manual sample preparation, but this variability should be minimised if operators adhere to strict laboratory standard operating procedures. The 11.3% of samples that had viral load values >log 0.5 copies/ml, indicate that samples with low viral load values are more readily quantified after manual extraction and that samples with high viral load values are more readily quantified after automated extraction. The automated extraction protocol offers the advantages of requiring fewer operational staff for a higher throughput of specimens and less chance for contamination (although contamination was not found to be an issue in this study). Manual extraction has the advantage of requiring less laboratory space, requiring less complex and expensive instrumentation, yet still generates reliable real time viral load results. Although potentially any RNA extraction method with sufficient yield and clean RNA product could be used for sample processing, the authors chose the Abbott Sample Preparation System since its CE marking ensures consistent, site-independent assay performance. Both automated and manual extraction methods provide accurate and reliable viral load results for patient management.
Acknowledgments
The authors wish to thank Lara Noble, Eveline Akkers and Kirthi Hira for their assistinace in sample and assay preparations conducted in Johannesburg, South Africa, the staff and patients of the Kilimanjaro Christian Medical Centre Infectious Diseases Clinic, and the staff and clients of Kikundi cha Wanawake Kilimanjaro Kupambana na UKIMWI (KIWAKKUKI; Women Against AIDS in Kilimanjaro) for their participation. The authors thank Ekyafyose E. Kimaro, Julitha Kimbi, Devotha Lyimo, and Editha Mushi for assistance with sample collection. This publication was made possible by the generous support of the American people through the US Agency for International Development. The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the US government. This research was supported by the Center for HIV/AIDS Vaccine Immunology, a United States National Institutes of Health (NIH) funded program (U01 AI067854). Authors received additional support from NIH awards International Studies of AIDS-associated Co-infections (ISAAC) (AI 062563, JAC and ABM), and the Duke Clinical Trials Unit and Clinical Research Sites (U01 AI069484-01, JAC). The authors are grateful to Abbott Laboratories for donating reagents for sample testing in Moshi and to Dr. Laurence Phillips from Abbott Molecular for assistance in addressing queries around manual extraction methods.
References
- Bland JM, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085–1087. doi: 10.1016/s0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- Crump JA, Scott LE, Msuya E, Morrissey AB, Kimaro EE, Shao JF, Stevens WS. Evaluation of the Abbott m2000rt RealTime HIV-1 assay with manual sample preparation compared with the ROCHE COBAS AmpliPrep/AMPLICOR HIV-1 MONITOR v1.5 using specimens from East Africa. J Virol Methods. 2009;162:218–222. doi: 10.1016/j.jviromet.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Salituro J, Tang N, Luk KC, Hackett J, Jr, Swanson P, Cloherty G, Mak WB, Robinson J, Abravaya K. Thermodynamically modulated partially double-stranded linear DNA probe design for homogeneous real-time PCR. Nucleic Acids Res. 2007;35:e101. doi: 10.1093/nar/gkm551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L, Carmona S, Stevens W. Performance of the new Roche Cobas AmpliPrep-Cobas TaqMan version 2.0 human immunodeficiency virus type 1 assay. J Clin Microbiol. 2009a;47:3400–3402. doi: 10.1128/JCM.00727-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LE, Galpin JS, Glencross DK. Multiple method comparison: statistical model using percentage similarity. Cytometry: Clin Commun. 2003;54B:46–53. doi: 10.1002/cyto.b.10016. [DOI] [PubMed] [Google Scholar]
- Scott LE, Noble LD, Moloi J, Erasmus L, Venter WD, Stevens W. Evaluation of the Abbott m2000 RealTime human immunodeficiency virus type 1 (HIV-1) assay for HIV load monitoring in South Africa compared to the Roche Cobas AmpliPrep-Cobas Amplicor, Roche Cobas AmpliPrep-Cobas Taq-Man HIV-1, and BioMerieux NucliSENS EasyQ HIV-1 assays. J Clin Microbiol. 2009b;47:2209–2217. doi: 10.1128/JCM.01761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz G. Diagnosis of HIV infection and laboratory monitoring of its therapy. J Clin Virol. 2001;21:187–196. doi: 10.1016/s1386-6532(01)00165-2. [DOI] [PubMed] [Google Scholar]


