Abstract
The shunt model predicts that small ORFs (sORFs) within the cauliflower mosaic virus (CaMV) 35S RNA leader and downstream ORF VII are translated by different mechanisms, that is, scanning–reinitiation and shunting, respectively. Wheat germ extract (WGE) and rabbit reticulocyte lysate (RRL) in vitro translation systems were used to discriminate between these two processes and to study the mechanism of ribosomal shunt. In both systems, expression downstream of the leader occurred via ribosomal shunt under the control of a stable stem and a small ORF preceding it. Shunting ribosomes were also able to initiate quite efficiently at non-AUG start codons just downstream of the shunt landing site in WGE but not in RRL. The short sORF MAGDIS from the mammalian AdoMetDC RNA, which conditionally suppresses reinitiation at a downstream ORF, prevented shunting if placed at the position of sORF A, the 5′-proximal ORF of the CaMV leader. We have demonstrated directly that sORF A is translated and that proper termination of translation at the 5′-proximal ORF is absolutely required for both shunting and linear ribosome migration. These findings strongly indicate that shunting is a special case of reinitiation.
Keywords: translational control, scanning, sORF, AdoMetDC, rabbit reticulocyte lysate, wheat germ extract
The 5′-leader sequence of an mRNA controls translation initiation in eukaryotes. In most cases, leaders are short and, unstructured, and direct initiation from the AUG codon nearest to the capped 5′ end by a linear scanning mechanism (Kozak 1999). Although most initiation events use the 5′-proximal AUG, about 10% of eukaryotic mRNAs (particularly those encoding growth-related factors, tumor suppressers, transcription factors, and proto-oncogenes), and many viral RNAs, contain one or more small open reading frames (sORFs) upstream of the main coding region. Such sORFs can modulate the translation process, depending on their location, length, and sequence. An sORF can either be bypassed if its start codon is in suboptimal context (leaky scanning) (Kozak 1989), or it can allow reinitiation after its own translation. Two types of reinitiation have been described: after translation of an upstream sORF, with reinitiation efficiency increasing with distance between the sORF and the true ORF (Kozak 1987; Fütterer and Hohn 1992; Luukkonen et al. 1995; Hinnebush 1997); and after translation of a large ORF, with a short distance between it and a second ORF in the presence of a nonclassical translation factor (TAV) found in caulimoviruses (Bonneville et al. 1989; Gowda et al. 1989; Fütterer and Hohn 1992).
In several cases, inhibition of downstream translation appears to depend on the amino acid sequence of the uORF, for example, in the transcript leaders of mammalian AdoMetDC (S-adenosyl-methionine-decarboxylase) (Hill and Morris 1992, 1993; Mize et al. 1998), human cytomegalovirus (CMV) gpUL4 (gp48) (Schleiss et al. 1991; Cao and Geballe 1996b, 1998), yeast CPA1 (Werner et al. 1987; Delbecq et al. 1994) and its homolog in Neurospora crassa, arg2 (Wang and Sachs 1997a,b). In all cases, missense mutations altering particular codons (especially at the carboxyl terminus of the peptide) derepress translation of the downstream ORF, whereas mutations in the same codons that retain the amino acid sequence preserve the inhibitory effect. Sequence-dependent sORFs can regulate downstream translation in response to environmental conditions (Werner et al. 1987; Hill and Morris 1992, 1993; Wang and Sachs 1997a). In all cases, the number of ribosomes associated with mRNA is reduced to 1–2 when inhibition occurs. The mechanism of action may involve peptide-specific ribosome arrest near the termination codon of the sORF (Geballe and Morris 1994). This has been directly demonstrated for the relatively long sORFs of the gp48 and arg-2 transcripts (24- and 22-amino acid sORFs, respectively; Cao and Geballe 1996a; Wang and Sachs 1997b), where toeprinting showed ribosome arrest near the sORF termination codon.
Another well-studied example of regulated translation is the yeast GCN4 mRNA (Hinnebusch 1997). In this case, the 5′-proximal sORF, ORF 1, seems to be translated and reinitiation at either the main ORF or at the competing uORF 4 depends on the availability of the eIF-2/GTP/Met-tRNAiMet ternary complex (Dever et al. 1992). The structural context of sORF 4 contributes to the inhibition of downstream reinitiation (Miller and Hinnebusch 1989). This is an example of a nucleotide sequence-dependent inhibitory sORF, whereas sORF 1 has a stimulatory role for GCN4 ORF expression.
At least two alternatives to linear scanning exist for mRNAs with long 5′-noncoding regions and multiple upstream AUGs: initiation directly at an internal ribosome entry site (IRES), for example, in the RNAs of picornovirus, hepatitis C virus, cellular Bip, or Drosophila antennapedia (Jackson 1996); or ribosomal shunting, as shown for the RNAs of CaMV (Fütterer et al. 1990, 1993), Sendai virus (Curran and Kolakofsky 1988; Latorre et al. 1998), rice tungro bacilliform virus (RTBV) (Fütterer et al. 1996), adenovirus (Yueh and Schneider 1996, 2000), and papillomavirus (Remm et al. 1999). In the shunt model, ribosomes start scanning from the capped 5′ end of the mRNA, bypass a large segment of 5′-UTR, which may include AUG codons and secondary structures inhibiting linear migration, and resume scanning further downstream (Hohn et al. 1998).
The cauliflower mosaic virus (CaMV) 35S RNA leader is unusually long, contains several sORFs, and folds into an extensive stem–loop structure (ΔG > −162 kcal/mol; Mfold, GCG) comprising three main stem sections (Hemmings-Mieszczak et al. 1997). The CaMV leader does not function as an IRES when placed between two cistrons (Dominguez et al. 1998). Rather, initiation of the first major viral ORF (ORF VII) downstream of the 35S RNA leader is cap dependent (Schmidt-Puchta et al. 1997) and occurs via ribosome shunt.
The ribosomal shunt in CaMV requires (1) the cap (Schmidt-Puchta et al. 1997), (2) a 5′-proximal sORF, sORF A (Dominguez et al. 1998; Pooggin et al. 1998), and (3) a stable stem (stem section 1) at a proper distance (Dominguez et al. 1998; Hemmings-Mieszczak et al. 1998). The leader hairpin structure promotes shunting by bringing the “shunt landing site” (Fütterer et al. 1993) upstream of the first CaMV ORF into close proximity with a shunt “take-off site” downstream of sORF A (Dominguez et al. 1998). Both the ribosome shunt mechanism itself and the crucial role of the 5′-proximal sORF in the process remained unclear.
In this study, we applied plant and animal in vitro translation systems and in vivo expression analysis to gain further insight into the mechanism of ribosomal shunt.
Results
Experimental design
The CAT reporter gene was placed downstream of the CaMV leader at the position of ORF VII. In the viral context, the last sORF within the leader (sORF F) partially overlapped the ORF VII (see Lm, Fig. 1B). However, we used a frameshift mutant fusing sORF F to the CAT ORF (LmF, Fig. 1A,B; Dominguez et al. 1998). The two AUG codons of sORF F (F and F′) and the AUG of the CAT reporter lay upstream and downstream of the shunt landing site, respectively (Fig. 1A). Ribosomes migrating through the central part of the leader and initiating at the start codon of sORF F or F′ should produce F::CAT or F′::CAT fusion polypeptides, whereas shunting ribosomes should initiate downstream of the shunt landing site, normally at the CAT AUG. High-resolution tricine/SDS–PAGE was used to separate these fusion polypeptides (differing sometimes by only a few amino acids) after translation in vitro.
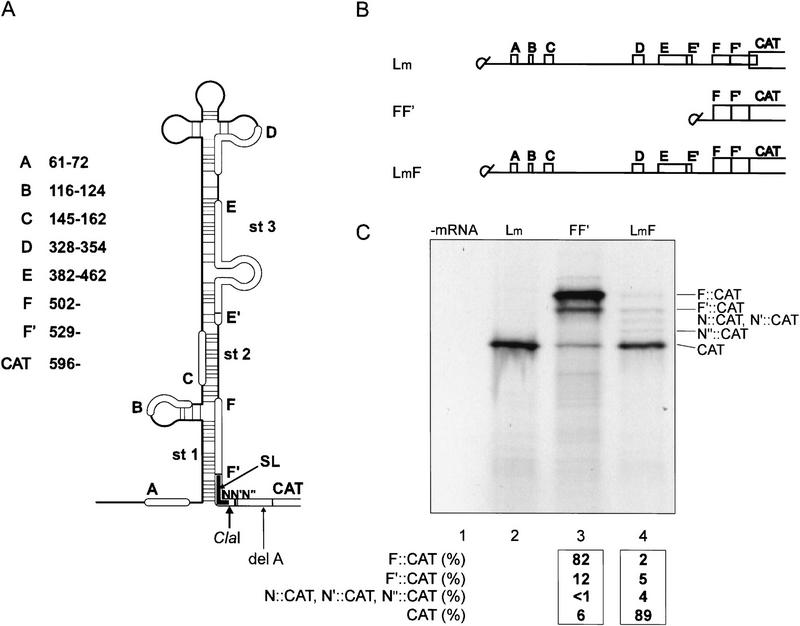
Figure 1.
The 35S RNA leader redirects expression to downstream AUG. (A) Schematic presentation of the predicted secondary structure (MFold) of the modified CaMV (strain S) leader (LmF) consisting of the three main stem sections (st1, st2, and st3) interrupted by bifurcations. The positions of sORFs (A–F′) are indicated by thick lines superimposed on the structure, and potential non-AUG initiation start sites, N, N′, and N′′, are shown. The shunt landing site (SL) is in black. The ClaI restriction site is indicated. (del A) Deletion of A579 to create an F::CAT fusion ORF. (B) Capped mRNA constructs used for translation in WGE. sORFs A–F′ are shown by boxes. In construct Lm, ORF F overlaps the CAT ORF as in the wild-type situation with ORF VII, whereas in FF′ and LmF it is in-frame with the CAT ORF. (C) Wheat germ translation reactions with either Lm (1.5 pmole), LmF (1.5 pmole), FF′ (0.75 pmole), or no added mRNA were resolved on a tricine/SDS–polyacrylamide gel. Positions of F::CAT, F′::CAT, N::CAT, N′::CAT, N′′::CAT, and CAT polypeptides are shown. The relative yields of CAT and CAT fusion polypeptides synthesized were calculated relative to the sum of the total level of translation. The values are the average from at least three independent experiments.
A truncated leader construct (FF′, Fig. 1B), lacking the region including sORFs A to E′, contained the AUG codons of F, F′, and CAT ORFs in moderate, weak, and strong initiation context, respectively (Kozak 1989), and correspondingly gave rise to three proteins (Fig. 1C, lane 3). In agreement with the scanning hypothesis (Kozak 1999), the first AUG encountered (F) was preferentially recognized by ribosomes and only a small fraction continued scanning to initiate at the AUGs of F′ or CAT ORFs (Fig. 1C, lane 3). In contrast, the full-length CaMV leader directed ribosomes to initiate mainly at the CAT AUG (Lm, LmF; Fig. 1C, lanes 2 and 4), indicative of shunting (see also Schmidt-Puchta et al. 1997). Additional fusion polypeptides larger than CAT but smaller than F′::CAT polypeptides were also detected in LmF, but not in FF′ or Lm (N::CAT, N′::CAT, and N′′::CAT polypeptides; Fig. 1C, lane 4).
Stem section 1 controls initiation downstream of the CaMV leader
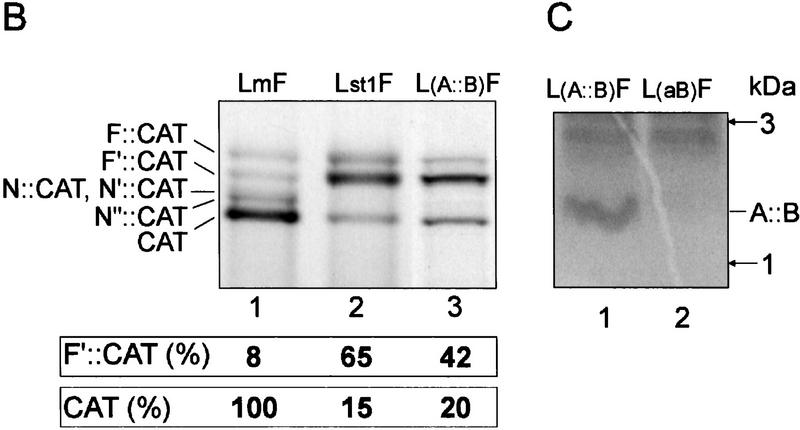
The CaMV leader with a destabilized stem section 1 does not promote shunting at the original take-off and landing sites (Dominguez et al. 1998). Physical disruption of stem section 1 by mutations in the left arm of the hairpin structure (Lst1F, Fig. 2A) resulted in preferential use of the F′ AUG (Fig. 2B, lane 2). A significant reduction of N::CAT, N′::CAT, and N′′::CAT ORF expression was also observed. A similar result was obtained with a further mutant, L(A::B)F, in which sORF A was elongated and fused to sORF B (Fig. 1A). Here, stem section 1 would be disrupted by ribosomes translating the A::B fusion ORF. The A::B peptide was detected (Fig. 2C, lane 1), showing that the AUG of sORF A was recognized and the sORF was translated. As an additional control, mutation of the latter AUG to UAG fully abolished A::B polypeptide synthesis (Fig. 2C, lane 2). Thus, destabilization of stem section 1 led to a significant reduction of CAT protein production by shunting ribosomes. The concomitant drop in the level of N::CAT, N′::CAT, and N′′::CAT polypeptides suggests that these proteins are also synthesized via shunting.
Figure 2.
Initiation downstream of the CaMV leader hairpin depends on stem section 1. (A) Schematic representation of secondary structures of constructs with mutated stem section 1 (Lst1F) or altered stop codon of sORF A [L(A::B)F] are shown. The A::B fusion ORF is indicated. (B) Representative WGE in vitro translation results. Yields of F′::CAT and CAT protein expressed as a percentage relative to the level of CAT from LmF, are given. (C) Low-molecular-weight polypeptide detected on translation of L(A::B)F (lane 1). The A::B ORF AUG codon is mutated to UAG in L(aB)F (lane 2). The position of the A::B fusion polypeptide on the gel is indicated (only the range from 1 to 3 kD is shown).
The F′ AUG was preferentially recognized in mutants with a destabilized stem section 1, despite the presence of stem sections 2 and 3 and additional ORFs D and E in the central part of the CaMV leader. These results are best explained by an alternative shunt event occurring just downstream of sORF B, which becomes apparent when the primary shunt is impaired by stem section 1 destabilization (L.A. Ryabova, M.M. Pooggin, D. Dominguez, and T. Hohn, in prep.).
The N::CAT, N′::CAT, and N′′::CAT polypeptides are derived from initiation events at non-AUG start codons downstream of the shunt landing site
We hypothesized that the N::CAT, N′::CAT, and N′′::CAT polypeptides, which appear only in the full-length leader where shunting is operating, are initiated from non-AUG start codons. Analysis of the Mg2+ dependence of their synthesis in wheat germ extract (WGE) gave an optimal value typical of non-AUG start codon recognition (2.0–2.2 mm), whereas at 1.5–1.7 mm Mg2+ (optimal for CAT synthesis) only 40% of this maximal expression was obtained (data not shown).
The shunt landing site is located just upstream of an AU-rich sequence (∼20 nucleotides long), which is conserved in all plant pararetroviruses (Pooggin et al. 1999). This region contains three potential non-AUG start codons, AUA553, AUC556, and AUA562, potentially able to initiate translation of the N::CAT, N′::CAT, and N′′::CAT polypeptides, respectively (Fig. 3A). The frameshift mutation in LmF also brought these codons into frame with AUG of CAT and allowed visualization of N::CAT and/or N′::CAT (N and N′ are too close to be well resolved) and N′′::CAT polypeptides, suggesting that these non-AUG start codons are recognized. The possibility that two additional non-AUG codons located at the end of the leader, AUU581 and AUC587, which were also in frame with the CAT ORF in our constructs, could be start sites for these extra polypeptides was excluded by knockout mutation of these codons in construct LΔN(last)F (Fig. 3B, cf. lanes 1 and 2). Insertion of two AUGs in perfect context in place of the N– and N′′–non-AUG start codons, while destroying the N′–non-AUG [LAUG(NN′′)F], showed that ribosomes were efficiently attracted at the position of the first non-AUG triplet, whereas initiation further downstream was blocked (Fig. 3B, lane 3).
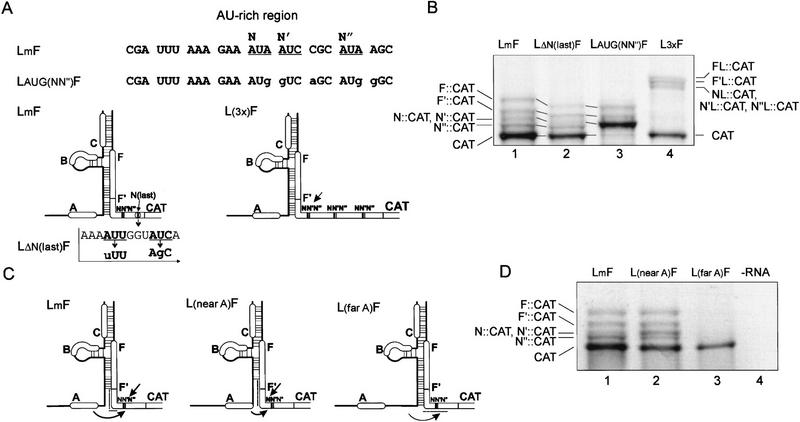
Figure 3.
Initiation at non-AUG triplets downstream of the shunt landing site. (A) Primary sequence of the 3′ unstructured region flanking the leader hairpin and secondary structure in mutant constructs used for translation in WGE are shown with reference to the LmF construct. Non-AUG codons in-frame with the CAT ORF are underlined and designated N, N′, N′′, and N(last). Mutations in these non-AUGs are shown in lower case. The arrow in L3xF denotes non-AUGs involved in translation initiation. (B) Representative translation in WGE with the constructs shown in A. Positions of F::CAT, F′::CAT, N::CAT, N′::CAT, N′′::CAT, and CAT polypeptides are indicated. Note that the mobility of F::CAT, F′::CAT, and the non-AUG::CAT fusions, but not CAT protein, in LΔN(last)F and LAUG(NN′′)F is slightly different because of the altered amino acid composition of the resulting fusion polypeptides [(Ile → Phe, Ile → Ser) and (Ile → Met, Ile → Val, Arg → Ser, Ile → Met, and Ser → Asp), respectively]. Note also that the mutant L3xF produces longer fusion products whose positions are also indicated (FL::CAT, F′L::CAT, NL::CAT, N′L::CAT, N′′L::CAT). (C) Schematic presentation of the secondary structures of the leader with wild-type and shifted positions of sORF A. The straight arrow indicates which non-AUG triplet is preferentially expressed, curved arrows indicate nonlinear migration of ribosomes. Broken lines indicate the position of the shunt landing site in LmF and its hypothetical positioning in L(nearA)F and L(farA)F. (D) Representative translation in WGE with constructs shown in C.
The unstructured region just 3′ of the shunt landing site and preceding the CAT AUG was triplicated (L3xF, Fig. 3A). Translation of this construct revealed the expected elongated F and F′::CAT products (Fig. 3B, lane 4). However, only one set of N::CAT, N′::CAT, and N′′::CAT polypeptides were produced. These originated from the first set of non-AUG start codons, whereas the second and third set were not recognized.
Because these non-AUG codons are preferentially recognized in shunt-mediated constructs and in close proximity to the shunt landing site, we conclude that shunting ribosomes can recognize start codons with lowered fidelity.
We took advantage of these multiple non-AUG start codons to study the effect of sORF A position on start codon recognition downstream of the shunt landing site. Wild-type sORF A is optimally positioned for maximal translation of a reporter ORF downstream of the leader (Dominguez et al. 1998). The non-AUG codon N′′, located 21 nucleotides downstream of the base of stem section 1, was recognized more efficiently than non-AUG codons N and N′, located 9 and 6 nucleotides closer to the base of this stem (Fig. 3D, lane 1). When the stop codon of sORF A directly abutted the base of stem section 1 (Fig. 3C) after deletion of 6 nucleotides downstream of the stop codon of sORF A [L(near A)F], CAT synthesis was slightly reduced, whereas recognition of the first two non-AUG codons (N or N′) was more efficient (Fig. 3D, cf. lanes 1 and 2). Increasing the distance between the sORF A stop codon and stem section 1 from 6 to 30 nucleotides [L(far A)F, Fig. 3C] completely abolished translation initiation events upstream of CAT, whereas CAT protein production was reduced but not eliminated (Fig. 3D, lane 3).
These results show that the position of the sORF A stop codon relative to the base of stem section 1 affects the sites and efficiencies of initiation events by shunting ribosomes downstream of the leader hairpin and we assume that this is caused by repositioning of the shunt landing site (as indicated in Fig. 3C). The position of 5′-proximal sORF contributes to ribosomal linear migration via leader hairpin to get to the F and F′ ORFs.
Ribosome shunting functions in a mammalian cell-free translation system
To investigate whether the shunt mechanism promoted by the CaMV leader can function in the absence of plant-specific translational factors, we used rabbit reticulocyte lysate (RRL) for in vitro translation experiments. The translational efficiency of the wild-type leader-containing RNA was compared to mRNAs with a strong stem either at the 5′ end or in the center of the leader (LC20, LCs2, LCs108, respectively). Consistent with previous results in WGE (Schmidt-Puchta et al. 1997), insertion of the −43.9 kcal/mole stem–loop structure at the 5′-end of the leader caused a strong repression of CAT reporter translation, whereas the same stem inserted within stem section 3 had no effect (Fig. 4A). Thus the 5′ end of the CaMV RNA leader is required for translation also in RRL, whereas the middle portion can be loaded with insertions without affecting translation.
Figure 4.
Stem section 1 and sORF A direct shunting in RRL. (A) Effect of stable stem-loop structures introduced into the CaMV leader on in vitro translation of the downstream CAT reporter in RRL. Constructs used for translation and the relative efficiencies of expression as determined by PhosphorImager quantification are shown. The values are the means from at least three independent experiments. (B) Representative translation in RRL of RNA constructs containing short (FF′) or full-sized leader (LmF and variants). The effect of stem section 1 destabilization (Lst1F), alteration of the 5′-proximal sORF [La(UAG)F, L(A::B)F, L(near A)F, and L(farA)F] and mutation of the non-AUG start codons [LAUG(NN′′)F] on CAT translation are shown. Reference translation in WGE of LmF RNA is shown at left. Yields of F′::CAT and CAT protein expressed as a percentage relative to the level of CAT detected on translation of LmF mRNA are shown below the gel.
Properties of the shunt process in RRL were examined in more detail using the above constructs in WGE. F::CAT, F′::CAT, and CAT polypeptides were expressed by linear and leaky scanning in decreasing order of efficiency from the short leader construct FF′ (Fig. 4B, lane 1; see Fig. 1B), showing that the 5′ proximal AUG of sORF F is preferentially recognized in RRL as in WGE. In the context of the complete leader sequence (LmF), CAT protein was mainly produced, as expected for shunting (Fig. 4B, lane 2).
Most of the other constructs tested behaved similarly in RRL and WGE: (1) Disruption of stem section 1 (Lst1F) greatly reduced CAT ORF translation but increased expression from the F′ AUG (Fig. 4B, lane 3). (2) Mutation of the AUG codon of sORF A to a stop codon, La(UAG)F, resulted in a 10-fold reduction in CAT ORF translation (Fig. 4B, lane 4; WGE: Dominquez et al. 1998). Shifting the position of sORF A back and forth relative to the base of stem section 1 [mutants L(near A)F and L(far A)F] reduced CAT polypeptide production three- to fourfold (Fig. 4B, lanes 6 and 7). (3) The position of sORF A also contributed to migration of ribosomes through the leader hairpin, as increasing the distance between the stop codon of sORF A and the base of stem section 1- to 30- nucleotides completely abolished F and F′::CAT fusion polypeptide production (Fig. 4B, lane 7). Thus, the shunting process in RRL was also under the control of sORF A and stem section 1 and there was no requirement for plant-specific factors in the shunt mechanism.
However, some special features of initiation differ between RRL and WGE: (1) N::CAT, N′::CAT, or N′′::CAT protein fusions were not detected in RRL (Fig. 4B, lane 2), showing that in this case the non-AUG start codons were poorly recognized. This was not due to a position effect, as AUGs placed at the position of the N and N′′ non-AUG codons were efficiently recognized, whereas CAT AUG recognition was strongly reduced (Fig. 4B, lane 8). (2) Disruption of stem section 1 by fusion of sORF A with sORF B [L(A::B)F] did not significantly switch the production of CAT polypeptides to the production of F′::CAT polypeptides (Fig. 4B, lane 5), as was the case with WGE (Fig. 2B, lane 3). These dissimilarities suggest that stem section 1 is more stable in RRL than in WGE and/or that reinitiation in RRL is less efficient. However, note that other secondary structure elements such as the iron-response element inhibits scanning more in WGE than in RRL (Paraskeva et al. 1999).
Control of shunting by an sORF derived from the mammalian AdoMetDC mRNA
To study the mechanistic details of how sORF acts on shunt-mediated translation, we took advantage of a specific uORF that conditionally suppresses reinitiation. The mammalian AdoMetDC mRNA sORF, encoding a hexapeptide with the amino acid sequence MAGDIS (Hill and Morris 1992), serves as a negative regulatory element by regulating reinitiation at the associated downstream cistron in a polyamine-responsive manner in T lymphocytes, HeLa cells, and yeast. Although the wild-type MAGDIS sORF represses reinitiation in vivo, the D4R mutation (MAGRIS sORF) alleviated this repression by 87% and deletion of the serine at position 6 (MAGDI sORF) completely abolished it (Mize et al. 1998). If this regulation also functions in vitro, the AdoMetDC sORF provides a means to study translational events at the sORF required for ribosomal shunt.
The AdoMetDC sORF variants described above were introduced in place of sORF A in the wild-type CaMV leader, LmF (LMAGDISF, LMAGRISF, LMAGDIF; Fig. 5A) or in a truncated derivative lacking the hairpin structure, TLsORF A (TLMAGDIS, TLMAGRIS, TLMAGDI; Fig. 5A), and translation assays were performed in WGE and RRL.
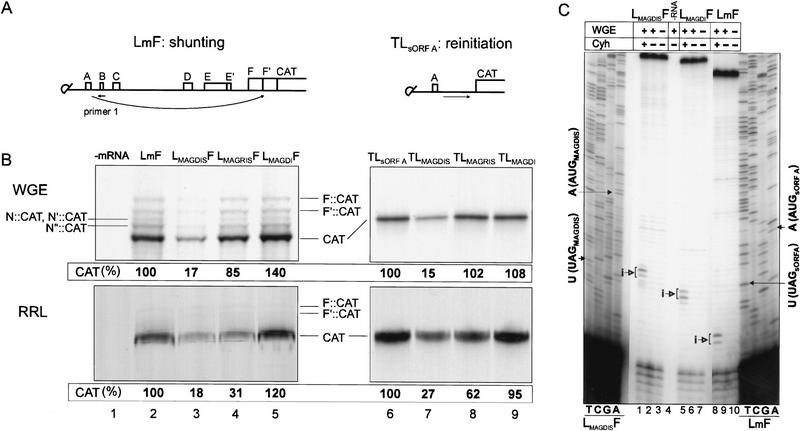
Figure 5.
Mammalian AdoMetDC sORF in place of sORF A represses ribosomal shunt- and linear migration-mediated expression in the CaMV leader. (A) Capped mRNA shunt (left) and reinitiation (right) supporting constructs with sORF A, as an example, used for translation. sORFs A–F′ are shown by boxes. Arrows indicate ribosome migration. Small arrow on LmF shows position of the primer for reverse transcription. (B) Analysis of [35S]methionine-labeled polypeptides produced by translation of synthetic RNA transcripts in WGE (top) and in RRL (bottom), with no added mRNA (lane 1), LmF (lane 2), LMAGDISF (lane 3), LMAGRISF (lane 4), LMAGDIF RNA (lane 5), TLsORF A (lane 6), TLMAGDIS (lane 7), TLMAGRIS (lane 8), TLMAGDI (lane 9). (C) Shunt-supporting RNAs (1.5 pmole) containing the wild type sORF A (LmF), or the wild-type or mutant AdoMetDC sORF (LMAGDISF and LMAGDIF) were translated for 20 min (27°C) in a 25-μl WGE reaction mixture containing Cyh where indicated. Radiolabeled primer (primer 1) was used for primer extension analyses (lanes 1–10) and for sequencing of LMAGDISF and LmF (left and right, respectively). Arrows indicate the positions of premature reverse transcription termination products (i) in the presence of Cyh, corresponding to ribosomes bound at the AUGs of MAGDIS sORF (lane 1), MAGDI sORF (lane 5), and sORF A (lane 8). The sequences read 3′ to 5′ from top to bottom.
With both sets of constructs and in both in vitro systems, the MAGDIS sORF significantly reduced CAT ORF translation compared with sORF A (Fig. 5B, cf. lanes 2 and 3 and 6 and 7). This inhibition was fully or partially reversed when the MAGRIS sORF was used and totally abolished by the MAGDI sORF (Fig. 5B, lanes 4,5,8,9).
Thus, the MAGDIS sORF inhibits reinitiation in vitro, both in a simple reinitiation context and in ribosome shunt-mediated translation. These results also show that the MAGRIS and MAGDI sORFs were perfectly able to replace sORF A and support efficient shunting (although MAGRIS sORF worked less well in RRL).
The comparable effects of MAGDIS sORF on both shunt- and reinitiation-supporting constructs strongly suggests that shunt-mediated translation is a variant of reinitiation. RNA stability did not appear to be a primary component of AdoMetDC sORF regulation in either extract, as most of the input RNA was recovered after the translation assay (data not shown) or reverse transcribed to the full-length cDNA after translation mixture incubation (see below, Fig. 5C).
Ribosomal complexes formed at sORF A (or at AdoMetDC sORFs) in the context of the full-length leader were also analyzed by a toeprinting (primer extension inhibition) assay in WGE (Fig. 5C). The leader hairpin had an inhibitory effect on reverse transcription primed with primer 2 complementary to a region downstream of the leader hairpin. Primer 1 complementary to the region of the sORF B loop between stem sections 1 and 2 of the leader hairpin was used to overcome this inhibitory effect. Reverse transcription in the absence of WGE yielded cDNA extension products predominantly corresponding to full-length transcripts (Fig. 5C, lanes 3, 7, and 10). Inhibition of translation elongation by cycloheximide (Cyh) revealed extension stop sites at positions corresponding to 17 and 18 bases downstream of base A of the sORF start sites [MAGDIS sORF (lane 1), MAGDI sORF (lane 5), and sORF A (lane 8)]. Thus, the AUG of sORF A (moderate context AUAAUGU) in front the CaMV leader hairpin was indeed recognized as efficiently as the AUGs of the MAGDIS and MAGDI sORFs (perfect context AUAAUGG), probably because of its position in front of a stem structure (Kozak 1990). This provides direct evidence that the initiation context of sORF A is not critical for recognition of sORF A in WGE.
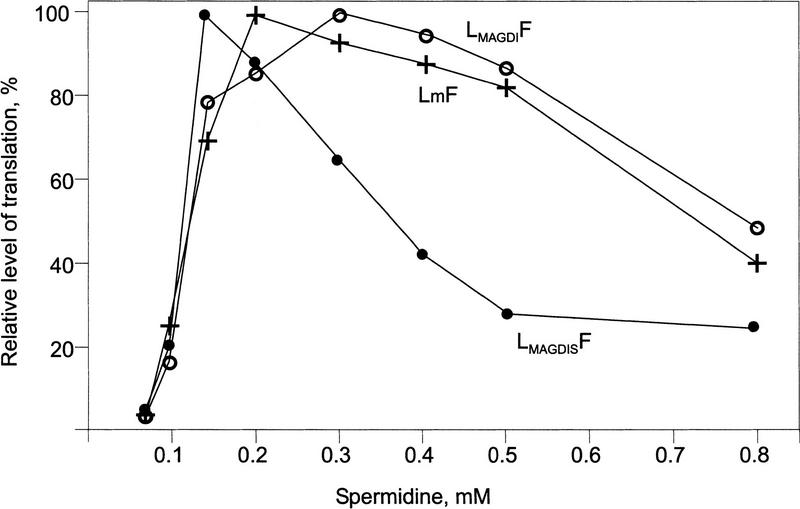
Standard RRL and WGE include polyamines. The standard concentrations of spermidine in WGE or RRL (0.6 and 0.5 mm, respectively) were already sufficient to downregulate CAT ORF translation programmed with LMAGDISF and TLMAGDIS RNAs (Fig. 5B, lanes 3 and 7). A decrease of spermidine or spermine concentrations in WGE strongly affected the total level of protein synthesis, whereas in RRL the level was decreased but not abolished. The effect of spermidine addition (0.1–0.8 mm) to a translation system containing gel-filtered RRL was further tested using three shunt-mediating constructs, LmF, LMAGDISF, and LMAGDIF (Fig. 6). Addition of 0.5 mm spermidine still allowed efficient translation of LMAGDIF RNA or LmF RNA, whereas translation of LMAGDISF RNA was strongly inhibited at this concentration. Similar results were obtained when shunting was not involved, that is, TLMAGDIS and TLMAGDI RNAs (data not shown).
Figure 6.
Effect of spermidine on translation of the AdoMetDC sORF-containing shunt-mediated constructs in RRL. Gel-filtered RRL reaction mixtures programmed with LmF (+), LMAGDIF (○), and LMAGDISF (●) RNAs were supplemented with different amounts of spermidine as indicated. Reactions were analyzed by tricine/SDS–PAGE. Radiolabeled bands were analyzed by PhosphorImager. The maximum level of translation for each mRNA (sum of CAT and CAT fusion polypeptides) was set as 100%. Yields at different spermidine concentrations are given as a percentage relative to this maximum.
Together, these results demonstrate that both the amino acid coding information of the AdoMetDC sORF and the presence of polyamines are required for suppression of downstream reinitiation and shunting in vitro.
Ribosomes stall at the AdoMetDC sORF stop codon, preventing reinitiation at a downstream ORF
The mechanism by which MAGDIS inhibits downstream translation has not been reported so far. Therefore, we employed the toeprinting assay to investigate the mechanism of AdoMetDC sORF function. The truncated CaMV RNA leader containing the MAGDIS sORF (or MAGDI sORF as a control) in place of sORF A (see Fig. 7A) was used because structure in the leader hairpin impedes reverse transcription. We used RRL as an homologous model system for the mammalian AdoMetDC ORF. In the absence of RRL, reverse transcription of both RNAs with a radiolabeled primer complementary to a region downstream of the AUG of the CAT ORF yielded long cDNA extension products predominantly corresponding to full-length transcripts (Fig. 7B, lanes 2 and 6). When translation elongation was inhibited by Cyh addition, a toeprint was observed 17 and 18 nucleotides downstream of base A of the sORF start codon (lanes 3 and 7, indicated by i). These products correspond to the inhibition of reverse transcription of the RNA template by ribosomes with the initiation codons in their P sites (Fig. 7C, top). The efficiency of AUG recognition in both constructs was similar (cf. lanes 3 and 7). Without Cyh, the toeprinting assay revealed ribosomes paused 14–15 nucleotides downstream of base U of the UAG stop codon with TLMAGDIS RNA, but not with TLMAGDI RNA (Fig. 7B, cf. lanes 4 and 8, indicated by t). These extension products correspond to ribosomes with the sORF termination codon located in their A sites (Fig. 7C, bottom). Increasing the spermidine concentration to 0.6 mm considerably increased the strength of this toeprint for the MAGDIS sORF (lane 5), whereas only a weak signal appeared in the case of MAGDI sORF (lane 9).
Figure 7.
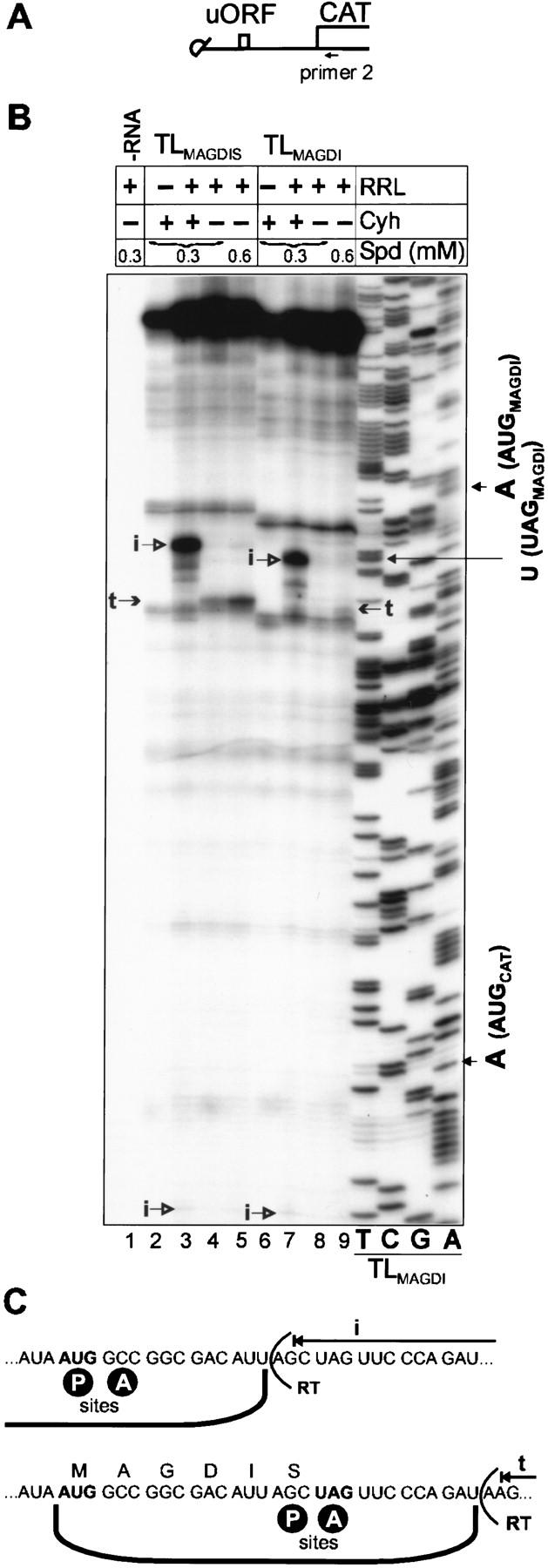
Toeprinting reveals stalling at the stop codon of the AdoMetDC sORF. (A) Capped reinitiation-supporting constructs used for toeprinting. Position of primer 2 is indicated. (B) Transcripts (1.5 pmole) containing the wild-type or mutant AdoMetDC sORF (TLMAGDIS and TLMAGDI RNA) upstream of the CAT ORF were translated for 20 min (30°C) in a 25-μl RRL reaction mixture. Spermidine (spd, 0.3 or 0.6 mm) and cycloheximide (Cyh, 0.9 mm) were included as indicated. Radiolabeled primer 2 was used for primer extension analyses (lanes 1–9) and for sequencing of the wild-type TLMAGDI template (the four righthand lanes). The sequence reads 3′ to 5′ from top to bottom. Arrows indicate the positions of premature reverse transcription termination products: (i) in the presence of Cyh, correspond to ribosomes bound at the AUGs of MAGDIS (lane 3) and MAGDI sORFs (lane 7); (t) correspond to ribosomes near the UAGs of MAGDIS (lanes 4,5) and MAGDI sORFs (lane 9). (C) 80S ribosome bound to mRNA with a start codon at the ribosomal P site causes toeprints 17–18 nucleotides downstream of base A of the start codon (Anthony and Merrick 1992) and ribosomes stalled near a stop codon at the ribosomal A-site of arg-2 ORF cause toeprints about 13 nucleotides downstream of base U of the stop codon (Wang and Sachs 1997b). (Top) A ribosome bound to TLMAGDIS RNA at the start codon of the MAGDIS sORF causing premature reverse transcriptase termination 17–18 nucleotides downstream of base A of the AUG. (Bottom) A ribosome stalled near the stop codon of the MAGDIS sORF with the sORF termination codon at its A site causes premature termination 14–15 nucleotides downstream of base U of the UAG. The position of reverse transcriptase (RT) is indicated; A and P ribosomal sites are also shown.
Thus, the mammalian AdoMetDC sORF mediates its inhibiting effect on downstream reinitiation by interfering with translation termination at its own stop codon. Depending on the environmental conditions, it is assumed that the nascent peptide interacts with the component of termination machinery, apparently blocking the step of peptidyl–tRNA hydrolysis as in the case of CMV gpUL4 (gp48) (Cao and Geballe 1998). Ribosome stalling has been demonstrated for long ORFs (>20 amino acids) (Cao and Geballe 1996a; Wang and Sachs 1997b). Here we show that a short nascent peptide (MAGDIS) causes the same type of stalling (Fig. 7B), indicating that termination is directly affected.
The AdoMetDC uORF also represses ribosomal shunting in vivo
The effect of sORF A substitution with the MAGDIS sORF and its derivatives on CAT ORF expression was also analyzed in plant protoplasts (Table 1). Consistent with the in vitro data, replacement of sORF A by MAGDIS sORF practically abolished expression. On the other hand, MAGRIS and MAGDI sORFs again supported shunting as efficiently as wild-type sORF A. We also tested the effect of the CaMV-derived translational transactivator TAV, which functions to promote reinitiation at the internal ORFs on polycistronic mRNA (Fütterer and Hohn 1991). TAV did not eliminate ribosomal stalling at the termination step of translation, but promoted a threefold increase in the initial level of ribosomal shunting. Thus, in vivo expression of the CAT ORF by shunting ribosomes requires translation of sORF A, shunting and subsequent reinitiation. TAV simply stimulates reinitiation at the AUG of the CAT ORF.
Table 1.
MAGDIS represses downstream CAT expression in plant protoplasts
| Constructs
|
Relative CAT expression (%)
|
||
|---|---|---|---|
| −TAV
|
+TAV
|
+TAV/−TAV
|
|
| LsORF A | 100 ± 8 | 329 ± 10 | 3.3 |
| LMAGDIS | 6 ± 1 | 18 ± 3 | 3.0 |
| LMAGRIS | 100 ± 9 | 299 ± 9 | 3.0 |
| LMAGDI | 102 ± 8 | 219 ± 10 | 2.1 |
Discussion
Three cis-acting elements are responsible for ribosome shunt-mediated translation downstream of the CaMV leader: the cap (Fütterer et al. 1993; Schmidt-Puchta et al. 1997), a 5′-proximal sORF (Dominguez et al. 1998; Pooggin et al. 1998), and a strong stem structure (Dominguez et al. 1998; Hemmings-Mieszczak et al. 1998). In this study we demonstrate that the sORF is translated, that this translation event allows ribosome shunt with subsequent reinitiation downstream of the shunt landing site, and that shunting does not depend on plant-specific factors.
At the beginning of our study, we envisaged four scenarios for shunting: (1) initiation transfer—an initiation complex, corresponding to complex II (Pestova et al. 1998), is transferred from the AUG of the sORF to the shunt landing site at the 3′ end of the leader; (2) hopping—the sORF is translated but not terminated and the 80S ribosome with the nascent peptide is transferred by a mechanism related to the hopping observed in prokaryotes (Herbst et al. 1994); (3) induced internal initiation—the sORF is translated and terminated, and partial melting of the stem creates an internal ribosome entry site; (4) reinitiation—the sORF is translated and terminated and then the 40S ribosome is shunted and used for reinitiation.
A key question is whether the sORF is translated and terminated before shunting occurs. The three-amino-acid peptide derived from sORF A is not easily detectable. Although we detected the 20-amino-acid peptide derived from the sORF A:B fusion (Fig. 2C), showing that a translation event at this position indeed occurs, this does not in itself prove that sORF-translating and shunting ribosomes are linked, as these could originate from different subpools.
More compelling evidence that the sORF is translated and that shunting occurs on the same RNA molecule comes from the inhibitory action on reinitiation and shunting of the short regulatory MAGDIS sORF. Sequence-dependent inhibition by short regulatory sORFs depends on their translation. Under conditions where reinitiation is blocked, the ribosome is stalled at the termination codon of the sORF. Thus, peptide sequence- and polyamine-dependent suppression of shunting by the MAGDIS polypeptide at the position of sORF A demonstrates that the 5′-proximal sORF is indeed translated by the majority of scanning ribosomes and that this translation leads to shunting.
This rules out both initiation transfer and hopping. Furthermore, the extended polypeptides expected from hopping would have been detected by the sensitive tricine/SDS–polyacrylamide gel system.
A structural change in the leader RNA might promote shunting by induced internal initiation. Such a change in fact occurs by melting of part of stem section 1 by the sORF-translating ribosome. However, ribosomes that have translated and terminated sORF A would be expected to unwind at least the first three G-C base pairs of stem section 1. Such a structural change could create an IRES. Our experiments show that MAGDIS sORF, terminating either at the exact position of sORF A (Fig. 5) or 3 and 6 nucleotides further into the stem (not shown), inhibits shunting. This rules out induced-IRES, in favor of the reinitiation, model (Fig. 8). In this model, the same ribosome that translates the 5′ proximal sORF would initiate downstream of the leader hairpin. The sORF A seems to be recognized by the majority of ribosomes that started scanning at the cap. The presence of the downstream strong stem could enhance sORF A recognition significantly. The fact that the sORF A initiation context is not critical supports this. The reinitiation model is further strengthened by the apparent promotion of shunting by the CaMV translational transactivator (Fütterer et al. 1993; Pooggin et al. 2000; Table 1), which specifically enhances reinitiation in linear constructs.
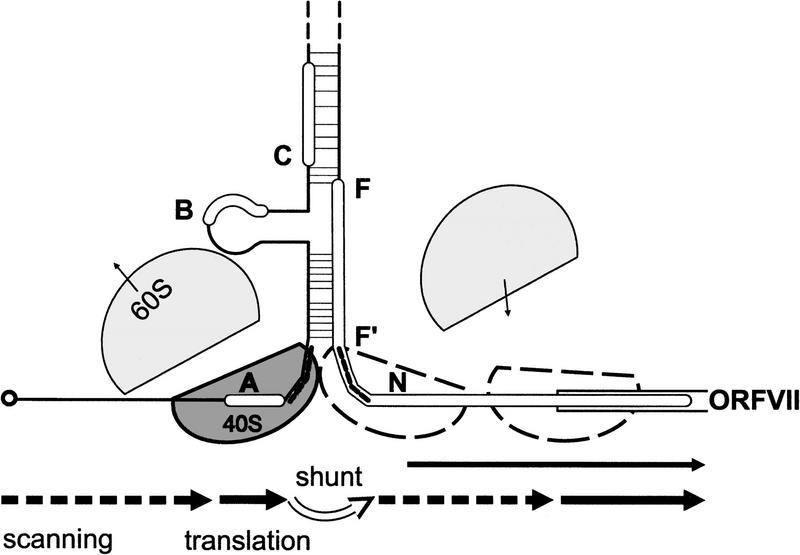
Figure 8.
Model for ribosome shunt. Schematic presentation of the secondary structure of the part of the 35S RNA leader with ORF VII is shown. Broken lines on the structure indicate the positions of the shunt takeoff and landing sites. Arrows show migration of ribosomes (40S and 60S subunits are shown in grey, with outlines representing the subsequent path of the same 40S subunit) by scanning (broken), translation (black), shunting (white). The small arrows show dissociation and association of 60S. Scanning ribosomes enter the 35S RNA at the capped 5′ end and scan until they reach the sORF A start codon. sORF A is translated and properly terminated by most of these ribosomes. The translation event at sORF A provides a specially modified shunt- and reinitiation-competent ribosome that bypasses stem section 1 and is ready to reinitiate just downstream of the shunt landing site. Reinitiation can occur at non-AUG codons (N), but the majority of postshunt ribosomes reinitiate at the AUG of ORF VII. The positive effect of TAV on ribosomal shunt strongly supports the reinitiation model for shunting in CaMV. Note that the translation event at sORF A also leads to partial melting of stem section 1, allowing some post-translation ribosomes to penetrate into stem section 1 and take the alternative route of linear scanning through the leader.
The position of the sORF affected shunting efficiency and determined the start site for reinitiation downstream of the leader hairpin (Fig. 3D). We suggest that it also affects the positions of both the take-off and landing sites with the take-off site related to the end of sORF A. Take-off and landing sites would be symmetrically arranged, that is, mirrored around the stem structure (Fig. 8). Moving the sORF towards the stem increased translation efficiency from non-AUG start codons close to the stem, whereas it was diminished by moving the sORF towards the cap. Upon termination of translation of a 5′-proximal ORF directly abutting the base of stem section 1, additional base pairs of this stem would be disrupted, apparently increasing exposure of the first set of non-AUG start codons for initiation (Fig. 3D). In contrast, when ribosomes terminate too far from the base of stem section 1 to melt it, the multiple non-AUG codons remain inaccessible to ribosomal binding.
An important observation is the shunt-mediated recognition of non-AUG start codons, provided they are positioned appropriately at the shunt-landing site. We assume that the fidelity of AUG recognition by shunting ribosomes is lowered near the shunt landing site, possibly because of temporary lack of “fidelity factors” through sORF A translation. This has been observed also for RTBV, where the ORF reached by shunting ribosomes starts naturally with an AUU codon situated just downstream of the leader hairpin (Fütterer et al. 1996). The efficient non-AUG recognition by post-shunting ribosomes may be due to local deficiency of certain eIFs required for ribosome progression after sORF A translation, for example, eIF 4A/4B (Jaramillo et al. 1991), such that more time for initiation at non-AUG start codons close to the shunt landing site is gained. Additionally or alternatively, eIFs required for maintaining accuracy of initiation may have been lost. Pestova et al. (1998) suggested a role for eIF1/1A in editing inappropriately formed initiation complexes.
Initiation at non-AUG start codons is also enhanced in a number of other interesting cases. In mammalian cells, a potentially different type of shunting found in Sendai virus C/P mRNA (Latorre et al. 1998) uses non-AUG start codons and their mutation to AUGs did not markedly enhance translation. Mutation of the authentic initiation codon used for internal initiation in hepatitis C virus (HCV) RNA (Reynolds et al. 1995) to a non-AUG start codon had little effect on initiation efficiency. Initiation at non-AUG start codons in WGE, as well as in plant protoplasts, seems to occur more efficiently than in mammalian systems (Fütterer and Hohn 1996).
sORF stem-mediated shunting seems not to be the only possibility, as other types of shunting could also exist. For instance, in the adenovirus tripartite leader shunting does not require an sORF but depends on sequences complementary to the 3′ end of the 18S ribosomal RNA (Yueh and Schneider 2000) and on partial depletion of eIF 4F (Yueh and Schneider 1996). A common denominator of both types of shunting may be the stalling event, that is, at the stem structure in CaMV and at the ribosomal RNA-binding site in adenovirus, combined with local or general factor depletion, respectively.
sORF A, as in this study and Pooggin et al. (1998), and stem section 1 of the CaMV RNA leader (Dominguez et al. 1998; Hemmings-Mieszczak and Hohn 1999) as well as both together (M. Hemmings-Mieszczak, T. Preiss, and T. Hohn, in prep.) can be exchanged for other natural or synthetic sORFs and stem structures, respectively, without greatly affecting the efficiency of shunting. Consequently, many, if not all, similar arrangements of sORF and stem structure in eukaryotic (and perhaps even prokaryotic) leaders may be shunt competent. The strength of the stem structure and the position and initiation context of the sORF will then regulate the efficiency of shunting. The amino acid composition of the sORF has little influence, with the important exception of sORFs derived from strongly regulated genes, as exemplified here by the MAGDIS sORF involved in feedback control of polyamine biosynthesis. An additional level of control may be provided by stabilization of the stem structure via protein.
Naturally occurring and artificial shunt structures could be exploited for translation of transgenes, allowing inclusion of secondary structure motifs in the unscanned part of the leader that act as signals, for example, for viral packaging. They can also be used to express dicistronic translation units (Fütterer et al. 1993) if the first ORF is located within a loop at the top of the stem structure and the second one is downstream of the stem. The strength of the stem structure would then determine the fraction of ribosomes translating the first ORF by stem melting or the second ORF by shunting.
Materials and methods
Constructs
Plasmid Lm containing the T7 promoter, the whole CaMV S strain leader (Franck et al. 1980) modified with new restriction sites (XhoI, NheI, SpeI) to facilitate the introduction of mutations, and the CAT reporter gene fused to the AUG of ORF VII was previously described as Lm-CAT (Dominguez et al. 1998). LmF, which carries a deletion of A579 to create F::CAT and F′::CAT fusion proteins was used as a reference construct and described as LmF–CAT (Dominguez et al. 1998). A series of derivatives lacking parts of the leader or having a mutated leader sequences is listed in Table 2. In FF′, most of the leader (sORF A to sORF F′) between the XhoI–ClaI sites in the reference construct was replaced with double-stranded oligonucleotides covering the region including the start codons of sORF F and sORF F′. Mutations around sORF A were introduced by the insertion of oligonucleotides of the desired sequence between the XhoI–NheI sites in LmF, resulting in clones La(UAG)F, L(A::B)F, L[a(UAG)B]F, L(near A)F, L(far A)F (Table 2, and see Dominquez et al. 1998). In Lst1F, stem section 1 is disrupted with several point mutations (Dominguez et al. 1998). Mutations around the shunt landing site were carried out by the insertion of double-stranded oligonucleotides between the ClaI–NcoI sites in LmF, yielding LAUG(N,N′′)F and LΔN(last)F. The insertion of a doubled ClaI–NcoI fragment with alterations of all stop codons into the ClaI site of LmF resulted in L3xF. Substitutions of sORF A with the original or mutated versions of the AdoMetDC sORF (Hill and Morris 1993) were achieved by the insertion of oligonucleotides of the desired sequence between the XhoI–NheI sites in LmF, resulting in clones LMAGDISF, LMAGRISF, LMAGDIF (Table 2). Plasmids TLsORF A, TLMAGDIS, TLMAGRIS, and TLMAGDI, truncated derivatives of LmF lacking the hairpin structure, carry a deletion of the central part of the 35S CaMV leader between the NheI–ClaI sites. Plasmids pSPLC20, pT3sLC2, and pSPLCs108 were described previously (Schmidt-Puchta et al. 1997).
Table 2.
Plasmids used
| Construct
|
Description of cloning
|
Sequence of original and mutated regions
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmF | ClaI/NcoI sequence | C | G | A | T | T | T | A | A | A | G | A | A | A | T | A | A | T | C | C | G | C | A | T | A | A | G | C | C | C | C | C | G | C | T | T | A | A | A | A | A | T | T | G | G | T | A | T | C | A | G | A | G | C | ||||||
| LAUG(NN“)F | ClaI/NcoI oligonucleotide | C | G | A | a | T | T | A | A | A | G | A | A | A | T | g | g | T | C | a | G | C | A | T | g | g | G | C | C | C | C | C | G | C | T | T | A | A | A | A | A | T | T | G | G | T | A | T | C | A | G | A | G | C | ||||||
| LΔN(last)F | ClaI/NcoI oligonucleotide | C | G | A | a | T | T | A | A | A | G | A | A | A | T | A | A | T | C | C | G | C | A | T | A | A | G | C | C | C | C | C | G | C | T | T | A | A | A | A | t | T | T | G | G | T | A | g | C | A | G | A | G | C | ||||||
| L3×F | oligonucleotide insertion in ClaI | C | G | A | a | T | T | c | A | A | G | A | A | A | T | c | A | T | C | C | G | C | A | T | c | A | G | C | C | C | C | C | G | C | T | T | c | A | A | A | A | A | T | T | G | G | t | A | T | C | A | G | ||||||||
| A | G | C | C | T | T | c | A | A | G | A | A | A | T | c | A | T | C | C | G | C | A | T | c | A | G | C | C | C | C | C | G | C | T | T | c | A | A | A | A | A | T | T | G | G | t | A | T | C | A | G | T | A | T | |||||||
| FF‘ | XhoI/ClaI oligonucleotide | T | C | G | A | G | A | T | A | A | (Δ61–517) | A | A | T | G | C | T | T | G | T | A | T | T | T | A | C | C | C | T | A | T | A | T | A | C | C | C | T | A | A | T | G | A | C | C | C | C | T | T | A | ||||||||||
| LmF | XhoI/NheI sequence | T | C | G | A | G | A | A | T | A | A | T | G | T | G | T | G | A | G | T | A | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T | A | G | G | G | T | T | T | C | G |
| La(UAG)F | XhoI/NheI oligonucleotide | T | C | G | A | G | A | A | T | A | t | a | G | T | G | T | G | A | G | T | A | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T | A | G | G | G | T | T | T | C | G |
| L(A∷B)F | XhoI/NheI oligonucleotide | T | C | G | A | G | A | A | T | A | A | T | G | T | G | T | G | A | G | T | c | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T | A | G | G | G | T | T | T | C | G |
| La(UAG)BF | XhoI/NheI oligonucleotide | T | C | G | A | G | A | A | T | A | t | a | G | T | G | T | G | A | G | T | c | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T | A | G | G | G | T | T | T | C | G |
| L(near A)F | XhoI/NheI oligonucleotide | T | C | G | A | G | A | A | T | A | t | a | G | T | G | T | G | A | G | T | A | G | – | – | – | – | – | – | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T | A | G | G | G | T | T | T | C | G |
| L(far A)F | XhoI/NheI oligonucleotide | T | C | G | A | G | a | t | g | t | g | t | g | a | g | t | a | g | t | t | c | c | c | a | g | a | A | A | T | A | t | T | G | T | G | T | G | A | G | T | A | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T |
| A | G | G | G | T | T | C | T | T | A | T | A | G | G | G | T | T | T | C | G | |||||||||||||||||||||||||||||||||||||||||
| LMAGDISF | XhoI/NheI oligonucleotide | T | C | G | A | G | A | A | T | A | A | T | G | g | c | c | g | g | c | g | a | c | a | t | t | a | g | c | T | A | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T |
| A | G | G | G | T | T | T | C | G | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| LMAGRISF | XhoI/NheI oligonucleotide | T | C | G | A | G | A | A | T | A | A | T | G | g | c | c | g | g | c | c | g | c | a | t | t | a | g | c | T | A | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T |
| A | G | G | G | T | T | T | C | G | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| LMAGDIF | XhoI/NheI oligonucleotide | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T | C | G | A | G | A | A | T | A | A | T | G | g | c | c | g | g | c | g | a | c | a | t | t | T | A | G | T | T | C | C | C | A | G | A | T | A | A | G | G | G | A | A | T | T | A | G | G | G | T | T | C | T | T | A | T | A | G | G | ||
| G | T | T | T | C | G | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
(Uppercase letters) Original CaMV leader sequence; (boldface letters) mutated nucleotide sequence introduced with double-stranded oligonucleotide; (lowercase letters) nucleotide chages; (dashes, Δ) deleted nucleotides.
Plasmids LsORF A, LMAGDIS, LMAGRIS, and LMAGDI used for transient infections in this study, containing the 35S RNA promoter, the complete leader with altered 5′-proximal sORF, a CAT reporter, and the CaMV terminator signal were subcloned from LmF, LMAGDISF, LMAGRISF, and LMAGDIF into pGC4.NS′ (Fütterer and Hohn 1992) using unique sites XhoI and PstI occurring downstream of the S1 region and downstream of the stop codon of the CAT gene, respectively.
RNA secondary structure analysis was performed using the MFOLD program (Wisconsin Package, version 6.0, Genetics Group, Madison, WI) at 25°C (Zuker 1989).
In vitro transcription
T7 plasmids were linearized with Acc65I and transcribed in the presence of the cap analog 7mGpppG (in sixfold excess over GTP) by incubation with T7 polymerase (Biofinex), according to the protocol of Gurevich (1996). Transcripts were purified by precipitation with 3 m lithium chloride, followed by precipitation with ethanol. The integrity of the synthesized transcripts was evaluated on a 6% denaturing polyacrylamide gel. RNA was quantified by measuring absorbance at 260 nm.
In vitro translation
WGE was prepared according to Roberts and Paterson (1973). RRL (gel-filtered) was from Roche Boehringer Mannheim Biochemica. FF′ transcripts (0.05 pmole/μl) and 0.1 pmole/μl each of other transcripts were used for in vitro translation in both extracts. The capped mRNAs used were relatively stable during 40 min of incubation in vitro.
A WGE translation reaction in 40 mm HEPES (pH 7.5), containing 3 μl of wheat germ extract, 1.5 mm MgAc2, 150 mm KAc, 10 mm creatine phosphate, 1 mm ATP, 0.2 mm GTP, 5 mm DTT, 0.6 mm spermidine, 1.5 μl of [35S]methionine (29.6 TBq/mmole; NEN, Boston, MA) and 50 μm each of the other 19 amino acids in a total volume of 15 μl was incubated at 28°C for 40 min. In 15 μl of rabbit reticulocyte standard translation reaction, MgAc2 and KAc were added to a final concentration of 1.5 mm and 100 mm, respectively, and the reaction was incubated at 30°C for 40 min. In experiments with different spd concentrations in gel-filtered RRL, the final concentration of MgAc2 was 1.5 mm (spd concentration as indicated in Fig. 6). Cycloheximide (Cyh, for toeprinting assays) was added into the reaction mixtures to bring the final concentration to 0.9 mm.
Translation reaction mixtures (3 μl) were resolved on high-cross-linking (16.5% T and 6% C) tricine/SDS-polyacrylamide gels as described in Schagger and von Jagow (1987). Gels were fixed in the presence of 10% glycerol, dried at 50°C overnight, and exposed to X-ray film (Fuji). Gels were also analyzed using a PhosphorImager PC (Molecular Dynamics, Sunnyvale, CA).
Primer extension inhibition assays
Primers (25 pmole each) for toeprinting and sequencing reactions were labeled with [γ-32P]ATP (>6000 Ci/mmole; NEN, Boston, MA) at their 5′ termini with T4 polynucleotide kinase (New England BioLabs). Primer 1 (ATGCTCAACACATGCTAGCGAAA) and primer 2 (AGGCCGTAATATCCAGCTGA) were purified over Sephadex G-25 and precipitated with ethanol. Oligonucleotides were dissolved in 50 μl water (0.5 pmole/μl). The primer extension inhibition (toeprinting) assay was done as described in Wang and Sachs (1997b) with slight modifications. The assay was performed by adding 5 μl of WGE or RRL translation mixtures (or pure RNA) to 45 μl of reverse transcription buffer on ice. This buffer contained 50 mm Tris-Ac (pH 7.5), 75 mm KAc, 5 mm MgAc2, 5 mm dithiothreitol, 0.3 mm spermidine, 0.5 mm cycloheximide when it was present in the translation reaction, 0.5 mm each dATP, dCTP, dGTP, dTTP, and 1000 units of RNase inhibitor (Promega). Samples were heated at 50°C for 2 min and then immediately placed on ice. 32P-Labeled primer (2 μl, ∼5 × 106 cpm Cherenkov counts) was added to each tube and annealed to the template at 37°C for 5 min. Superscript II RNase H-reverse transcriptase (100 units) (GIBCO BRL) was added. Incubation was carried out for 25 min at 37°C. Reactions were terminated by phenol–chloroform extractions. The aqueous phase was mixed with an equal volume of formamide dye. Samples were heated and loaded on 9% (for primer 1) or 6% (for primer 2) RNA denaturation sequencing polyacrylamide gels. The gels were dried and exposed to film (Fuji) for 1 or 2 days at −80°C. DNA sequencing markers were obtained using the labeled primers used for toeprinting to sequence the corresponding plasmids.
Protoplast transfections and reporter gene assays
Protoplasts were prepared from suspension cultures of Orychophragmus violaceus and transfected with plasmid DNA by electroporation as described by Fütterer et al. (1989). Protoplasts (0.7 ml; ∼2 million) were mixed with plasmids on ice, transferred to 0.4-cm electrode gap cuvettes, and electroporated by discharging 960 μF at 450 V and 200 Ω using a BioRad Gene Pulser. After 20–24 hr incubation at 27°C, protoplasts were harvested and protein extracts prepared by three cycles of freezing and thawing in 200 μl of GUS extraction buffer. Aliquots (30 μl) were immediately used for reporter gene assays. CAT expression levels were determined using the CAT ELISA kit (Roche Molecular Biochemicals) and GUS activity was measured by the MUG fluorimetric assay.
LsORF A or (LMAGDIS, or LMAGRIS, or LMAGDI) plasmid (10 μg) was always cotransfected with 2.5 μg of GUS coexpressing plasmid (pV594) to serve as an internal standard of transfection efficiency. For transactivation, 5 μg of plasmid pHELP7 (Bonneville et al. 1989) expressing the TAV protein was also added. For each CAT construct, transfections were repeated three times in duplicate (±TAV) in independent batches of protoplasts. CAT expression was always calculated relative to the GUS activity of internal controls measured as described by Fütterer and Hohn (1991).
Acknowledgments
We thank David Kirk for technical help, P. Müller for the provision of oligonucleotides, and Mike Rothnie for graphical work. We gratefully acknowledge the active interest and help of our colleagues, especially M. Pooggin, J. Fütterer, D. Dominguez, and H. Rothnie. Special thanks to Helen Rothnie for critical reading of the manuscript and stimulating discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Hohn@fmi.ch; FAX 4161-697 39 76.
References
- Anthony DD, Merrick WC. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- Bonneville J-M, Sanfacon H, Fütterer J, Hohn T. Posttranscriptional transactivation in cauliflower mosaic virus. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- Cao J, Geballe AP. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996a;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996b;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA. 1998;4:181–188. [PMC free article] [PubMed] [Google Scholar]
- Curran J, Kolakofsky D. Scanning independent ribosomal initiation of the sendai virus X protein. EMBO J. 1988;7:2869–2874. doi: 10.1002/j.1460-2075.1988.tb03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbecq P, Werner M, Feller A, Filipkowski RK, Messenguy F, Piérard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Dominguez DI, Ryabova LA, Pooggin MM, Schmidt-Puchta W, Fütterer J, Hohn T. Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J Biol Chem. 1998;273:3669–3678. doi: 10.1074/jbc.273.6.3669. [DOI] [PubMed] [Google Scholar]
- Franck A, Guilley H, Jonard G, Richards KE, Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980;21:285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Fütterer J, Hohn T. Translation of a polycistronic mRNA in presence of the cauliflower mosaic virus transactivator protein. EMBO J. 1991;10:3887–3896. doi: 10.1002/j.1460-2075.1991.tb04958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Role of an upstream open reading frame in the translation of polycistronic mRNA in plant cells. Nucleic Acids Res. 1992;20:3851–3857. doi: 10.1093/nar/20.15.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Translation in plants—rules and exceptions. Plant Mol Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Gordon K, Pfeiffer P, Sanfaçon H, Pisan B, Bonneville J, Hohn T. Differential inhibition of downstream gene expression by the cauliflower mosaic virus 35S RNA leader. Virus Genes. 1989;3:45–55. doi: 10.1007/BF00301986. [DOI] [PubMed] [Google Scholar]
- Fütterer J, Gordon K, Sanfaçon H, Bonneville JM, Hohn T. Positive and negative control of translation by the leader of cauliflower mosaic virus pregenomic 35S RNA. EMBO J. 1990;9:1697–1707. doi: 10.1002/j.1460-2075.1990.tb08293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Kiss-László Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- Fütterer J, Potrykus I, Bao Y, Li L, Burns TM, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus Rice Tungro Bacilliform Virus translation. J Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe AP, Morris DR. Initiation codons within 5′-leaders of mRNAs as regulators of translation. Trends Biochem Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- Gowda S, Wu FC, Schlthof HB, Shepherd RJ. Gene VI of figwort mosaic virus (caulimovirus group) functions in posttranscriptional expression of genes on the full-length RNA transcript. Proc Natl Acad Sci. 1989;86:9203–9207. doi: 10.1073/pnas.86.23.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV. Use of bacteriophage RNA polymerase in RNA synthesis. Methods Enzymol. 1996;275:382–397. doi: 10.1016/s0076-6879(96)75023-1. [DOI] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M, Hohn T. A stable hairpin preceded by a short ORF promotes nonlinear ribosome migration on a synthetic mRNA leader. RNA. 1999;5:1149–1157. doi: 10.1017/s1355838299990325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M, Steger G, Hohn T. Alternative structures of the cauliflower mosaic virus 35 S RNA leader: Implications for viral expression and replication. J Mol Biol. 1997;267:1075–1088. doi: 10.1006/jmbi.1997.0929. [DOI] [PubMed] [Google Scholar]
- ————— Regulation of CaMV 35 S RNA translation is mediated by a stable hairpin in the leader. RNA. 1998;4:101–111. [PMC free article] [PubMed] [Google Scholar]
- Herbst KL, Nichols LM, Gesteland RF, Weiss RB. A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc Natl Acad Sci. 1994;91:12525–12529. doi: 10.1073/pnas.91.26.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JR, Morris DR. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA. J Biol Chem. 1992;267:21886–21893. [PubMed] [Google Scholar]
- ————— Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. J Biol Chem. 1993;268:726–731. [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Hohn T, Dominguez DI, Schärer-Hernandez N, Pooggin MM, Schmidt-Puchta W, Hemmings-Mieszczak M, Fütterer J. Ribosome shunting in eukaryotes: What the viruses tell me. In: Bailey-Serres J, Gallie DR, editors. A look beyond transcription: Mechanisms determining mRNA stability and translation in plants. Rockville, MD: American Society of Plant Physiologists; 1998. pp. 84–95. [Google Scholar]
- Jackson RJ. A comparative view of initiation site selection mechanisms. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 71–112. [Google Scholar]
- Jaramillo M, Dever TE, Merrick WC, Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF- 4B. Mol Cell Biol. 1991;11:5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eukaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Context effects and inefficient initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PF, Hinnebusch AG. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes & Dev. 1989;3:1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- Mize GJ, Ruan H, Low JJ, Morris DR. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- Paraskeva E, Gray NK, Schlager B, Wehr K, Hentze MW. Ribosomal pausing and scanning arrest as mechanisms of translation regulation from cap-distal iron-responsive elements. Mol Cell Biol. 1999;19:807–816. doi: 10.1128/mcb.19.1.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pooggin MM, Hohn T, Fütterer J. Forced evolution reveals the importance of short open reading frame A and secondary structure in the cauliflower mosaic virus 35S RNA leader. J Virol. 1998;72:4157–4169. doi: 10.1128/jvi.72.5.4157-4169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin MM, Fütterer J, Skryabin KG, Hohn T. A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J Gen Virol. 1999;80:2217–2228. doi: 10.1099/0022-1317-80-8-2217. [DOI] [PubMed] [Google Scholar]
- Pooggin, M.M., T. Hohn, and J. Fütterer. 2000. Role of a short ORF in ribosome shunt on the CaMV RNA leader. J. Biol. Chem. (in press). [DOI] [PubMed]
- Remm M, Remm A, Ustav M. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J Virol. 1999;73:3062–3070. doi: 10.1128/jvi.73.4.3062-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BE, Paterson BM. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc Natl Acad Sci. 1973;70:2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schleiss MR, Degnin CR, Geballe AP. Translational control of human cytomegalovirus gp48 expression. J Virol. 1991;65:6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Puchta W, Dominguez D, Lewetag D, Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854–2860. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sachs MS. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J Biol Chem. 1997a;272:255–261. doi: 10.1074/jbc.272.1.255. [DOI] [PubMed] [Google Scholar]
- ————— Ribosomal stalling is responsible for arginine-specific translation attenuation in Neurospora crassa. J Biol Chem. 1997b;272:255–261. [Google Scholar]
- Werner M, Feller A, Messenguy F, Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987;49:805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ. Selective translation initiation by ribosome jumping in adenovirus-infected and heat shocked cells. Genes & Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- ————— Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes & Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]