Abstract
Harmonin is a scaffolding protein required for normal mechanosensory function in hair cells. Here, we describe a novel presynaptic association of harmonin and Cav1.3 Ca2+ channels at the mouse inner hair cell synapse, which limits channel availability through a ubiquitin-dependent pathway.
Voltage-gated Cav1.3 (L-type) Ca2+ channels mediate Ca2+ influx and exocytosis at the inner hair cell (IHC) synapse and are required for hearing1,2. The C-terminus of the Cav1.3 α1 subunit (α11.3) binds to PDZ (PSD-95 (Postsynaptic density-95)/Discs large/ZO-1 (Zona occludens-1) domains, which affect neuronal Cav1.3 localization and function3. In screening for PDZ-proteins that interact with Cav1 α1-subunits, we discovered that harmonin (variant a), a PDZ-protein expressed in IHCs4, binds to α11.3 via the second of its three PDZ domains (Fig. 1a,b).
Figure 1.
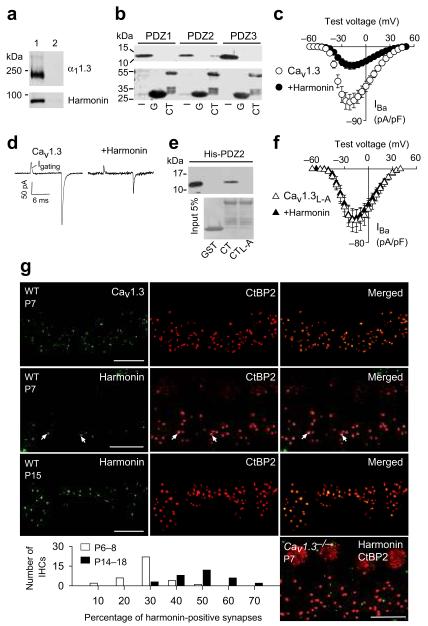
Harmonin inhibits Cav1.3 channels and is localized at inner hair cell synapses.
(a) Myc-harmonin coimmunoprecipitates with FLAG-α11.3 in cotransfected HEK293T cells (lane 1) but not in cells transfected with myc-harmonin alone (lane 2). (b) Western blot (upper panel) from pull-down assay shows GST-1.3CT (CT) but not GST (G) binds his-tagged PDZ2 of harmonin. Input (I) represents ~5% protein His-PDZ protein input. Ponceau staining (lower panel) shows amounts of GST-proteins used. Full length blots are presented in Supplementary Fig.5. (c) Harmonin inhibits Cav1.3 IBa density in transfected HEK293T cells. Cav1.3 alone, n=11; +harmonin, n=11. (d) Representative traces showing gating currents (Igating) of Cav1.3 ± harmonin measured at the IBa reversal potential (+60 mV). (e) His-PDZ2 of harmonin binds to α11.3 CT but not with L-A mutation. Pull-down assay was done as in b. (f) No effect of harmonin on IBa density for Cav1.3 with L-A mutation (Cav1.3L-A). Cav1.3L-A alone, n=8; Cav1.3+harmonin, n=7. (g) Confocal projections of whole mounts of Organ of Corti from wild-type (WT, P7 or P15) or Cav1.3−/− mice (P7) double-labeled for CtBP2 (red) and Cav1.3 (green) or harmonin (green). Synaptic labeling of CtBP2 appears as spots basal to the nucleus, which is also labeled by these antibodies. Areas of colocalization (arrows) are yellow in the merged images. Scale bars, 10 μm. Graph shows distribution of harmonin-positive synapses in individual IHCs from P6–8 and P14–16 IHCs. In c and f, error bars represent s.e.m.
Like the PDZ-protein erbin5, harmonin promoted voltage-dependent facilitation of Cav1.3 in transfected HEK293T cells (not shown). However, harmonin also significantly suppressed peak Cav1.3 Ba2+ current (IBa) density (~66%, p<0.001; Fig. 1c) without major effects on voltage-dependent activation (Supplementary table 1). Harmonin significantly decreased maximal “on” gating charge (Qon) of IBa and tail current amplitudes at the reversal potential (Fig. 1d, Supplementary Fig. 1), which suggests that harmonin reduces the number of available Cav1.3 channels rather than the open probability. Disrupting the type I PDZ binding sequence of α11.3 by substituting the C-terminal leucine for alanine (L-A) prevented harmonin binding (Fig. 1e) and inhibition of Cav1.3 peak current density (p=0.93; Fig. 1f).
As described previously 6-8, harmonin is strongly localized in the apical hair bundles of IHCs (Supplementary Fig.2). However, closer scrutiny revealed harmonin clusters at the base of IHCs, which colocalized with the ribbon synapse protein ribeye/CtBP2 at a subset of synapses (Fig. 1g). Comparisons before (P6–8) and after (P14–16) hearing onset revealed a developmental increase in the number of harmonin-positive IHC synapses (Fig. 1g). In most immature IHCs (30 of 35), harmonin was localized at ~10–30% of synapses per IHC. However, in mature IHCs (18 of 31), ~40–60% of synapses per IHC were harmonin-positive (Fig. 1g). Cav1.3 channels may be necessary for the synaptic localization of harmonin: the fraction of harmonin-positive synapses was significantly reduced in IHCs from Cav1.3−/− mice (~26% of harmonin-positive synapses in WT, P6-8, n=84 IHCs, vs. ~10% in Cav1.3−/−, P6-8, n=82 IHCs, p<0.001; Fig. 1g). Harmonin also coimmunoprecipitated with Cav1.3 from cochlear extracts (Supplementary Fig. 2). These findings support a presynaptic association of harmonin with Cav1.3, primarily in mature IHCs.
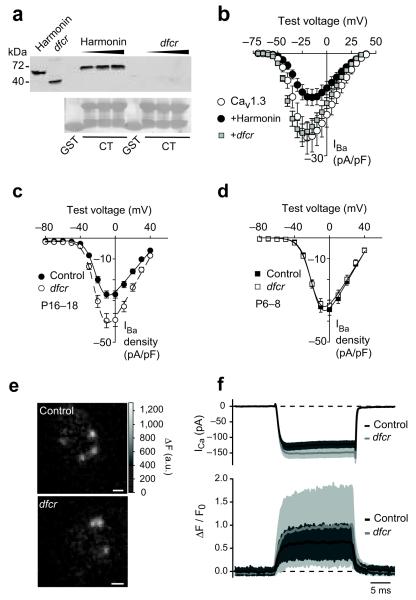
To determine if harmonin regulates presynaptic Cav1.3 channels in IHCs, we analyzed “deaf-circler” mice (dfcr)9. While the mutant (dfcr harmonin) retains PDZ2 that interacts with α11.3 (Fig. 1b), an internal deletion removes the coiled-coil domain and causes abnormal proximity of the PDZ domains. Dfcr harmonin did not bind to α11.3 (Fig. 2a) or affect peak Cav1.3 IBa density in transfected HEK293T cells (p=0.10; Fig. 2b), which demonstrates that dfcr harmonin does not functionally interact with α11.3. In agreement with a normally inhibitory role for harmonin, whole cell IBa density was greater in P16–18 dfcr IHCs than in control IHCs (~33%, p<0.01; Fig. 2c), despite otherwise normal Cav1.3 activation properties (Supplementary Table 2). This difference was not observed at P6-8 (p=0.59, Fig. 2d), consistent with sparse synaptic localization of harmonin at this age (Fig. 1g). Ca2+ microdomains, which primarily reflect presynaptic Ca2+ influx10, had higher amplitudes in mature dfcr than control IHCs (~53%, p=0.02; Fig. 2e, f). Taken together, these results show that harmonin directly inhibits synaptic Cav1.3 channel density in IHCs, which is disrupted by the dfcr mutation.
Figure 2.
Dfcr mutation prevents functional interaction of harmonin and Cav1.3
(a) Pull-down assays with GST or GST-α11.3 CT and increasing amounts of lysates from HEK293T cells transfected with myc-harmonin or myc-dfcr harmonin, as indicated. Left 2 lanes show input (~5%) and Ponceau staining (lower panel) shows GST or GST-α11.3 CT used in the assay. Full length blots are presented in Supplementary Fig.5. (b) Same protocol as in Fig.1c except in cells transfected with Cav1.3 alone, n=7, +harmonin, n=9, or +dfcr-harmonin, n=8. (c,d) Plots of IBa density vs. test voltage from whole-cell patch clamp recordings at P16-18 (c) and P6-8 (d) from control (+/−; n=23 in c, n=45 in d) or dfcr (−/−; n=31 in c, n=42 in d) IHCs. (e,f) Simultaneous whole-cell patch clamp recordings and fluorescent measurement of presynaptic Ca2+ microdomains. (e) Representative hotspots of Fluo-5N fluorescence in control (+/+) and dfcr (−/−) IHCs during depolarization. Scale bar, 1 μm. (f) Top, Whole-cell ICa recorded in control (black line and dark grey area, n = 14) and dfcr (grey line and light grey area, n = 16) IHCs. Bottom, Background-subtracted and normalized Fluo-5N fluorescence change (ΔF/F0, mean±SD) in the center of the Ca2+ microdomain (41 domains in 14 control IHCs and 41 domains in 16 dfcr IHCs). In b-d, error bars represent s.e.m. Protocols for the use of mice were approved by the Institutional Animal Care and Use Committee at U. Iowa and U. Gottingen.
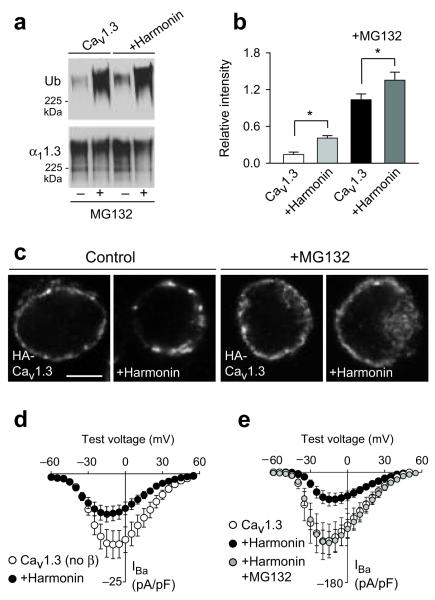
Cell surface density of ion channels can be regulated by ubiquitination and targeting for proteosomal or lysosomal degradation11. Thus, we tested if harmonin enhanced ubiquitination of Cav1.3 in HEK293T cells. Following immunoprecipitation with α11.3 antibodies, western blotting revealed higher levels of ubiquitinated channels cotransfected with harmonin, which was intensified by the proteosomal inhibitor MG132 (Fig. 3a,b). In addition, harmonin altered the distribution of cell-surface Cav1.3 channels immunofluorescently labeled through an extracellular hemagglutinin epitope. While Cav1.3 channels alone showed a fairly continuous distribution, harmonin caused Cav1.3 channels to form large clusters in the plasma membrane (Fig. 3c, Supplementary Fig. 3). The patchier cell-surface distribution of Cav1.3 channels caused by harmonin was reversed by MG132 (Fig. 3c). Given that MG132 broadly affects the ubiquitin system and may affect both proteosomal and lysosomal protein pathways, the altered distribution of Cav1.3 channels caused by harmonin may result from improper trafficking and/or degradation of ubiquitinated channels. Although Cavβ subunits inhibit ubiquitination of Cav channels12,13, the inhibition of Cav1.3 current density by harmonin did not require Cavβ (Fig. 3d) but was blocked by MG132 (Fig. 3e; MG132 did not affect current density in cells with Cav1.3 alone, Supplementary Fig. 4). We conclude that harmonin tags Cav1.3 channels for ubiquitination and alters their functional levels at the cell surface. This may constrain the number of available presynaptic Cav1.3 channels in IHCs, a parameter that varies between IHC synapses10 and during development14. Together with evidence that harmonin regulates mechanotransduction currents8,15, our findings reveal harmonin as not simply a scaffold, but an integral component of ion channel complexes and fundamental regulator of electrical and Ca2+ signaling in auditory hair cells.
Figure 3.
Harmonin inhibits plasma membrane Cav1.3 channel density through a ubiquitin-dependent pathway
(a,b) Ubiquitination of Cav1.3 channels in HEK293T cells transfected with Cav1.3±harmonin. Cav1.3 channels were immunoprecipitated with α11.3 antibodies and western blotting was performed with anti-ubiquitin (Ub, top panel) or α11.3 antibodies (bottom panel). Cells were maintained with (+) or without (−) MG132 (5 μM, 12 h) prior to lysis. Ubiquitination of Cav1.3 was quantitated and plotted as relative intensities (b); *, p<0.05. Full length blots are presented in Supplementary Fig.5. (c) Confocal micrographs showing immunofluorescence of cell-surface labeled HA-Cav1.3 channels transfected alone or with harmonin. Pretreatment with MG132 was as in (a). Scale bars, 5 μm. (d,e) IBa density recorded as in Fig.1c in HEK293T cells cotransfected without Cavβ (d; n=12 for Cav1.3, n=11 for +harmonin) or with Cavβ (e; n=12 for Cav1.3 alone, n=14 for +harmonin, n=11 for +harmonin+MG132). In (e), cells were treated with MG132 (5 μM) 1-3 h before recording. In b, d, e, error bars represent s.e.m.
Supplementary Material
Acknowledgments
This work was supported by the NIH (DC009433, HL087120, DC 10382 (AL); DC008417 (ICJ); DA015040, K12/GM000680 (FDG); Deafness Research Foundation (AL, ICJ); DFG (Center for Molecular Physiology of the Brain, FZT-103), the BMBF (Bernstein Center for Computational Neuroscience Goettingen: 01GQ1005A) to TM; and Alexander von Humboldt fellowship to TP. The authors thank J.Striessnig for Cav1.3 KO mice; U. Mueller and U. Wolfrum for cDNAs and antibodies; Q. Zheng for harmonin KO tissue; H.Couchoux, A. Inagaki for contributing data; B. Fritzsch, C.Harata, M.Stamnes, R.Piper for advice and discussion; and J. Diamond for comments on the manuscript.
Footnotes
Declaration of competing financial interests
References
- 1.Platzer J, et al. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 2.Brandt A, et al. J Neurosci. 2005;25:11577–11585. doi: 10.1523/JNEUROSCI.3411-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin-Jageman I, Lee A. J Neurochem. 2008;105:573–583. doi: 10.1111/j.1471-4159.2008.05286.x. [DOI] [PubMed] [Google Scholar]
- 4.Verpy E, et al. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 5.Calin-Jageman I, et al. J Neurosci. 2007;27:1374–1385. doi: 10.1523/JNEUROSCI.5191-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiners J, et al. Invest Ophthalmol Vis Sci. 2003;44:5006–5015. doi: 10.1167/iovs.03-0483. [DOI] [PubMed] [Google Scholar]
- 7.Boeda B, et al. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grillet N, et al. Neuron. 2009;62:375–387. doi: 10.1016/j.neuron.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson KR, et al. Hum Mol Genet. 2003;12:3075–3086. doi: 10.1093/hmg/ddg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank T, et al. Proc Natl Acad Sci U S A. 2009 [Google Scholar]
- 11.Abriel H, Staub O. Physiology (Bethesda) 2005;20:398–407. doi: 10.1152/physiol.00033.2005. [DOI] [PubMed] [Google Scholar]
- 12.Waithe D, et al. J. Biol. Chem. 2011;286:9598–9611. doi: 10.1074/jbc.M110.195909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altier C, et al. Nature neuroscience. 2011;14:173–180. doi: 10.1038/nn.2712. [DOI] [PubMed] [Google Scholar]
- 14.Beutner D, Moser T. J Neurosci. 2001;21:4593–4599. doi: 10.1523/JNEUROSCI.21-13-04593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michalski N, et al. Pflugers Arch. 2009;459:115–130. doi: 10.1007/s00424-009-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.