Abstract
In this paper we describe an application called peFinder for document-level classification of CT pulmonary angiography reports. peFinder is based on a generalized version of the ConText algorithm, a simple text processing algorithm for identifying features in clinical report documents. peFinder was used to answer questions about the disease state (pulmonary emboli present or absent), the certainty state of the diagnosis (uncertainty present or absent), the temporal state of an identified pulmonary embolus (acute or chronic), and the technical quality state of the exam (diagnostic or not diagnostic). Gold standard answers for each question were determined from the consensus classifications of three human annotators. peFinder results were compared to naive Bayes’ classifiers using unigrams and bigrams. The sensitivities (and positive predictive values) for peFinder were 0.98(0.83), 0.86(0.96), 0.94(0.93), and 0.60(0.90) for disease state, quality state, certainty state, and temporal state respectively, compared to 0.68(0.77), 0.67(0.87), 0.62(0.82), and 0.04(0.25) for the naive Bayes’ classifier using unigrams, and 0.75(0.79), 0.52(0.69), 0.59(0.84), and 0.04(0.25) for the naive Bayes’ classifier using bigrams.
Keywords: medical language processing, ConText, pulmonary emboli, computed tomography, CTPA
1. Introduction
Thromboembolic disease is a vascular disease with a high incidence in the United States, with estimates of 398,000 cases of deep venous thrombosis (DVT) and 347,000 cases of pulmonary embolus (PE) per year. Deaths due to PE are estimated to be about 235,000 per year. The mortality due to untreated, clinically apparent PE is approximately 30%. However, if correctly diagnosed and anticoagulant therapy is initiated, mortality drops to below 3% [16]. A large proportion of deaths due to PE are believed to be due to missed diagnoses rather than failure of therapies. Currently, contrast-enhanced CT pulmonary angiography (CTPA) is the standard diagnostic procedure to rule out suspected PE. CTPA exams can provide exquisite detail but are also liable to artifacts, such as respiratory motion and beam harding, that can make image interpretation uncertain or effectively impossible, depending on the severity of the artifacts. Improvements to acquisition hardware and protocols and development of post-processing schemes are continually being made and evaluated. Such technological developments and evaluations require the ability to identify and characterize a large number of CTPA exams.
Currently at our institution, identification of cases requires an honest broker to manually review the radiology information system to identify exams ordered to rule out PE and then to access and review the dictated report to determine the characteristics of the resulting exam. This human bottleneck increases the cost and slows the rate of case identification. Text processing tools offer the promise of semi-automating this process so that large numbers of relevant radiology exams can be identified rapidly with less cost. In this paper, we describe our initial efforts to develop a text processing application called peFinder for document-level classification of CTPA reports for exams that were ordered to rule out PE.
peFinder relies on identification of features within the report regarding whether the radiologist suspects a PE, whether a suspected PE is new, and the quality of the radiology exam. To generate the features, we decided to apply an existing algorithm called ConText [22]. However, to apply ConText to this new task, we developed a more generalized implementation of the ConText application and extended the algorithm with properties relevant to identifying patients with an acute PE in high quality CTPA exams. Con-Text determines whether a clinical condition is absent or present, whether the condition is historical, recent, or in the future, and whether the experiencer of the condition is the patient or someone else. In a previous study [22], Harkema and colleagues showed that ConText, which was developed for emergency department notes, performed similarly on discharge summaries but did not perform as well on radiology reports. We believe that determining whether a new finding is present or absent is a more complex task in radiology reports than it is in emergency department reports and discharge summaries. Identifying indications of a new radiology finding, such as a PE, from a radiology report involves integrating information about whether the finding was seen on the exam, whether the finding is new or has been seen previously, how certain the radiologist is that the opacity indicates the finding, and the quality of the radiology study, which if poor could compromise the radiologist’s impression of the image. Moreover, it is important to be able to distinguish between mentions of the finding as an observation (which indicates potential presence of the finding) and mentions of the finding as the reason for exam (which does not indicate presence or absence of the finding).
We extended the contextual properties assigned by ConText to account for these features in a more generalized Python implementation of the algorithm called pyConText.
First, we provide a brief background on ConText and the modications we made in the pyConText implementation. Second, we describe how we built peFinder using features generated by pyConText. Then we evaluate how well peFinder can classify documents to answer the following questions: 1) Is a PE present in the exam? 2) Is there uncertainty related to the disease state? 3) If a PE is present, is it chronic or acute? and 4) Does the exam exhibit notable quality limitations? We compared peFinder classifications against classifications made by domain experts and against n-gram document-level classifiers.
2. Background
For some tasks, the challenge in identifying relevant patients is accounting for the myriad of ways the case definition can be described in text. For example, finding patients with chest pain may require dozens of keywords or phrases, such as “angina,” “chest discomfort,” and “pain when I press on the lower part of the sternum.” In contrast, only a few lexical variants are commonly used to describe pulmonary embolism in a CTPA exam: for example, “pulmonary embolism,” “embolus/emboli,” or “PE.” The challenge in identifying PE cases from CTPA exams is not the lexical variation in describing the condition itself but the ability to identify the contextual modifiers that determine whether the case represents a new PE. Once we locate a mention of PE in the exam, we need to know whether the PE is present or absent, whether the PE is new or pre-existing, how certain the radiologist is of the presence of a PE, and how reliable the imaging study was in relation to the diagnosis. We hypothesized that an algorithm accounting for lexical cues surrounding the findings of interest could successfully identify the modifying information we need.
ConText [22], an extension of the NegEx algorithm [5], is a simple algorithm that looks for lexical cues in the context of mentions of signs, symptoms, and diseases. If a clinical condition falls within the scope of a lexical cue, the condition is assigned the property represented by that cue. For instance, PE in the sentence “No evidence of PE” falls within the scope of the negation cue “no” and would therefore be assigned the property negated. The current version of ConText, as described in [22], assigns the following properties: existence (affirmed, negated); temporal (historical, recent, future/non-specific); and experiencer (patient, other). We generalized ConText in this study by allowing it to assign arbitrary, user-defined properties as well as arbitrary, user-defined relationships between properties.
2.1. Related Work
Others have developed applications similar to ConText for characterizing clinical named entities. Several algorithms exist for negation detection [30, 15, 24]. Recently, machine learning classifiers have been applied to the problem of assertion detection in clinical reports, determining whether a concept is asserted, negated, or uncertain [36], and we are developing a feature-based machine learning version of ConText. In addition, there is current work on temporal annotation of clinical text [43, 33, 28]. ConText differs from many of these other applications in its simplicity, which makes it easy to apply to a variety of problems but often less accurate than more specialized applications that do not rely only on lexical patterns. The properties identified by ConText and similar algorithms can be extremely useful for accurate case detection or characterization from clinical reports.
Clinical research often relies on identifying patients with specified case definitions from the EMR. Many studies use claims data to identify relevant patients; however, studies relying solely on structured data often generate inaccurate disease or screening rates [17, 21]. Free-text clinical documents provide more accurate and complete clinical detail for identifying patients with a specific case definition. Therefore many studies rely on chart review to identify relevant cases. Dublin et al. identified 1,410 people with newly-recognized atrial fibrillation from ICD-9 codes and validated cases by review of medical records to examine the association of diabetes with risk of atrial fibrillation [14]. Over a two-year period, Nelson et al. manually reviewed 70,000 chest radiograph reports to study the impact of the introduction of pneumococcal conjugate vaccine on rates of community acquired pneumonia [31]. The expense involved in chart review is massive [27, 25]. Text processing methods can be applied to increase the efficiency of chart review.
Various approaches have been used to classify or retrieve textual documents that match a specific query or case definition. Medline and Google are two examples of classification systems using a query submitted by a user. Text classification approaches vary in accuracy and in sophistication. Boolean keyword-based approaches return a relevant document if the document includes words or word variants from the query. Statistical retrieval techniques attempt to differentiate documents from each other based on the words and their frequencies in relevant and irrelevant document. The tf/idf weight (term frequency inverse document frequency) [26] is a standard statistical measure that weights a word’s ability to discriminate relevant and irrelevant documents in a corpus. A word’s weight increases proportionally to the number of times a word appears in the document but is offset by the frequency of the word in the entire corpus. Similarly, a Bayesian text classification algorithm may use the positive or negative likelihood ratios of a word or a group of words to quantify the word’s ability to discriminate relevant and irrelevant documents [2]. There are also a number of symbolic NLP applications designed for or applied to clinical texts, including MedLEE [19], MPLUS [10], MetaMap [3], Hitex [42], CTAKES (http://sourceforge.net/projects/ohnlp/files/), Multi-threaded Clinical Vocabulary Server (MCVS), and ONYX [9]. Symbolic text classification systems use linguistic techniques, such as part-of-speech tagging, parsing, entity recognition, and relation identification, to identify relevant information in a document. The relevant information can then be reasoned with using rules or machine learning algorithms to classify a document. Statistical, symbolic, and hybrids have been applied to text classification of clinical reports [32, 6, 35, 7, 18, 38].
Statistical techniques are simple to apply but may not accurately capture the relationships among words in a document. For example, a bag-of-words classifier using n-grams as features may accurately classify reports describing a PE, but may not do as well at determining whether the PE was negated: the assumption that two words occuring in the same document are related is often an inaccurate assumption, as in “Positive for PE. No other abnormality.” Symbolic techniques attempt to identify modifying information for clinical conditions, but clinical NLP applications using symbolic techniques can be cumbersome to implement and use outside of the institution where the application was developed. Moreover, only recently are researchers making such NLP applications available for open use by others [42]. Similar to what South and colleagues found [34], when a task involves a limited number of named entities (as in detection of influenza or of pulmonary emboli) and relies strongly on contextual properties for successful detection, a more targeted algorithm like ConText can perform as well as more sophisticated, general-purpose NLP applications like MedLEE.
3. Methods
The objectives of this study were to create a CTPA report classifier based on a more generalizable and extensible version of ConText. All research done for this study was approved by the University of Pittsburgh IRB.
The CTPA report classifier (peFinder) was used to answer the following questions: Was the patient positive for a pulmonary embolism? Was the pulmonary embolism acute or chronic? Was there uncertainty regarding the diagnosis of PE? Was the exam of diagnostic quality?
To evaluate the performance of peFinder at answering these questions, we compared its output to answers provided by human annotators who read the same reports. We also compared peFinder document classification performance to that of two bag-of-word naive Bayesian classifiers. Bag-of-words classifiers are frequently used as a baseline comparison for document-level classification, in large part due to their simplicity. In spite of their simplicity, bag-of-words classifiers are actually often difficult to beat. [12] Although an n-gram classifier at the sentence level may have been more accurate than our document-level classifier, the annotation effort required for a baseline sentence-level classifier would exceed the effort involved in configuring py-ConText for this task. In this section, we describe selection of our dataset, manual report annotation, the adaptations we made to ConText, and our evaluation process and outcome measures.
3.1 Data Set
We retrospectively identified 4,657 CTPA reports that were ordered to rule out pulmonary emboli. The reports were identified using radiology procedure codes that are included in the header of the dictated reports archived in MARS [41]. The reports were all de-identified in a fully HIPAA-compliant manner using the De-ID software [20]. Using the NegEx algorithm to process the Impression section of the reports, we identified 449 potentially positive cases in the set and randomly selected 11% of the remaining reports, for potentially negative cases. We randomly split the 928 reports into three sets. The first 20 reports were used to train human annotators on the task. A set of 250 reports was used to develop our automated classifiers, and the remaining set of 658 reports was held out as a test set. Ultimately, 40 reports were discarded because no user annotations were obtained for them; two failures were due to anomalies in the Impression section, as will be discussed later, the remaining 38 failures were due to our annotation software skipping the reports. Our final set consisted of 202 development reports and 656 test reports.
3.2. Report Annotation
Three second-year medical students independently annotated the reports using a GUI interface to a database that we developed. Users were not allowed to edit their annotations. For each annotator, the first twenty reports were used to train the annotators on the annotation task. Developers reviewed the annotations and discussed them with the annotators. The annotation schema for DISEASE/CERTAINTY STATE and QUALITY STATE are presented in Tables 1 and 2, respectively. We initially captured the TEMPORAL STATE informally by asking the annotators to provide a free-text comment if there was a discussion of a chronic PE.
Table 1.
DISEASE/CERTAINTY STATE Annotation Schema for CTPA Reports
| Value | Example Criteria |
|---|---|
| definitely negative | Absence of pulmonary embolism is explicitly stated, as in “No PE” |
| probably negative | Mention of PE is modified as in “No evidence,” “Not excluded,” “No PE identified/seen” |
| Indeterminate | Pulmonary embolism not discussed (e.g. exam described as non-diagnostic) |
| probably positive | Mention of PE modified by terms such as “consis- tent with,” “worrisome for,” “likely” |
| definitely positive | Presence of pulmonary embolism explicitly stated, as in “PE” |
Table 2.
QUALITY STATE Annotation Schema for CTPA Reports
| Value | Example Criteria |
|---|---|
| diagnostic | No description of exam |
| limited | Discussion of one of the following factors as influ- encing evaluation of the study: motion, artifact, bolus timing, other specific limitation |
| non-diagnostic | A term meaning “non-diagnostic” explicitly used |
In peFinder, we chose to annotate DISEASE and CERTAINTY STATES separately. Binary DISEASE and CERTAINTY STATES were generated from the user annotations as follows: probably positive and definitely positive were collapsed to positive; probably negative, indeterminate, and definitely negative were considered negative; definitely negative and definitely positive were considered certain; and probably negative, inderminate, and probably positive were considered uncertain. After collapsing annotations to binary values, we generated consensus states for each report by a majority vote of the annotators.
Free-text entries describing the TEMPORAL STATE were often missing; therefore, we asked annotators to explicitly annotate the TEMPORAL STATE of the consensus disease-positive cases. The annotation scheme for the TEMPORAL STATE is described in Table 3. Because of the unavailability of two of the original annotators, two physicians performed the temporal annotations along with one of the original medical students.
Table 3.
TEMPORAL STATE Annotation Schema
| Value | Example Criteria |
|---|---|
| new | Mention of previously undocumented acute PE |
| old | Mention of previously documented PE; mention of resolved PE; description of chronic PE |
| mixed | Both old and new PE mentioned |
3.3. Generalization of ConText
ConText was developed for the task of identifying the presence of acute clinical conditions from ED reports. The task of identifying acute PEs seems similar to the task ConText was initially developed for; however, the implementation of the ConText algorithm would not accommodate all of the types of knowledge required to perform our task. We hypothesized that lexical surface patterns may be sufficient for accomplishing our task of identifying acute pulmonary emboli in CTPA reports, so we developed a new version of ConText. We implemented the algorithm using the Python programming language in a package named pyConText. We populated the knowledge structures of the algorithm with all existing ConText surface cues and supplemented it with a number of new cues relevant for our task.
The core component of the pyConText algorithm is the domain and syntactic knowledge incorporated into four-tuples called items. Each item consists of the following elements: 1) a literal, 2) a category, 3) a regular expression, and 4) a rule.
The items are used by the pyConText algorithm to create tagObjects on a sentence-by-sentence basis. Each tagObject receives the four attributes of the item used to define it and adds the following attributes: 1) the actual matched phrase from the text, 2) the character span within the sentence of the matched phrase, 3) the scope within the sentence of the tagObject, 4) a collection of other tagObjects modified by the current tagObject, and 5) a collection of other tagObjects modifying the current tagObject. The relationships between tagObjects defined within a sentence form the basis for textual inferencing with the objects.
The details of items are described in Section 3.3.1 and the details of tagObjects are described in 3.3.2.
3.3.1. Description of items
The details of the four-tuple components of items are as follows:
The literal is a lexical cue (word or phrase) relevant for the domain problem.
-
The category defines what the item represents, for example a finding or an uncertainty term. The categories from the previous implementations of ConText included finding, negated existence term, conjunction, pseudo-negated existence term, and temporality term. In pyConText categories are user-defined and there are no limits to the number of categories that can be used.
As an example, for our PE identification task, we added additional categories exam feature, exam quality descriptor, and indication for exam. In the previous version of ConText, all literals (except those with the category finding) act as modifiers for literals with the category finding. In pyConText, the user can define modifier relationships among literals in whatever way the task at hand requires.
-
The regular expression is what is used in the program to actually capture the literal in the text. If a regular expression is not provided in the definition of the item, the literal is used to directly generate the regular expression.
The previous implementations of ConText attempted to be comprehensive in the list of literals (e.g., including “rule him out for,” “rule her out for,” “rule patient out for,” etc.). The use of regular expressions reduces the proliferation of literals and increases comprehensiveness in matching variant phrases in the text.
The rule states how the tagObjects generated from the item will interact with other tagObjects generated within the sentence. For our task we applied the rules forward and backward, as in the previous implementation of ConText, and we added the rule bidirectional. The rule forward means the tagObject generated from the item can only modify a tagObject to the right of itself in the sentence. The rule backward is similar but in the opposite direction. A literal with the rule terminate terminates the scope of another tagObject. Examples of items with a rule terminate are conjunctions such as “though” and “but.”
pyConText comes with a large number of pre-defined items, based both on literals listed on the NegEx website (http://code.google.com/p/negex/) as well as additional items we defined for this project. However, items can easily be created by a user for a specific application.
3.3.2. tagObject Description
For each item, the regular expression associated with the item is used to identify any literals in the sentence. When a literal is identified, a tagObject is created. Attributes of the tagObject include the following:
the literal for which the tagObject was created (defined by the generating item)
the actual phrase matched by the regular expression in the text
the character span of the matched phrase
the category of the tagObject (defined by the generating item)
the rule of the tagObject (defined by the generating item)
the scope of the tagObject’s influence (i.e., the character range in the sentence over which the tag can operate)
an empty collection of tagObjects modified by the current tagObject and an empty collection of tagObjects modifying the current tagObject; these collections are filled in based on the scope of other tagObjects.
The generation and modification of tagObjects created within a sentence consists of a four step process: 1) Create tagObjects; 2) Prune tagObjects; 3) Update scope of the tagObjects; 4) Assign modifiers to targets. Each of these steps are now described and illustrated in detail.
-
Create tagObjects
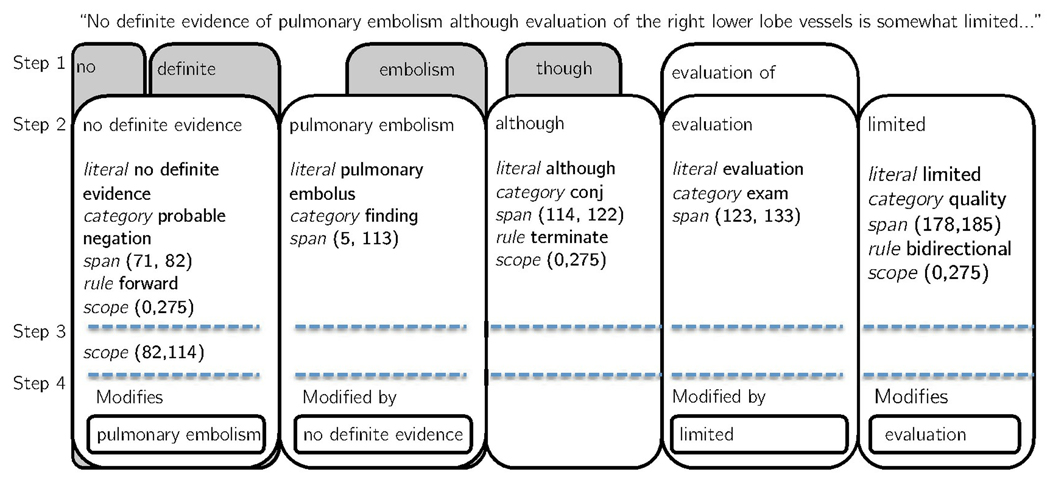
Figure 1 shows the tagObjects generated for the sentence “No definite evidence of pulmonary embolism although evaluation of the right lower lobe vessels is somewhat limited…” In Step 1, 10 tagObjects were created for the literals in the sentence. The tagObjects in grey were pruned in Step 2, as described next.
-
Prune tagObjects
A tagObject can be a subset of another tagObject. For example, “embolism” and “pulmonary embolism” both generated tagObjects in the sentence shown in Fig. 1. Step 2 in pyConText removes any tagObjects that are encompassed by a larger tagObject. The pruning is applied to all categories of literals.
-
Update Scope of tagObjects
A tagObject can have a scope: tagObjects with a rule of bidirectional have a default scope of the entire sentence; tagObjects with a rule of forward have a scope from the end of the tagObject to the end of the sentence; and tagObjects with a rule of backward have a scope from the start of the tagObject to the beginning of the sentence. However, the default scope can be modified by other tagObjects within the sentence. In Step 3, ConText loops through all the tagObjects in the sentence and updates the scope for each tagObject based on the rule of the other tagObjects in the sentence. For instance, in the sentence shown in Fig. 1 the scope of the tagObject with the literal “no definite” extends to the end of the sentence (character 275). However, because the rule for the literal “although” is terminate, the scope of the tagObject with the literal “no definite” is adjusted in Step 3 to terminate at “although” (character 114).
-
Assign tagObjects as targets or modifiers
In the last step, the remaining tagObjects are assigned as target and modifier objects. In the previous version of ConText, the algorithm assigns modifiers to pre-annotated targets that are assumed to be clinical conditions. In pyConText, the user can specify which category of tagObjects are targets, which are modifiers, and the desired relationship between targets and modifiers. Any tagObject that is identified as a target whose matched phrase falls within the scope of a modifier tagObject is modified by that tagObject. For instance, in the sentence shown in Fig. 1, the tagObject with the literal “pulmonary embolus” is identified as a target (as are all tagObjects with category finding), and the tagObject with the literal “no definite” is identified as a modifier. Since “pulmonary embolism” lies within the scope of “no definite,” the tagObject for “no definite” modifies the “pulmonary embolism” tabObject. Because the value for the modifier is probably negated existence, we know that the pulmonary embolism is believed to be absent with some uncertainty.
Figure 1.
After the pruning stage, six of the ten tagObjects generated by pyConText for the example sentence were kept (white boxes in figure). The tagObject with the literal “although” terminates the scope of “no definite evidence.” However, “no definite evidence” modifies “pulmonary embolism,” which is still within its scope, indicating that the pulmonary embolism is probably negated. The tagObject for “evaluation” is modified by “limited.” A tagObject was created for “evaluation of,” but the shorter, overlapping one (“evaluation”) was not pruned, because the tagObjects have different categories.
3.4. peFinder: a pyConText-based Classifier for CTPA Reports
To identify CTPA reports that describe the presence of an acute pulmonary embolism from an exam of high diagnostic quality, we developed a document-level classifier called peFinder. peFinder uses pyConText annotations to perform rule-based classification of CTPA reports. We implemented peFinder in three steps:
-
Define CTPA-specific items in pyConText:
For peFinder, we defined several items with a number of new categories to describe the certainty or uncertainty of existence. Specifically, we added categories of definite negated existence, probable negated existence, and probable existence (to existing categories of negated and affirmed and removed possible.) Our creating separate categories for positive and negative uncertainty was motivated by our observation that different literals are often used to indicate uncertainty of existence when the polarity is negative than when polarity is positive. We also supplemented existing items with literals and regular expressions that occurred in our development reports. For instance, we added literals such as “documented,” for probable affirmation and “subacute” and “resolution of” for historical. Finally, we created items with the following new categories, which were necessary to accomplish our task:
pe finding: these items represent the PE and include literals such as “pe,” “embolism,” and “pulmonary embolus.”
exam: these items represent the terms used by the radiologist to describe the CT exam and the features of the exam. The literals include terms such as “bolus timing,” “scan,” “evaluation,” and “study,”
quality feature: these items represent the adjectives used by the radiologist to describe limited or non-diagnostic quality of a CT exam. The literals include phrases such as “suboptimal,” “degraded,” “non-diagnostic,” and “limited.”
artifact: these items represent terms used by radiologist to describe artifacts in the CT exam. The literals include “respiratory motion,” “bulk motion,” and “artifact.”
exam indication: these items represent terms used by radiologist to describe the reason for exam, such as “evaluate for,” or “rule out.” findings modified by a tagObject with the category exam indication can be understood to not be actual findings or observations by the radiologist and are ignored by peFinder.
-
Apply pyConText to CTPA Reports
We applied pyConText to the Impression sections of CTPA reports, assuming that the Impression section would contain a relevant summary of the information regarding the disease state, uncertainty, and exam quality. For peFinder, our targets included tagObjects with category finding, exam, and artifact. All other categories could be assigned as modifiers to any of the targets within their scope, with the exception of quality feature, that could only modify exam (e.g., “suboptimal evaluation”).
-
Query pyConText Annotations to Answer PE Specific Questions:
pyConText performs sentence-level analyses, providing a collection of sentence-level annotations of targets and modifiers in each sentence. Document-level reasoning in peFinder was achieved by aggregating the sentence-level pyConText annotations into a virtual database and querying the annotations to answer the following questions:
DISEASE STATE: Was the patient positive for a pulmonary embolism?
TEMPORAL STATE: Was the pulmonary embolism new (acute) or pre-existing (chronic)?
CERTAINTY STATE: Was there uncertainty regarding the diagnosis of PE?
QUALITY STATE: Was the exam quality diagnostic?
The DISEASE STATE for the report was considered positive if there was at least one finding tagObject in the report that was either unmodified or was modified by probable affirmation or definite affirmation. If there were multiple finding tagObjects, as long as one of them was modified by probable or definite affirmation, DISEASE STATE was considered positive. finding tagObjects modified by exam indication were ignored.
The TEMPORAL STATE for the report was considered acute if at least one positive finding tagObject was unmodified by historical. If all positive finding tagObjects were modified by historical, the TEM-PORAL STATE was considered chronic.
The CERTAINTY STATE for the patient was considered uncertain if any of the following occurred in the report: an exam tagObject was modified by a quality feature tagObject; or the report contained no tagObject, indicating that no mention was made of pulmonary emboli; or a finding tagObject was modified by a tagObject with category probable affirmation or probable negation. probable affirmation and probable negation concepts were modeled after the instructions given to the annotators in Table 1, and were designed to be symmetric (e.g. “not seen” and “seen” expressed uncertain negation and uncertain affirmation respectively).
The QUALITY STATE was considered non-diagnostic if the report contained an artifact tagObject or contained an exam tag Object modified by a quality feature tagObject. We used separate pyCon- Text objects to answer the disease questions (existence, uncertainty, temporality) and the exam quality questions.
The values for DISEASE STATE, TEMPORAL STATE, CERTAINTY STATE, and QUALITY STATE can be combined in various ways by a specific application, depending on the needs of the application. For instance, a screening application might want to capture all CTPA exams that address pulmonary embolism, regardless of certainty, temporality, or quality of the exam. For our purpose—retrieving high quality images of positive, acute pulmonary emboli for imaging studies—we may allow some uncertainty but would want to assure that the exam was high quality and that the pulmonary embolus was acute.
3.5. Evaluation
We compared peFinder’s classifications of the four patient states against consensus manual annotations that were generated from raw annotations described in Tables 1 and 2 as follows:
DISEASE STATE: Patients with reference standard annotations of probably positive and definitely positive were considered positive
TEMPORAL STATE: Reference standard annotations for patients as new or mixed were considered acute
CERTAINTY STATE: Reference standard annotations of probably positive and probably negative were considered uncertain
QUALITY STATE: Reference standard annotations of limited and non-diagnostic were considered non-diagnostic
From standard contingency tables, we calculated the following outcome values:
sensitivity: TP/(TP + FN)
specificity: TN/(TN + FP)
positive predictive value (PPV): TP/(TP + FP)
accuracy:(TP + TN)/(TP + FP+TN + FN)
3.6 Baseline Comparison
As a baseline comparison, for every patient state we implemented a naive Bayesian document classifier trained on unigrams and a document classifier trained on bigrams from the training set, using the Python package Orange (http://www.ailab.si/orange/). To preserve generalizability, we limited the number of features to one-tenth of the number of cases in the training set. The most informative attributes were pre-selected using the Orange filter orngFSS.FilterBestNAtts.
4. Results
4.1. Rater Agreement
To assess rater agreement we computed the complete agreement fraction (i.e., agreement among all three raters) and partial agreement fraction (i.e., agreement between two of the three raters) for each of our annotation states. We computed Fleiss’ Kappa to assess the overall multi-reader agreement as well as Cohen’s Weighted Kappa Coefficient [11] with squared weighting for each pair of raters. Kappa values were computed in R (http://www.r-project.org/). TEMPORAL STATE annotations were performed by a different set of annotators than the DISEASE and QUALITY STATE annotations; one annotator (Rater 1) was common between both groups.
The Fleiss’ Kappa coefficients shown in Table 5 were all between 0.76 and 0.87 with p-values less than 0.00455, indicating very good overall annotator agreement. Pairwise Cohen’s Weighted Kappa coefficients are shown in Table 6. Again the Kappa coefficients are high with p-values less than 0.05, indicating very good rater agreement.
Table 5.
Multi-reader Agreement
| State | Kappa |
|---|---|
| Disease | 0.847 |
| Quality | 0.87 |
| Uncertainty | 0.787 |
| Temporal | 0.762 |
Table 6.
Inter-rater Agreement
| DISEASE | CERTAINTY | QUALITY | TEMPORALITY | |
|---|---|---|---|---|
| Rater 1 and Rater 2 | 0.958 | 0.916 | 0.937 | NA |
| Rater 1 and Rater 3 | 0.944 | 0.840 | 0.860 | NA |
| Rater 2 and Rater 3 | 0.944 | 0.886 | 0.903 | NA |
| Rater 1 and Rater 4 | NA | NA | NA | 0.753 |
| Rater 1 and Rater 5 | NA | NA | NA | 0.706 |
| Rater 4 and Rater 5 | NA | NA | NA | 0.87 |
4.2. Baseline Classifier Performance
The combined corpus of the training and testing sets contained 2,511 unique unigrams and 10,078 unique bigrams. In order to reduce over-training, we limited the number unigrams or bigrams used for each model to be one tenth the number of cases in the training set.
Through feature selection, the most informative unigrams or bigrams were selected prior to training the baseline classifier and are shown in Tables 7–9.
Table 7.
Twenty Selected Features for Baseline Classifier: DISEASE STATE
| Unigrams | Bigrams |
|---|---|
| “at” “atelectasis” “con- veyed” “ct” “discussed” “embolism” “evidence” “findings” “large” “left” “lobe” “lower” “mid- dle” “negative” “no” “positive” “time” “to” “upper” “were” |
“at the” “bilateral pul- monary” “conveyed to” “discussed with” “evi- dence of” “left lower” “lobe pulmonary” “lower lobe” “no pulmonary” “positive pulmonary” “pulmonary emboli” “pulmonary embolism” “right lower” “right middle” “the left” “the posterior” “the time” “time of” “to the” “were discussed” |
Table 9.
Twenty Selected Features for Baseline Classifier: QUALITY STATE
| Unigrams | Bigrams |
|---|---|
| “airspace” “artifact” “be” “but” “central” “definite” “due” “em- boli” “evaluation” “large” “left” “may” “mild” “motion” “no” “patient” “scan” “study” “suboptimal” “to” |
“but no” “cannot be” “central pulmonary” “definite evidence” “due to” “emboli are” “evaluation for” “faxed to” “for pulmonary” “highly suspicious” “motion and” “motion artifact” “no definite” “pulmonary emboli” “pulmonary embolism” “study due” “subopti- mal due” “suboptimal study” “suspicious for” “the left” |
Tables 10 and 11 show the performance measures for the baseline classifiers.
Table 10.
Naive Bayes Unigram Baseline Classifier Performance on the Test Set
| State | PPV | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| DISEASE | 0.77 (155/202) | 0.68 (155/228) | 0.89 (381/428) | 0.82 (536/656) |
| QUALITY | 0.87 (88/101) | 0.67 (88/131) | 0.98 (512/525) | 0.91 (600/656) |
| CERTAINTY | 0.82 (207/253) | 0.62 (207/333) | 0.86 (277/323) | 0.74 (484/656) |
| TEMPORAL | 0.25 (1/4) | 0.04 (1/25) | 0.99 (200/203) | 0.88 (201/228) |
Table 11.
Naive Bayes Bigram Baseline Classifier Performance on the Test Set
| State | PPV | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| DISEASE | 0.79 (170/214) | 0.75 (170/228) | 0.90 (384/428) | 0.84 (554/656) |
| QUALITY | 0.69 (68/98) | 0.52 (68/131) | 0.94 (495/525) | 0.86 (563/656) |
| UNCERTAINTY | 0.84 (195/232) | 0.59 (195/333) | 0.89 (286/323) | 0.73 (481/656) |
| TEMPORAL | 0.25 (1/4) | 0.04 (1/25) | 0.99 (200/203) | 0.88 (201/228) |
4.3. peFinder Performance
The performance of peFinder on the test set is shown in Table 12. A true positiveindicates correctly classifying the patient as follows: DISEASE STATE positive; TEMPORAL STATE acute; CERTAINTY STATE uncertain; and QUALITY STATE non-diagnostic.
Table 12.
peFinder Classification Performance on the Test Set
| State | PPV | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| DISEASE | 0.83 (223/269) | 0.98 (223/228) | 0.89 (382/428) | 0.92 (605/656) |
| QUALITY | 0.96 (113/118) | 0.86 (113 /131) | 0.99 (520/525) | 0.96 (633/ 656) |
| CERTAINTY | 0.93 (313/ 336) | 0.94 (313/ 333) | 0.93 (300/ 323) | 0.93 (613 /656) |
| TEMPORAL | 0.90 (18/20) | 0.60 (18/ 30) | 0.99 (196 /198) | 0.94 (214/228) |
We also measured agreement between peFinder and consensus annotations for the combined DISEASE and CERTAINTY STATE. Table 6 shows the contingency table for classifications of pulmonary embolism definitely negative, probably negative, probably positive, and definitely positive. Rows indicate reference standard annotations, and columns indicate peFinder annotations. Similar to the inter-rater agreement analysis in Section 4.1, we calculated weighted (squared) Cohen’s kappa coefficients using R. The weighted kappa was 0.932 (z=23.4, p-value=0). The close clustering along the diagonal indicates that a main source of error in the DISEASE STATE of present or absent shown in Table 13 was differences in interpretation of uncertainty, such as the 33 cases classified as probably negative by the reference standard and probably positive by peFinder.
Table 13.
Agreement on DISEASE/CERTAINTY STATE Combinations
| def. neg. | prob. neg. | prob. pos. | def. pos. | |
|---|---|---|---|---|
| def. neg. | 130 | 2 | 0 | 1 |
| prob. neg. | 6 | 224 | 33 | 6 |
| prob. pos. | 0 | 3 | 30 | 5 |
| def. pos. | 0 | 2 | 19 | 169 |
4.4. Error Analysis
Six of the DISEASE STATE errors were due to typographical errors resulting in a missed negation, such as “noevidence of PE.” Most other errors were due to missing literals: Seven errors resulted from not including “resolved” as a literal with the category negation; three were due to not including “clinical history” as a literal with the category exam indication.
The majority (n = 23) of errors in CERTAINTY STATE were due to annotators not following the guidelines for annotating certainty, such as classifying a case as definitely positive rather than classifying it as probably positive. Errors by peFinder were largely due to missing literals for indicating uncertainty (n = 13) (i.e., “appreciated,” “is difficult to completely exclude,” and “possible”). Another source of CERTAINTY STATE errors was mistakes in other modifiers that indicate uncertainty during the document-level reasoning. For instance, peFinder did not create a tagObject with category quality for the sentence “though the timing of the bolus was not ideal” and therefore did not classify the document as uncertain.
Annotators did not always follow guidelines for QUALITY STATE annotations either, accounting for six errors. But again the majority (n = 16) of QUALITY STATE errors were due to missing literals for indicating limited or non-diagnostic quality, such as “technical error,” “nondiagnostic,” “compromised,” and “quality is diminished.” Twelve of fourteen TEMPORAL STATE errors were failures to identify the positive pulmonary embolus as chronic. Nine of these were due to missing literals such as “interval progression” or “again noted.” Our development set only had six mentions of chronic pulmonary embolus, whereas the test set had 30. Other errors were due to spelling errors and difficult cases such as “It is not clear cut whether these are acute or whether they may be old althoughthere [sic] are features favoring chronic rather than acute.”
5. Discussion
We developed a text processing application (peFinder) for classifying CTPA reports with respect to pulmonary embolism. This classification task was built upon a more generalizable implementation of ConText that we called pyConText. pyConText can facilitate an unlimited set of user-defined features and supports user-defined relationships between features. As such, for peFinder we defined contextual properties related to CTPA such as pulmonary embolism findings, uncertainty, and exam quality. peFinder showed promising results as a document classifier compared to consensus annotations from human readers and compared to simple bag-of-words document classifiers.
Our comparison of the peFinder algorithm to naive Bayes’ classifiers based on unigrams and bigrams shows the vast improvement that can be obtained by incorporating knowledge about the context in which the words appear. Similar to other studies based on ConText, our poorest performance occurred with identifying the temporal state, which was largely due to the small number of cases in our training set and consequently poor inclusion of relevant literals. Identifying more literals that indicate a finding is chronic is likely to succeed in addressing the poor performance for identifying temporal state in radiology reports, whereas identifying chronic conditions in other free-text reports, such as discharge summaries, is more complex and can only partially be addressed by adding more literals [29].
One of the most challenging aspects of this study was modeling uncertainty. Radiology reports have been criticized for being vague or not definitive [4], and the amount of uncertainty expressed in a report has been shown to be associated with lack of clarity in the report [8]. In addition to uncertainty due to poor expression by the radiologist, there is uncertainty intrinsic to the radiological exam being interpreted: first, there may be uncertainty in the relationship between a finding in the image and the underlying pathology generating the finding; second, there may be uncertainty regarding whether a finding is real or simply an artifact of the acquisition. Our guidelines to the human annotators included both types of uncertainty (see Table 1). However, annotators were not consistent in applying the guidelines. In retrospect, having reviewers explicitly annotate VISUALIZATION STATE may have been a better way to capture the uncertainty that accompanies the inability to visualize a finding. A schema used in a data structuring and visualization system for neuro-oncology at the UCLA Medical Imaging Informatics Group (http://www.mii.ucla.edu/r01-neuro-oncology/) annotates for visualization state with possible values Clearly seen, Appears, and Difficult to Visualize. We suspect that inter-annotator agreement on uncertainty would have increased if we had separated certainty from visualization ability.
Another issue related to identification of uncertainty is due to the relationship between negation and uncertainty. It was not clear whether negation and uncertainty should be combined into a single feature (e.g., probably absent) or whether the two properties should be represented individually and their relationship considered after annotation of the individual properties. In our application of ConText, modifiers interact with findings but not with each other. However, sometimes one modifier has scope over another modifier. For example, in our illustrative sentence (see Fig. 1 “No definite evidence of PE”) “no” indicates negation but does not directly negate the finding “PE;” instead, “no” negates the assertion of certainty (“definite evidence of”). In other words the certainty that a PE exists is negated, which indicates that the PE does not exist. Instead of specifying in pyConText that certain types of modifiers have scope over other types of modifiers, we remained consistent with the original version of ConText and included a literal for the complete phrase “no definite evidence of” that resulted in a classification of probably negated. A more elegant solution would be to specify interactions between modifiers. The current implementation of pyConText allows modifiers and targets to be specified and combined in any way the user desires; therefore, this change would only require splitting apart compound phrases, such as “no definite evidence of” and allowing uncertainty modifiers to be targets of negation modifiers. This type of extension would also help address double negatives, such as “cannot rule out,” in a more principled way.
peFinder does not account for any document structure. Since we were only processing the Impression section of radiology reports, this limitation did not affect our results; however, for processing more complex reports, such as history and physical exam reports, the structure of the report can influence the values assigned to the contextual properties. We are currently developing a statistical version of ConText that accounts for the lexical features and scope in the same way as the current version but that can incorporate additional information, such as the report structure, syntactic relations of modifiers and findings, and temporal expressions. Uzuner and colleagues [37] compared NegEx against a machine-learning-based classifier at assertion classification. Similar to Uzuner’s findings, we expect incorporating additional knowledge will improve ConText’s performance; however, we also believe that ConText’s performance is respectable in spite of the lack of sophisticated linguistic modeling. The simplicity and intuitiveness of the algorithm makes it appealing to apply, especially for developers without extensive training in natural language processing. And the extension we developed for this study will allow ConText to be applied to a variety of tasks, increasing its usefulness for assigning properties to annotated concepts.
There are several limitations to this study. We only processed the Impression section of the report. Consequently, we missed some information about pulmonary emboli. For example, we reviewed the complete reports for the 31 cases that had a consensus disease state classification of Indeterminate. Thirteen of the 31 cases were technically non-diagnostic exams. Of the remaining 18 cases, six discussed PE in the findings section but made no mention of PE in the Impression. One report made no mention of PE in any part of the report, and one was stated as being indeterminate as to whether the filling defects in the exam were embolic events or tumor processes. Unfortunately, we found that for ten of the cases, our captured Impression sections were missing the first sentence of the actual Impression section where the PE disease state had been mentioned. This failure to capture the first sentence was due to variant typesetting that conflicted with the text processing script used by the honest broker to capture the Impression section.
There was some inhomogeneity in our annotators. Three of the five annotators were second-year medical students, whereas the remaining two were experienced physicians (a pathologist and infectious disease specialist). The disease, uncertainty, and quality annotations were done by the medical students. The temporal annotations were done by one of the medical students and the two physicians. For the temporal annotation, agreement between the medical student and the physicians was notably lower than agreement between the two physicians; this may indicate the medical students used less domain knowledge for the annotations.
Abujudeh, et al. [1] reviewed over 2000 reports for CTPA exams to rule out PE and found that 62% of all reports mentioned motion artifact as a limitation in image quality and 28% of reports mentioned contrast enhancement as a limitation in image quality. Despite the high incidence of technical limitations, conclusive diagnoses were still frequently made. In the accompanying editorial, Hatabu and Hunsaker [23] conclude that radiologist tend to over-describe quality limitations, even when they do not seem to impact the certainty of diagnosis. Since our analysis of image quality lumped together “limited” and “non-diagnostic” quality descriptors and inferred diagnostic uncertainty in the presence of sub-optimal quality, our analysis likely overestimates the number of cases with uncertainty.
Our selection of unigram and bigram naive Bayes’ classifiers may be criticized for their simplicity, since neither sentence boundary information nor sentence contextual information is used. Nonetheless, bag-of-words classifiers are common baseline document-level classifiers and perform remarkably well, in general, as has been pointed out by experts in the field (e.g. “The story that it’s hard to beat an n-gram language model is fairly ubiquitous.” http://nlpers.blogspot.com/2006/06/beating-n-gram.html). Trigram models often perform better than bigram and unigram models, but require large training sets to avoid sparse feature vectors. Finally, we did not pre-process our text. There were a large number of concatenated words in our corpus. We were able to work through some of these errors by using regular expressions, but running a spell-checker prior to processing would be a preferred solution.
pyConText provides a flexible framework for processing information in free-text reports and showed high sensitivity and specificity for identifying disease state, uncertainty, and technical quality of the exam related to pulmonary embolic disease. peFinder is currently being used to identify exams for a research project developing algorithms for computer-aided detection of PE depicted in CTPA studies. We also have interest in using peFinder as a quality assurance tool to assess changes in incidence of limited or nondiagnostic scans as improved technologies (e.g. intelligent power injectors, faster CT scanners) are introduced into our hospital system. The pyConText framework is flexible, and we were able to quickly customize it to build an application in an entirely different domain: to identify personal and family history of mesothelioma and ancillary cancers in history and physical exam reports [39, 40]. This customization essentially consisted of creating new items relevant for the new domain, including additional negation terms and cancer terms to be used as findings, and defining new categories such as anatomic location to be used as modifiers.
We believe pyConText is sufficiently general that many tasks involving identification of targets and their modifiers could be accomplished using pyConText without further generalizations to the implementation. To apply pyConText to another task, one would add new values to the lexicons of targets and modifiers, as we did with the mesothelioma study. For example, to apply pyConText to assessing colorectal cancer screening rates [13] one would create a new target item for a colorectal exam and new categories for modifiers, such as patient consent for an exam (e.g., “patient refused colorectal exam”) and when an exam was scheduled (e.g., “Exam scheduled for next Wednesday”). Unlike the previous version of ConText, which was specific to a few particular targets and modifiers, the task of adapting pyConText to other problems is now one of creating specialized lexicons and implementing them within an existing general framework. The ultimate success of the application of the algorithm to any particular problem depends partly on the coverage of the lexicon created by the user and partly on the suitability of the algorithm itself, which is based on regular expressions without consideration of deeper linguistic features.
6. Conclusion
We developed an application called peFinder that performed well at identifying diagnostic CTPA reports that describe pulmonary emboli. peFinder’s success is contingent on the ability to identify whether a PE is present or absent, to determine whether there is uncertainty about the presence of a PE, to conclude whether the PE is acute or chronic, to ignore mentions of PE in the context of the reason for exam, and to understand whether the exam was of diagnostic quality. To assess this information, peFinder implemented pyConText for assigning modifiers to targeted concepts and then performed queries based on their relationships. pyConText provides an extension of the existing ConText algorithm. The extension accommodates user specification of the modifier types and can provide a more accurate model of the relationship between a modifier and the concept it modifies, which promises to make ConText more useful for a variety of text processing tasks.
The pyConText code can be obtained from the NegEx website http://code.google.com/p/negex/.
Table 4.
Complete and Partial Agreement Fractions: DISEASE STATE, CERTAINTY STATE, QUALITY STATE, and TEMPORAL STATE
| DISEASE | CERTAINTY | QUALITY | TEMPORALITY | |
|---|---|---|---|---|
| Full Agreement | 0.847 | 0.938 | 0.906 | 0.915 |
| Partial Agreement | 0.142 | 0.06164 | 0.0930 | 0.0785 |
Table 8.
Twenty Selected Features for Baseline Classifier: CERTAINTY STATE
| Unigrams | Bigrams |
|---|---|
| “a” “and” “as” “at” “embolism” “evidence” “for” “groundglass” “infiltrate” “is” “me- diastinal” “negative” “new” “no” “of” “pul- monary” “right” “the” “upper” “with” |
“a new” “a pulmonary” “at the” “cannot be” “ct evidence” “emboli can- not” “evidence of” “for pulmonary” “mild inter- stitial” “new pulmonary” “no evidence” “no pul- monary” “of a” “of pul- monary” “of the” “pul- monary emboli” “right hilar” “scan suggested” “suggested after” “very limited” |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abujudeh HH, Kaewlai R, Farsad K, Orr E, Gilman M, Shepard JA. Computed tomography pulmonary angiography: an assessment of the radiology report. Acad Radiol. 2009 Nov;16:1309–1315. doi: 10.1016/j.acra.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Aronis JM, Cooper GF, Kayaalp M, Buchanan BG. Identifying patient subgroups with simple Bayes’. Proc AMIA Symp. 1999:658–662. [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Berlin L. Pitfalls of the vague radiology report. AJR Am J Roentgenol. 2000 Jun;174:1511–1518. doi: 10.2214/ajr.174.6.1741511. [DOI] [PubMed] [Google Scholar]
- 5.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 6.Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, Olszewski RT. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med. 2005 Jan;33:31–40. doi: 10.1016/j.artmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Chapman WW, Cooper GF, Hanbury P, Chapman BE, Harrison LH, Wagner MM. Creating a text classifier to detect radiology reports describing mediastinal findings associated with inhalational anthrax and other disorders. J Am Med Inform Assoc. 2003;10:494–503. doi: 10.1197/jamia.M1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman WW, Fiszman M, Frederick PR, Chapman BE, Haug PJ. Quantifying the characteristics of unambiguous chest radiography reports in the context of pneumonia. Acad Radiol. 2001 Jan;8:57–66. doi: 10.1016/S1076-6332(03)80744-4. [DOI] [PubMed] [Google Scholar]
- 9.Christensen LM, Harkema H, Haug PJ, Irwin JY, Chapman WW. Onyx: a system for the semantic analysis of clinical text; BioNLP ’09: Proceedings of the Workshop on BioNLP; Morristown, NJ, USA: Association for Computational Linguistics; 2009. pp. 19–27. [Google Scholar]
- 10.Christensen LM, Haug PJ, Fiszman M. Mplus: a probabilistic medical language understanding system; Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain; Morristown, NJ, USA: Association for Computational Linguistics; 2002. pp. 29–36. [Google Scholar]
- 11.Cohen J. Weighted kappa: nominal scale agreement with provision for scaled disagreement or partial credit. Psychol Bull. 1968 Oct;70:213–220. doi: 10.1037/h0026256. [DOI] [PubMed] [Google Scholar]
- 12.Daume H. Beating an n-gram. 2006 http://nlpers.blogspot.com/2006/06/beating-n-gram.html.
- 13.Denny JC, Peterson JF, Choma NN, Xu H, Miller RA, Bas-tarache L, Peterson NB. Development of a natural language processing system to identify timing and status of colonoscopy testing in electronic medical records; AMIA Annu Symp Proc; 2009. p. 141. [PMC free article] [PubMed] [Google Scholar]
- 14.Dublin S, Glazer NL, Smith NL, Psaty BM, Lumley T, Wiggins KL, Page RL, Heckbert SR. Diabetes Mellitus, Glycemic Control, and Risk of Atrial Fibrillation. J Gen Intern Med. 2010 Apr; doi: 10.1007/s11606-010-1340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkin PL, Brown SH, Bauer BA, Husser CS, Carruth W, Bergstrom LR, Wahner-Roedler DL. A controlled trial of automated classification of negation from clinical notes. BMC Med Inform Decis Mak. 2005;5:13. doi: 10.1186/1472-6947-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedullo PF, Tapson VF. Clinical practice. The evaluation of suspected pulmonary embolism. N. Engl. J. Med. 2003 Sep;349:1247–1256. doi: 10.1056/NEJMcp035442. [DOI] [PubMed] [Google Scholar]
- 17.Fisher ES, Whaley FS, Krushat WM, Malenka DJ, Fleming C, Baron JA, Hsia DC. The accuracy of Medicare’s hospital claims data: progress has been made, but problems remain. Am J Public Health. 1992 Feb;82:243–248. doi: 10.2105/ajph.82.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automaticdetection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman C. A broad-coverage natural language processing system; Proc AMIA Symp; 2000. pp. 270–274. [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta D, Saul M, Gillbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am. J. Clin. Pathol. 2004 Feb;121:176–186. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- 21.Haque R, Chiu V, Mehta KR, Geiger AM. An automated data algorithm to distinguish screening and diagnostic colorectal cancer endoscopy exams. J. Natl. Cancer Inst. Monographs. 2005:116–118. doi: 10.1093/jncimonographs/lgi049. [DOI] [PubMed] [Google Scholar]
- 22.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009 Oct;42:839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatabu H, Hunsaker AR. The cost and consequence of ”uncertainty”. Acad Radiol. 2009 Nov;16:1307–1308. doi: 10.1016/j.acra.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Lowe H. A grammar-based classification of negations in clinical radiology reports; Proc AMIA Annu Fall Symp; 2005. p. 988. [PMC free article] [PubMed] [Google Scholar]
- 25.Jha AK, Kuperman GJ, Teich JM, Leape L, Shea B, Ritten-berg E, Burdick E, Seger DL, Vander Vliet M, Bates DW. Identifying adverse drug events: development of a computer-based monitor and comparison with chart review and stimulated voluntary report. J Am Med Inform Assoc. 1998;5:305–314. doi: 10.1136/jamia.1998.0050305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones KS. A statistical interpretation of term specificity and its application in retrieval. Journal of Documentation. 1972;28:11–21. [Google Scholar]
- 27.Kaushal R, Bates DW, Franz C, Soukup JR, Rothschild JM. Costs of adverse events in intensive care units. Crit. Care Med. 2007 Nov;35:2479–2483. doi: 10.1097/01.CCM.0000284510.04248.66. [DOI] [PubMed] [Google Scholar]
- 28.Mowery D, Harkema H, Chapman W. Temporal annotation of clinical text; BioNLP Workshop of the 46th Annual Meeting of the Association of Computational Linguistics; 2008. p. (in press). [Google Scholar]
- 29.Mowery DL, Harkema H, Dowling JN, Lustgarten JL, Chapman WW. Distinguishing historical from current problems in clinical reports: which textual features help?; BioNLP ’09: Proceedings of the Workshop on BioNLP; Morristown, NJ, USA: Association for Computational Linguistics; 2009. pp. 10–18. [Google Scholar]
- 30.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc. 2001;8:598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson JC, Jackson M, Yu O, Whitney CG, Bounds L, Bittner R, Zavitkovsky A, Jackson LA. Impact of the introduction of pneumococcal conjugate vaccine on rates of community acquired pneumonia in children and adults. Vaccine. 2008 Sep;26:4947–4954. doi: 10.1016/j.vaccine.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Pakhomov S, Bjornsen S, Hanson P, Smith SS. Quality perfor-mance measurement using the text of electronic medical records. Med Decis Making. 2008;28:462–470. doi: 10.1177/0272989X08315253. [DOI] [PubMed] [Google Scholar]
- 33.Savova G, Bethard S, Styler W, Martin J, Palmer M, Masanz J, Ward W. Towards temporal relation discovery from the clinical narrative; AMIA Annu Symp Proc; 2009. pp. 568–572. [PMC free article] [PubMed] [Google Scholar]
- 34.South BRSP, Swaminathan AD, Anthony J, Delisle S, Perl T, Samore MH. Text-processing of va clinical notes to improve case detection models for influenza-like illness. Advances in Disease Surveillance. 2007;2:28. [Google Scholar]
- 35.Trick WE, Chapman WW, Wisniewski MF, Peterson BJ, Solomon SL, Weinstein RA. Electronic interpretation of chest radiograph reports to detect central venous catheters. Infect Control Hosp Epidemiol. 2003 Dec;24:950–954. doi: 10.1086/502165. [DOI] [PubMed] [Google Scholar]
- 36.Uzuner Ö, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. Journal of the American Medical Informatics Association. 2009;16(1):109–115. doi: 10.1197/jamia.M2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uzuner O, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. J Am Med Inform Assoc. 2009;16:109–115. doi: 10.1197/jamia.M2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahner-Roedler DL, Welsh GA, Trusko BE, Froehling DA, Froehling DA, Temesgen Z, Elkin PL. Using natural language processing for identification of pneumonia cases from clinical records of patients with serologically proven influenza; AMIA Annu Symp Proc; 2008. p. 1165. [PubMed] [Google Scholar]
- 39.Wilson RA. Master’s thesis. University of Pittsburgh; 2010. Automated ancillary cancer history classification for mesothelioma patients from free-text clinical reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson RA, Chapman WW, DeFries SJ, Becich MJ, Chap man BE. Automated ancillary cancer history classification for mesothelioma patients from free-text clinical reports. J Pathol Inform TBD. 2010 doi: 10.4103/2153-3539.71065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yount R, Vries JCDC. The medical archival system: An information retrieval system based on distributed parallel processing. Information Processing & Management. 1991;27:379–391. [Google Scholar]
- 42.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L, Melton GB, Parsons S, Hripcsak G. A temporal constraint structure for extracting temporal information from clinical narrative. J Biomed Inform. 2005 Sep 15; doi: 10.1016/j.jbi.2005.07.002. [DOI] [PubMed] [Google Scholar]